Abstract
The optimal hepatitis B virus (HBV) DNA quantitative assay for clinical use remains to be determined. We examined the sensitivity, linearity, and variability of a novel second-generation antibody capture solution hybridization assay, the Digene Hybrid Capture II assay (HCII), and compared it with another widely used solution hybridization assay, the branched-DNA (bDNA) assay (Quantiplex; Chiron Corp.). Our results showed similar and satisfactory assay linearity values, as well as interassay and intra-assay variability values, for both HCII and bDNA assays across different ranges of HBV DNA. Ninety-one percent of 102 serum samples from hepatitis B surface antigen-positive patients showed concordant results with the two assays. The HCII assay was more sensitive than the bDNA assay by 1 dilution, with the lowest reading being 0.9 pg/ml (3.8 pg/ml by bDNA assay). The HBV DNA seropositivity rates for the 102 samples were 58, 67, and 97% by bDNA, HCII, and nested PCR, respectively. While the relationship between results obtained with the bDNA assay and those with the HCII assay was nonlinear, with the bDNA assay yielding values 2.83 ± 0.92-fold higher than those of the HCII assay, especially at high HBV DNA levels, a linear relationship was observed between the two sets of data after logarithmic conversion. The formula for interassay conversion of results was derived as follows: HBV DNA by HCII (picograms per milliliter) = 3.19 × [HBV DNA by bDNA (megaequivalents per milliliter)]0.866. The HCII assay was technically less complex and required a shorter assay time (4 h) than the bDNA assay (24 h). We conclude that the HCII assay compares favorably with the bDNA assay and offers the additional advantages of increased sensitivity and shorter assay time. The increased sensitivity should be particularly useful in monitoring the efficacy of antiviral therapies and detecting the emergence of drug-resistant HBV mutants.
Hepatitis B virus (HBV) infection is a major cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma and accounts for 1 million deaths annually (5, 13). Information on the virus load and the replicative activity of HBV is of paramount importance in the management of patients with chronic HBV infection, especially after the recent advent of medications which can effectively suppress HBV replication, such as the nucleoside analogues. Serological parameters which have been used in this regard include the hepatitis B e antigen (HBeAg) status, DNA polymerase, HBV DNA status by qualitative methods such as PCR or semiquantitative dot blot hybridization, and more recently HBV DNA quantitation by solution hybridization (8, 11, 13). The direct and quantitative nature of the latter makes it a useful clinical test to monitor serially the efficacy of antiviral therapy (7, 12, 14).
Solution hybridization techniques for HBV DNA quantitation can utilize radioactive (e.g., Genostics; Abbott Laboratories), antibody capture (e.g., Hybrid-Capture; Murex Diagnostics Ltd.), or branched-DNA (bDNA) signal detection systems (e.g., Quantiplex; Chiron Corp.). Comparisons among these assays are complicated by variations in methodology, standards, and units. The bDNA assay is the most sensitive and precise among the three (1, 2, 16). Since a universal standard is lacking and the bDNA assay both is technically intricate and requires a long assay time, the optimal choice of quantitative HBV DNA assay for clinical use remains to be defined.
Recently, a novel (second-generation) antibody capture solution hybridization HBV DNA quantitative assay (Digene Hybrid Capture II assay [HCII]; Digene Corp., Beltsville, Md.) has offered sensitivity improved over that of the bDNA assay. In this study, we evaluated the sensitivity, specificity, linearity, and technical complexity of this novel HCII assay and compared it with the bDNA assay. Correlations between results obtained with the HCII and those with the bDNA assay were examined, and an equation was derived for the conversion of results between the two quantitative assays.
MATERIALS AND METHODS
Serum samples from 102 hepatitis B surface antigen (HBsAg)-positive patients and 22 HBsAg-negative controls were assayed for HBV DNA by (i) HCII assay, (ii) bDNA assay, and (iii) an in-house nested PCR (nPCR) assay. The patients were randomly included from known chronic HBV carriers attending regular follow-up for serial monitoring of their liver status. Blood samples were centrifuged within 4 h to obtain the serum fractions, which were then aliquoted and kept at −80°C before assay.
HCII assay.
The Digene HCII (standard) assay (Digene Corp.) quantitates HBV DNA by solution hybridization, followed by immunocapture and chemiluminescent signal detection. The assay protocol was according to the manufacturer’s instructions. Briefly, 30 μl of denaturing reagent was added to each microplate well containing 30 μl of test samples or HBV DNA standards (0 to 6,000 pg/ml). The plate was incubated at 65°C for 30 min for the lysis of HBV and DNA denaturation. RNA-DNA hybridization was achieved by adding 30 μl of RNA probe (specific for HBV ad and ay strains) to each well and incubating the wells at 65°C for 60 min. Seventy-five microliters of the hybrid-containing solution was then transferred into RNA-DNA capture wells and shaken (Thermolyne Maxi-Mix III) at 1,100 rpm at room temperature for 60 min. The solution in the wells was then removed by aspiration. Seventy-five microliters of alkaline phosphatase-conjugated antibodies to RNA-DNA hybrids was added to each well and incubated at room temperature for 30 min. After six washings, 75 μl of chemiluminescent substrate was added, and light emission after 15 min was measured by a chemiluminometer (DML 2000 luminometer; Digene Corp.). Results were expressed in picograms per milliliter according to the plot of standards. The sensitivity according to the manufacturer was 0.5 pg/ml or 0.142 × 106 copies/ml.
bDNA assay.
The bDNA assay (Quantiplex; Chiron Corp.) is a sandwich nucleic acid hybridization assay. A set of oligonucleotide probes bind single-stranded HBV DNA to a solid phase, which is detected by a second set of oligonucleotide probes. bDNA then serves to amplify the signal, which is generated by adding alkaline phosphatase-conjugated probes for bDNA and dioxetane as substrate (6). The assay protocol was according to the manufacturer’s instructions. Briefly, 10 μl of test samples or HBV DNA-positive standards (0.5 to 4,400 MEq/ml) was added to 10 μl of lysis reagent on a microwell plate coated with oligonucleotide probes and incubated for 30 min at 63°C to release HBV DNA from the viral particles. Ten microliters of the second set of probes in denaturing buffer was then added to the wells and incubated for 30 min at 63°C. Ten microliters of neutralizing reagent was then added, and the hybridization process continued for 16 h. After washing, 40 μl of bDNA amplifiers was added and incubated for 30 min at 53°C. Forty microliters of alkaline phosphatase-conjugated probes was then added to each well and incubated for 15 min at 53°C. After washing, 30 μl of dioxetane substrate was added. The plate was incubated for 25 min at 37°C in the Chiron luminometer, and light emission was measured for each well. The result of HBV DNA level, expressed as megaequivalents per milliliter, was generated by software supplied by the manufacturer. The sensitivity according to the manufacturer was 0.7 MEq/ml (0.7 × 106 copies/ml) or 2.5 pg/ml (1 MEq/ml = 3.53 pg/ml [9]).
nPCR assay.
The nPCR was performed according to a previously published protocol (3). The primer sets for the HBV core region were GGAGTGTGGATTCGCACTCCTCC (map positions 2269 to 2288) and ATACTAACATTGAGATTCCC (2457 to 2438) for the first-round PCR and AGACCACCAAATGCCCCTAT (2299 to 2318) and GATCTTCTGCGACGCGGCGA (2429 to 2410) for the second round. Briefly, 10 μl of serum was mixed with 8 μl of 0.2 M NaOH solution and incubated at 37°C for 1 h and then at 98°C for 5 min. Two microliters of 0.5 M Tris-HCl was added to bring the pH to 8. Eighty microliters of the PCR solution, with 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 200 μM (each) deoxyribonucleoside triphosphate, 1 μM (each) first-round primers, and 1.25 μl of Taq polymerase, was then added. The mixture was heated to 95°C for 5 min, followed by 30 PCR cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min in a thermal cycler (PTC-100; MJ Research, Watertown, Mass.). For the second-round PCR, 5 μl of the first-round PCR product was added to 40 μl of the second-round reaction mixture prepared as described above except that the second set of primers was used. The nPCR product was electrophoresed in a polyacrylamide gel and visualized under UV light after staining with ethidium bromide. Detection of a DNA band of 130 bp denoted positivity. All the samples were tested in duplicate, and positive and negative controls were included in each run. Our in-house nPCR assay had a sensitivity of 1 fg/ml (300 copies/ml).
Statistics.
Correlation between HBV DNA results obtained with the HCII and those with bDNA assays was examined by Pearson’s method.
RESULTS
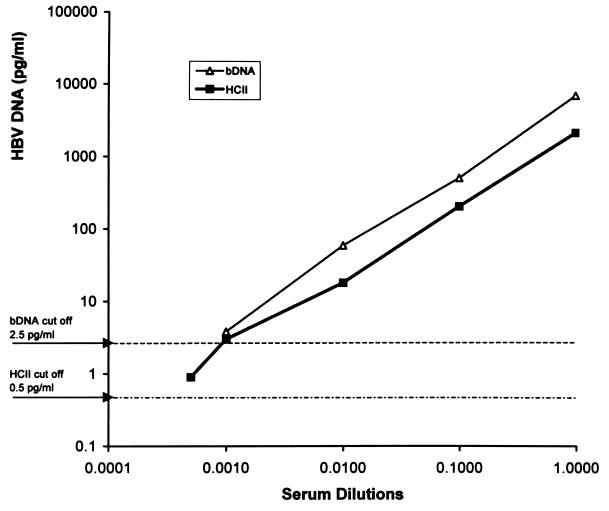
A serum sample with a high HBV DNA concentration (7,081 pg/ml by bDNA assay and 2,164 pg/ml by HCII assay) was tested upon serial dilution with normal human serum at 1/10, 1/100, 1/1,000, and 1/2,000 dilutions in order to determine and compare the sensitivity and performance linearity values of the bDNA and HCII assays. Repeated testing of this dilution series three times yielded consistent results. The sensitivity limit for the bDNA assay was at a 1/1,000 dilution, giving an assay result of 3.8 pg/ml, while that for the HCII assay was at a dilution of 1/2,000, yielding an assay result of 0.9 pg/ml. Linearity values of the two assays were similar (Fig. 1).
FIG. 1.
Graph showing the linearity of the HCII and bDNA assay results as determined by testing a standard serum sample with a high HBV DNA concentration at serial dilutions.
To compare the precision of the bDNA and HCII assays, three serum samples with HBV DNA levels previously determined to be at the low (L) (2.5 to <40 pg/ml), medium (M) (40 to 3,900 pg/ml), and high (H) (>3,900 to 17,000 pg/ml) levels according to the standard ranges of bDNA assay (actual readings of 37.5, 909.2, and 10,525.3 pg/ml, respectively) were tested six times with both assays. The bDNA assay yielded HBV DNA levels 1.7 to 3.6 times higher than those obtained with the HCII assay (Table 1). The intra-assay and interassay coefficients of variation (CV) were similar for the two assays (P value not significant).
TABLE 1.
Comparison of corresponding values and intra- and interassay variations when identical serum samples with L, M, and H levels of HBV DNA were tested with the HCII and the bDNA assays
| Variation and specimen identification | HCII
|
bDNA
|
||
|---|---|---|---|---|
| Mean ± SD | CV (%) | Mean ± SD | CV (%) | |
| Intra-assay (n = 6) | ||||
| L | 22.4 ± 1.3 | 5.8 | 37.5 ± 2.1 | 5.6 |
| M | 355.9 ± 24.3 | 6.8 | 909.2 ± 64.6 | 7.1 |
| H | 2,884.3 ± 96.6 | 6.8 | 10,525.3 ± 292.0 | 2.8 |
| Interassay (n = 4) | ||||
| L | 20.8 ± 2.3 | 11.1 | 36.3 ± 3.0 | 8.3 |
| M | 337.4 ± 26.0 | 7.7 | 858.5 ± 82.2 | 9.6 |
| H | 2,871.0 ± 177.7 | 6.1 | 10,061.1 ± 656.4 | 6.5 |
The prevalence of HBV DNA as determined by bDNA, HCII, and nPCR methods in the 102 HBsAg-positive samples and 22 HBsAg-negative samples is given in Table 2. The HBV DNA seropositivity rate was highest with the nPCR assay, followed by the HCII assay and then the bDNA assay, irrespective of the HBeAg-antibody status. The two quantitative (bDNA and HCII) assays gave 59 concordant positive and 34 concordant negative results, giving an overall concordance rate of 91% (93 of 102). All the bDNA-positive samples tested positive with the HCII assay. In contrast, nine samples were HCII positive but bDNA negative, giving a sensitivity gain of 13% (9 of 68) for the HCII assay. The HBV DNA concentration of the nine serum samples was 3.8 ± 4.9 pg/ml (range, 0.9 to 14.9 pg/ml). None of the three assays detected HBV DNA in the 22 HBsAg-negative serum samples.
TABLE 2.
Prevalence of HBV DNA as detected by bDNA, HCII, and nPCR methods in relation to their HBeAg and anti-HBe status in 102 HBsAg-positive samples and in 22 HBsAg-negative samples
| Assay | No. of HBV DNA-positive samples (%)
|
||||
|---|---|---|---|---|---|
| HBsAg+
|
HBsAg−(n = 22) | ||||
| HBeAg+ and anti-HBe−(n = 53) | HBeAg− and anti-HBe+(n = 46) | HBeAg− and anti-HBe−(n = 3) | Total (n = 102) | ||
| bDNA | 46 (87) | 12 (26) | 1 | 59 (58) | 0 |
| HCII | 50 (94) | 17 (37) | 1 | 68 (67) | 0 |
| nPCR | 53 (100) | 43 (93) | 3 | 99 (97) | 0 |
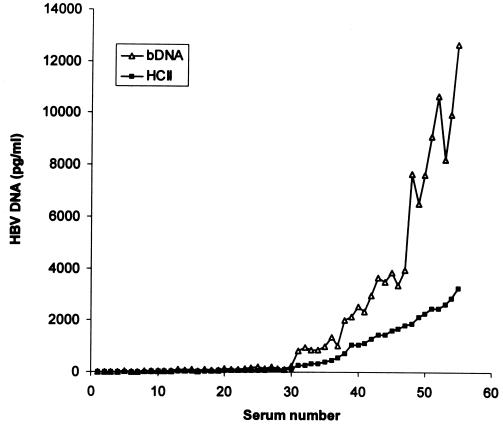
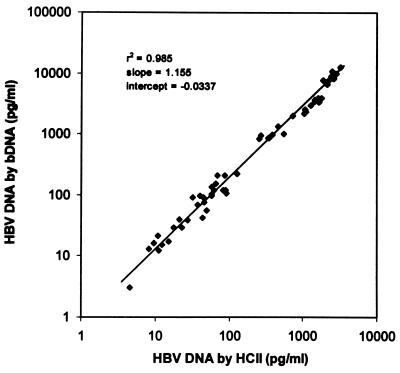
HBV DNA levels measured with the HCII assay were compared to corresponding values determined with the bDNA assay in 55 of the 59 concordant positive samples (Fig. 2). Four samples yielded readings higher than the detection range of both assays and were not included in this comparison. The bDNA assay yielded increasingly higher values relative to the HCII readings as the HBV DNA concentration increased. The ratio of corresponding bDNA to HCII results (in picograms per milliliter) was 2.83 ± 0.92 (range, 0.65 to 4.31). A good linear relationship was observed when the logarithmic conversions of HBV DNA levels determined by the two assays were plotted against each other (r2 = 0.985, slope = 1.155) (Fig. 3). The following formulae were derived accordingly for the interassay conversion of results: log [HBV DNA by bDNA (picograms per milliliter)] = 1.155 × log [HBV DNA by HCII (picograms per milliliter)] − 0.0337 or simplified as HBV DNA by bDNA (megaequivalents per milliliter) = (1) 0.262 × [HBV DNA by HCII (picograms per milliliter)]1.155 HBV DNA by HCII (picograms per milliliter) = 3.19 × (2)[HBV DNA by bDNA (megaequivalents per milliliter)]0.866
FIG. 2.
Comparison of corresponding HBV DNA levels as determined by the HCII or the bDNA assay in 55 concordant positive serum samples.
FIG. 3.
A log-log plot of HBV DNA concentrations in 55 concordant positive serum samples as determined by the bDNA and HCII assays.
With regard to the complexity of assay procedures, the HCII assay was technically simpler and yielded results faster than the bDNA assay since it involved fewer steps and did not require many freshly prepared agents (Table 3).
TABLE 3.
Comparison of the technical complexities of the HCII and bDNA HBV DNA assays
| Characteristic | HCII | bDNA |
|---|---|---|
| Sample vol (μl) | 30 | 10 |
| No. of reagents used (no. that need to be freshly prepared) | 4 (1) | 6 (4) |
| No. of incubations | 5 | 9 |
| No. of washes | 1 | 3 |
| Color indicator | Yes | No |
| Assay time (h) | 4 | 24 |
DISCUSSION
Although the novel HCII assay claimed to offer improved sensitivity compared to other quantitative solution hybridization HBV DNA assays currently in use, these assays have not been directly compared against one other. Due to the lack of a universal HBV DNA standard (1, 2, 16), the identical blood sample can yield markedly different HBV DNA levels when tested by different assays. This represents a major difficulty in the interpretation of data in the literature. In this study, we not only examined the reliability of the HCII assay but also compared its results with those obtained by the bDNA assay and derived formulae for interconversion. In addition, the technical aspects and the assay duration of these two quantitative assays were compared, since these properties have a major bearing on their usefulness in the clinical management of patients. Our results showed satisfactory and similar linearity, intra-assay variability, and interassay variability values for the HCII and bDNA assays, which were applicable across different ranges of HBV DNA.
We demonstrated an increase in the sensitivity for HBV DNA detection for the HCII assay compared to the bDNA assay, from a 1/1,000 to a 1/2,000 dilution, with corresponding HBV DNA levels going down from 3.8 to 0.9 pg/ml. The increased sensitivity may relate to the higher starting sample volume used in the HCII assay, which was 30 μl compared to the 10 μl used in the bDNA assay. Despite the apparently small (twofold) difference in sensitivity, testing of serum samples from HBsAg-positive patients showed a 13% gain in HBV DNA detection rate with the HCII assay. The difference in HBV DNA detection rates between the two assays is influenced by the proportion of tested samples with low viremia levels, which may test positive by the HCII assay but negative by the bDNA assay. In this regard, our study population was randomly chosen from HBsAg-positive patients being monitored for serial liver status. From a clinical perspective, the increased sensitivity of the HCII assay at low HBV DNA levels would be a significant advantage in monitoring patients to confirm efficacy of antiviral therapy and to detect the emergence of drug-resistant mutants.
Although the nPCR assay is most sensitive, its nonquantitative nature and the technical complexity hinder its routine clinical use. Another commercially available HBV DNA assay, the Amplicor HBV Monitor (Roche Diagnostic Systems, Basel, Switzerland), quantitates serum HBV DNA by competitive PCR with a quantitation standard and a microwell plate nonradioactive hybridization and detection system. The Amplicor assay has been reported to be reliable and sensitive, with a detection limit of 102 to 103 copies of HBV DNA per ml (4, 10). We have not included the Amplicor assay in the present study since our main objective was to compare the solution hybridization quantitative assays with regard to their assay performance and clinical utility. In this context, the Amplicor assay is also more labor-intensive, with a longer assay duration, and about three times more expensive than the HCII assay. In view of the inherent technical and financial implications, it would be difficult to establish the Amplicor assay as a routine test in clinical practice.
Comparison of concordant samples showed that the bDNA assay yielded higher values compared to corresponding readings with the HCII assay. The difference was more marked at high HBV DNA levels. This nonlinear relationship between the two sets of readings was exemplified by the conversion factors ranging from 0.65 to 4.31. We observed a good linear correlation when the results by the two assays were plotted against each other after logarithmic conversion. The ensuing formulae for interconversion of results should be helpful in the interpretation and comparison of data generated with these assays.
Apart from precision and reliability, the cost, technical complexity, and result turnaround time have important bearings on the usefulness of an assay in the clinical management of patients. In this regard, the HCII assay offers the advantages of being technically simpler and faster in yielding results than the bDNA assay. In conclusion, our results show that the new HCII assay is a sensitive and reliable assay for HBV DNA quantitation. Its enhanced sensitivity, reduced technical complexity, and shortened assay time compared to other quantitative HBV DNA assays make it a useful test in the management of patients with HBV infection.
ACKNOWLEDGMENTS
This study was supported by the CRCG Research Grant (10202135/20428/20600/323/01) and the Lee Wing Tat Renal Research Fund of The University of Hong Kong.
REFERENCES
- 1.Aspinall S, Mphahlele M J, Peenze I, Steele A D. Detection and quantitation of hepatitis B virus DNA: comparison of two commercial hybridization assays with polymerase chain reaction. J Viral Hepatitis. 1995;2:107–111. doi: 10.1111/j.1365-2893.1995.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 2.Butterworth L A, Prior S L, Buda P J, Faoagali J L, Cooksley W G E. Comparison of four methods for quantitative measurement of hepatitis B viral DNA. J Hepatol. 1996;24:686–691. doi: 10.1016/s0168-8278(96)80264-9. [DOI] [PubMed] [Google Scholar]
- 3.Chung H T, Lok A S F, Lai C L. Re-evaluation of α-interferon treatment of chronic hepatitis B using polymerase chain reaction. J Hepatol. 1993;17:208–214. doi: 10.1016/s0168-8278(05)80040-6. [DOI] [PubMed] [Google Scholar]
- 4.Gerken G, Gomes J, Lampertico P, Colombo M, Rothaar T, Trippler M, Colucci G. Clinical evaluation and applications of the Amplicor HBV Monitor test, a quantitative HBV DNA PCR assay. J Virol Methods. 1998;74:155–165. doi: 10.1016/s0166-0934(98)00081-0. [DOI] [PubMed] [Google Scholar]
- 5.Gitlin N. Hepatitis B: diagnosis, prevention, and treatment. Clin Chem. 1997;43:1500–1506. [PubMed] [Google Scholar]
- 6.Hendricks D A, Stowe B J, Hoo B S, Kolberg J, Irvine B D, Neuwald P D, Urdea M S, Perillo R P. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am J Clin Pathol. 1995;104:537–546. doi: 10.1093/ajcp/104.5.537. [DOI] [PubMed] [Google Scholar]
- 7.Hoofnagle J H. Alpha-interferon therapy of chronic hepatitis B. Current status and recommendations. J Hepatol. 1990;11:S100–S107. doi: 10.1016/0168-8278(90)90173-o. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko S, Miller R H, Di Bisceglie A M, Feinstone S M, Hoofnagle J H, Purcell R H. Detection of hepatitis B virus DNA in serum by polymerase chain reaction. Application for clinical diagnosis. Gastroenterology. 1990;99:799–804. doi: 10.1016/0016-5085(90)90971-3. [DOI] [PubMed] [Google Scholar]
- 9.Kapke G F, Watson G, Sheffler S, Hunt D, Frederick C. Comparison of the Chiron Quantiplex branched DNA (bDNA) assay and the Abbott Genostics solution hybridization assay for quantification of hepatitis B viral DNA. J Viral Hepatitis. 1997;4:67–75. doi: 10.1046/j.1365-2893.1997.00127.x. [DOI] [PubMed] [Google Scholar]
- 10.Kessler H H, Pierer K, Dragon E, Lackner H, Santner B, Stunzner D, Stelzl E, Waitzl B, Marth E. Evaluation of a new assay for HBV DNA quantitation in patients with chronic hepatitis B. Clin Diagn Virol. 1998;9:37–43. doi: 10.1016/s0928-0197(97)10008-3. [DOI] [PubMed] [Google Scholar]
- 11.Kuhns M C, McNamara A L, Perrillo R P, Cabal C M, Campbell C R. Quantitation of hepatitis B viral DNA by solution hybridization: comparison with DNA polymerase and hepatitis B antigen during antiviral therapy. J Med Virol. 1989;27:274–281. doi: 10.1002/jmv.1890270404. [DOI] [PubMed] [Google Scholar]
- 12.Lai C L, Chien R N, Leung N W, Chang T T, Guan R, Tai D I, Ng K Y, Wu P C, Dent J C, Barber J, Stephenson S L, Gray D F. A one-year trial of lamivudine for chronic hepatitis B. Asia hepatitis lamivudine study group. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 13.Lee W M. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 14.Lok A S F, Lai C L, Wu P C, Leung E K Y. Long-term follow-up in a randomized controlled trial of recombinant α2-interferon in Chinese patients with chronic hepatitis B infection. Lancet. 1988;ii:298–302. doi: 10.1016/s0140-6736(88)92355-0. [DOI] [PubMed] [Google Scholar]
- 15.Scotto J, Hadchouel M, Hery C, Yvart J, Tiollais P, Brechot C. Detection of hepatitis B virus DNA in serum by a simple spot hybridization technique: comparison with results for other viral markers. Hepatology. 1983;3:279–284. doi: 10.1002/hep.1840030301. [DOI] [PubMed] [Google Scholar]
- 16.Zaaijer H L, ter-Borg F, Cuypers H T, Hermus M C, Lelie P N. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 1994;32:2088–2091. doi: 10.1128/jcm.32.9.2088-2091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]