ABSTRACT
This study describes the epidemiology of listeriosis in New Zealand between 1999 and 2018 as well as the retrospective whole-genome sequencing (WGS) of 453 Listeria monocytogenes isolates corresponding to 95% of the human cases within this period. The average notified rate of listeriosis was 0.5 cases per 100,000 population, and non-pregnancy-associated cases were more prevalent than pregnancy-associated cases (averages of 19 and 5 cases per annum, respectively). WGS data was assessed using multilocus sequencing typing (MLST), including core-genome and whole-genome MLST (cgMLST and wgMLST, respectively) and single-nucleotide polymorphism (SNP) analysis. Thirty-nine sequence types (STs) were identified, with the most common being ST1 (21.9%), ST4 (13.2%), ST2 (11.3%), ST120 (6.1%), and ST155 (6.4%). A total of 291 different cgMLST types were identified, with the majority (n = 243) of types observed as a single isolate, consistent with the observation that listeriosis is predominately sporadic. Among the 49 cgMLST types containing two or more isolates, 18 cgMLST types were found with 2 to 4 isolates each (50 isolates in total, including three outbreak-associated isolates) that shared low genetic diversity (0 to 2 whole-genome alleles), some of which were dispersed in time or geographical regions. SNP analysis also produced results comparable to those from wgMLST. The low genetic diversity within these clusters suggests a potential common source, but incomplete epidemiological data impaired retrospective epidemiological investigations. Prospective use of WGS analysis together with thorough exposure information from cases could potentially identify future outbreaks more rapidly, including those that may have been undetected for some time over different geographical regions.
KEYWORDS: Listeria monocytogenes, listeriosis, clinical, whole-genome sequencing, Listeria, genotyping
INTRODUCTION
Listeria monocytogenes is a Gram-positive bacterium that can cause disease in humans (listeriosis), with symptoms ranging from mild self-limiting gastroenteritis and fever in healthy individuals to severe sepsis, meningitis, and even death in immunocompromised persons, pregnant women, and the elderly (1). In 2013, expert elicitation was used to estimate the proportion of human cases of specific microbial diseases, including listeriosis in New Zealand (NZ), as well as the proportion of the foodborne burden that was due to the transmission by specific foods (2). Expert elicitation refers to a systematic approach of obtaining and synthesizing subjective judgement from experts on a subject where there is uncertainty as a result of insufficient data. For listeriosis, it was estimated that 88% (95th percentile credible interval, 58% to 99%) of listeriosis incidences in New Zealand were due to foodborne transmission, which was comparable to that in other countries (2).
L. monocytogenes is ubiquitous in the environment, in animals, and in humans and can contaminate all raw foods and ingredients (1). Foods of most concern for listeriosis are those in which L. monocytogenes can multiply. Ready-to-eat (RTE) foods present a significant risk as these products do not undergo further listeriocidal treatment such as cooking before consumption, and growth of L. monocytogenes can occur during refrigerated storage (3, 4). Consumption of contaminated foods such as unpasteurized milk or cheese, contaminated pasteurized soft cheeses, contaminated vegetables, deli meats and pâté, or shellfish have been major sources of infection internationally (1, 5).
In New Zealand, listeriosis has been a notifiable disease since 1983 and is a disease that is often sporadic, with outbreaks rarely reported. The sources of contamination can be difficult to identify due to the variable and often long incubation periods (ranging from 1 to 70 days) and the condition of patients (typically severely ill), thus affecting the quality and completeness of food histories (6, 7). All listeriosis cases are administered a standardized questionnaire by public health authorities to identify key risk factors. Additional food exposure information may also be obtained by local authorities. Common food exposures between cases can prompt an extended questionnaire and multiagency investigation with the New Zealand Food Safety, Ministry for Primary Industries (MPI), the regulator for the New Zealand food industry. A guidance document on L. monocytogenes management for food producers is available through MPI which includes the microbiological limits for L. monocytogenes in food products, management of contamination, hazard analysis, shelf-life information, and food safety documentation (8). There is currently no mandatory requirement for industry to perform molecular characterization of L. monocytogenes identified within their factory environments.
In New Zealand, L. monocytogenes isolates from invasive listeriosis cases are sent to the National Reference Laboratory at the Institute of Environmental Science and Research (ESR) for typing. Since 2017, New Zealand has transitioned from typing L. monocytogenes isolates using pulsed-field gel electrophoresis (PFGE) to using whole-genome sequencing (WGS). Internationally, the implementation of WGS has resulted in the identification of an increased number of clusters and outbreaks with fewer cases and has enabled linking of human illness to specific foods or production environments with greater confidence than ever before (6, 9).
In this work, the epidemiology of listeriosis in New Zealand over a 20-year period is described along with a retrospective WGS analysis on 453 L. monocytogenes clinical isolates in an effort to identify what is required to enable successful real-time surveillance of listeriosis in New Zealand.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All L. monocytogenes strains were recovered from −80°C storage by streak plating onto Columbia blood agar (CBA) incubating the plates at 37°C for 24 to 48 h. A single colony was inoculated into 10 ml tryptone soya broth and incubated at 37°C for 18 h prior to DNA extraction. All L. monocytogenes isolates are listed in Data Set S1 in the supplemental material.
DNA extraction, library preparation, and genome sequencing.
For each L. monocytogenes isolate, 1 ml of broth culture was used for DNA extraction using the Qiagen DNeasy blood and tissue kit (Qiagen, Hilden, Germany) with modifications. Cell lysis using 20 mg/ml lysozyme was performed at 37°C for 2 h, and final elutions were performed in 50 μl RNase-free water. DNA quality and concentration were assessed using a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA), Qubit, and PicoGreen (Quant-iT; Thermo Fisher Scientific). Sequencing libraries containing 1 ng of DNA were prepared using Nextera XT chemistry (Illumina, San Diego, CA, USA) for 250 bp or 150-bp pair-end sequencing on an Illumina MiSeq or NextSeq sequencer, respectively, according to the manufacturer’s recommendations (Illumina).
Whole-genome sequencing analysis.
Initial sequence quality and species identification were determined using the Nullarbor pipeline (10). Genome assemblies were performed using SPAdes within BioNumerics version 7.6.3 (Applied Maths NV, Belgium). Assembly-based allele calls were performed within BioNumerics using multilocus sequence typing (MLST) (7 loci [11]), core genome MLST (cgMLST) (1,748 loci [12]), and whole-genome MLST (wgMLST) (4,797 loci; using the scheme within BioNumerics). MLST profiles were classified into sequence types (STs) and grouped into clonal complexes (CCs) as previously described (11). cgMLST profiles were grouped into cgMLST types (CTs) and sublineages (SLs), using the cutoffs of 7 and 150 allelic mismatches, respectively, as previously described (12). Profiles and types were defined by using international nomenclature (12), available at BIGSdb-Lm (https://bigsdb.pasteur.fr/listeria/). Whole-genome single-nucleotide polymorphism (SNP) analysis was performed within BioNumerics using strict SNP filtering options. Separate SNP analyses were performed for selected cgMLST types, where the isolate corresponding to the first clinical case of that cgMLST type was selected as the reference sequence for the group (13, 14). Cluster analyses were performed using cgMLST, wgMLST, and/or SNP data (categorical data values) and the single-linkage algorithm.
Pulsed-field gel electrophoresis.
A total of 371 isolates outlined in Data Set S1 were previously PFGE typed using AscI and ApaI restriction enzymes as per standard protocols outlined by the Centers for Disease Control and Prevention, Atlanta, GA, PulseNet and analyzed using procedures previously outlined (15). The discriminatory power of cgMLST and PFGE was compared using Simpson’s index of diversity (16).
Epidemiological case information.
ESR undertakes the surveillance of notifiable diseases in New Zealand, including listeriosis, on behalf of the Ministry of Health. Data used in this study were obtained from New Zealand’s national notifiable disease database (EpiSurv). The data used to generate Fig. 1 to 3 were extracted from EpiSurv on 25 August 2020. A confirmed case requires definitive laboratory evidence (isolation of L. monocytogenes or detection of L. monocytogenes nucleic acid) in conjunction with a clinically compatible illness (17). Cases were classified as pregnancy-associated if illness occurred in a pregnant woman, fetus, or infant aged ≤28 days. Cases were followed up promptly by health protection staff in local public health units upon notification, but protocols may have differed between public health units.
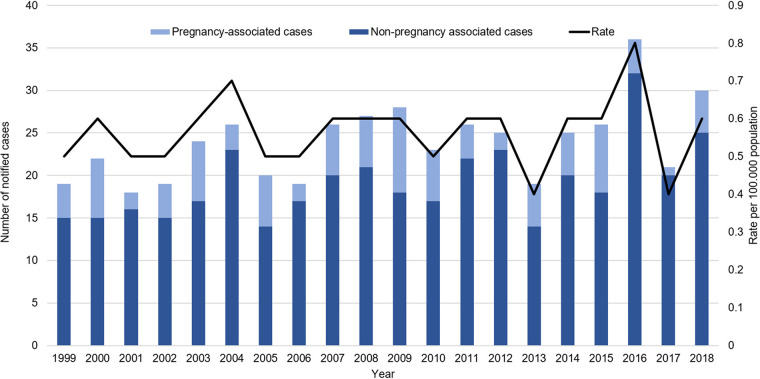
FIG 1.
Distribution of numbers of notified listeriosis cases and rates per 100,000 population between 1999 and 2018. Rate was calculated as the number of cases per 100,000 population, based on the Statistics New Zealand mid-year population estimates.
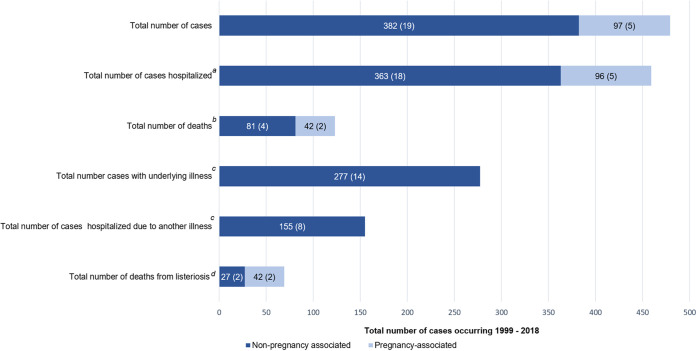
FIG 2.
Total numbers of pregnancy- and non-pregnancy-associated cases (average per annum) in New Zealand between 1999 and 2018 as recorded in EpiSurv. Numbers on the bars are the total numbers of pregnancy- or non-pregnancy-associated cases (and the average numbers of cases per annum) for each outcome recorded. a, total of 377 non-pregnancy-associated cases (average 19 cases per annum) with hospitalization status recorded; b, deaths occurring during a period after the reporting date may not be captured in EpiSurv. For pregnancy-associated deaths, one case each in 2004 and 2006 was an adult, and all other deaths were fetal. c, data not available for pregnancy-associated cases; d, data not collected in EpiSurv prior to 2007, all pregnancy-associated deaths were associated with the disease. For non-pregnancy-associated cases, 250 cases were recorded with this information, and the average number of cases was calculated using values available for 14 years.
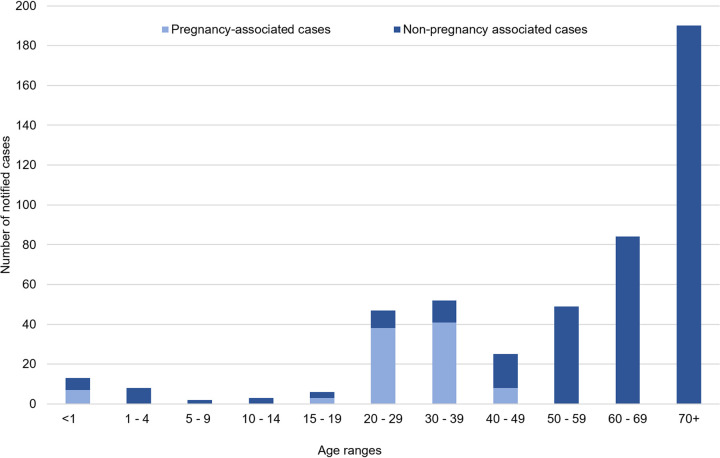
FIG 3.
Distribution of numbers of notified listeriosis cases between 1999 and 2018 according to case age. Cases are classified as pregnancy-associated if illness occurred in a pregnant woman, fetus, or infant aged ≤28 days.
Data availability.
Raw sequence files for all isolates analyzed in this study are available on National Centre for Biotechnology Information (NCBI) short read archive (SRA) with BioProject number PRJNA741099 (Data Set S1). A total of 20 isolates were sequenced as a part of the University of California, Davis, 100K Pathogen Genome Project (18), with SRA accession numbers outlined in Data Set S1.
RESULTS
Epidemiology of human listeriosis in New Zealand 1999 to 2018.
Between 1999 and 2018, the notification rate of listeriosis in New Zealand ranged between 0.4 and 0.8 per 100,000 population (Fig. 1). The majority of notified cases were non-pregnancy-associated listeriosis (64 to 95%; average of 19 cases per annum) (Fig. 1 and 2). The hospitalization status was available for 99% (377 of 382) of non-pregnancy-associated cases, of which 96% (363 of 377; average of 18 cases per annum) were hospitalized (Fig. 2). However, of those hospitalized cases, 43% (155 of 363; average of 8 cases per annum) were hospitalized due to another illness. Since 2007, when data of deaths attributed to listeriosis were recorded, 27 of 250 non-pregnancy-associated cases with this information (11%; average of 2 cases per annum) were for patients that died due to listeriosis. Approximately 50% of the total number of non-pregnancy-associated cases (190 of 382) between 1999 and 2018 were for patients 70 years or older (Fig. 3).
The average number of notified pregnancy-associated listeriosis cases between 1999 and 2018 was five cases per annum (5% to 36% of the total number of cases per annum) (Fig. 2). With the exception of one case where the hospitalization status was unknown, all pregnancy-associated cases were hospitalized. All pregnancy-associated deaths were recorded as being attributed to listeriosis, where 43% (42 of 97; average of 2 cases per annum) of cases resulted in death of the fetus and two of these cases reported the death of the mother.
Molecular diversity of NZ clinical L. monocytogenes.
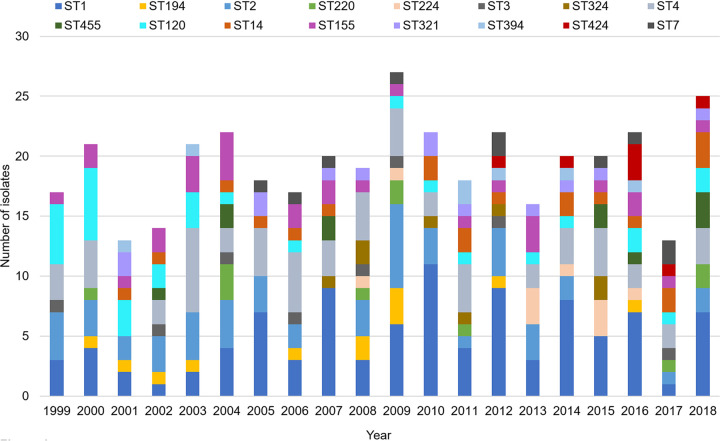
Whole-genome sequencing was performed on 453 laboratory-confirmed L. monocytogenes clinical cases in New Zealand between 1999 and 2018, representing 95% of total notified cases. Quality control statistics for all genomes used in the study are outlined in Data Set S1 in the supplemental material. This data set includes 15 isolates associated with three recognized national listeriosis outbreaks and one suspected cluster of five pregnancy-associated cases in 2009 (5, 19, 20). Sixty-three percent of isolates (n = 286) belonged to phylogenetic lineage I, while the remaining 35% (n = 157) and 2% (n = 10) belonged to phylogenetic lineages II and III, respectively. Thirty-nine STs were identified among the NZ data set. The five most frequent STs identified were ST1 (n = 99; 21.9%), ST4 (n = 60; 13.2%), ST2 (n = 51; 11.3%), ST120 (n = 31; 6.1%), and ST155 (n = 29, 6.4%) (Fig. 4). ST1 has been observed every year between 1999 and 2018, while ST2, ST4, ST120, and ST155 appeared for 17 to 19 years between 1999 and 2018. ST324, ST224, and ST424 were observed from 2007, 2008, or 2012 onwards, respectively. At the level of core genome, 291 different cgMLST types were identified. The majority of cgMLST types assigned (243 of 292 [83%]) contained a single isolate, whereas 49 cgMLST types contained two or more isolates (Tables 1 and 2). The most prevalent cgMLST types detected were L1-SL4-ST4-CT187 (n = 24; 5%), L2-SL8-ST120-CT786 (n = 23; 5%), and L1-SL1-ST1-CT4574 (n = 10; 2%).
FIG 4.
Distribution of sequence types (STs) among L. monocytogenes isolates between 1999 and 2018. ST inferred using whole-genome sequence and multilocus sequence typing.
TABLE 1.
Core-genome multilocus sequence type groups containing two isolates
| cgMLST typea | Region(s)b | Yr(s) | Pulsotype(s)c | Maximum no. of differencesd |
|
|---|---|---|---|---|---|
| cgMLST | wgMLST | ||||
| L1-SL2-ST2-CT4548 | L, M | 2012, 2013 | Asc0023:Apa0031, NT | 0 | 0 |
| L2-SL321-ST321-CT5870 | B, C | 2001 | Asc0051:Apa0032 | 0 | 0 |
| L1-SL4-ST4-CT5903 | F | 2009 | Asc0021:Apa0023 | 0 | 0 |
| L1-SL1-ST1-CT5954 | B, C | 2005, 2005 | Asc0058:Apa0005 | 0 | 0 |
| L1-SL220-ST220-CT5993 | F, I | 2009 | Asc0036:Apa0039 | 0 | 0 |
| L1-SL1-ST1-CT3416 | S | 2010, 2011 | Asc0045:Apa007 | 0 | 1 |
| L1-SL4-ST4-CT4534 | C, E | 2011 | Asc0021:Apa0023 | 0 | 2 |
| L1-SL1-ST1-CT5963 | S | 2001 | Asc0078:Apa0006 | 0 | 2 |
| L1-SL1-ST1-CT5950 | C, D | 2007 | Asc0045:Apa0152 | 1 | 1 |
| L2-SL8-ST120-CT5874 | Q | 2001 | Asc0002:Apa0002 | 1 | 2 |
| L2-SL7-ST7-CT5986 | B, D | 2006, 2007 | Asc0015:Apa0026 | 1 | 2 |
| L1-SL2-ST2-CT3414 | H | 2012, 2013 | Asc0023:Apa0031 | 1 | 4 |
| L2-SL37-ST37-CT4583 | D | 2011, 2017 | Asc0055a:Apa0057, NT | 3 | 5 |
| L2-SL155-ST155-CT2607 | E, G | 2003, 2004 | Asc0126:Apa0041 | 3 | 11 |
| L1-SL1-ST1-CT4552 | B, D | 2011, 2012 | Asc0040:Apa0045, Asc0040:Apa, NT | 4 | 6 |
| L1-SL2-ST2-CT5917 | C, F | 2002 | Asc0023:0031 | 5 | 8 |
| L1-SL2-ST2-CT5936 | C, J | 2004, 2008 | Asc0063:Apa0012, Asc0077:Apa0012 | 5 | 12 |
| L1-SL220-ST220-CT4597 | B, D | 2000, 2011 | Asc0036:Apa0039 | 5 | 13 |
| L2-SL155-ST155-CT4611 | E, F | 2011 | Asc0055a:Apa0057, Asc0044:Apa0045 | 7 | 9 |
| L2-SL14-ST14-CT4622 | E, G | 2011, 2012 | Asc0050:Apa0046, Asc0059:Apa0046a | 7 | 11 |
| L2-SL155-ST155-CT4608 | O, Q | 2015, 2016 | Asc0037:Apa0044, NT | 9 | 16 |
| L1-SL2-ST2-CT5934 | B, C | 1999, 2009 | Asc0023:Apa0071, Asc0082:Apa0129 | 9 | 23 |
| L2-SL321-ST321-CT4591 | B | 2007, 2014 | Asc00001:Apa0055 | 6 | 8 |
| L2-SL14-ST14-CT6025 | F, T | 2004, 2010 | Asc0038:Apa0046 | 6 | 10 |
| L2-SL101-ST101-CT6007 | D, NS | 2000, 2001 | Asc007:Apa0042, Asc0006:Apa0056 | 6 | 12 |
| L2-SL204-ST204-CT638 | B, R | 2006, 2013 | Asc0055:Apa0096, Asc0055:Apa0107 | 13 | 28 |
The lineage (L)-sublineage (SL)-multilocus sequence type (ST)-core genome multilocus sequence types (CTs) were inferred from WGS data and defined by using international nomenclature (13).
Regions within New Zealand that have been anonymized. NS, no region stated for the case.
Pulsotype assigned as a part of the PulseNet New Zealand/Aotearoa database. NT, not tested.
Maximum allele differences observed for isolates within a cgMLST type using cgMLST and whole-genome MLST.
TABLE 2.
Core-genome multilocus sequence type groups containing more than two isolates
| cgMLST typea | No. (%) of isolates in CTb | Region(s)c | Yrs | Pulstotyped | No. of allele differences (maximum and minumum)e |
||
|---|---|---|---|---|---|---|---|
| cgMLST | wgMLST | SNP | |||||
| L1-SL2-ST2-CT5919 | 3f (0.7) | B, D, K | 2000, 2004, 2005 | Asc0030:Apa0025 | 0–2 | 7–10 | 9–10 |
| L1-SL1-ST1-CT2373 | 3 (0.7) | D, K, S | 2007, 2010 | Asc0058:Apa0005 | 0–2 | 0–2 | 0–2 |
| L1-SL224-ST224-CT3426 | 7 (1.6) | A, B, C, D, E, F, M | 2012, 2013, 2014, 2015, 2016 | Asc0060:Apa0124a, Asc0021:Apa0023, Asc0119:Apa0104, NT | 0–2 | 0–4 | 0–5 |
| L2-SL155-ST155-CT4612 | 8 (1.7) | E, F, G, L, K, M, N, S | 2000, 2003, 2004, 2006, 2007, 2013 | Asc0037:Apa0044 | 0–4 | 0–12 | 0–16 |
| L1-SL315-ST194-CT5908 | 5 (1.1) | S | 2008, 2009 | Asc00018:Apa006 | 0–6 | 1–8 | 1–10 |
| L2-SL8-ST120-CT786f | 23 (5.1) | A, B, C, D, F, K, L, M, O, Q, R, S | 1999, 2000, 2001, 2003, 2004, 2006, 2004, 2010, 2011, 2016, 2017, 2018 | Asc0011:Apa0002, Asc0069:Apa0002, Asc0002:Apa0002, Asc0028:Apa0002, Asc0023:Apa0002 | 0–9 | 0–14 | 0–15 |
| L1-SL1-ST1-CT4553 | 4 (0.9) | D, T | 2008, 2010, 2012, 2015 | Asc0045:Apa0013 | 1–4 | 5–12 | 5–7 |
| L1-SL1-ST1-CT4576 | 8 (1.7) | C, D, K, Q | 2003, 2005, 2009, 2011, 2014, 2016 | Asc0003:Apa0063, Asc0003:Apa0063a | 1–7 | 2–15 | 2–19 |
| L1-SL2-ST2-CT5935 | 5 (1.1) | B, C, G, K, Q, S | 1999, 2000, 2002, 2004, 2005, 2009 | Asc0023:Apa0065, Asc0023:Apa0071, Asc0063:Apa0012 | 1–7 | 7–15 | 8–15 |
| L1-SL4-ST4-CT5890 | 5 (1.1) | A, B | 2000, 2006, 2007 | Asc0021a:Apa0023 | 1–8 | 3–20 | 2–21 |
| L1-SL315-ST194-CT4541 | 6 (1.3) | B, C, D, E | 2001, 2002, 2003, 2006, 2012, 2016 | Asc0072:Apa0028 | 2–7 | 2–15 | 2–15 |
| L1-SL4-ST4-CT187 | 24 (5.3) | A, B, C, D, E, J, K, L, M, N, O, P, Q, S, NS | 1999, 2000, 2002, 2003, 2005, 2006 | Asc00021:Apa0023, Asc0021a:Apa0023, Asc0037:Apa0044, NT | 2–9 | 3–17 | 3–22 |
| L2-SL321-ST321-CT691 | 6 (1.3) | C, D, F, G, K, O | 2005, 2008, 2010, 2011, 2018, | Asc0128:Apa0001, Asc0031:Apa0001, Asc0001:Apa0001, Asc0074:Apa0001 | 3–7 | 8–17 | 8–16 |
| L2-SL155-ST155-CT852 | 6 (1.3) | B, D, K, F | 2006, 2009, 2012, 2017 | Asc0037:Apa0044, NT | 3–10 | 4–29 | 6–80 |
| L2-SL18-ST18-CT2990 | 3 (0.7) | A, S | 2003, 2014, 2018 | Asc0047:Apa0117, NT | 4–7 | 12–18 | 13–23 |
| L1-SL1-ST1-CT4574 | 10 (2.2) | B, C, D, L, M, Q | 2008, 2009, 2013, 2014, 2015, 2018 | Asc0088:Apa0126, Asc0081:Apa0063, Asc0068:Apa0064, NT | 4–7 | 9–11 | 6–20 |
| L2-SL9-ST9-CT4539 | 3 (0.7) | L, S | 2014, 2016, 2018 | Asc0046:Apa0058, NT | 5 | 11–15 | 11–14 |
| L1-SL1-ST1-CT4562 | 3 (0.7) | B, E, Q | 1999, 2004, 2011 | Asc0010:Apa0008 | 5 | 9–12 | 13–17 |
| L1-SL1-ST1-CT2810 | 4 (0.9) | B, C, L, N | 1999, 2003, 2005, 2015, | Asc0083:Apa0013, Asc0067:Apa0021, NT | 5–7 | 11–27 | 11–103 |
| L2-SL7-ST7-CT3420 | 3 (0.7) | E | 2005, 2012, 2015 | Asc0043:Apa0026, Asc0043a:Apa0026 | 5–7 | 13–19 | 16–24 |
| L1-SL324-ST324-CT4618 | 5 (1.1) | C, D, E | 2008, 2011, 2012 2015 | Asc0161:Apa0019, Asc0161a, Apa0119, Asc0161:Apa0132 | 6–11 | 12–21 | 12–27 |
| L2-SL20-ST424-CT2456 | 3 (0.7) | E, F | 2012, 2016, 2018 | Asc0094:Apa0104, Asc0119:Apa0104, NT | 9–11 | 27–36 | ND |
| L2-SL14-ST14-CT956 | 10 (2.2) | B, C, E, F, N, Q, R, | 2002, 2005, 2007, 2010, 2011, 2014, 2015, 2017, 2018, | Asc0059:Apa0046a, Asc0005:Apa0046a, Asc0038:Apa0046 | 10–12 | 11–38 | ND |
The lineage (L)-sublineage (SL)-multilocus sequence type (ST)-core genome multilocus sequence types (CTs) were inferred from WGS data and defined by using international nomenclature (13).
Percentage of isolates within the cgMLST type from total number of isolates in study (n = 453).
Regions within New Zealand that have been anonymized. NS, no region stated for the case.
Pulsotype assigned as a part of the PulseNet Aotearoa/New Zealand database; NT, not tested.
Minimum and maximum allele differences observed for isolates within a cgMLST type using cgMLST and whole-genome MLST; single-nucleotide polymorphism (SNP) analysis was performed for cgMLST groups that had fewer than 7 core genome MLSTs.
Single case from CT5919 included in an outbreak that also involved three cases identified within L2-SL8-ST120-CT786.
A comparison between cgMLST and PFGE typing (pulsotypes previously assigned are outlined in Data Set S1) resulted in Simpson’s indices of 0.991 and 0.978, respectively. A total of 152 pulsotypes were identified among the 371 isolates with PFGE typing data, but 281 cgMLST types were identified among the same isolates. Only six pulsotypes accounted for 30% of isolates (n = 111), but 63 cgMLST types were identified among these same isolates. Twenty pulsotypes accounted for 50% of the isolates (n = 188), but 109 cgMLST types were identified among the same isolates.
Retrospective genomic analysis of outbreak-related isolates.
(i) Invasive listeriosis cases linked to a febrile listeriosis outbreak attributed to RTE meat, 2000. An outbreak of 31 febrile noninvasive gastrointestinal listeriosis cases was attributed to RTE corned meat product from a single manufacturer (20). During the outbreak, PFGE typing using SmaI and ApaI was performed by an Australian laboratory, and all clinical and RTE meat isolates associated with the outbreak were reported to have indistinguishable PFGE profiles.
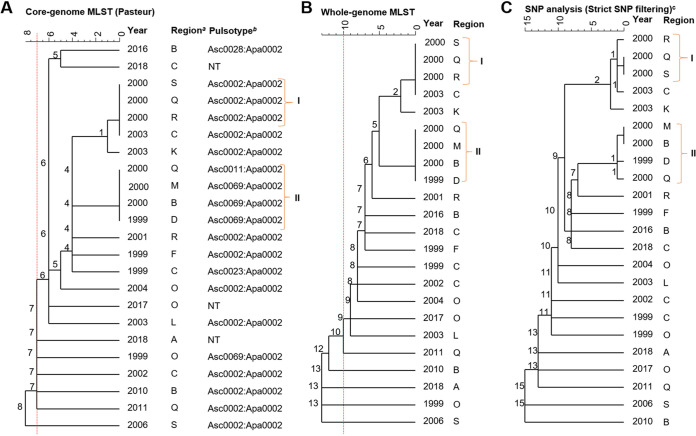
Four invasive listeriosis cases in the same year were epidemiologically attributed to consumption of an RTE corned meat product from the same manufacturer identified for the febrile gastrointestinal listeriosis cases. Historical records indicate that the isolates obtained from the invasive listeriosis cases were not PFGE typed by the Australian laboratory at the time of the outbreak. In New Zealand, PFGE typing was implemented in 2002 and performed retrospectively on an annual basis; therefore, it was unlikely to have been used to inform epidemiological investigations of these invasive listeriosis cases at the time of the outbreak. Retrospective PFGE typing performed in New Zealand showed that three of the four isolates were of the same pulsotype (Asc0002:Apa0002). Using retrospective WGS analysis, these same three isolates were also identified as the same cgMLST type, L2-SL8-ST120-CT786, and shared no cgMLST or wgMLST allelic differences and 0 to 1 SNP differences (Fig. 5, cluster I), confirming their close genetic relationship.
FIG 5.
Dendrograms (single linkage) representing the genetic relationship between L. monocytogenes isolates identified as L2-SL8-ST120-CT786 by using core genome multilocus sequence typing (cgMLST) (A), whole-genome MLST (wgMLST) (B), and single-nucleotide polymorphism (SNP) (C). Numbers on the branches indicate the allele/SNP differences between isolates. Isolates indicated with I are confirmed outbreak isolates. Those isolates indicated with II had low genetic diversity and occurred at approximately the same time as the outbreak isolates (I). a, regions within New Zealand have been anonymized; b, pulsotype assigned as a part of the PulseNet Aotearoa/New Zealand database; NT, not tested; c, SNP analysis was performed using the first clinical case (1999) for L2-SL8-ST120-CT786 as the reference. Red dotted lines represent cutoffs of 7 and 10 core and whole-genome allele differences, respectively (A and B), as previously proposed (6, 13).
Retrospective WGS analysis showed that there were an additional four isolates associated with invasive listeriosis cases from 1999 to 2000 that identified as L2-SL8-ST120-CT786 and shared 0 to 1 core and whole-genome allele and SNP differences between them (Fig. 5, cluster II). Cluster II isolates were not linked to the outbreak at the time due to the limited epidemiological data, and no additional links were identified between the cases. Pulsotype Asc0002:Apa0002 identified for the cluster I outbreak isolates was one of the most frequently observed pulsotypes in the PulseNet New Zealand/Aotearoa database. Cluster II isolates were identified as one of two pulsotypes different from those in the cluster I outbreak isolates, but these pulsotypes only differed by a single restriction enzyme fragment for the AscI PFGE pattern (data not shown).
A fourth invasive listeriosis case was previously included in this outbreak due to consumption of a corned beef product. However, retrospective PFGE and cgMLST analysis identified this isolate as a different genotype (Apa0025:Asc0030) and cgMLST type (L1-SL2-ST2-CT5919) compared to those of the other three epidemiologically linked isolates, suggesting this case may not have had the same infection source. Two other isolates from 2004 and 2005 were also identified as cgMLST type L1-SL2-ST2-CT5919 and differed from the 2000 isolate by 0 to 2 cgMLST and 7 to 10 wgMLST alleles and SNPs (Fig. 6 and 7; Table 2). However, a retrospective analysis did not find any associations between the cases. The food and clinical isolates associated with the febrile listeriosis cases were no longer available for retrospective WGS analysis. Therefore, it was not possible to genetically confirm the epidemiological link between the invasive cases and the febrile outbreak cases.
FIG 6.
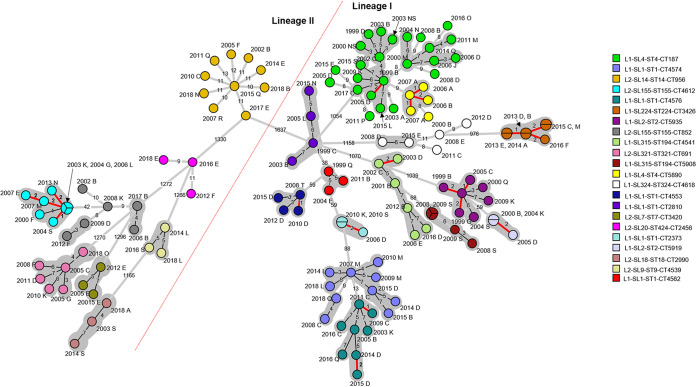
Minimum spanning tree based on core genome multilocus sequence typing of all L. monocytogenes within core genome multilocus sequence types containing more than two isolates (excluding L2-SL8-ST120-CT786). Branch length scaling is logarithmic. The numbers of allele differences between isolates are shown on the branches. The year and region code are displayed next to each isolate. NS is used where a region was not stated. Isolates within a gray-shaded area share seven or less core genome allele differences between them, as previously proposed as a threshold (13). Isolates within a pie have zero core differences, and isolates that share 1 to 2 allele differences are highlighted with a red branch. A red dotted line separates lineages I and II.
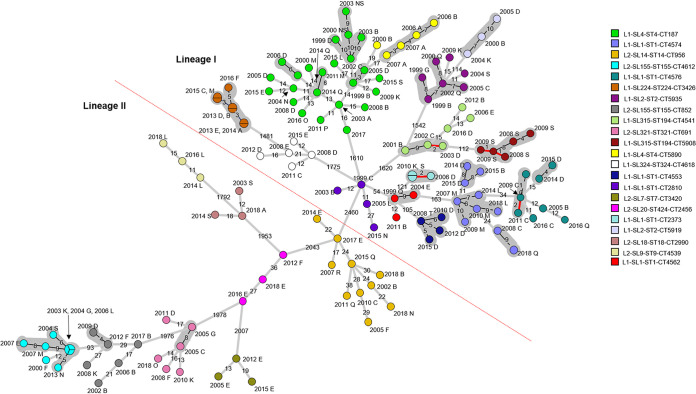
FIG 7.
Minimum spanning tree based on whole-genome multilocus sequence typing of all L. monocytogenes within core genome multilocus sequence types containing more than two isolates (excluding L2-SL8-ST120-CT786). Branch length scaling is logarithmic. The numbers of allele differences between isolates are shown on the branches. The year and region code are displayed next to each isolate. NS is used where a region was not stated. Isolates within a gray-shaded area share 10 or less whole-genome allele differences between them, as previously proposed as a threshold (6). Isolates within a pie have zero core differences, and isolates that share 1 to 2 allele differences are highlighted with a red branch. A red dotted line separates lineages I and II.
(ii) Smoked fish multistrain outbreak, 2009.
This outbreak involved two cases (including one pregnancy-associated case) implicating smoked fish based on epidemiological information. Retrospective PFGE typing of the isolates resulted in two different pulsotypes: Asc0078:Apa0006, which was previously observed for six other clinical isolates between 2001 and 2009, and Asc0030:Apa0023a, which was not previously observed (Data Set S1). Retrospective WGS analysis of the two isolates identified two different cgMLST types: L1-SL2-ST2-CT5914 and L1-SL1-ST1-CT5964, both of which were the sole isolates of that cgMLST type in the data set (Data Set S1). These two isolates had significant core genome and whole-genome (1,119 and 1,682, respectively) differences between them. L. monocytogenes was isolated from the food producer implicated at the time of the outbreak, but PFGE typing results for these food isolates were not identified. The food isolates were not retained and therefore not available for retrospective WGS analysis.
(iii) Suspected pregnancy-associated cluster, 2009.
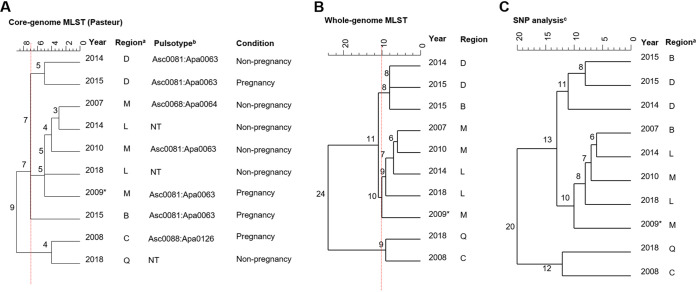
A retrospective study describing the epidemiology of pregnancy-associated listeriosis in New Zealand identified a cluster of five pregnancy-associated cases in early-to-mid-September 2009 (19). Retrospective PFGE typing results showed four different pulsotypes (Data Set S1) among the five isolates associated with the cases, but no epidemiological link between cases was identified at the time. Retrospective cgMLST analysis of the five isolates identified four different cgMLST types, two of which (L1-SL1-ST1262-CT3418 [1 isolate] and L1-SL4-ST4-CT5903 [2 isolates] were the sole isolates identified for their respective cgMLST types (Data Set S1). The two isolates identified as L1-SL4-ST4-CT5903 shared no core genome or whole-genome allelic differences (Table 1). The case investigation findings noted that the cases were possibly linked, but no common food source was identified (21).
One isolate within this pregnancy-associated cluster identified as cgMLST type L1-SL220-ST220-CT5993 had no core genome or whole-genome allele differences compared to the isolate from a non-pregnancy associated case that occurred 3 months later in a different region in New Zealand (Table 1). These two isolates are the only two isolates identified for this cgMLST type within the data set, and retrospective analysis of the cases did not find any further associations between the cases.
The remaining isolate within this pregnancy-associated cluster was identified as cgMLST type L1-SL1-ST1-CT4574. This cgMLST type was identified for nine other isolates within the data set, including three other pregnancy-associated cases occurring in different years and regions of New Zealand (Fig. 8). This 2009 pregnancy-associated isolate shared 5 to 10 core- and whole-genome allele and SNP differences with an isolate from a non-pregnancy associated case in 2018.
FIG 8.
Dendrograms (single linkage) representing the genetic relationship between L. monocytogenes isolates identified as L1-SL1-ST1-CT4574 using core genome multilocus sequence typing (cgMLST) (A), whole-genome MLST (wgMLST) (B), and single-nucleotide polymorphism (SNP) (C). Number on the branches indicate the allele/SNP differences between isolates. a, regions within New Zealand have been anonymized; b, pulsotype assigned as a part of the PulseNet New Zealand/Aotearoa database; NT, not tested; c, SNP analysis was performed using the first clinical case (2007) for L1-SL1-ST1-CT4574 as the reference. Red dotted lines represent cutoffs of 7 and 10 core- and whole-genome allele differences, respectively (A and B), as previously proposed as thresholds (6, 13). *, isolate corresponding to case within the suspected pregnancy-associated cluster, 2009.
(iv) RTE meat outbreak, 2012.
This outbreak involved four cases, including two deaths, and was attributed to RTE meats supplied to a hospital from a single manufacturer (5). A retrospective study compared the PFGE typing and retrospective WGS analysis for this outbreak (5). All four isolates from cases were different cgMLST types (L1-SL9-ST9-CT3406, L2-SL101-ST101-CT3423, L2-SL7-ST7-CT732, and L1-SL1-ST1-CT3417) and were the sole isolates for each of their cgMLST types in the data set (Data Set S1).
Retrospective genomic analysis highlights previously unidentified clusters.
Table 1 outlines the 26 cgMLST types that contained only two isolates each. Eleven of the 26 cgMLST types were observed to have isolates that shared very low levels of core genome and whole-genome MLST (0 to 2) allele differences between them. While the cases in seven of these cgMLST types were from different regions, they were geographically close.
Table 2 outlines the cgMLST types that contained more than two isolates and the ranges of cgMLST, wgMLST, and SNP differences between isolates within each cgMLST type. Cluster diagrams representing core genome and whole-genome MLST analysis for all cgMLST types containing more than two isolates (except for L2-SL8-ST120-CT786 [Fig. 5]) are also shown in Fig. 6 and 7, respectively. It was observed that within 12 of these cgMLST types, including L2-SL8-ST120-CT786, there were between 2 and 7 isolates that shared a very low level (0 to 2) of core genome allele differences between them (Fig. 6). Seven of these cgMLST types, including L2-SL8-ST120-CT786 (Fig. 5), were also observed to have very low levels (0 to 2) of whole-genome allelic differences (Fig. 7). SNP analysis produced results similar to those of wgMLST, but eight cgMLST types, including those outlined in Fig. 9 and L2-SL8-ST120-CT786 shown in Fig. 5 and L1-SL1-ST1-CT2373 (Table 2), were observed to have isolates with very low-level (0 to 2) SNP differences between them.
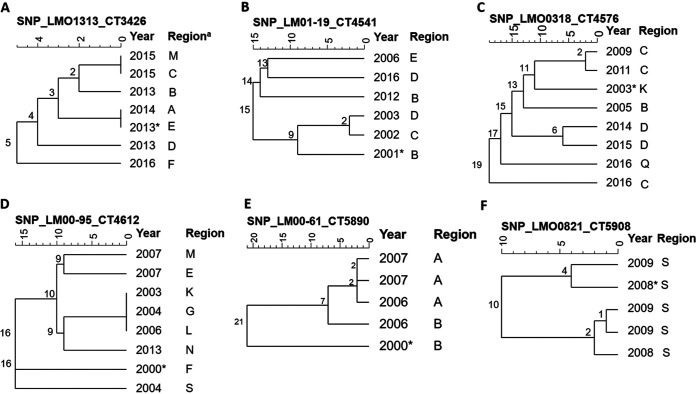
FIG 9.
Dendrograms (single linkage) representing the genetic relationship between L. monocytogenes isolates identified as cgMLST types L1-SL224-ST224-CT3426 (A), L1-SL315-ST194-CT4541 (B), L1-SL1-ST1-CT4576 (C), L2-SL155-ST155-CT4612 (D), L1-SL4-ST1624-CT5890 (E), and L1-SL315-ST194-CT5908 (F) using single-nucleotide polymorphism (SNP) analysis. Numbers on the branches indicate the SNP differences between isolates. These cgMLST types had more than two isolates and were observed to have isolates that shared very low levels (0 to 2) of SNP differences between them. a, regions within New Zealand have been anonymized; *, first clinical case for the cgMLST type, which was used as reference for the remaining isolates in the cgMLST type.
With the exception of isolates within L1-SL4-ST4-CT5903 and L2-SL8-ST120-CT786 (associated with the suspected pregnancy cluster or febrile listeriosis outbreak described above), retrospective analysis of the epidemiological information for the remaining cases was limited, and no sources or additional links between cases was identified.
DISCUSSION
Listeriosis is rare in New Zealand, with an average of 0.5 cases per 100,000 population between 1998 and 2018. This incidence is, however, higher than those recently reported for the United States, Canada, Australia, and the European Union and European Economic Area (EU/EEA) (0.23 to 0.42 cases per 100,000 population) (22–25). The NZ listeriosis rate has remained relatively stable during 1998 to 2018, with no clear trends in the incidence of disease (1997 to 2016) previously reported for pregnancy-associated cases in New Zealand (19). In New Zealand, the majority (64% to 95%) of listeriosis cases between 1999 and 2018 were non-pregnancy associated, and as observed in other countries, listeriosis is the cause of a large proportion of severe cases and deaths in susceptible populations such as the elderly and immunocompromised persons (26). The detection of cases with L. monocytogenes infection in New Zealand is biased toward detecting cases of invasive listeriosis, as fecal testing for Listeria spp. is not normally performed by laboratories (27). As a result, febrile gastroenteritis may be contributing to a portion of the undiagnosed sporadic gastroenteritis disease reported each year and is more likely to be detected if it is a part of an outbreak (20). Including Listeria species testing of fecal samples from gastrointestinal cases and surveillance may potentially highlight linkages between noninvasive and invasive listeriosis cases in New Zealand. In turn, this may provide greater opportunities to identify food and environmental sources of L. monocytogenes more quickly and improve the overall public heath burden of listeriosis in New Zealand.
Retrospective whole-genome sequence analysis demonstrated that more than 50% of NZ clinical isolates belonged to internationally spread MLST sequence types (ST1, ST4, ST2, and ST155) (28–33). Further discrimination between isolates within a ST was achieved using cgMLST, with similar relationships also observed using wgMLST and/or SNP analysis within cgMLST types, supporting the use of multiple WGS analytical methods for closely related isolates (6, 31). The use of cgMLST demonstrated that the majority of the L. monocytogenes types associated with clinical cases between 1998 and 2018 were the sole isolate identified for a cgMLST type, confirming that listeriosis cases are mostly sporadic, which is also observed internationally (19, 34).
A cutoff of ≤7 core genome alleles, as proposed by Moura et al. (12), appears to be useful for identifying potential clusters of isolates in New Zealand, with prioritization of effort thereafter based on decreasing allelic differences. In the present study, 18 cgMLST types contained isolates (50 isolates in total, including three outbreak-associated isolates) that shared low genetic diversity (0 to 2 whole-genome allele differences), suggesting a possible common source between cases. SNP analysis also produced results comparable to those of wgMLST. Other outbreak investigations using the same cgMLST scheme have also reported that clinical isolates were highly related to nonclinical isolates with four allele and <10 SNP differences (35, 36). This observation was consistent with the 2012 RTE meat outbreak in New Zealand, where clinical and food isolates had 0 to 1 core genome allele and SNP differences between them, and in concordance with epidemiological data (5). However, the lack of epidemiological data on food consumption of cases not already accounted for in a recognized outbreak impaired retrospective epidemiological investigations. It is plausible that the cgMLST types with low genetic diversity (0 to 2 whole-genome alleles/SNPs) represent small monoclonal outbreaks that have remained undetected, even over a number of years.
For many pathogens associated with foodborne outbreaks, low genetic diversity may denote a recent transmission or a common source. However, several studies have reported that the genetic diversity of L. monocytogenes may exceed currently proposed thresholds, in particular, in the long-term outbreaks and cases in which contamination may originate from different sources and/or sources with a genetically diverse population (37–41). This observation was also observed during the 2012 RTE outbreak in New Zealand, where isolates from RTE products implicated during the outbreak resulted in nine core genome MLST differences and between 10 and 96 SNP differences from those in clinical cases (5). This highlights that epidemiological concordance (i.e., food consumption data) remains the key factor in outbreak investigation. This is especially important for multigenotype outbreaks, for which genotyping can provide support or confirmation alongside food testing (6, 42).
The assignment of cgMLST types was performed using the BIGSdb-Lm database (12), which incorporates a number of international isolates. It was observed in the present study that there were isolates assigned a cgMLST type that exceeded the seven-allele threshold. This is due to the presence of intermediary isolates in the database that share fewer than seven allele differences with isolates within this current data set. For example, L1-SL1-ST1-CT4574 (Fig. 8) has two isolates with nine core allele differences with the other clinical isolates in the database but was previously identified in clinical isolates from the United States (data not shown). Inclusion of international isolates sharing the same cgMLST type may provide greater context and contribute to identifying potential imported food sources.
Indeed, there is increasing evidence that linking patient information with genotyping data from food and/or food processing environments can facilitate rapid hypothesis generation through case-case comparison of outbreak-related and sporadic cases (43). In the United States, a multiagency collaboration undertakes real-time WGS on all L. monocytogenes isolates from clinical cases, food, and the environment, including from food manufacturing. Combining epidemiological and product trace-back data with WGS data has resulted in the detection of more listeriosis clusters and has solved more outbreaks (6). Integrated WGS surveillance systems also exist within Europe and have been advantageous in identifying cross-border listeriosis outbreaks (35, 44–46). A similar approach of introducing a comprehensive WGS-based typing of food and human isolates in real-time in New Zealand could expedite the identification of food sources and assist officials in undertaking regulatory actions early to prevent further illnesses.
The inclusion of the current WGS data of historical strains will provide a valuable resource that can assist with the identification of prospective outbreaks that may have been undetected for some time (13). As an example, in the present study, L2-SL8-ST120-CT786, containing outbreak-associated isolates, appeared to have additional isolates (with low genetic diversity) that span across 20 years and different geographical regions in New Zealand. PFGE, the typing tool of choice prior to WGS, showed five different AscI pulsotypes for this cgMLST group, one of which was among the most common pulsotypes (Asc0002:Apa0002) observed in New Zealand. This pulsotype was also observed to include seven other cgMLST types. Misleading variability in pulsotype and insufficient true discrimination using PFGE were also observed for other common pulsotypes and undoubtedly impeded previous linking of cases. Studies using WGS analysis have observed that these AscI PFGE differences are due to the loss or gain of prophages over time (33). As demonstrated internationally, the unprecedented discriminatory power of WGS analysis, together with thorough exposure information from cases will be able to better link cases in future outbreaks more rapidly and possibly those that have been undetected for some time over different geographical regions (35, 38, 44–46).
In conclusion, listeriosis rates have remained relatively stable in New Zealand between 1999 and 2018, and retrospective WGS analysis supports the observation that the majority of listeriosis cases in New Zealand are sporadic. WGS analysis indicating a close genetic relationship between many groups of isolates within cgMLST types suggests potential common sources or outbreaks. However, it was not possible to confirm these sources retrospectively due to incomplete epidemiological data. Currently in New Zealand, WGS analysis of key pathogens, including L. monocytogenes, occurs routinely when isolates from invasive listeriosis cases arise and predominately supports outbreak investigations. Improvements in the real-time integration of genomic and epidemiological information is ongoing for timely detection of outbreaks. The inclusion of L. monocytogenes isolates from food sources obtained from regulatory testing is already shown to be advantageous in outbreak situations in New Zealand (5), but more routine testing and the development of a system that can integrate these data together with epidemiological information are required to improve the overall surveillance practice in New Zealand.
ACKNOWLEDGMENTS
We thank the Ministry of Health for supporting this work, including review of the manuscript and use of the epidemiological data available on EpiSurv. We thank the New Zealand Public Health Units for the collection of case information and the staff in diagnostic laboratories and at ESR that conducted the laboratory investigations. We also thank Sarah Jefferies and Angela Cornelius for reviewing the manuscript and Jing Wang for her bioinformatic assistance with uploading genomes to NCBI. Finally, we thank Helen Withers and Tanya Soboleva from NZ Food Safety, MPI for their review of the manuscript.
The Ministry of Health provided funding for the whole-genome sequencing of isolates. Funds to prepare this publication were also provided by the ESR Science Strategic Investment Fund of the Ministry of Business, Innovation and Employment.
Footnotes
Supplemental material is available online only.
Contributor Information
Lucia Rivas, Email: lucia.rivas@esr.cri.nz.
Daniel J. Diekema, University of Iowa College of Medicine
REFERENCES
- 1.Jordan K, McAuliffe O. 2018. Listeria monocytogenes in Foods. Adv Food Nutr Res 86:181–213. 10.1016/bs.afnr.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Cressey PJ, Lake RJ, Thornley C, Campbell D. 2019. Expert elicitation for estimation of the proportion foodborne for selected microbial pathogens in New Zealand. Foodborne Pathog Dis 16:543–549. 10.1089/fpd.2018.2576. [DOI] [PubMed] [Google Scholar]
- 3.Tompkin RB. 2002. Control of Listeria monocytogenes in the food-processing environment. J Food Prot 65:709–725. 10.4315/0362-028x-65.4.709. [DOI] [PubMed] [Google Scholar]
- 4.Kurpas M, Wieczorek K, Osek J. 2018. Ready-to-eat meat products as a source of Listeria monocytogenes. J Vet Res 62:49–55. 10.1515/jvetres-2018-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivas L, Dupont PY, Wilson M, Rohleder M, Gilpin B. 2019. An outbreak of multiple genotypes of Listeria monocytogenes in New Zealand linked to contaminated ready-to-eat meats-a retrospective analysis using whole-genome sequencing. Lett Appl Microbiol 69:392–398. 10.1111/lam.13227. [DOI] [PubMed] [Google Scholar]
- 6.Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P. 2016. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis 63:380–386. 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaminathan B, Gerner-Smidt P. 2007. The epidemiology of human listeriosis. Microbes Infect 9:1236–1243. 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Ministry for Primary Industries. 2017. Guidance for the control of Listeria monocytogenes in ready-to-eat foods. Part 3: monitoring activities. Ministry for Primary Industries (MPI), Wellington, New Zealand. [Google Scholar]
- 9.Besser JM, Carleton HA, Trees E, Stroika SG, Hise K, Wise M, Gerner-Smidt P. 2019. Interpretation of whole-genome sequencing for enteric disease surveillance and outbreak investigation. Foodborne Pathog Dis 16:504–512. 10.1089/fpd.2019.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seemann T, Goncalves da Silva A, Bulach DM, Schultz MB, Kwong JC, Howden BP. Nullarbor GitHub. https://github.com/tseemann/nullarbor. Accessed 6 September 2019.
- 11.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, Bjorkman JT, Dallman T, Reimer A, Enouf V, Larsonneur E, Carleton H, Bracq-Dieye H, Katz LS, Jones L, Touchon M, Tourdjman M, Walker M, Stroika S, Cantinelli T, Chenal-Francisque V, Kucerova Z, Rocha EP, Nadon C, Grant K, Nielsen EM, Pot B, Gerner-Smidt P, Lecuit M, Brisse S. 2016. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2:16185. 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, Bulach DM, Stinear TP, Seemann T, Howden BP. 2016. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 54:333–342. 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahl V, Sundqvist L, Hedenstrom I, Lofdahl M, Alm E, Ringberg H, Lindblad M, Wallensten A, Thisted Lambertz S, Jernberg C. 2017. A nationwide outbreak of listeriosis associated with cold-cuts, Sweden 2013–2014. Infect Ecol Epidemiol 7:1324232. 10.1080/20008686.2017.1324232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas L, Horn B, Cook R, Castle M. 2017. Microbiological survey of packaged ready-to-eat red meats at retail in New Zealand. J Food Prot 80:1806–1814. 10.4315/0362-028X.JFP-17-179. [DOI] [PubMed] [Google Scholar]
- 16.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.New Zealand Ministry of Health. 2018. Communicable disease control manual: listeriosis. New Zealand Ministry of Health, Wellington, New Zealand. [Google Scholar]
- 18.Chen P, Kong N, Huang B, Thao K, Ng W, Storey DB, Arabyan N, Foutouhi A, Foutouhi S, Weimer BC. 2017. 100K pathogen genome project: 306 Listeria draft genome sequences for food safety and public health. Genome Announc 5:e00967-16. 10.1128/genomeA.00967-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffs E, Williman J, Brunton C, Gullam J, Walls T. 2020. The epidemiology of listeriosis in pregnant women and children in New Zealand from 1997 to 2016: an observational study. BMC Public Health 20:116. 10.1186/s12889-020-8221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sim J, Hood D, Finnie L, Wilson M, Graham C, Brett M, Hudson JA. 2002. Series of incidents of Listeria monocytogenes non-invasive febrile gastroenteritis involving ready-to-eat meats. Lett Appl Microbiol 35:409–413. 10.1046/j.1472-765x.2002.01207.x. [DOI] [PubMed] [Google Scholar]
- 21.Institute of Environmental Science and Environment. 2009. New Zealand Public Health surveillance report. December 2009: covering July to September 2009. Institute of Environmental Science and Environment, Wellington, New Zealand. [Google Scholar]
- 22.Tack DM, Ray L, Giffin PM, Cieslak PR, Dunn J, Rissman T, Jervis R, Lathrop S, Muse A, Duwell M, Smith K, Tobin-D'Angelo M, Vugia G, Zablotsky Kufel J, Wolpert BJ, Tauxe R, Payne DC. 2020. Preliminary incidence and trends of infections with pathogens transmitted commonly through food - foodborne diseases active surveillance network, 10 U.S. Sites, 2016–2019. MMWR Morb Mortal Wkly Rep 69:509–514. 10.15585/mmwr.mm6917a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foodnet Canada. 2019. Annual report 2018. Public Health Agency of Canada, Ottawa, Canada. [Google Scholar]
- 24.New South Wales Health. 2017. Listeriosis control guideline for public health units. New South Wales Health, St Leonards, Australia. [Google Scholar]
- 25.European Centre for Disease Prevention and Control. 2020. Listeriosis: annual epidemiological report for 2017. European Centre for Disease Prevention and Control, Stockholm, Swedend. [Google Scholar]
- 26.de Noordhout CM, Devleesschauwer B, Angulo FJ, Verbeke G, Haagsma J, Kirk M, Havelaar A, Speybroeck N. 2014. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect Dis 14:1073–1082. 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicol C, King N, Pirie R, Dufour M. 2010. Diagnostic and public health management practices of foodborne bacterial diseases. Institute of Environmental Science and Research, Wellington, New Zealand. [Google Scholar]
- 28.Caruso M, Fraccalvieri R, Pasquali F, Santagada G, Latorre LM, Difato LM, Miccolupo A, Normanno G, Parisi A. 2020. Antimicrobial susceptibility and multi-locus sequence typing of Listeria monocytogenes isolated over 11 years from food, humans, and the environment in Italy. Foodborne Pathog Dis 17:284–294. 10.1089/fpd.2019.2723. [DOI] [PubMed] [Google Scholar]
- 29.Toledo V, den Bakker HC, Hormazabal JC, Gonzalez-Rocha G, Bello-Toledo H, Toro M, Moreno-Switt AI. 2018. Genomic diversity of Listeria monocytogenes isolated from clinical and non-clinical samples in Chile. Genes (Basel) 9:396. 10.3390/genes9080396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camargo AC, Moura A, Avillan J, Herman N, McFarland AP, Sreevatsan S, Call DR, Woodward JJ, Lecuit M, Nero LA. 2019. Whole-genome sequencing reveals Listeria monocytogenes diversity and allows identification of long-term persistent strains in Brazil. Environ Microbiol 21:4478–4487. 10.1111/1462-2920.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halbedel S, Prager R, Fuchs S, Trost E, Werner G, Flieger A. 2018. Whole-genome sequencing of recent Listeria monocytogenes isolates from Germany reveals population structure and disease clusters. J Clin Microbiol 56:e00119-19. 10.1128/JCM.00119-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knabel SJ, Reimer A, Verghese B, Lok M, Ziegler J, Farber J, Pagotto F, Graham M, Nadon CA, Gilmour MW, Canadian Public Health Laboratory Network (CPHLN). 2012. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J Clin Microbiol 50:1748–1751. 10.1128/JCM.06185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desai AN, Anyoha A, Madoff LC, Lassmann B. 2019. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: a review of ProMED reports from 1996 to 2018. Int J Infect Dis 84:48–53. 10.1016/j.ijid.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Walle I, Bjorkman JT, Cormican M, Dallman T, Mossong J, Moura A, Pietzka A, Ruppitsch W, Takkinen J, European Listeria Wgs Typing Group. 2018. Retrospective validation of whole genome sequencing-enhanced surveillance of listeriosis in Europe, 2010 to 2015. Euro Surveill 23:1700798. 10.2807/1560-7917.ES.2018.23.33.1700798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith AM, Tau NP, Smouse SL, Allam M, Ismail A, Ramalwa NR, Disenyeng B, Ngomane M, Thomas J. 2019. Outbreak of Listeria monocytogenes in South Africa, 2017–2018: laboratory activities and experiences associated with whole-genome sequencing analysis of isolates. Foodborne Pathog Dis 16:524–530. 10.1089/fpd.2018.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Gonzalez-Escalona N, Hammack TS, Allard MW, Strain EA, Brown EW. 2016. Core genome multi-locus sequence typing for identification of globally distributed clonal groups and differentiation of outbreak strains of Listeria monocytogenes. Appl Environ Microbiol 82:6258–6272. 10.1128/AEM.01532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Luo Y, Curry P, Timme R, Melka D, Doyle M, Parish M, Hammack TS, Allard MW, Brown EW, Strain EA. 2017. Assessing the genome level diversity of Listeria monocytogenes from contaminated ice cream and environmental samples linked to a listeriosis outbreak in the United States. PLoS One 12:e0171389. 10.1371/journal.pone.0171389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pightling AW, Pettengill JB, Luo Y, Baugher JD, Rand H, Strain E. 2018. Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front Microbiol 9:1482. 10.3389/fmicb.2018.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papic B, Kusar D, Zdovc I, Golob M, Pate M. 2020. Retrospective investigation of listeriosis outbreaks in small ruminants using different analytical approaches for whole genome sequencing-based typing of Listeria monocytogenes. Infect Genet Evol 77:104047. 10.1016/j.meegid.2019.104047. [DOI] [PubMed] [Google Scholar]
- 41.Reimer A, Weedmark K, Petkau A, Peterson CL, Walker M, Knox N, Kent H, Mabon P, Berry C, Tyler S, Tschetter L, Jerome M, Allen V, Hoang L, Bekal S, Clark C, Nadon C, Van Domselaar G, Pagotto F, Graham M, Farber J, Gilmour M. 2019. Shared genome analyses of notable listeriosis outbreaks, highlighting the critical importance of epidemiological evidence, input datasets and interpretation criteria. Microb Genom 5:e000237. 10.1099/mgen.0.000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerner-Smidt P, Besser J, Concepcion-Acevedo J, Folster JP, Huffman J, Joseph LA, Kucerova Z, Nichols MC, Schwensohn CA, Tolar B. 2019. Whole genome sequencing: bridging One-Health surveillance of foodborne diseases. Front Public Health 7:172. 10.3389/fpubh.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O'Connor KA, Cosgrove S, Gossack JP, Parachini SS, Jain NS, Ettestad P, Ibraheem M, Cantu V, Joshi M, DuVernoy T, Fogg NW, Jr, Gorny JR, Mogen KM, Spires C, Teitell P, Joseph LA, Tarr CL, Imanishi M, Neil KP, Tauxe RV, Mahon BE. 2013. Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med 369:944–953. 10.1056/NEJMoa1215837. [DOI] [PubMed] [Google Scholar]
- 44.Moura A, Tourdjman M, Leclercq A, Hamelin E, Laurent E, Fredriksen N, Van Cauteren D, Bracq-Dieye H, Thouvenot P, Vales G, Tessaud-Rita N, Maury MM, Alexandru A, Criscuolo A, Quevillon E, Donguy MP, Enouf V, de Valk H, Brisse S, Lecuit M. 2017. Real-time whole-genome sequencing for surveillance of Listeria monocytogenes. Emerg Infect Dis 23:1462–1470. 10.3201/eid2309.170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schjorring S, Gillesberg Lassen S, Jensen T, Moura A, Kjeldgaard JS, Muller L, Thielke S, Leclercq A, Maury MM, Tourdjman M, Donguy MP, Lecuit M, Ethelberg S, Nielsen EM. 2017. Cross-border outbreak of listeriosis caused by cold-smoked salmon, revealed by integrated surveillance and whole genome sequencing (WGS), Denmark and France, 2015 to 2017. Euro Surveill 22:17-00762. 10.2807/1560-7917.ES.2017.22.50.17-00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleta S, Hammerl JA, Dieckmann R, Malorny B, Borowiak M, Halbedel S, Prager R, Trost E, Flieger A, Wilking H, Vygen-Bonnet S, Busch U, Messelhäußer U, Horlacher S, Schönberger K, Lohr D, Aichinger E, Luber P, Hensel A, Al Dahouk S. 2017. Molecular tracing to find source of protracted invasive listeriosis outbreak, Southern Germany, 2012–2016. Emerg Infect Dis 23:1680–1683. 10.3201/eid2310.161623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download JCM.00849-21-s0001.xlsx, XLSX file, 0.06 MB (62.5KB, xlsx)
Data Availability Statement
Raw sequence files for all isolates analyzed in this study are available on National Centre for Biotechnology Information (NCBI) short read archive (SRA) with BioProject number PRJNA741099 (Data Set S1). A total of 20 isolates were sequenced as a part of the University of California, Davis, 100K Pathogen Genome Project (18), with SRA accession numbers outlined in Data Set S1.