Abstract
There is a critical need to develop high throughput assays to identify compounds that offer therapy for individuals suffering from neurodegenerative diseases. Most brain disorders, including neurodegenerative diseases, share the common neuropathology of mitochondria dysfunction, which can lead to apoptosis of neurons, over-production of reactive oxygen species (ROS), and other cellular neuropathologies characteristic of these diseases. Human iPSCs with a stable genomic insertion of the neurogenin-2 transcription factor under the control of the TetOn promoter can be differentiated into excitatory human neurons (i3Neurons) within 3 days of exposure to doxycycline. These neurons have been used to develop and validate a live cell assay for parameters of mitochondrial dynamics and function using two compounds known to promote mitochondrial elongation in mouse neurons, 4-hydroxychalcone and 2,4-dihyrdroxychalcone. The assay involves plating the neurons in 384 well microtiter plates, treating them with known or unknown substances, and then capturing morphological information for the neuronal mitochondria using a lentivirus vector to express a mitochondrial-targeted fluorescence reporter. The i3Neuron cultures exposed to these two compounds for 24hr exhibit significantly decreased circularity and significantly increased length compared to controls, two morphological parameters correlated with increased mitochondrial health. The assay is rapid, with results obtained after a one week-long i3Neuron culture or one month if neurons are co-cultured with astrocytes. This live cell, mitochondrial phenotypic assay can be used for high-throughput screening or as an orthogonal assay for compounds obtained via other high throughput screening campaigns.
Keywords: iPSC, mitochondria, human neuron, high throughput assay
Introduction:
The progress in bringing potential therapeutics for neurodegenerative diseases from the research bench to the clinic has been extraordinarily slow. In fact, only 8% of drugs targeted to treat central nervous system diseases become approved after clinical trials 1. Compared to drug discovery for other metabolic or autoimmune diseases, these diseases present difficult target identification and validation, animal models with poor face validity, and a lack of biomarkers 1–4. However, phenotypic high throughput compound screens offer promise by providing an unbiased method for uncovering novel targets for neurodegenerative diseases 5.
Mitochondrial dysfunction is a hallmark of most neurodegenerative diseases including Alzheimer’s Disease (AD), Parkinson’s Disease (PD), Huntington’s Disease (HD) and Amyotrophic lateral sclerosis (ALS) 6, 7. Mitochondria perform numerous biochemical functions in cells beyond the typically recognized functions of ATP generation and calcium buffering, including the regulation of apoptosis, generation of reactive oxygen species (ROS), lipid and pyrimidine biosynthesis and others 8–11. When mitochondria become compromised in neurons due to aging or other factors, the precise regulation of these functions lead to the pathologies observed in many neurodegenerative diseases 7. With the advent of iPSC technology 12, we can now employ human derived neurons in high throughput screens to search for compounds that improve mitochondrial health and function.
iPSC derived human neurons are now being effectively used in high throughput screening platforms. Two recent examples include the search for compounds that reactivate the silenced Fmr I gene in Fragile X to compounds that promote neurite outgrowth and branching for potential nerve regeneration 4, 13, 14. iPSCs exposed to the neurogenin-2 (Ngn-2) transcription factor via lentivirus transduction differentiate into glutamatergic neurons 15. Fernandopulle et al 16 improved upon this system by stably integrating the Ngn-2 gene under TetOn promoter 17 regulation in a human iPSC line so that virtually all cells treated with doxycycline differentiate into excitatory human neurons (i3Neurons). The i3Neurons express classic cortical neuron markers such as MAP2, TAU and Tuj1 (Beta-III Tubulin) after differentiation 16. When the i3Neurons are co-cultured with astrocytes, they exhibit spontaneous excitatory currents that can be inhibited by glutamate receptor antagonists 16.
Here, we have combined the advantages offered by i3Neurons with a novel and effective high content assay we developed to extract morphological features of mitochondria in mouse neurons as proxies for parameters of mitochondrial health and function 18. Structure and function of mitochondria are highly related. For instance, when mitochondria become defective, they assume a small and rounded morphology which can be measured by a circularity index. Moreover, 90% (45/50) of the compounds identified to promote mitochondrial length in mouse neurons in this high content assay caused a corresponding increase in ATP generation 18. i3Neurons are generated rapidly and in large batches that are ideal for reducing variability in high throughput assays and screens. In addition, virtually all of the iPSC differentiate into excitatory neurons, which reduces variability from the multiple cell types found in primary neuron cultures established from brain tissue. The assay generates results in a week’s time from the plating of cryopreserved i3Neurons to imaging, allowing for the rapid accumulation of data. The assay as described for measuring mitochondrial features in neurons can be adapted to any cellular target for which a fluorescent reporter can be fashioned.
Materials and Methods:
iPSC maintenance, i3Neuron differentiation and cryopreservation
All reagents were obtained from Thermo Fisher Scientific, Waltham, MA, USA unless otherwise noted. iPSCs and i3Neurons were maintained and differentiated according to protocols outlined in Fernandopulle et al 16. In short, iPSCs were thawed and plated in Essential 8 media with 1X RevitaCell™ Supplement in a 6 well dish coated with 0.5μg/cm2 vitronectin, and then incubated at 37°C with 5% CO2. Media was changed daily using only Essential 8 media after the first day. iPSCs were clump passaged twice with 0.5mM EDTA in 1X DPBS before being plated in a 15cm2 dish coated with a 1:100 dilution of growth factor reduced matrigel in DMEM/F12 media. When the 15cm2 dish was 70% confluent, the iPSCs were single cell passaged with 7mL of Accutase. Ten million iPSCs were plated per 15cm2 dish coated with matrigel in induction media: 1X N2 supplement, 1X NEAA, 1X L-glutamine with a DMEM/F12 and HEPES base supplemented with 1X RevitaCell Supplement and 2 ug/ml doxycycline (Millipore Sigma, Burlington, MA, USA). We consider this as cell day 0 for all experimental timelines described below. On days 1 and 2, 100% media changes were performed with induction media supplemented with only doxycycline (minus 1X RevitaCell Supplement). On day 3, i3Neurons were cryopreserved in CTS™ PSC cryomedium and stored at −80°C. In all succeeding experiments with i3Neurons, each plate is derived from an individual cryovial of previously differentiated cells and can therefore be considered to be independent of other plates used.
i3Neuron lentiviral MOI titration assay
i3Neurons, now considered to be at cell day 4, were revived and plated in cortical neuron media: 1X B27 supplement, 10ng/mL BDNF (PeproTech, Rocky Hill, NJ, USA), 10ng/mL NT-3 (PeproTech), 1ug/mL laminin with a BrainPhys™ Neuronal Medium base (StemCell Technologies, Vancouver, Canada) in 384 well plates coated with PLO and laminin (Aurora Microplates, Whitefish, MT, USA). i3Neurons were plated at either 7,500 or 15,000 cells per well in 40μL of cortical neuron media. CAG>mtTagGFP2 lentivirus (packaged by Vigene Biosciences, Rockville, MD, USA) was mixed with the cell suspension prior to plating at the following MOIs (multiplicity of infections): 1, 3, 5, 7 and 10. On day 5, 20μL of media with lentivirus was removed and 60μL of fresh cortical neuron media was added. On cell day 7, the i3Neuron 384 well plates were imaged on the IN Cell Analyzer 6000 confocal microscope (GE Healthcare Life Sciences, Marlborough, MA, USA) capturing the TagGFP2 signal in three slices ∆Z =0.7μm; aperture = 1.0 AU; λexc = 488nm; λem = 515 to 535nm using a 60X objective as described in Varkuti et al 18. Four field images were collected per well. The imaging data were analyzed as described below to determine the optimal cell density and MOI to use for further experiments for each batch of i3Neurons prepared.
I3Neuron immunocytochemical (ICC) studies
iPSCs were dissociated into single cells and plated in 96 well plates at a cell density of 24,000 cells per well. On the second day after plating, cells were fixed for 15 minutes with a final concentration of 4% paraformaldehyde in each well, permeabilized with 0.5% triton X-100 in 1X DPBS for 15 minutes and blocked with 3% bovine serum albumin in 1X DPBS for 1 hour at room temperature. Wells were washed twice with 1X DPBS between all steps of the protocol. The following primary antibodies were added at a 1:500 dilution in blocking buffer (3% BSA in 1X DPBS) overnight at 4°C: polyclonal rabbit Oct4 (Abcam, Cambridge, MA, USA cat# ab18976), monoclonal mouse Sox2 (Abcam cat# ab79351), polyclonal rabbit MAP2 (Abcam cat# ab32454), and monoclonal mouse Tuj1 (Abcam cat# ab78078). The following secondary antibodies were added at a 1:1000 dilution in blocking buffer to the appropriate wells and incubated for 1 hour in the dark at room temperature: Alexa Fluor 568 goat anti rabbit (cat# A11011) and Alexa Fluor 488 goat anti mouse (cat# A11029). Wells were washed twice with 1X DPBS and in the final wash, NucBlue Live Stain ReadyProbes reagent was added at a concentration of 2 drops per 1mL of 1X DPBS. Wells were imaged using a 20X objective in 2D after a 5 minute incubation using the IN Cell Analyzer 6000 confocal microscope (GE Healthcare Life Sciences, Marlborough, MA, USA). I3Neuron ICC studies were performed similarly with exceptions described below. iPSCs were dissociated into single cells and plated on 384 well plates coated with matrigel at a cell density of 3,000 cells per well. The cells were then exposed to doxycycline in induction media to induce differentiation for 3 days and ICC studies were commenced at this time. Day 3 i3Neuron ICC images were taken with a 60X objective. I3Neuron day 7 and day 10 images were obtained by reviving and plating previously cryopreserved cells at a density of 7,500 cells per well on 384 well poly-L-ornithine (PLO) and laminin dual coated plates in cortical neuron media. Once plated, i3Neurons were aged until appropriate time point and the ICC procedure outlined above was commenced. I3Neuron day 28 images were obtained by co-culturing human astrocytes (iCell Astrocytes, Cellular Dynamics, Madison, WI, USA) on PLO and laminin dual coated plates in cortical neuron media. Human astrocytes were plated at a cell density of 5,000 cells per well 1 day prior to i3Neuron plating (cell density 10,000 cells per well). Cells were then aged to appropriate time point and ICC was commenced with images taken using a 60X objective.
i3Neuron high throughput mitochondrial assay
The i3Neurons were revived and immediately plated with CAG>mtTagGFP2 lentivirus at a MOI of 5 in 384 well plates coated with PLO and laminin at a cell density of 15,000 i3Neurons in 40μL of cortical neuron media/well. On cell day 5, 20μL of media with lentivirus was removed per well and 60μL of fresh cortical neuron media was added. On cell day 6, 20μL of media was removed per well and compounds solubilized in DMSO were added at the appropriate concentrations so that DMSO concentration remained constant in all wells of the plate at 0.125%. On cell day 7 (24hr after exposure to the compounds), i3Neuron plates were imaged as described above. The i3Neuron plates were also imaged on cell day 8 to obtain 48hr exposure data. For experiments using 4-hydroxychalcone (Indofine Chemical Co Inc, Hillsborough, NJ, USA) and 2,4-dihydroxychalcone (Millipore Sigma), two independent plates were assayed in parallel.
Astrocyte extended culture assay
i3Neurons to be used in extended culture assays were prepared as described above except that CAG>mtTagGFP2 lentivirus was added to the differentiating i3Neurons on day 2 of differentiation rather than cell day 4 after being revived from cryopreservation. 700,000 single cell passaged iPSCs were added to each well of a matrigel coated 6 well dish on day 0. On day 2 (during doxycycline induction), the differentiating neurons in one well were dissociated to determine the cell count to be used for MOI calculation. CAG>mtTagGFP2 lentivirus was added at a MOI = 5 to the media of each i3Neuron well to be transduced. On day 3, the transduced i3Neurons were dissociated as single cells and cryopreserved. One day prior to plating i3Neurons, human astrocytes (Cellular Dynamics) were plated at a cell density of 5,000 astrocytes per well in 40μL of cortical neuron media/well. After 24hr, 10,000 i3Neurons per well were then plated with the human astrocytes in an additional 40μL of cortical neuron media/well (bringing the total volume to 80μL of cortical neuron media/well). Three identical plates of i3Neurons and human astrocytes were plated and maintained in culture. 75% media changes were performed with cortical neuron media one time each week as the experiments progressed. 4-hydroxychalcone and 2,4-dihydroxychalcone were added to the plates as described above at cell days 13, 20 and 27 and imaged 24hr later at cell days 14, 21 and 28.
Data Analysis
Each 60X image stack was merged into a projection image using a custom macro supplied with the GE Developer Toolbox (1.9.2, build 2415) software (GE Healthcare Life Sciences). These final images were segmented into individual mitochondria as described in Varkuti et al 18, masking the somatic mitochondria to yield images containing only axonal and dendritic mitochondria, which were analyzed together. Supplemental Figure 1 contains images that illustrate how the projection images were segmented. The following data was generated for each image collected: cell body count, mitochondrial count, sum of the total mitochondria area (CA), the median circularity of the mitochondria per field, and the median length of the mitochondria per field. Since four images were taken per well, the parameter values for each image within a well were averaged together and reported as a well value. These well values were then analyzed along with the corresponding values from other wells. If an experiment consisted of multiple plates assayed in parallel, robust z-scores were calculated from the well data using the DMSO well data on the same assay plate.
Results:
Human iPSC differentiation into homogenous excitatory neurons
iPSCs with a stably integrated Ngn-2 gene in the genome under the regulation of a TetOn promoter can produce large quantities of high quality human excitatory neurons upon the addition of doxycycline to induce expression of the transcription factor 16. After only three days of Ngn-2 expression, the human iPSCs visibly shift from compact colonies of cells with large nuclei and little cytoplasm (Figure 1A) to young excitatory neurons with distinct somas and neuritic extensions (Figure 1B). At this time point, the i3Neurons exhibit expression of the neuronal markers MAP2 and Tuj1 (beta-III tubulin) (Supplemental Figure 2). At day 3 of differentiation, i3Neurons must be re-plated in the desired assay format or cryopreserved because the neurites become so extensive that passaging them produces damage. The i3Neurons survive on PLO laminin coated plates for approximately 2 weeks (Figure 1C); however, after cell day 10, the cells begin to clump and their overall health begins to deteriorate.
Figure 1.
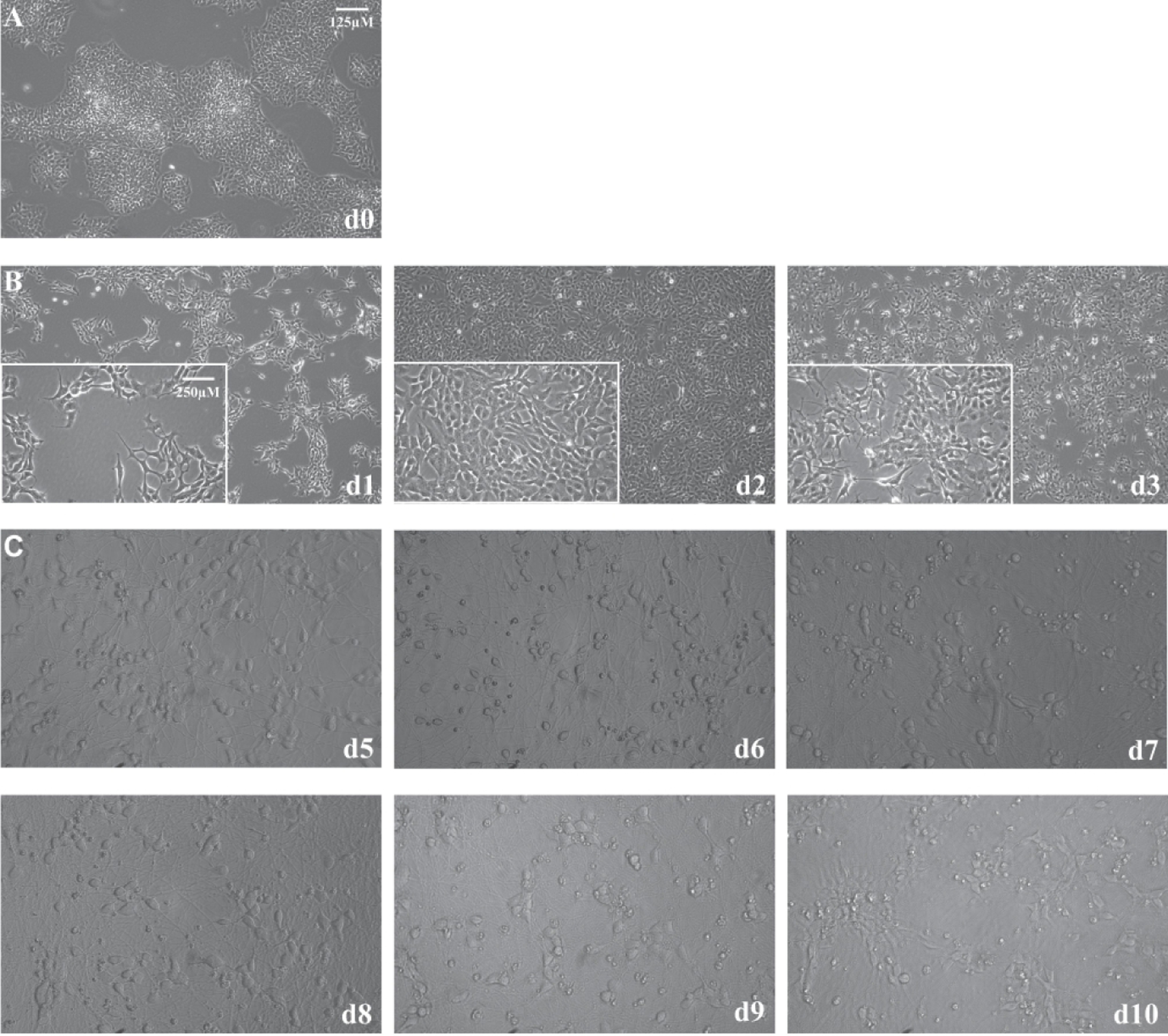
A) i3Neuron differentiation. At day 0, 10X image of iPSCs that were dissociated into single cells and plated in 15cm matrigel coated dishes for differentiation into i3Neurons. B) 10X images of i3Neurons days 1–3 (inset images are at 40X). Doxycycline was added to the media to induce the expression of neurogenin-2, encoded by a stable genomic insertion under the control of a TetOn promoter. By day 3, neuritic extensions were observed (see 40X insert image d3). On day 3, i3Neurons were dissociated as single cells for re-plating or for cryopreservation. C) 10X images of i3Neurons at cell days 5 through 10. These images show i3Neurons plated in PLO laminin coated 384 well plates at 15,000 cells/well. The i3Neurons make extensive networks over these 6 days.
Development of a high content, mitochondrial screening platform for human neurons
Varkuti et al (2020) describe a high content assay and screen to search for compounds that increase mitochondrial content, health, and/or function using mouse cortical neurons 18. This assay employed a mitochondrial targeted version of the TagGFP2 reporter engineered into the Rosa26 locus behind a conditional promoter in order to sparsely label mitochondria in the cultured neurons. Expression of the reporter is dependent on treating the cultures with AAV expressing Cre recombinase. A similar scheme was used to develop an assay for human neurons differentiated from iPSCs. First, a lentivirus (Figure 2A) was created to express mitochondrial targeted TagGFP2 in the human neurons under the control of the strong, ubiquitous CAG promoter which contains the CMV early enhancer element, a minimal promoter, the first exon and intron of the chicken beta-actin gene and the splice acceptor of the rabbit beta-globin gene 19, 20. iPSCs and iPSC derived cells have been found to silence gene expression from certain promoters via methylation and histone modification 21, 22. Therefore, it was critical to use a robust promoter that was not susceptible to these silencing processes.
Figure 2.
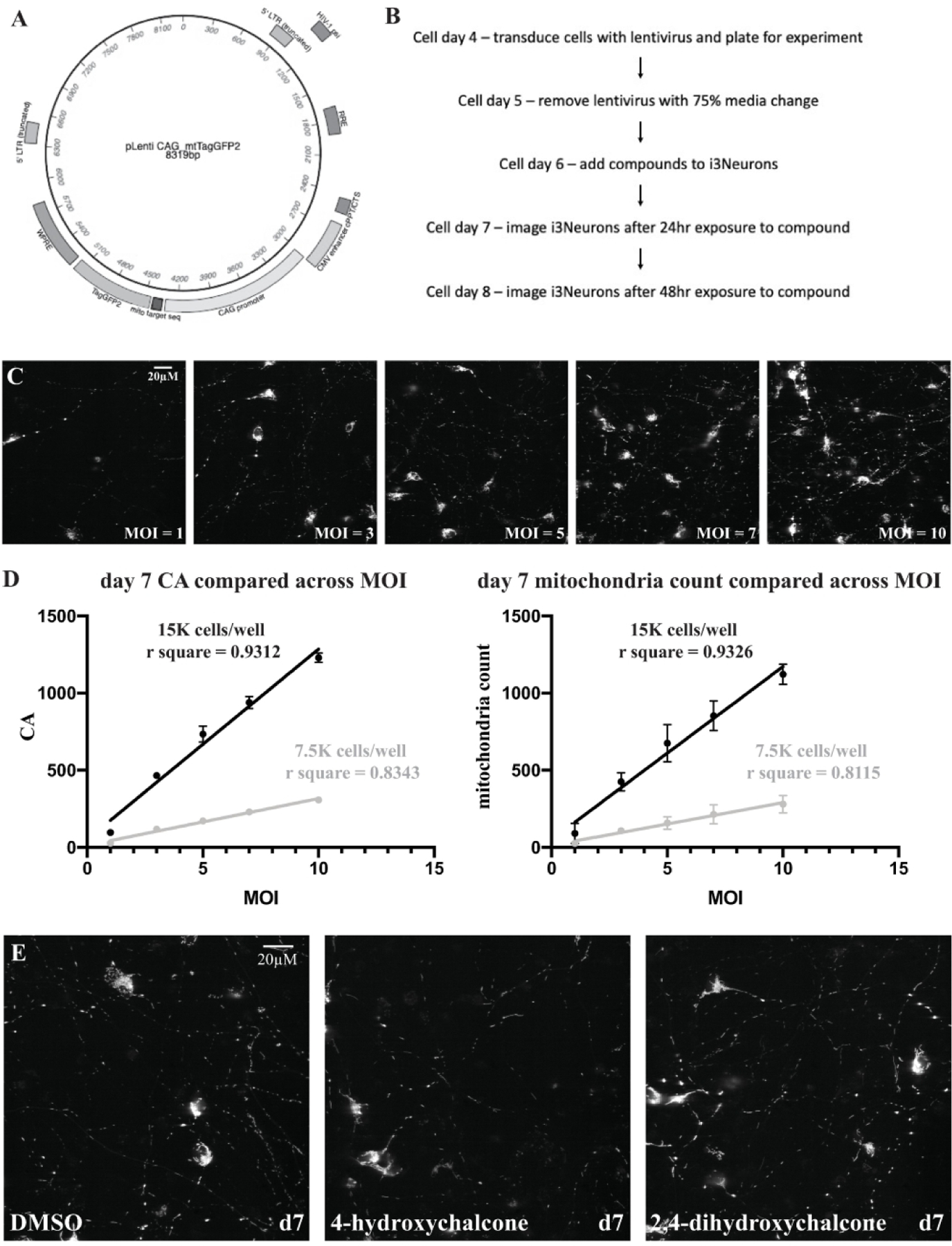
A) Map of the pLenti CAG>mtTagGFP2 plasmid used for lentivirus production. B) Timeline of the multiplexed, high content assay of i3Neurons. C) 60X representative images of a MOI titration experiment using CAG>mtTagGFP2 at cell day 7. 15,000 cells per well were infected with increasing MOI from left to right. D) MOI plotted against CA and mitochondria count on cell day 7 for both 7,500 and 15,000 cells per well densities. These two parameters are most informative in determining how accurately an i3Neuron image is segmented for individual mitochondria. The average±SEM values for each parameter at each MOI are plotted on the graphs (many of the SEMs are too small to be seen on the graph). MOIs between 3 and 7 for the 15,000 cells per well density illustrated a linear relationship with sufficient signal to be accurately segmented. E) 60X representative images of i3Neurons with TagGFP2 labeled mitochondria. The compounds, 4-hydroxychalcone and 2,4-dihydroxychalcone, when exposed to the i3Neurons at a concentration of 3 μM, cause visible elongation of mitochondria without a loss in overall CA (mitochondrial content) compared to the DMSO control.
We next turned our attention to finding the optimum cell density per well and MOI that would offer sparse but robust signals from mitochondria required for accurate segmentation. We found from experience that images obtained from random fields containing too few objects tend to increase the variability of the assay. Too many objects within a field, in contrast, leads to overlapping objects and inaccuracy in the assay. We analyzed a single plate with either 7,500 or 15,000 i3Neurons per well with varying MOIs of CAG>mtTagGFP2 lentivirus: 1,3,5,7 and 10 on cell day 4 and imaged the neurons at cell day 7 and cell day 8. Tables 1 and 2 show the results obtained from the experiment using 15,000 cells per well at the various MOIs for CA (cumulative area: sum of the areas of all mitochondria) and mitochondrial count, the two critical parameters for determining optimal segmentation of neuritic mitochondria. Supplemental Tables 1 and 2 show the results obtained from the 7,500 cells per well assay. Plotting the MOI vs CA or mitochondrial count demonstrated high linearity across the 1–10 MOI range for the 15,000 cells per well data, with perhaps the best fit using MOI of between 3 and 7 (Figure 2D). Thus, we chose to use 15,000 cells per well for the assay, and while MOIs between 3 and 7 could be used; we chose a MOI of 5 to conserve on use of the lentivirus.
Table 1.
i3Neuron CAG>mtTagGFP2 lentivirus MOI titration day 7 results for 15,000 cells per well cell density
| parameter | MOI | day | average* | SEM | ANOVA** |
|---|---|---|---|---|---|
| cell body count | 1 | 7 | 1.2 | 0.3 | p value = 0.0002 |
| cell body count | 3 | 7 | 5.3 | 0.6 | NS |
| cell body count | 5 | 7 | 7.5 | 0.4 | |
| cell body count | 7 | 7 | 11.0 | 0.3 | p value = 0.0012 |
| cell body count | 10 | 7 | 12.0 | 0.9 | p value < 0.0001 |
|
| |||||
| CA | 1 | 7 | 96 | 26 | p value < 0.0001 |
| CA | 3 | 7 | 465 | 24 | p value < 0.0001 |
| CA | 5 | 7 | 735 | 52 | |
| CA | 7 | 7 | 940 | 39 | p value = 0.0030 |
| CA | 10 | 7 | 1230 | 31 | p value < 0.0001 |
|
| |||||
| mito count | 1 | 7 | 90 | 25 | p value < 0.0001 |
| mito count | 3 | 7 | 425 | 22 | p value < 0.0001 |
| mito count | 5 | 7 | 676 | 46 | |
| mito count | 7 | 7 | 853 | 36 | p value = 0.0041 |
| mito count | 10 | 7 | 1122 | 25 | p value < 0.0001 |
|
| |||||
| median circularity | 1 | 7 | 0.59 | 0.15 | NS |
| median circularity | 3 | 7 | 0.79 | 0.00 | NS |
| median circularity | 5 | 7 | 0.78 | 0.00 | |
| median circularity | 7 | 7 | 0.77 | 0.00 | NS |
| median circularity | 10 | 7 | 0.77 | 0.00 | NS |
|
| |||||
| median length | 1 | 7 | 1.03 | 0.27 | p value = 0.0349 |
| median length | 3 | 7 | 1.55 | 0.01 | NS |
| median length | 5 | 7 | 1.55 | 0.01 | |
| median length | 7 | 7 | 1.58 | 0.02 | NS |
| median length | 10 | 7 | 1.59 | 0.01 | NS |
CA = sum of the total mitochondrial area; NS = not significant; MOI = multiplicity of infection.
4 fields were imaged per well and 7 independent wells were tested per MOI; field data was collapsed into an average well value for each variable tested; the 7 well values were averaged together to provide the data in the table above.
ANOVA results illustrate whether the parameter value at a particular MOI was significantly different from that same parameter value at a MOI=5.
Table 2.
i3Neuron CAG>mtTagGFP2 lentivirus MOI titration day 8 results for 15,000 cells per well cell density
| parameter | MOI | day | average* | SEM | ANOVA** |
|---|---|---|---|---|---|
| cell body count | 1 | 8 | 1.5 | 0.4 | p value < 0.0001 |
| cell body count | 3 | 8 | 6.1 | 0.4 | p value = 0.0440 |
| cell body count | 5 | 8 | 8.9 | 0.8 | |
| cell body count | 7 | 8 | 11.8 | 0.6 | p value = 0.0283 |
| cell body count | 10 | 8 | 13.1 | 1.0 | p value = 0.0009 |
|
| |||||
| CA | 1 | 8 | 114 | 31 | p value < 0.0001 |
| CA | 3 | 8 | 591 | 30 | p value = 0.0002 |
| CA | 5 | 8 | 953 | 73 | |
| CA | 7 | 8 | 1247 | 56 | p value = 0.0029 |
| CA | 10 | 8 | 1686 | 54 | p value < 0.0001 |
|
| |||||
| mito count | 1 | 8 | 111 | 31 | p value < 0.0001 |
| mito count | 3 | 8 | 561 | 29 | p value = 0.0001 |
| mito count | 5 | 8 | 916 | 69 | |
| mito count | 7 | 8 | 1166 | 54 | p value = 0.0078 |
| mito count | 10 | 8 | 1605 | 47 | p value < 0.0001 |
|
| |||||
| median circularity | 1 | 8 | 0.58 | 0.15 | NS |
| median circularity | 3 | 8 | 0.79 | 0.00 | NS |
| median circularity | 5 | 8 | 0.79 | 0.00 | |
| median circularity | 7 | 8 | 0.78 | 0.00 | NS |
| median circularity | 10 | 8 | 0.78 | 0.00 | NS |
|
| |||||
| median length | 1 | 8 | 1.06 | 0.27 | NS |
| median length | 3 | 8 | 1.54 | 0.01 | NS |
| median length | 5 | 8 | 1.53 | 0.01 | |
| median length | 7 | 8 | 1.56 | 0.01 | NS |
| median length | 10 | 8 | 1.55 | 0.01 | NS |
CA = sum of the total mitochondrial area; NS = not significant; MOI = multiplicity of infection.
4 fields were imaged per well and 7 independent wells were tested per MOI; field data was collapsed into an average well value for each variable tested; the 7 well values were averaged together to provide the data in the table above.
ANOVA results illustrate whether the parameter value at a particular MOI was significantly different from that same parameter value at a MOI=5
As anticipated, increasing the MOIs led to significantly increased cell body count, CA, and mitochondrial count compared to a MOI = 5 for the data obtained using 15,000 cells per well (Figure 2C, Table 1 and 2). Median circularity and median length parameters were not expected to differ when varying the MOIs. On cell day 7, the median circularity parameter was not significantly different between the various MOIs; however, MOI = 1 had a significantly lower median length than MOI = 5 (Table 1). By day 8, there were no significant differences between the median circularity or length values. The difference observed can be attributed to low signal intensity that is resolved by day 8. The CA values (Table 1 and 2, Supplemental Table 1 and 2) using 7,500 cells per well were approximately 25% the values observed when using 15,000 cells per well, rather than the expected 50%. This could be due to increased growth-factor stimulated generation of neurite growth from the higher cell density. These data indicate that the assay needs to be carefully optimized for cell density and virus MOI in order to obtain consistent results. We chose to use 15,000 cells per well for subsequent assays.
i3Neurons reproduce mitochondrial phenotypes observed in mouse cortical neurons
Our prior study using mouse neurons identified multiple compounds named MnMs (Modulators of neuronal Mitostasis) that influence mitochondrial content, elongation and circularity. We chose to use two of the compounds, 4-hydroxychalcone and 2,4-dihydroxychalcone, to develop the high content assay using i3Neurons. These compounds were found to reduce median circularity and dramatically increase median mitochondria length 18. The i3Neurons were plated on cell day 4 followed by compound addition on cell day 6 with imaging on cell day 7 (24hr after compound exposure) and cell day 8 (48hr after compound exposure) (Figure 2B). Each compound was solubilized in DMSO and assayed over a concentration range of 1 to 10 μM with each concentration being assayed in 4 wells. DMSO alone was also placed in 20 wells of the plate as a control at a final concentration of 0.125%. Two independent plates were assayed in parallel and robust z-scores were calculated using the DMSO wells on each plate as the control population for other wells on that plate. Figure 2E shows representative images for the DMSO control and both chalcone compounds at a concentration of 3 μM, illustrating that these compounds dramatically lengthen mitochondria to the point of being visually apparent. At this concentration, there was no significant difference in mitochondrial content as calculated by CA (data not shown) between the chalcones and DMSO.
Dose response curves were constructed for each compound for the median circularity and median length parameters (Figure 3). The EC50 values after 24 hr of exposure to 4-hydroxychalcone were 1.9 μM for both mitochondrial median circularity and median length. These values remained fairly constant after 48hr of exposure to the compound, increasing slightly to 2.6 μM and 2.4 μM respectively. The EC50 values after 24 hr of exposure to 2,4-dihydroxychalcone were similar to 4-hydroxchalcone at 2.0 μM for median circularity and 2.1 μM for median length, increasing slightly with 48hr of exposure to 3.1 μM and 3.2 μM, respectively.
Figure 3.
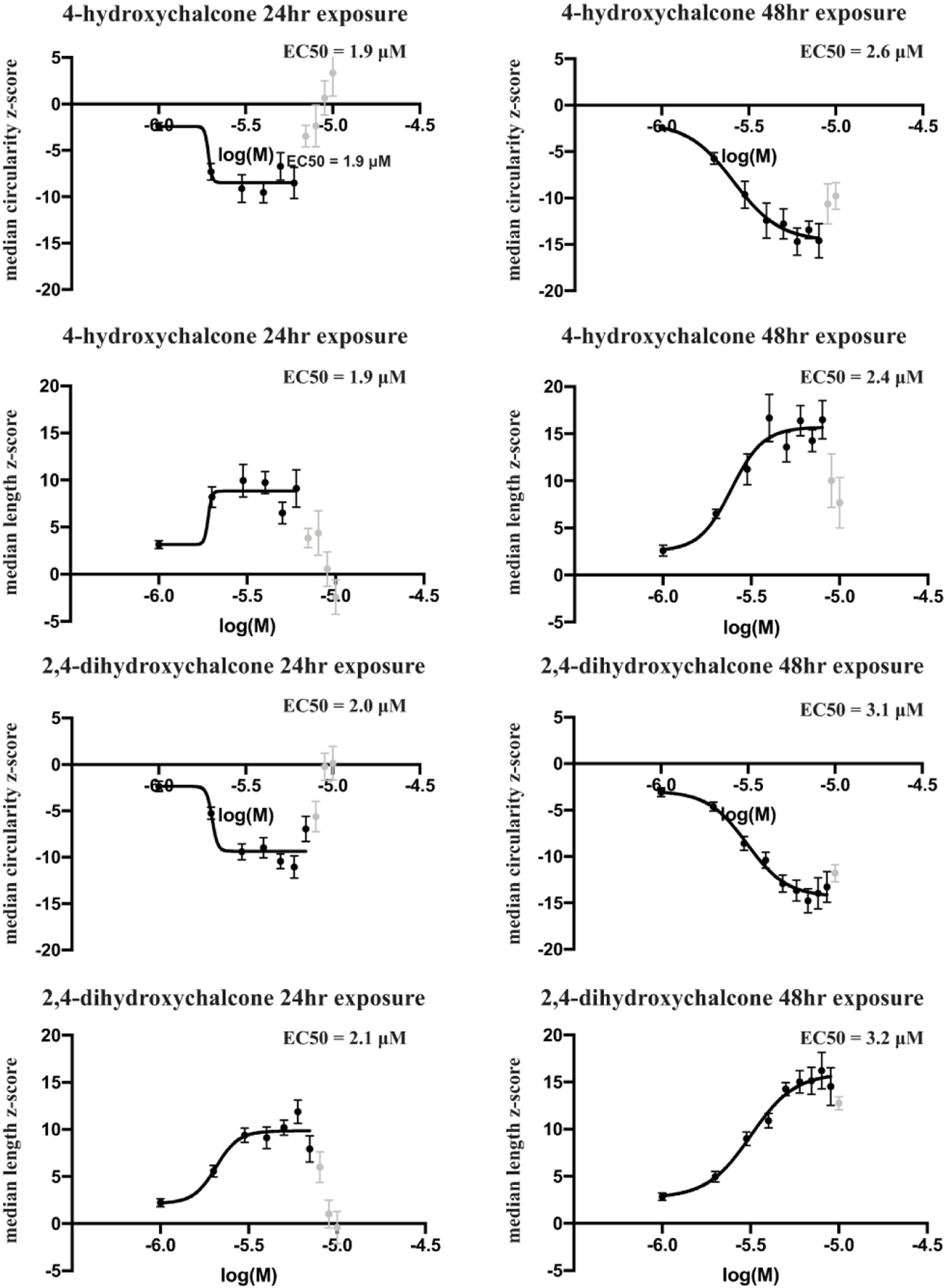
Dose response curves for mitochondrial median circularity and length using 4-hydroxychalcone and 2,4-dihydroxychalcone. Each compound was assayed over a concentration range from 1 to 10 μM on two duplicate plates. Robust z-scores were calculated for mitochondrial median circularity and length values of the neuritic mitochondria for each well and were plotted against the log10 of the molar concentrations of the compounds. Each point on the plot represents the robust z-score average±SEM of 8 independent wells between the two independent plates. High concentrations of each compound caused toxicity in the wells as illustrated by increased and/or decreased z-scores (light grey points) where appropriate. These values were excluded for calculating the EC50 values.
i3Neurons respond to compounds in similar ways under extended culture conditions
The i3Neurons can be maintained in a healthy state in culture for over 1 month by co-culturing the neurons with human astrocytes. Figures 4A and 4B illustrate how i3Neuron mitochondrial content changes over a 4-week culture period as axonal and dendritic networks become more developed. Figure 4C illustrates how the CA (sum of the total mitochondria area) remains rather constant through 14 days of culture, but then significantly increases at cell days 21 and 28. The 4-hydroxychalcone and 2,4-dihydroxychalcone dose response experiments were repeated using the extended culture conditions in which media was changed every 7 days until the compound was added on cell days 13, 20 and 27 for imaging on cell days 14, 21 and 28 (Figure 5). Again, CA values did not differ significantly from the DMSO control (data not shown). However, a significant dose response was observed to both 4-hydroxychalcone and 2,4-dihydroxchalcone for the parameters of median circularity and median length at each weekly time point tested, similar to that observed at cell days 7 and 8 under the basal culturing conditions. The EC50 obtained using the extended culture conditions were very similar to those obtained using the simpler basal culturing conditions without human astrocytes. One noteable difference is the reduced toxicity of the compounds at high concentrations using the extended vs basal culturing conditions, which presumably reflects increased health of the neurons when astrocytes are present.
Figure 4.
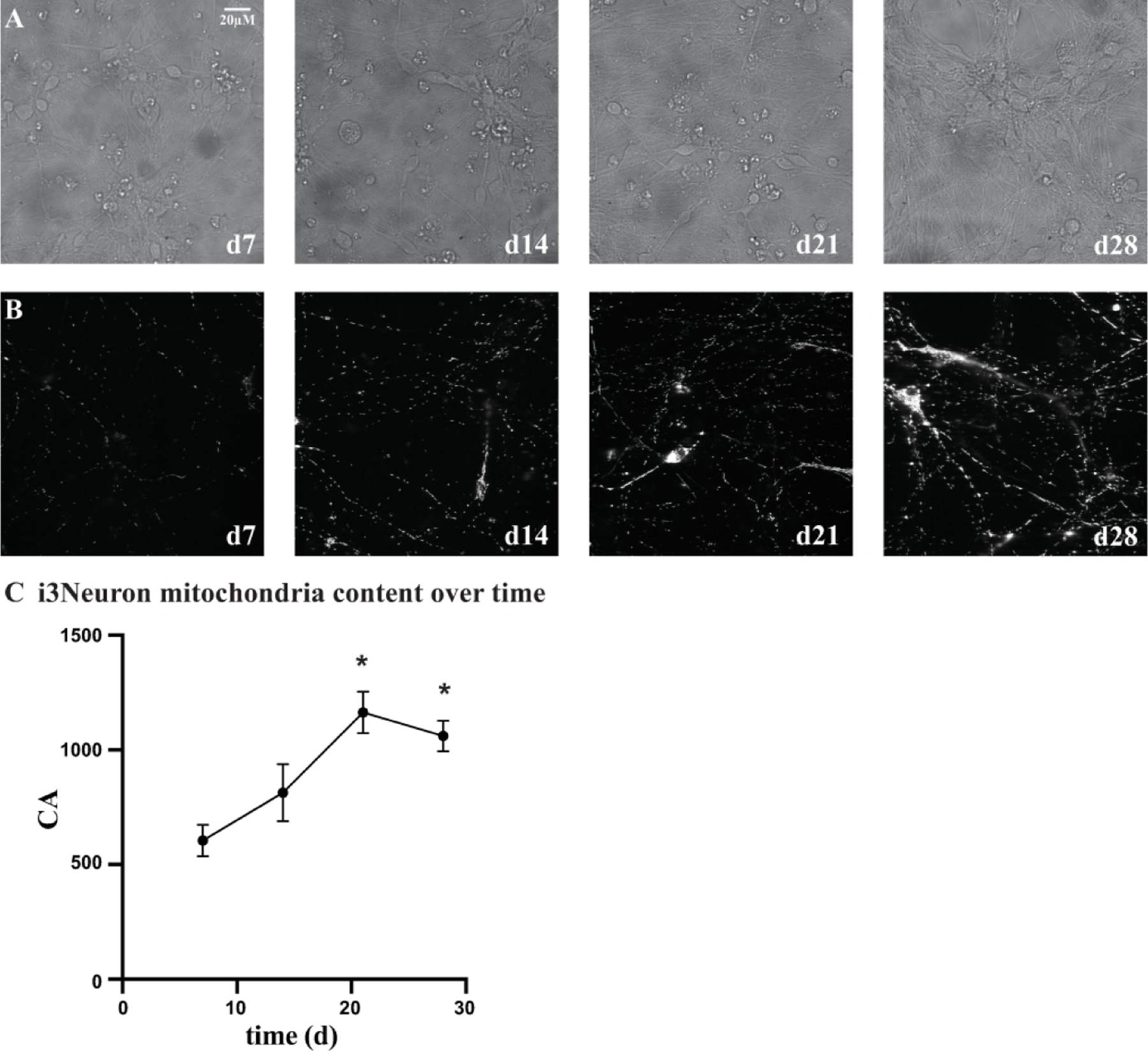
i3Neuron health and viability extended with astrocyte co-culture. A) 60X brightfield images of i3Neurons transduced with CAG>mtTagGFP2 lentivirus at day 2. i3Neurons were cryopreserved on day 3 and revived to be co-plated with human astrocytes at a 2:1 ratio in each well of a 384 well plate (10,000 i3Neurons/5,000 human astrocytes). B) Corresponding 60X TagGFP2 images of i3Neurons. The number of neuritic processes and mitochondria increased with time in culture. C) Quantification of i3Neuron mitochondrial content over time. The average CA values for i3Neuron mitochondria when co-cultured with astrocytes are plotted over the 4-week time culturing period. While the CA values are not significantly different between cell day 7 and day 14, they significantly increase (asterisk) by cell day 21 and day 28 (ANOVA with Tukey multiple comparison. p value = 0.0004 and p value = 0.0048 for cell day 21 and cell day 28, respectively. compared to cell day 7). Values are the average±SEM of 32 i3Neuron wells cultured in cortical neuron media.
Figure 5.
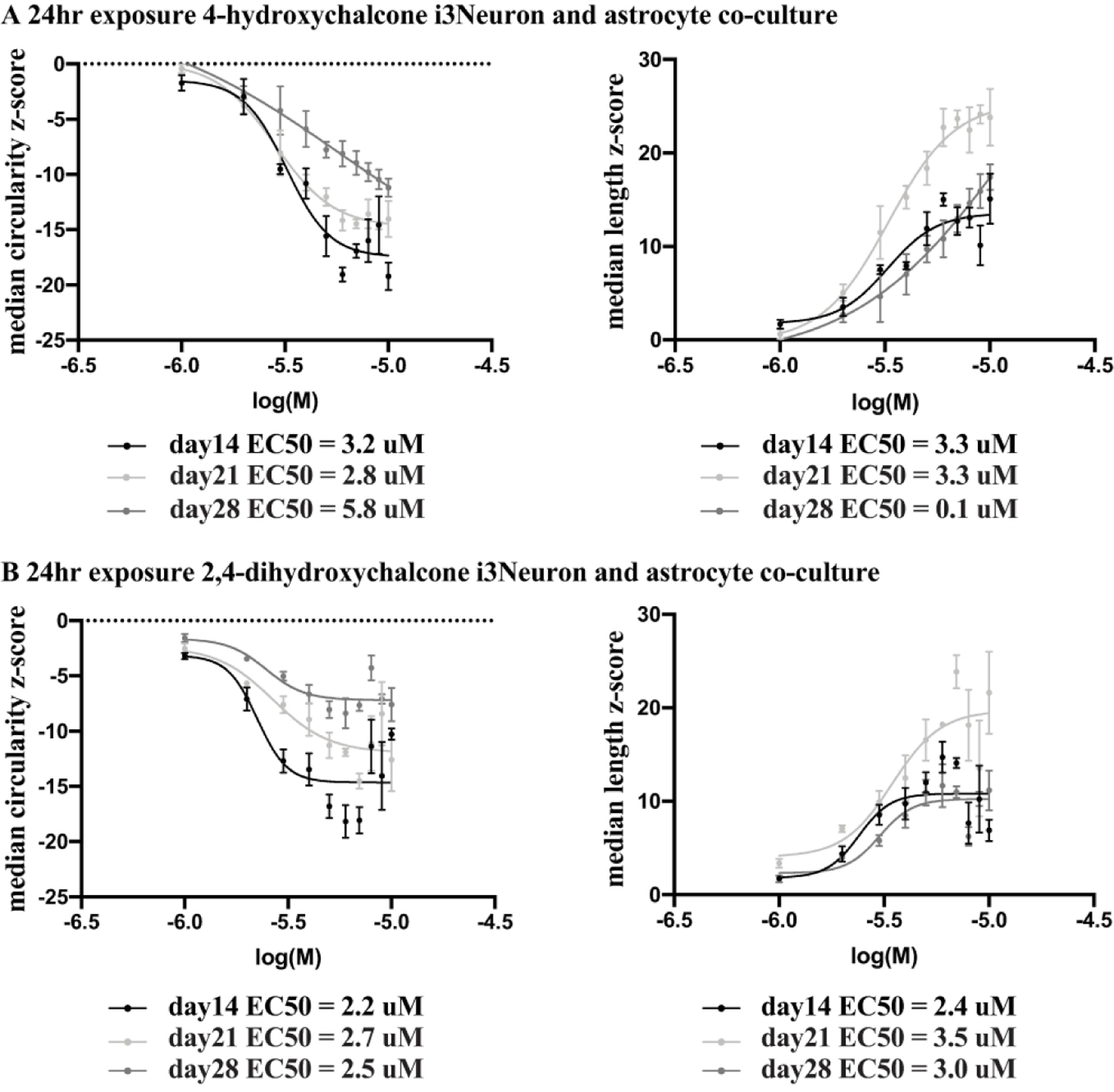
Dose response curves for i3Neurons co-cultured with human astrocytes over a three-week culture period. A) i3Neurons co-cultured with human astrocytes were exposed to 4-hydroxychalcone for a 24hr period prior to imaging at cell days 14, 21 and 28. Robust z-scores were calculated against DMSO control values at each time point, plotted as dose response curves, and resulting EC50 values are shown beneath each graph for the mitochondrial median circularity and median length parameters. Each point on the plot represents the robust z-score average±SEM of 4 independent wells on the single plate at each weekly time point. The day 14 and day 21 EC50 values remain consistent with the day 14 values having the largest span values for median circularity and the day 21 values having the largest span values for median length. B) i3Neurons co-cultured with human astrocytes were exposed to 2,4-dihydroxychalcone for a 24hr period prior to imaging and dose response curves were graphed as described above. For this compound, the EC50 values at all three time points remained relatively consistent with the day 14 values having the largest span values for mitochondrial median circularity and the day 21 values having the largest span values for median length.
Discussion:
Mitochondrial dysfunction is a hallmark of aging for many different diseases, including neurodegenerative diseases and other neuropsychiatric disorders. For some genetic forms of PD, ALS, and Charcot-Marie Tooth disease, an insult to mitochondria is the precipitating factor for the disease 23–25. For others, including AD, mitochondrial dysfunction remains as a very prominent aspect of the neuropathology. However, a hypothesis of increasing influence – the mitochondrial cascade hypothesis for sporadic Alzheimer’s disease – envisions mitochondrial dysfunction as an initiator or very near the top of the hierarchy of system failures involved in disease progression 26, 27. The prominence of impairments in mitochondrial dysfunction across these diseases commands the development of new assays that may offer strategies for obtaining the needed therapeutics. To date, very few high throughput assays have been developed that focus on mitochondrial phenotypic readouts to aid in the search for compound targets that improve mitochondria health and function 18, 28–30. The assay presented here has great potential for mitotherapeutic discovery.
This i3Neuron mitochondrial phenotypic assay offers many advantages: (1) It is performed using a homogenous population of the target cells of interest. (2) It is based in human biology rather than that of a rodent. (3) It relies on live imaging of the mitochondria in culture such that multiple imaging sessions can be performed across time. (4) The cells can be produced quickly and in large quantities. (5) The assay can be accomplished within one week. (6) The assay is adaptable to other types of cells in the CNS involved in disease pathology. However, the lentivirus multiplicity of infection is batch dependent and must be optimized for each new production of i3Neurons. Careful planning of cell based screening assays will help ensure that the same assay optimizations can be used throughout the length of the screen.
Most assays used in the past were developed using cell systems that are easy to maintain such as HEK293 cells. While these cells are derived from a human source, they may not offer the required environment for discovering effective therapeutics targeting neurons, given the unique anatomy and physiology of the neuron. Neuroscience based drug discovery in the past has relied on rodent model systems. There have been many failed clinical trials for compounds passed through the rodent drug development pipelines, giving pause as to whether they can be considered reliable for mimicking human neuronal environments 31. Substantial efforts in prior neurodegeneration research has relied on fixed, frozen tissue. Fixation procedures distort cellular morphology while live cell imaging circumvents these issues. High throughput screens based on primary neuron cultures obtained from the mouse can be plagued from a lack of starting material at a critical time, given that cultures are inevitably started from embryonic or early postnatal tissues. The i3Neurons can be produced in large quantities and cryopreserved for all experiments for a screen. Finally, the response to tested compounds using the basal or the extended culture conditions appear to remain stable; therefore, one can obtain reliable results after only 1 week of culture. Thus, the assay developed has many important advantages in screening for potential mitotherapeutics compared to previously developed assays using rodent neurons.
We validated the assay primarily using two potent positive compounds, 4-hydroxychalcone and 2,4-dihydroxychalcone, obtained from our prior screens using rodent assays (18), In addition, several other compounds identified initially from our screens using rodent neurons have exhibited significant responses using this assay that are currently being studied. However, not all of the compounds identified from the rodent screens exhibit strong potency using the i3Neuron assay. To date, we have screened 26 compounds found to improve mitochondrial health and/or count using mouse primary neurons. Of these compounds, 8 (including 4-hydroxychalcone and 2,4-dihydroxychalcone) have shown evidence of improving mitochondrial health. This observation is notable for three major reasons. First, identifying negative compounds during assay development is an important part of the validation of the assay. Second, the difference in efficacy may reveal authentic differences in the biology of rodent vs human neurons, commanding greater emphasis on the use of human neurons to discover new therapeutics. Finally, the disparity may be due to the simple differences in the two assays. For instance, the rodent assays employ a heterogenous set of neurons isolated from the forebrain, whereas the i3Neuron assay uses homogenous glutamatergic neurons.
This i3Neuron mitochondrial phenotypic assay by itself can only suggest novel compounds for future pre-clinical development. Additional assays that help select the most promising compounds will complement these studies. Assays that mimic the insults to mitochondria that neurodgenerative diseases present include increasing oxidative stress, adding glutamate to mimic glutamate toxicity, or treating cultures with toxic peptides 18 All three of these assay examples are amenable to the i3Neuron assay. In addition, there are options for pursuing studies to reveal mechanism of action, including RNA-seq and proteomics approaches that may define the pathways in which selected compounds alter human mitochondrial dynamics. In sum, the phenotypic and high throughput assay using iPSC derived human neurons to select compounds altering mitochondrial dynamics has the potential to quickly identify key compounds for further research and development.
Supplementary Material
Acknowledgement:
We would like to thank Michael E. Ward of the National Institute of Neurological Disorders and Stroke, National Institutes of Health for providing the Ngn-2 iPSC line used to differentiate i3Neurons.
Funding:
This research was supported by NIH grant R01MH109957 and 3R01MH109957–03S1 to RLD.
Abbreviations:
- AD
Alzheimer’s Disease
- PD
Parkinson’s Disease
- HD
Huntington’s Disease
- ALS
Amyotrophic lateral sclerosis
- iPSC
induced pluripotent stem cells
- Ngn-2
neurogenin-2
- EDTA
ethylenediaminetetraacetic acid
- DPBS
Dulbecco’s phosphate buffered saline
- NEAA
non-essential amino acids
- PLO
poly-L-ornithine
- MOI
multiplicity of infection
- BDNF
brain-derived neurotrophic factor
- NT-3
neurotrophin 3
- PDL
poly-D-lysine
- DMSO
dimethylsulfoxide
- FBS
fetal bovine serum
- AAV
adeno-associated virus
- ICC
immunocytochemistry
- Tuj1
beta-III tubulin
Footnotes
Author disclosure statement:
The authors declare no conflict of interest.
References:
- 1.Miller G. Is pharma running out of brainy ideas? Science 2010, 329, 502–4. [DOI] [PubMed] [Google Scholar]
- 2.Heilker R; Traub S; Reinhardt P.; et al. iPS cell derived neuronal cells for drug discovery. Trends Pharmacol Sci 2014, 35, 510–9. [DOI] [PubMed] [Google Scholar]
- 3.Pankevich DE; Altevogt BM; Dunlop J.; et al. Improving and accelerating drug development for nervous system disorders. Neuron 2014, 84, 546–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherman SP; Bang AG High-throughput screen for compounds that modulate neurite growth of human induced pluripotent stem cell-derived neurons. Dis Model Mech 2018, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown DG; Wobst HJ Opportunities and Challenges in Phenotypic Screening for Neurodegenerative Disease Research. J Med Chem 2020, 63, 1823–1840. [DOI] [PubMed] [Google Scholar]
- 6.Johri A; Beal MF Mitochondrial dysfunction in neurodegenerative diseases. J Pharmacol Exp Ther 2012, 342, 619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y; Chen M; Jiang J. Mitochondrial dysfunction in neurodegenerative diseases and drug targets via apoptotic signaling. Mitochondrion 2019, 49, 35–45. [DOI] [PubMed] [Google Scholar]
- 8.Murphy MP How mitochondria produce reactive oxygen species. Biochem J 2009, 417, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nunnari J; Suomalainen A. Mitochondria: in sickness and in health. Cell 2012, 148, 1145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tait SW; Green DR Mitochondrial regulation of cell death. Cold Spring Harb Perspect Biol 2013, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lou G; Palikaras K; Lautrup S.; et al. Mitophagy and Neuroprotection. Trends Mol Med 2020, 26, 8–20. [DOI] [PubMed] [Google Scholar]
- 12.Stadtfeld M; Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev 2010, 24, 2239–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirenko O; Hesley J; Rusyn I.; et al. High-content high-throughput assays for characterizing the viability and morphology of human iPSC-derived neuronal cultures. Assay Drug Dev Technol 2014, 12, 536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufmann M; Schuffenhauer A; Fruh I.; et al. High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen 2015, 20, 1101–11. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y; Pak C; Han Y.; et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013, 78, 785–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandopulle MS; Prestil R; Grunseich C.; et al. Transcription Factor-Mediated Differentiation of Human iPSCs into Neurons. Curr Protoc Cell Biol 2018, 79, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gossen M; Freundlieb S; Bender G.; et al. Transcriptional activation by tetracyclines in mammalian cells. Science 1995, 268, 1766–9. [DOI] [PubMed] [Google Scholar]
- 18.Varkuti BH; Kepiro M; Liu Z.; et al. Neuron-based high-content assay and screen for CNS active mitotherapeutics. Sci Adv 2020, 6, eaaw8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niwa H; Yamamura K; Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991, 108, 193–9. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki J; Takaki S; Araki K.; et al. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene 1989, 79, 269–77. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann D; Schott JW; Geis FK; et al. Detailed comparison of retroviral vectors and promoter configurations for stable and high transgene expression in human induced pluripotent stem cells. Gene Ther 2017, 24, 298–307. [DOI] [PubMed] [Google Scholar]
- 22.Pfaff N; Lachmann N; Ackermann M.; et al. A ubiquitous chromatin opening element prevents transgene silencing in pluripotent stem cells and their differentiated progeny. Stem Cells 2013, 31, 488–99. [DOI] [PubMed] [Google Scholar]
- 23.Dupuis L; Gonzalez de Aguilar JL; Oudart H.; et al. Mitochondria in amyotrophic lateral sclerosis: a trigger and a target. Neurodegener Dis 2004, 1, 245–54. [DOI] [PubMed] [Google Scholar]
- 24.Haelterman NA; Yoon WH; Sandoval H.; et al. A mitocentric view of Parkinson’s disease. Annu Rev Neurosci 2014, 37, 137–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burte F; Carelli V; Chinnery PF; et al. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat Rev Neurol 2015, 11, 11–24. [DOI] [PubMed] [Google Scholar]
- 26.Swerdlow RH; Burns JM; Khan SM The Alzheimer’s disease mitochondrial cascade hypothesis: progress and perspectives. Biochim Biophys Acta 2014, 1842, 1219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez Ortiz JM; Swerdlow RH Mitochondrial dysfunction in Alzheimer’s disease: Role in pathogenesis and novel therapeutic opportunities. Br J Pharmacol 2019, 176, 3489–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efthymiou A; Shaltouki A; Steiner JP; et al. Functional screening assays with neurons generated from pluripotent stem cell-derived neural stem cells. J Biomol Screen 2014, 19, 32–43. [DOI] [PubMed] [Google Scholar]
- 29.Little D; Luft C; Mosaku O.; et al. A single cell high content assay detects mitochondrial dysfunction in iPSC-derived neurons with mutations in SNCA. Sci Rep 2018, 8, 9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varkuti BH; Liu Z; Kepiro M.; et al. High-Throughput Small Molecule Screen Identifies Modulators of Mitochondrial Function in Neurons. iScience 2020, 23, 100931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shanks N; Greek R; Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med 2009, 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


