ABSTRACT
The U.S. Food and Drug Administration (FDA) regulates manufacturing and testing of advanced therapeutic medicinal products (ATMPs) to ensure the safety of each product for human use. Gold-standard sterility testing (USP<71>) and alternative blood culture systems have major limitations for the detection of fungal contaminants. In this study, we evaluated the performance of iLYM (lactic acid-fermenting organisms, yeasts, and mold) medium (designed originally for the food and beverage industry) to assess its potential for use in the biopharmaceutical field for ATMP sterility testing. We conducted a parallel evaluation of four different test systems (USP<71>, BacT/Alert, Bactec, and Sabouraud dextrose agar [SDA] culture), three different bottle media formulations (iLYM, iFA Plus, and Myco/F Lytic), and two incubation temperatures (22.5°C and 32.5 to 35°C) using a diverse set of fungi (n = 51) isolated from NIH cleanroom environments and previous product contaminants. Additionally, we evaluated the effect of agitation versus delayed-entry static preincubation on test sensitivity and time to detection (TTD). Overall, delayed entry of bottles onto the BacT/Alert or Bactec instruments (with agitation) did not improve test performance. USP<71> and SDA culture continued to significantly outperform each automated culture condition alone. However, we show, for the first time, that a closed-system, dual-bottle combination of iLYM 22.5°C and iFA Plus 32.5°C can provide high fungal sensitivity, statistically comparable to USP<71>, when tested against a diverse range of environmental fungi. Our study fills a much-needed gap in biopharmaceutical testing and offers a favorable testing algorithm that maximizes bacterial and fungal test sensitivity while minimizing risk of product contamination associated with laboratory handling.
KEYWORDS: cell therapy, BacT/Alert, mold, sterility testing, gene therapy, advanced therapeutic medicinal products, fungi, iLYM, Myco/F Lytic, Bactec, ATMPs
INTRODUCTION
Advanced therapeutic medicinal products (ATMPs) which include cell, gene, and immunotherapies have become a critical tool for the targeted and personalized treatment of certain malignancies and genetic diseases. Recently, several ATMPs have been approved by the United States Food and Drug Administration (FDA) for commercial use (https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/approved-cellular-and-gene-therapy-products). The majority of ATMPs, however, are labeled as “investigational new drugs” (INDs) that are typically under study at major academic medical centers in the form of clinical trials. As such, clinical microbiology laboratories are often asked to assist with ATMP-related testing such as sterility testing and/or environmental monitoring as noted in two recent review articles (1, 2) and numerous discussions of this topic on the American Society for Microbiology’s clinical microbiology Division C and ClinMicroNet listservs.
Sterility testing of ATMPs is an important component to assess the absence of viable microbial contamination to ensure patient safety prior to product infusion. The gold standard for sterility testing in the United States is USP<71> published by the U.S. Pharmacopeia (3). Introduced in 1932, USP<71> was designed originally for the testing of traditional, bulk-manufactured sterile pharmaceuticals and consists of at least 14 days of culture incubation with manual observation for turbidity. The FDA has acknowledged that the USP<71> method may be unsuitable for sterility testing of ATMPs due to the inherent turbidity and short shelf-life of cellular therapy products (4). Respiration technology (i.e., blood culture systems) that can provide automated and objective detection of microbial growth have become favorable alternatives for ATMP sterility testing to overcome the limitations identified with USP<71> (1, 5, 6).
Studies comparing various blood culture systems with USP<71> have been reported, but most focus on bacterial contaminants and/or media formulations that trigger detection algorithms (on the instruments) designed for clinical bloodstream infections (7–12). Very few studies have reported on the performance of “industry” bottles that activate detection algorithms designed specifically for environmental organisms targeted for sterile drug product testing (5, 13). Furthermore, a major limitation in previous works has been the narrow spectrum of organisms evaluated, with most reports focusing on system performance using the six USP<71> quality control (QC) type strains only (Staphylococcus aureus ATCC 6538, Pseudomonas aeruginosa ATCC 9027, Bacillus subtilis ATCC 6633, Clostridium sporogenes ATCC 19404, Candida albicans ATCC 10231, and Aspergillus brasiliensis ATCC 16404) (1, 3). These isolates are clearly not reflective of typical environmental flora (particularly fungi) and can allude to a false sense of security regarding system performance due to the failure to rigorously challenge the system(s) beyond QC isolates. In contrast to smaller studies, we recently reported suboptimal performance of the Bactec (clinical bottles) and BacT/Alert (industry bottles) compared with USP<71> using a wider and more diverse organism challenge set (n = 118), thereby demonstrating that rigorous testing using in-house isolates, in addition to QC strains, is critically important for a system validation that is reflective of the daily environment and microbial contamination risk (5). We showed that use of aerobic and anaerobic BacT/Alert bottles alone were insufficient for the detection of fungal contaminants, but that addition of a Sabouraud dextrose agar (SDA) culture paired with the BacT/Alert industry bottles, however, resulted in an overall detection of bacterial and fungal isolates that was statistically equivalent to USP<71> (5). From this study, a testing algorithm consisting of BacT/Alert (iFA Plus and iFN Plus bottles, inoculated by the ATMP manufacturing personnel and incubated at 32.5°C) with SDA culture (inoculated by laboratory personnel and incubated at 22.5°C) has been used in our laboratory since 2019 to support the sterility testing of a variety of cellular and gene therapies, positron emission tomography (PET)-radiolabeled drugs, and viral vectors manufactured on the NIH campus.
While addition of the SDA culture has significantly improved our test sensitivity for fungal contaminants, the risk of “drop-in” culture contaminants caused by laboratory handling is high and can lead to challenging risk-benefit decisions as to whether to proceed with product infusion for a critically ill patient (14). In this study, we aimed to evaluate a closed-system alternative to the SDA culture to optimize fungal detection in ATMPs, focusing on the BacT/Alert iLYM aerobic bottle (lactic acid-fermenting organisms, yeasts, and mold; used commonly in the food and beverage industry). Additionally, we chose to evaluate the performance of the Bactec FX Myco/F Lytic (MFL) bottle, as it had been excluded from our original proof-of-principle study (5), and its formulary is intended for the recovery of mycobacteria, yeast, and other fungi from blood and sterile body fluids. In this study, a large and diverse set of 51 fungi were evaluated over seven culture conditions, including various combinations of test systems (USP<71>, BacT/Alert, Bactec, and SDA culture), media formulations (iLYM, iFA Plus, and MFL), and incubation temperatures (22.5°C and 32.5 to 35°C). “Delayed-entry” studies, whereby Bactec and BacT/Alert bottles were incubated offline prior to placing on the instrument, were also conducted to evaluate whether automated bottle agitation on the instruments could disrupt mycelial development, thereby impacting test sensitivity and delaying detection.
MATERIALS AND METHODS
Fungal isolates.
A total of 51 fungal isolates representing 44 distinct organisms were evaluated (Table 1). Forty-nine isolates were cultured from previously contaminated products or from the NIH cGMP environment. Two USP<71> fungal QC isolates, Candida albicans (ATCC 10231) and Aspergillus brasiliensis (ATCC 16404), were prepared from a commercial standardized inoculum (Microbiologics, St. Cloud, MN).
TABLE 1.
Forty-four distinct fungal isolates representing the NIH cGMP environment and previous product contaminants (n = 49), along with two USP<71> QC organisms (C. albicans and A. brasiliensis)
| Isolate |
|---|
| Yeast and yeast-like organisms (n = 8) |
| Aureobasidium sp. |
| Candida albicans (n = 2) |
| Candida parapsilosis |
| Cryptococcus sp. |
| Rhodotorula mucilaginosa |
| Sporobolomyces sp. |
| Trichosporon inkin |
| Mucorales (n = 2) |
| Rhizopus oryzae |
| Syncephalastrum sp. |
| Hyaline molds (n = 24) |
| Arthrinium sp. |
| Aspergillus brasiliensis |
| Aspergillus fumigatus |
| Aspergillus niger |
| Aspergillus terreusa (n = 2) |
| Aspergillus versicolor |
| Byssochlamys spectabilis |
| Byssomerulius corium |
| Fusarium incarnatum-equiseti complex |
| Hamigera sp. |
| Hypocrea sp. |
| Irpex sp. |
| Neosartorya hiratsukae |
| Paecilomyces variotii |
| Penicillium adametzioides |
| Penicillium capsulatum |
| Penicillium chrysogenum |
| Penicillium sp. (n = 3) |
| Purpureocillium lilacinum |
| Sistotrema sp. |
| Trametes sp. |
| Melanized molds (n = 17) |
| Alternaria sp. |
| Botrytis sp. |
| Chaetomium globosuma |
| Cladosporium domesticum |
| Cladosporium halotolerans |
| Cladosporium sp. (n = 3) |
| Curvularia sp. |
| Epicoccum nigrum |
| Epicoccum sp. |
| Ochroconis sp.a |
| Pithomyces chartarum |
| Pithomyces sp. |
| Ramularia sp. |
| Verrucladosporium sp.a |
Isolation from a biopharmaceutical product.
Culture methods and colony counts.
Non-QC isolates were subcultured from frozen stock onto SDA and incubated at 22.5°C. Following a second passage, a suspension was created to form a concentration of 10 to 99 CFU per 100 μl. Briefly, for each test isolate, a moistened cotton swab was rolled gently over the surface of the mold, transferred into sterile water, and set aside to allow large particulate matter to settle. The supernatant was used to create a fungal suspension with 80 to 82% transmittance (%T) at 530 nm (Genesys 20 visible spectrophotometer; Thermo Fisher Scientific, Waltham, MA). Serial dilutions ranging from 10−1 to 10−4 were cultured onto SDA in duplicate. Initial experiments consisted only of serial colony count dilution studies to establish a target dilution that resulted in 10 to 99 CFU/100 μl for each isolate. Fungal suspensions were cultured immediately upon preparation. After the target dilution had been determined for each isolate, fresh fungal suspensions were prepared for subsequent experimental growth promotion studies. Each experimental isolate was accompanied by a colony count plate to verify inoculum concentration. The inoculum of <100 CFU is defined by USP<71> (3).
Pilot delayed-entry static preincubation time point study.
Ten organisms consisting of two QC strains (C. albicans, A. brasiliensis) and eight environmental isolates (C. albicans, Cryptococcus sp., Rhodotorula sp., Aspergillus terreus, Irpex sp., Penicillium sp., Purpureocillium lilacinum, and Syncephalastrum sp.) were evaluated in an initial pilot study to determine the optimal delayed-entry static preincubation time point in hours for further study. Four delayed-entry static preincubation time frames (2 to 8 h, 16 to 24 h, 24 to 36 h, or >48 h) were tested alongside a time zero (0 h) control. Each organism was inoculated to a SDA plate and into tryptic soy broth (TSB; Hardy Diagnostics), iFA Plus bottles (iFA Plus; bioMérieux Industry, Marcy-l’Étoile, France), iLYM bottles (bioMérieux Industry), and Myco/F Lytic bottles (MFL; Becton, Dickinson, Franklin Lakes, NJ). iFA Plus and iLYM were incubated on the BacT/Alert Dual-T instrument (bioMérieux Industry). MFL bottles were incubated on the Bactec FX instrument (Becton, Dickinson). Test combinations are illustrated in Fig. 1, with all cultures monitored until growth was detected or for up to 360 h to account for full 14-day sterility testing incubation time frame as defined in USP<71>. Terminal visual inspection of the automated bottles was also conducted at the end of incubation to look for growth that may have gone undetected by the instrument (5). Culture bottles with no fungal growth were finalized on day 15 as not detected (ND). Sensitivity and days to detection were used to assess the optimal delayed-entry time point for further study.
FIG 1.
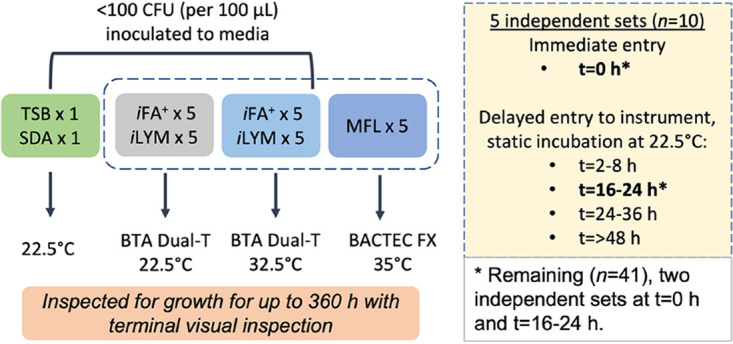
Test combination design.
Media evaluation study.
Based on results from the delayed-entry time point study, the remaining 41 isolates were set up with 0-h and 16- to 24-h time points only. For each isolate, the same set of inoculated medium was evaluated as described in Fig. 1, with each BacT/Alert and Bactec bottle tested without (0 h) and with (16 to 24 h) static preincubation at 22.5°C.
Acceptance criteria.
In accordance with USP<71>, a test was considered acceptable if fungal bioburden was <100 CFU, and growth was detected within 5 days of incubation (growth promotion qualification testing). Current practice reflects monitoring cultures for at least 14 days (as indicated for product sterility testing). Therefore, test comparisons across culture conditions were conducted within the context of growth detected up to 360 h of incubation (to account for the full 14 days of incubation).
Statistical analysis.
Qualitative analysis of sensitivity was performed using Fisher’s exact test (GraphPad, online resource). A log-rank test (GraphPad Prism 9) was used for statistical analysis of time to positivity between culture conditions.
RESULTS
Delayed-entry static preincubation does not improve test sensitivity or time to detection.
The pilot study performed using 10 isolates showed that delayed-entry static preincubation did not demonstrate a consistent pattern and did not significantly improve time to detection or test sensitivity (Table S1 in the supplemental material). Given these findings, only a single static preincubation time point of 16 to 24 h was chosen for further study on the remaining 41 isolates, as this time frame was most practical for laboratory workflow. Subsequently, with the expanded organism challenge set, no statistical difference in test sensitivity nor time to detection was observed between time (t) of 0 h and t of 16 to 24 h (Fig. 2A to E). Table S2 displays the days to detection for all 51 fungal isolates across the different culture conditions at t of 0 h and delayed entry t of 16 to 24 h.
FIG 2.
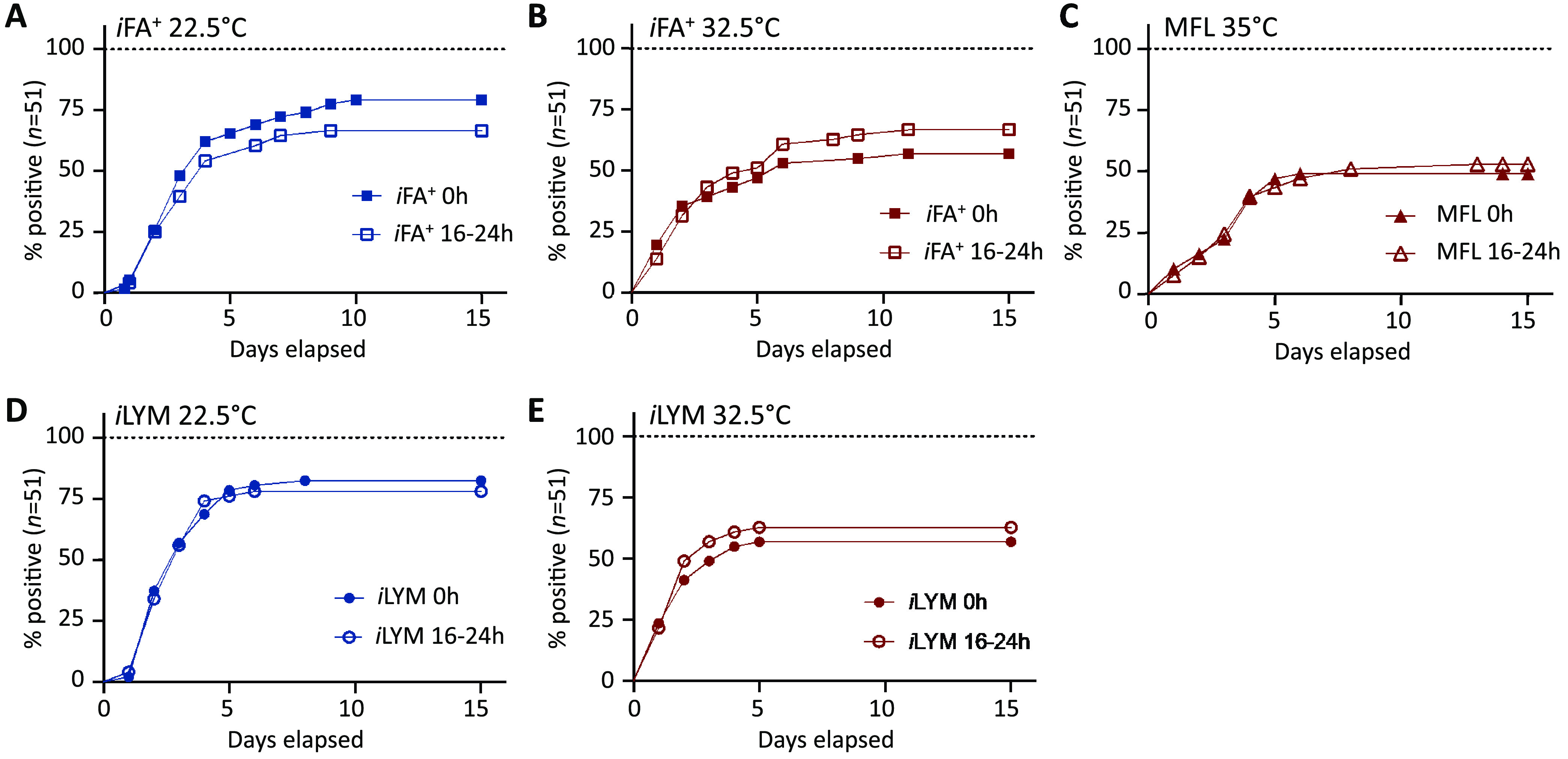
Delayed entry of bottles did not improve time to detection. The percentage of positive cultures (n = 51) for each automated bottle test combination is plotted over time. Fungal detection over 14 days is compared for 0-h and 16- to 24-h preincubation conditions for iFA Plus 22.5°C (A), iFA Plus 32.5°C (B), MFL 35°C (C), iLYM 22.5°C (D), and iLYM 32.5°C (E). A log-rank test showed that delayed entry had no statistical impact on time to detection.
Wide test variability across isolates and culture conditions.
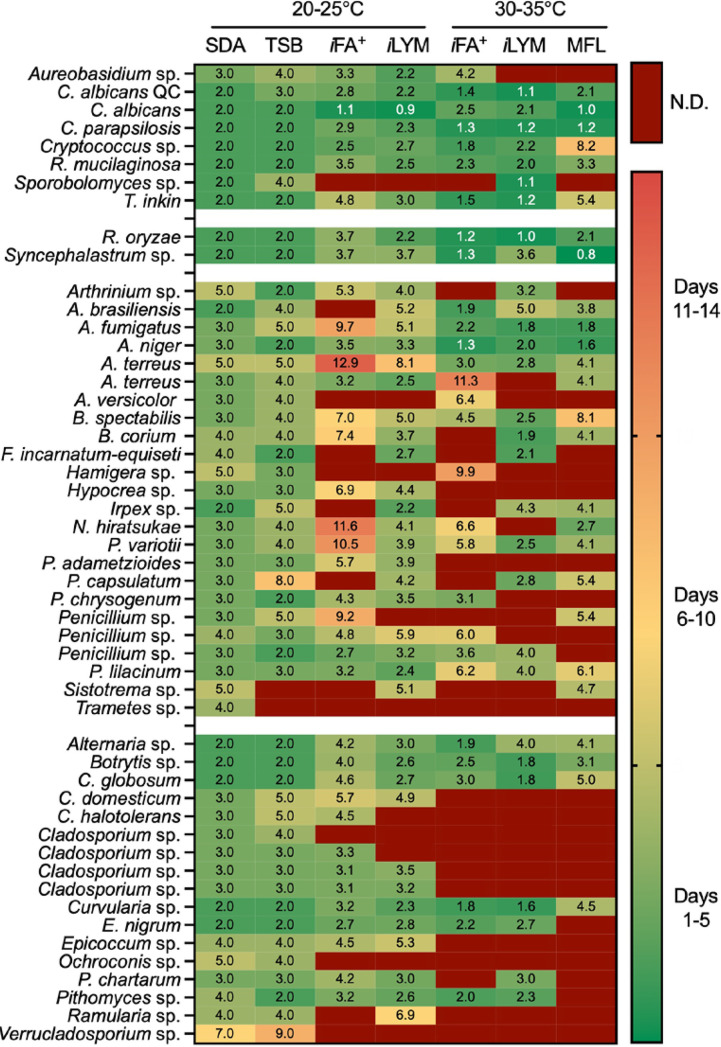
Figure 3 is a heatmap summarizing time to detection for the 51 fungal isolates studied at 0 h. Data are categorized by organism type (yeast, Mucorales, hyaline, and melanized molds, respectively) with one isolate per row and culture conditions represented by each column. Fungal detection was statistically equivalent between SDA and TSB at 25°C (USP<71>), which aligns with our previous findings (5). Automated culture systems (t = 0 h) performed better for yeast detection (34/40, 85%) across all conditions compared with mold (131/215, 61%, P = 0.0036; Fig. 3). Additionally, fungal isolates were significantly more likely to be detected by automated culture systems at the lower temperature incubation of 22.5°C (iFA Plus and iLYM; 68% to 78%) than with 32.5°C (iFA Plus and iLYM; 52 to 57%) and 35°C (MFL; 45 to 51%) when applying either the 5-day or 14-day incubation thresholds (Fig. 4A). Unfortunately, the Bactec FX instrument is manufactured only for clinical use and is limited to 35 ± 1.5°C as opposed to the BacT/Alert Dual-T instrument that contains both a regular incubator and a separate refrigerated unit capable of providing stable incubation of bottles down to 20°C. The SDA culture statistically outperformed all BacT/Alert and Bactec bottle media regardless of incubation temperature and/or delayed-entry condition (Fig. 3 and Fig. 5A).
FIG 3.
Time to detection varies widely across environmental fungi regardless of culture condition. Days to positivity are specified in each box for the corresponding organism (rows; n = 51) and culture condition (columns). Green represents a positive culture detected within 5 days, yellow shades represent detection between 6 and 10 days, and orange signifies late detection of 11 to 14 days. Cultures with undetectable growth within 14 days are dark red.
FIG 4.
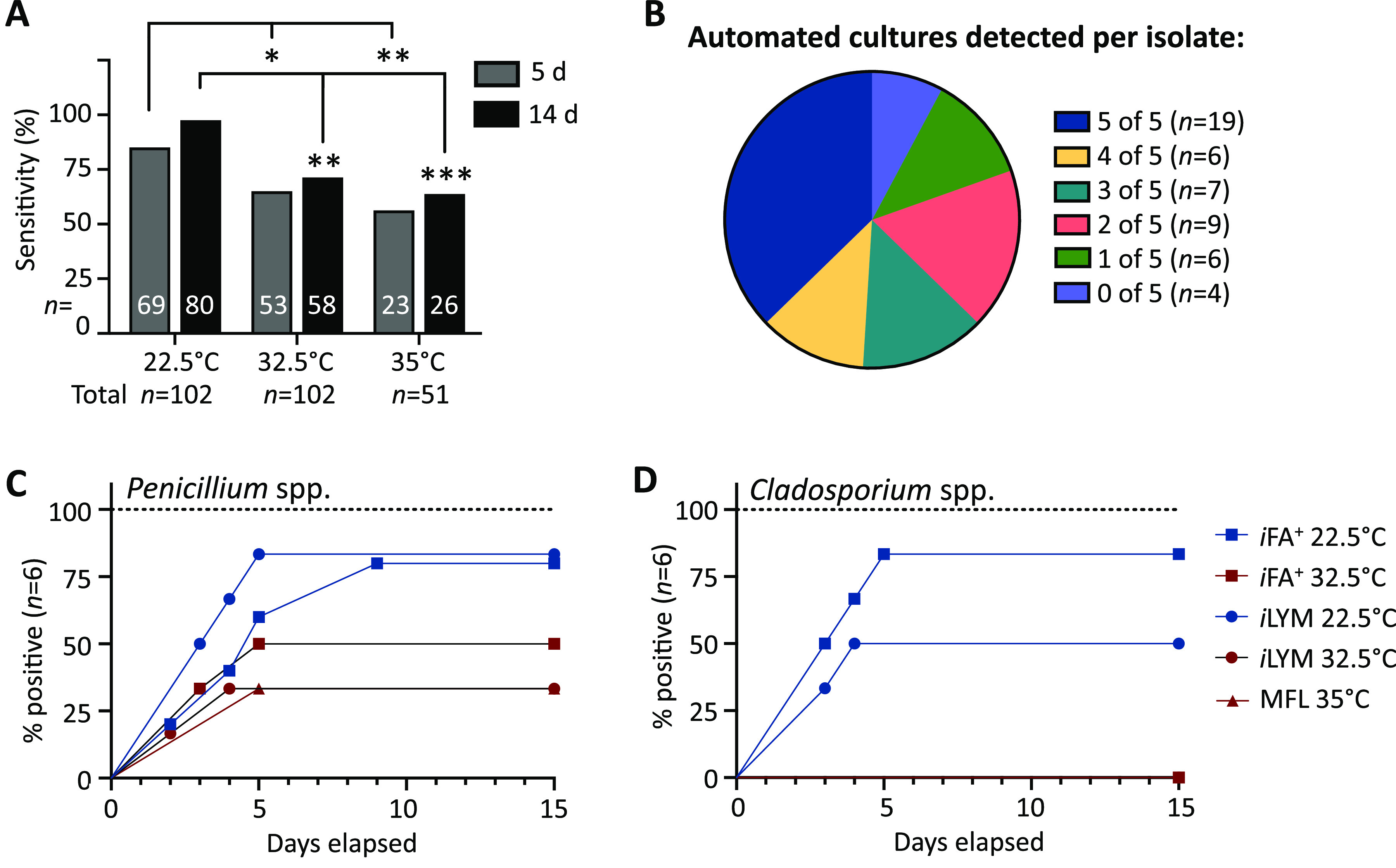
Test performance is organism and culture condition dependent. (A) Detection of fungal cultures within 5 and 14 days comparing automated culture bottles incubated at 22.5°C (n = 102; iFA Plus, iLYM), 32.5°C (n = 102; iFA Plus, iLYM), and 35°C (n = 51; MFL) using Fisher’s exact test. *, P < 0.05; **, P < 0.01; ***, P < 0.0001. (B) Each fungal isolate (n = 51) was categorized by how many automated culture bottles had growth detected. (C and D) Different culture conditions for Penicillium spp. (n = 6; P = 0.2088) (C) and Cladosporium spp. (n = 6; P = 0.0006) (D) were compared by log-rank test.
FIG 5.
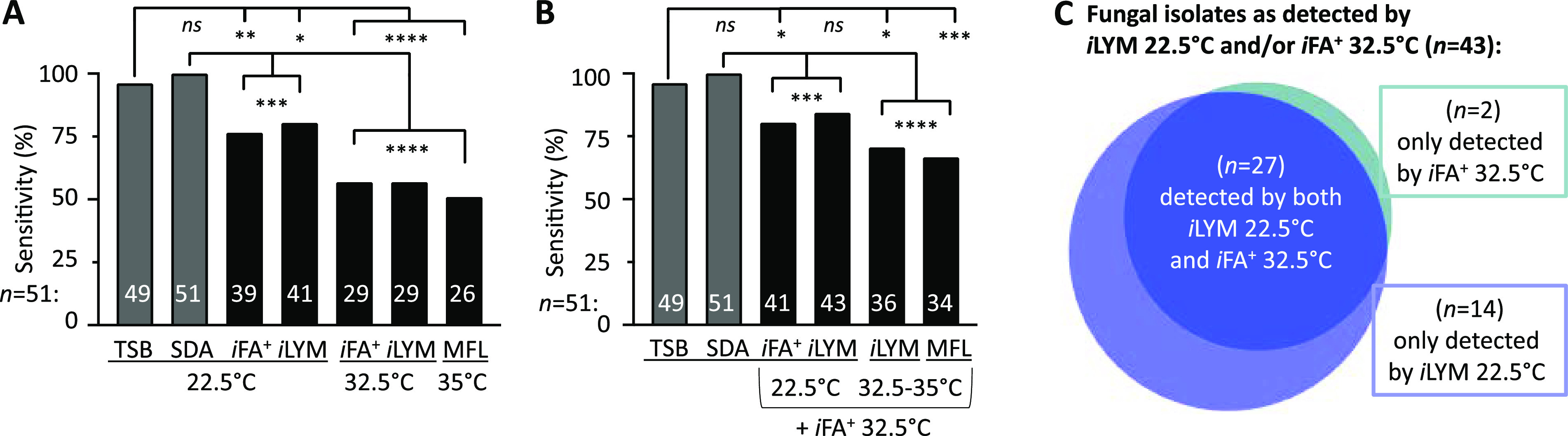
Combination of iLYM 22.5°C and iFA Plus 32.5°C has equivalent sensitivity to USP<71>. Fungal sensitivity after 14 days incubation is shown for each individual culture condition (A) or for all dual-culture conditions when combined with iFA Plus 32.5°C (B), as labeled on the x axis. A Fisher’s exact test compared sensitivity between TSB 22.5°C (USP<71>) or SDA 22.5°C with each individual culture (A) and with all culture conditions in combination with iFA Plus 32.5°C (B). *, P < 0.05; **, P < 0.01; ***, P < 0.0001; ****, P ≤ 0.0001; ns, not significant. (C) The area-proportional Venn diagram (generated with BioVenn [© 2007-2021 Tim Hulsen]) describes detection characteristics of fungal isolates (n = 43) detected by dual culture conditions iLYM 22.5°C and iFA Plus 32.5°C.
Figure 4B illustrates that only 19/51 (37%) isolates were detected across all five automated blood culture bottle conditions at 0 h, while the remaining 32 had at least one condition that failed detection. Only four (8%) isolates failed detection across all five automated test culture combinations (Fig. 4B). No fungal categories (yeast, Mucorales, hyaline, or melanized molds) had perfect detection within 14 days incubation (Fig. 3). Common environmental fungi, including Penicillium isolates, demonstrated marked strain variability in detection, denoted by a wide range in sensitivity (33% to 83%) among culture conditions (Fig. 4C). Cladosporium isolates also showed strain-dependent variability and, notably, were unable to thrive at 35°C, leading to significant differences in test sensitivity and time to detection (Fig. 4D). No single automated culture condition alone exhibited statistical equivalence to the USP<71> gold standard, TSB 22.5°C (Fig. 5A).
Combination of iLYM 22.5°C and iFA Plus 32.5°C improved fungal sensitivity and can reduce risk of false-positive SDA cultures caused by laboratory handling.
A two-bottle closed-system approach, combining iFA Plus 32.5°C and iLYM 22.5°C (n = 43, 84%) was significantly more sensitive than iFA Plus 32.5°C alone (n = 29, 57%; P = 0.0043). Furthermore, sensitivity was not statistically different from USP<71> (n = 49, 96%; P = 0.092; Fig. 5B). Importantly, 14/43 (33%) isolates were detected by iLYM 22.5°C alone, highlighting the substantial improvement in test sensitivity associated with iLYM media formulation and lower temperature incubation (Fig. 5C). Using a completely automated testing algorithm, iLYM 22.5°C increased fungal detection from 57% with iFA Plus 32.5°C alone to 84% for this dual-bottle combination for the 51 isolates tested (Fig. 5A and B).
DISCUSSION
Since 2015, the NIH sterility testing algorithm has included a supplemental SDA plate to improve fungal detection. The SDA culture was initiated following two contamination events whereby gross mold contamination failed to be detected by the Bactec system used at the time (2, 15, 16). A comprehensive study conducted in 2019, consisting of 118 aerobic and anaerobic bacteria, yeast, and mold representing environmental flora from the NIH manufacturing facilities as well as previous product contaminants, showed that the BacT/Alert system was significantly superior to the Bactec and that statistically equivalent results were obtained with USP<71> if the BacT/Alert (iFA Plus and iFN Plus at 32.5°C) was paired with an SDA culture at 22.5°C (5). However, between October 2015 and May 2021, 34/54 (63%) positive SDA cultures in our laboratory have been attributed to laboratory contamination (our unpublished data), each separately leading to a challenging risk assessment regarding product safety and release to a critically ill patient (14). Thus, the aim of this study was to reevaluate if a closed-system alternative was available for improved fungal detection. In this study, we evaluated a diverse set of 51 fungal isolates against a total of 7 different culture conditions (media formulation, incubation temperature) and test systems (USP<71>, BacT/Alert, Bactec, and SDA) to identify an alternative, closed-system, reduced-risk method that may replace the SDA culture while still allowing for optimized and highly sensitive detection of microbial contaminants.
Figure 3 and Fig. 5A clearly show that no test system alone is perfect, including the compendial USP<71> method, which was outcompeted by SDA culture. Similar to our previous report (5), we confirmed that the iFA Plus alone at 32.5°C had poor sensitivity for the detection of fungi (57%), while iFA Plus alone at 22.5°C had much improved sensitivity (76%) (Fig. 5A). Interestingly, BacT/Alert and Bactec media formulations (iLYM and MFL, respectively) targeted specifically for fungi performed poorly at higher temperatures (Fig. 4A). Performance of iLYM greatly improved when the incubation temperature was lowered to 22.5°C (80%; Fig. 5A). The current NIH testing algorithm, consisting of BacT/Alert iFA Plus and iFN Plus bottles at 32.5°C with SDA culture at 22.5°C, has been optimized for the detection of bacterial environmental contaminants (5) yet relies on the manual SDA culture (prone to contamination due to laboratory handling) to maximize fungal sensitivity. Here, we show, for the first time, the potential to replace SDA 22.5°C with the automated iLYM 22.5°C, to establish a highly sensitive, automated, closed-system algorithm for ATMP sterility testing at the NIH. Using a diverse fungal challenge set, we have demonstrated that a two-bottle combination of iFA Plus 32.5°C and iLYM 22.5°C was statistically equivalent to USP<71>. Using this bottle combination, only eight isolates (16%) failed to be detected. These isolates included Cladosporium sp. (n = 3), Ochroconis sp. (n = 1), Penicillium sp. (n = 1), Sporobolomyces sp. (n = 1), Trametes sp. (n = 1), and Verrucladosporium sp. (n = 1). So, while not perfect in sensitivity (as is normal for most test systems), the combined use of iFA Plus 32.5°C and iLYM 22.5°C provides an automated, closed-system option that would significantly reduce the risk of product contamination currently associated with SDA culture laboratory handling. As expected by the FDA, each institution would need to assess their own environmental flora and evaluate the risk associated with potentially missing certain environmental isolates such as the eight that failed to be detected in this study. This proof-of-principle study supports replacement of the SDA culture with iLYM 22.5°C at our facility, thereby resulting in an all-inclusive closed-system sterility testing algorithm consisting of iFA Plus 32.5°C, iFN Plus 32.5°C, and iLYM 22.5°C, all inoculated by manufacturing personnel prior to sample submission to the laboratory. Further testing and validation in the presence of various product matrices is required, but method suitability testing conducted thus far suggests minimal interference on test sensitivity and time to detection against a wide range of product matrices (17, 18).
Although we attempted to evaluate a wide breadth of fungal isolates, ultimately, this study was limited to environmental strains specific to cGMP manufacturing and testing facilities on the NIH campus, with purposeful overrepresentation of Cladosporium sp. and Penicillium sp. given their prominence in our facilities. Other limitations included the lack of technical replicates to assess for reproducibility and ruggedness. It is possible that the optimal sterility culture conditions used at the NIH as presented here and in England et al. (5) may not be applicable to other manufacturing sites that have different environmental strains. Therefore, it is extremely important that alternative methods for sterility testing are validated specifically at each site using facility-specific environmental flora in addition to the six USP<71> defined QC isolates. Nevertheless, the results from this study have filled a much-needed gap in the literature by challenging test system performance with a variety of fungi beyond the two USP<71> fungal QC strains.
Importantly, the industry portion of the BacT/Alert, defined by an “i” at the beginning of the bottled media, was evaluated. This industry media utilizes an algorithm designed for the detection of slower growing environmental contaminants and it differs from the detection algorithm applied to clinical blood cultures collected in routine clinical aerobic (FA) and anaerobic media (FN). Furthermore, the performance of iLYM media for product sterility testing has not been rigorously studied, with only one previous report that was limited to the six USP<71> QC organisms (19). The success of iLYM 22.5°C combined with iFA Plus 32.5°C to detect fungal growth in our study suggests that there may be potential to apply this culture combination toward the detection of fungemia, which has generally low sensitivity (20). A study comparing performance of iLYM 22.5°C and iFA Plus 32.5°C against the preferred fungal isolator lysis centrifugation may identify a potential clinical application for iLYM beyond the food and pharmaceutical industries, and, as with all culture tests, sensitivity is dependent upon the amount of organism bioburden. Molecular methods such as next-generation sequencing (NGS) and targeted pan-fungal PCRs have been used widely in the clinical setting to provide sensitive detection of organisms typically from sterile fluids (21–24). The application of NGS or targeted pan-fungal and pan-bacterial PCRs in the cGMP field, however, has not been widely studied due to the highly regulated area of cGMP with regard to use of alternative non-USP<71> sterility tests and concerns regarding detection of nonviable organisms, which can lead to challenging risk-based decisions as to the safety and quality of the product for release and patient infusion. In contrast to the cGMP field, where the focus remains on product safety and quality alone, clinical laboratories generally have more flexibility in being able to discern and interpret results from highly sensitive molecular assays because results can be assessed alongside patient signs and symptoms and other supporting diagnostic tests. To our knowledge, no laboratory currently provides NGS, pan-fungal, or pan-bacterial testing in replacement of sterility testing culture for product release.
This study is important to share with the clinical community because many academic centers may already be conducting some form of cell therapy testing in the clinical microbiology laboratory, sometimes unknowingly due to inherited test systems. Inoculation of various cell products into blood culture bottles for incubation in the clinical microbiology lab has become common practice over the last 20 years. It is critical that clinical laboratorians conducting ATMP testing are aware of the different FDA regulatory oversight governing cell therapy product release testing (1, 2), and the risks associated with poor fungal sensitivity if additional cultures, beyond routine aerobic and anaerobic bottles, are not included in the testing algorithm.
In summary, ATMP sterility testing is continuing to adapt to advanced technologies for quick and accurate detection of product contaminants. The additional SDA culture used in the NIH testing algorithm considerably increased fungal sensitivity and was a justified approach given the poor sensitivity of the Bactec system used at the time. However, continued use of the SDA culture also led to several challenging risk-assessments regarding product safety following positive cultures that were likely attributable to laboratory handling. Our findings fill a significant gap in the mycology field that can impact both the pharmaceutical and clinical industries. We show here, for the first time, that an automated, closed-system, dual-bottle combination of iLYM 22.5°C and iFA Plus 32.5°C can provide highly sensitive fungal detection, statistically comparable to USP<71>, when tested against a diverse range of environmental fungal isolates. Furthermore, the proposed NIH algorithm of BacT/Alert iFA Plus 32.5°C and iFN Plus 32.5°C (current culture setup), with iLYM 22.5°C (replacing the manual SDA culture) maintains optimal sensitivity for the detection of bacterial contaminants as shown previously by England et al. (5). Notably, assessment of non-NIH environmental isolates was not evaluated here, so it is possible that higher incubation temperatures may be favored by some environmental organisms in studies conducted at other institutions. Individual site-specific validations are required for any laboratory applying non-USP<71> alternative sterility testing methods for ATMP quality control and release. Optimization of sterility testing culture conditions should include facility-specific flora in addition to the six QC organisms listed in USP<71>. Further validation testing in the presence of different product matrices is required at the NIH, but these proof-of-principle data are exciting and have potential to revolutionize biopharmaceutical testing.
ACKNOWLEDGMENTS
We thank Karen Frank and James Gebo for critical review of the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health. The content is solely our responsibility and does not represent the official views of the National Institutes of Health.
Footnotes
Supplemental material is available online only.
Contributor Information
Anna F. Lau, Email: anna.lau@nih.gov.
Kimberly E. Hanson, University of Utah
REFERENCES
- 1.Gebo JET, Lau AF. 2020. Sterility testing for cellular therapies: what is the role of the clinical microbiology laboratory? J Clin Microbiol 58:e01492-19. 10.1128/JCM.01492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gebo JET, East AD, Lau AF. 2021. A side-by-side comparison of clinical vs cGMP microbiology laboratory requirements for sterility testing of cellular and gene therapy products. Clin Microbiol Newsl, in press. [Google Scholar]
- 3.U.S. Pharmacopeia-National Formulary. 2020. USP<71> sterility tests in USP43-NF38 2S. U.S. Pharmacopeia, Rockville, MD. [Google Scholar]
- 4.Food and Drug Administration. 2020. Chemistry, manufacturing, and control (CMC) information for human gene therapy investigational new drug applications (INDs) - guidance for industry. U.S. Department of Health and Human Services, Food and Drug Administration, Washington, DC. [Google Scholar]
- 5.England MR, Stock F, Gebo JET, Frank KM, Lau AF. 2019. Comprehensive evaluation of compendial USP<71>, BacT/Alert Dual-T, and Bactec FX for detection of product sterility testing contaminants. J Clin Microbiol 57:e01548-18. 10.1128/JCM.01548-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cundell T, Drummond S, Ford I, Reber D, Singer D, members of the Pharmaceutical Microbiology Expert Discussion Group. 2020. Risk assessment approach to microbiological controls of cell therapies. PDA J Pharm Sci Technol 74:229–248. 10.5731/pdajpst.2019.010546. [DOI] [PubMed] [Google Scholar]
- 7.Golay J, Pedrini O, Capelli C, Gotti E, Borleri G, Magri M, Vailati F, Passera M, Farina C, Rambaldi A, Introna M. 2018. Utility of routine evaluation of sterility of cellular therapy products with or without extensive manipulation: best practices and clinical significance. Cytotherapy 20:262–270. 10.1016/j.jcyt.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Khuu HM, Patel N, Carter CS, Murray PR, Read EJ. 2006. Sterility testing of cell therapy products: parallel comparison of automated methods with a CFR-compliant method. Transfusion 46:2071–2082. 10.1111/j.1537-2995.2006.01041.x. [DOI] [PubMed] [Google Scholar]
- 9.Khuu HM, Stock F, McGann M, Carter CS, Atkins JW, Murray PR, Read EJ. 2004. Comparison of automated culture systems with a CFR/USP-compliant method for sterility testing of cell-therapy products. Cytotherapy 6:183–195. 10.1080/14653240410005997. [DOI] [PubMed] [Google Scholar]
- 10.Mastronardi C, Perkins H, Derksen P, denAdmirant M, Ramirez-Arcos S. 2010. Evaluation of the BacT/ALERT 3D system for the implementation of in-house quality control sterility testing at Canadian Blood Services. Clin Chem Lab Med 48:1179–1187. 10.1515/CCLM.2010.240. [DOI] [PubMed] [Google Scholar]
- 11.Mastronardi C, Yang L, Halpenny M, Toye B, Ramirez-Arcos S. 2012. Evaluation of the sterility testing process of hematopoietic stem cells at Canadian Blood Services. Transfusion 52:1778–1784. 10.1111/j.1537-2995.2011.03530.x. [DOI] [PubMed] [Google Scholar]
- 12.Ramirez-Arcos S, Evans S, McIntyre T, Pang C, Yi QL, DiFranco C, Goldman M. 2020. Extension of platelet shelf life with an improved bacterial testing algorithm. Transfusion 60:2918–2928. 10.1111/trf.16112. [DOI] [PubMed] [Google Scholar]
- 13.Arlt N, Rothe R, Sielaff S, Juretzek T, Peltroche H, Moog R. 2018. Sterility release testing of peripheral blood stem cells for transplantation: impact of culture bottles and incubation temperature. Transfusion 58:2918–2923. 10.1111/trf.14910. [DOI] [PubMed] [Google Scholar]
- 14.Panch SR, Bikkani T, Vargas V, Procter J, Atkins JW, Guptill V, Frank KM, Lau AF, Stroncek DF. 2019. Prospective evaluation of a practical guideline for managing positive sterility test results in cell therapy products. Biol Blood Marrow Transplant 25:172–178. 10.1016/j.bbmt.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reardon S. 2015. Contamination shuts down NIH pharmacy centre. Nature 10.1038/nature.2015.17703. [DOI] [Google Scholar]
- 16.Gandhi TK. 2016. Safety lessons from the NIH clinical center. N Engl J Med 375:1705–1707. 10.1056/NEJMp1609208. [DOI] [PubMed] [Google Scholar]
- 17.Adams A, Tuznik K, Gebo JET, East AD, Lau AF. 2021. Evaluation of different product matrices and test systems for the detection of microbial contaminants. Abstr ASM/FEMS World Microbe Forum [Google Scholar]
- 18.Lotfi R, Rojewski MT, Zeplin PH, Funk W, Pullig O, Noth U, Schrezenmeier H. 2020. Validation of microbiological testing of cellular medicinal products containing antibiotics. Transfus Med Hemother 47:144–151. 10.1159/000501284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bugno A, Saes DPS, Almodovar AAB, Dua K, Awasthi R, Ghisleni DDM, Hirota MT, de Oliveira WA, de Jesus Andreoli Pinto T. 2018. Performance survey and comparison between rapid sterility testing method and pharmacopoeia sterility test. J Pharm Innov 13:27–35. 10.1007/s12247-017-9303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campigotto A, Richardson SE, Sebert M, McElvania TeKippe E, Chakravarty A, Doern CD. 2016. Low utility of pediatric isolator blood culture system for detection of fungemia in children: a 10-year review. J Clin Microbiol 54:2284–2287. 10.1128/JCM.00578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G, Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology, Microbiology Resource Committee of the College of American Pathologists. 2017. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med 141:776–786. 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell SL, Simner PJ. 2019. Next-generation sequencing in clinical microbiology: are we there yet? Clin Lab Med 39:405–418. 10.1016/j.cll.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Wilson MR, Sample HA, Zorn KC, Arevalo S, Yu G, Neuhaus J, Federman S, Stryke D, Briggs B, Langelier C, Berger A, Douglas V, Josephson SA, Chow FC, Fulton BD, DeRisi JL, Gelfand JM, Naccache SN, Bender J, Dien Bard J, Murkey J, Carlson M, Vespa PM, Vijayan T, Allyn PR, Campeau S, Humphries RM, Klausner JD, Ganzon CD, Memar F, Ocampo NA, Zimmermann LL, Cohen SH, Polage CR, DeBiasi RL, Haller B, Dallas R, Maron G, Hayden R, Messacar K, Dominguez SR, Miller S, Chiu CY. 2019. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med 380:2327–2340. 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau A, Chen S, Sorrell T, Carter D, Malik R, Martin P, Halliday C. 2007. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J Clin Microbiol 45:380–385. 10.1128/JCM.01862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download JCM.01357-21-s0001.pdf, PDF file, 0.2 MB (187.9KB, pdf)
Table S2. Download JCM.01357-21-s0002.xlsx, XLSX file, 0.01 MB (14.5KB, xlsx)