Summary
Premature infants are at substantial risk for suffering from perinatal white matter injury. Though the gut microbiota has been implicated in early-life development, a detailed understanding of the gut-microbiota-immune-brain axis in premature neonates is lacking. Here, we profiled the gut microbiota, immunological, and neurophysiological development of 60 extremely premature infants, which received standard hospital care including antibiotics and probiotics. We found that maturation of electrocortical activity is suppressed in infants with severe brain damage. This is accompanied by elevated γδ T cell levels and increased T cell secretion of vascular endothelial growth factor and reduced secretion of neuroprotectants. Notably, Klebsiella overgrowth in the gut is highly predictive for brain damage and is associated with a pro-inflammatory immunological tone. These results suggest that aberrant development of the gut-microbiota-immune-brain axis may drive or exacerbate brain injury in extremely premature neonates and represents a promising target for novel intervention strategies.
Keywords: perinatal white matter injury, extremely premature infants, gut-immune-brain axis, T cell ontogeny, microbiome, neurophysiology, early-life development, Klebsiella
Graphical abstract
Highlights
-
•
Microbiota, immune, and neurophysiological profile of 60 extremely premature infants
-
•
Pro-inflammatory T cell response correlates with suppressed electrocortical maturation
-
•
γδ T cells have central implications for the pathogenesis of brain injury
-
•
Klebsiella overgrowth in the gastrointestinal tract is predictive for brain damage
Seki et al. performed a comprehensive, time-resolved analysis of the gut microbiota-immune-brain axis in extremely premature infants. This analysis links early-life microbiome establishment to immunological and neurological development, identifying candidate biomarkers of perinatal brain injury and promising targets for personalized treatments to alleviate, or even prevent, life-long neurological morbidities.
Introduction
The incidence of premature birth has been rising worldwide and is one of the leading causes of perinatal morbidity and mortality (Tommiska et al., 2007). Although recent advances in neonatal intensive care have increased the survival of extremely premature infants (gestational age < 28 weeks), the number of survivors with severe morbidity and life-long neurodevelopmental impairment remains high (Adams-Chapman et al., 2018). The third trimester of pregnancy is a critical period for the establishment, refinement, and maturation of human brain connectivity, which determines subsequent cognitive potential (Matthews et al., 2018). Extremely premature infants are born on the verge of the third trimester and their neural circuitry is therefore established in the face of manifold environmental cues and insults associated with preterm extra-uterine life (Stoecklein et al., 2020).
One inevitable environmental cue is the immediate post-natal colonization of the body by microorganisms. The establishing gut microbiota is in contact with a plurality of neurons of the enteric nervous system (Cryan et al., 2019). Increasing evidence suggests that during the period of early-life, enteric microorganisms participate in bidirectional signaling between the gastrointestinal tract (GIT) and the brain (Warner, 2019). This reciprocal communication between prokaryotes and the human central nervous system (CNS) is established via the gut-brain axis, whereby parasympathetic fibers from the vagus nerve are directly exposed to bacteria and their metabolites (Bonaz et al., 2018), while the immune (Rooks and Garret, 2017) and endocrine system mediate indirect crosstalk (Keunen et al., 2015). The developing brain of premature infants is at especially high risk of suffering from perinatal white matter injury (PWMI), which can manifest as intraventricular hemorrhage (IVH), periventricular leukomalacia (PVL), or diffuse white matter injury (DWMI) (Volpe, 2009). It is now widely accepted that perinatal infection and associated inflammation are involved in the pathophysiology of PWMI and can worsen the outcome of brain injuries (Niño et al., 2018), resulting in life-long morbidities such as cognitive and motor delay, cerebral palsy, neurosensory impairments, and behavioral abnormalities such as attention deficit hyperactivity disorder and learning disabilities (Woodward et al., 2006).
A balanced relationship between the developing immune system and gut microbiota is crucial to prevent excessive inflammation (Mazmanian et al., 2008). However, the establishment of a mutualistic relationship between the gut microbiota and its host seems to be impaired in extremely premature infants, despite daily administration of probiotics in many neo-intensive care units (NICUs) (Alcon-Giner et al., 2020). It remains unclear if this impairment in early-life development is due to the intensive medical care give to this vulnerable patient cohort, or if certain expected and required immunological cues are missing. T lymphocytes are key mediators of sepsis-induced brain injury (Benakis et al., 2016). γδ T cells, in particular, are abundant in the neonatal gut and are unique among T cells due to the flexibility of their receptor chains (Ferreira, 2013). This flexibility allows them to respond directly to antigens without requiring prior antigen-processing and major-histocompatibility-complex presentation for their activation. They have also been detected in brain lesions of premature infants with white matter injury (Albertsson et al., 2018), suggesting that they may be important mediators of gut-brain axis communication.
Here, we report a systems-level analysis of the gut microbiota, immune system, and neurophysiological development during hospitalization of 60 extremely premature infants. We conducted 16S rRNA gene amplicon sequencing in combination with qPCR for quantitative microbiome profiling, flow cytometry for T cell phenotyping, multiplex cytokine and chemokine quantification, fecal metabolite analysis, neurophysiological measurements, and neuroimaging to gain a holistic understanding of the developing gut microbiota-immune-brain-axis in early life. We found that brain development in extremely premature infants is comprised three major phases: (1) an initial quiescent phase, (2) a period of neurophysiological maturation, and (3) a term-equivalent phase. We identified features of the microbiota and immune system in the quiescent phase that may initiate an inflammatory cascade that extends through the neurophysiological maturation phase and ultimately aggravates brain injury.
Results
White matter injury is preceded by distinct neurophysiological signatures
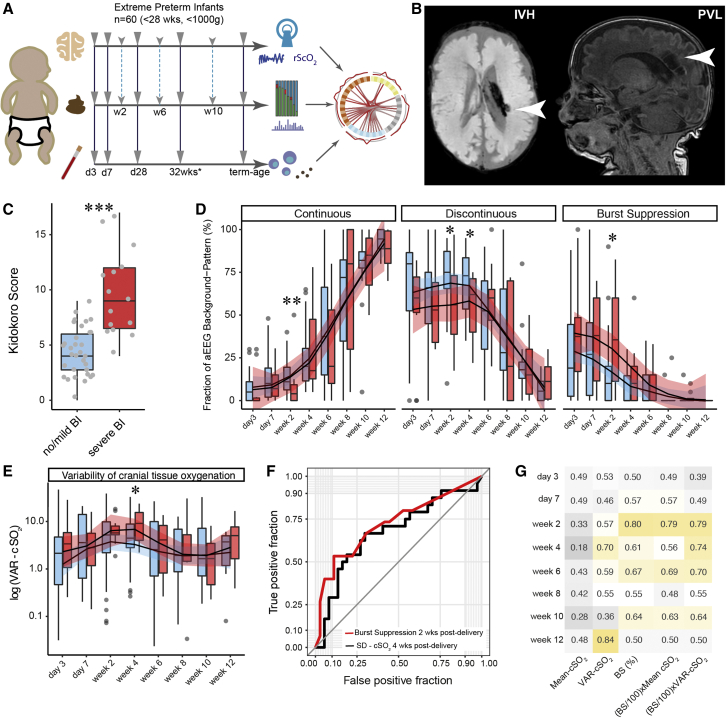
Sixty extremely premature infants were enrolled between September 2017 and June 2019 at the Medical University of Vienna. Characteristics of the study cohort can be found in Tables 1 and S1. Over the course of hospitalization, neurophysiological development was monitored, stool samples were collected, and blood was drawn at multiple time points (Figure 1A). Brain injuries were identified by combined assessment of ultrasound imaging at multiple time points during hospitalization as well as by magnetic resonance imaging (MRI) at term-equivalent age. Out of the 60 infants, seven died, 38 displayed age-adequate MRIs or only mild brain injuries, and 15 infants were diagnosed with severe pathological brain injuries. Observed injuries included periventricular hemorrhagic infarctions (n = 5), intraventricular hemorrhages (IVH grade > 2, n = 13), cerebellar hemorrhages (n = 7), and periventricular leukomalacia (PVL, n = 4). Many of these pathologies were accompanied by a reduction of brain volume and an enlargement of the subarachnoid spaces. Representative MRI images for IVH and PVL are shown in Figure 1B. In order to quantify brain injury, the Kidokoro score was used as a standardized MRI assessment tool of the nature and extent of brain injury as well as impaired brain growth (Kidokoro et al., 2013). As expected, the Kidokoro score was significantly higher in infants with severe brain injuries (Figure 1C; t test; p = 0.0001). Although gestational age (GA) has been suggested to be an important predictor of cerebral injury, we found no significant association between GA at birth and neuroimaging outcome (t test; p = 0.4371).
Table 1.
Subject cohort demographics
| Characteristic | Extremely premature infants without or with mild brain injury | Extremely premature infants with severe brain injury | p value (t test and Fisher’s exact test) |
|---|---|---|---|
| Individuals, number | 38 | 15 | |
| GA at birth, weeks | 25.5 ± 1.2 | 24.8 ± 1.3 | 0.100 |
| Birth weight, grams | 774 ± 137 | 718 ± 156 | 0.213 |
| Birth head circumference, cm | 23.2 ± 1.4 | 22.7 ± 1.6 | 0.234 |
| Birth length, cm | 32.9 ± 2.3 | 32.4 ± 2.0 | 0.520 |
| Female gender, % | 63% | 53% | 0.546 |
| Spontaneous birth, % | 11% | 53% | 0.001 |
| IUGR, % | 8% | 13% | 0.614 |
| pPROM, % | 63% | 33% | 0.068 |
| Received lung maturation, % | 97% | 93% | 0.489 |
| Apgar score at 5 min | 8.4 ± 0.9 | 8.4 ± 1 | 0.874 |
| Apgar score at 10 min | 8.7 ± 0.8 | 8.9 ± 0.2 | 0.468 |
| CSI, % | 32% | 47% | 0.351 |
| Culture-proven EOS, % | 5% | 0% | 1 |
| Clinical LOS, % | 16% | 33% | 0.257 |
| Culture-proven LOS, % | 29% | 20% | 0.731 |
| Hospitalization, days | 91.82 ± 37.32 | 91.53 ± 24.29 | 0.884 |
| NEC, % | 8% | 0% | 1 |
| ROP, % | 47% | 100% | 0.0003 |
| BPD, % | 27% | 20% | 0.733 |
| PDA, % | 37% | 60% | 0.217 |
| Antibiotic intervention, days | 15.89 ± 11.5 | 18.00 ± 13.4 | 0.598 |
| Age at discharge, weeks | 39.2 ± 4.5 | 38.7 ± 2.0 | 0.693 |
| Weight at discharge, grams | 2,929 ± 956 | 2,788 ± 325 | 0.600 |
| Length at discharge, cm | 47.0 ± 5.2 | 46.4 ± 2.6 | 0.699 |
| Head circumference at discharge, cm | 33.0 ± 3.1 | 32.8 ± 2.0 | 0.855 |
| Milk formula fortifier administered shortly before discharge | 77% | 77% | 1 |
IUGR, intrauterine growth restriction; pPROM, preterm premature rupture of membranes; CSI, clinical-suspected inflammation; EOS, early-onset sepsis; LOS, late-onset sepsis; BPD, bronchopulmonary dysplasia; NEC, necrotizing enterocolitis; ROP, retinopathy of prematurity; PDA, persistent ductus arteriosus. Data are presented as mean ± SD or percentage (%).
Figure 1.
Neurophysiological development of extremely premature infants with and without severe brain injuries
Blue color for age-adequate cranial magnetic resonance imaging (cMRI) results or mild brain injury (BI). Red color for severe BI.
(A) Schematic illustration of study design. Solid arrows indicate sampling time points with blood drawings on day 3, day 7, day 28 post-delivery, 32 weeks gestational age, and at term-equivalent age. Dashed lines indicate time points for stool sampling and neurophysiological assessment (day 3, day 7, and day 14, followed by biweekly sampling until discharge).
(B) Representative cMRI images at term-equivalent age for IVH > 2 (left) and PVL (right).
(C) Comparison of Kidokoro scores between infants with and without severe BI.
(D) Comparison of aEEG background patterns (1) continuous voltage (CV), (2) discontinuous voltage (DV), (3) burst suppression (BS).
(E) Comparison of the variability of cranial oxygen supply (VAR-cSO2) (fraction of deviation beyond 10% from the mean of the total NIRS measurement) between infants with and without BI.
(F) Receiver operating characteristic (ROC) curve visualizing the predictive potential of cranial oxygen fluctuation (VAR-cSO2) 4 weeks post-delivery, and BS signatures 2 weeks post-delivery for brain damage.
(G) Summary of area under the curve (AUC) scores of neurophysiological parameters at given time points. Color gradient ranges from dark gray (low AUC score) to dark yellow (high AUC score). Box plots show group median and interquartile range. Smoothed lines result from locally estimated scatterplot smoothing (LOESS) and indicate trends of neurophysiological development. Asterisks represent p values: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In order to characterize neurophysiological development in our patients, we employed the amplitude-integrated electroencephalography (aEEG) sum score (Hellstro m-Westas et al., 2006; Klebermass et al., 2011), which quantifies four distinct patterns of electrocortical activity: (1) continuous voltage (CV), (2) discontinuous voltage (DV), (3) burst suppression (BS), and (4) flat trace (FT). These patterns reflect the cortical activity of the patient at the time of measurement. We observed a consistent trend of neurophysiological maturation that was characterized by an initial quiescent phase, in which DV was the dominant aEEG background pattern (Figures 1D, S1A, and S1B). This was followed by a period of neurophysiological maturation (maturation phase) in which CV continuously increased and eventually plateaued by term-equivalent age (term-equivalent phase). Generally, infants with brain injuries experienced a delayed onset of the maturation phase, with lower CV and higher BS 2 weeks post-delivery (Wilcoxon; p = 0.008 and 0.028, respectively). Also, DV was significantly lower in infants with brain injuries 2 and 4 weeks post-delivery (Wilcoxon; p = 0.038 and 0.045, respectively) (Figure 1D). Neurophysiological maturation patterns converged by 4 weeks post-delivery, after which both groups continued to follow a similar course.
We next evaluated brain tissue oxygenation using near-infrared spectroscopy (NIRS). In order to quantify potentially harmful instabilities in brain tissue oxygenation, we calculated the prevalence of extreme fluctuations in brain tissue oxygenation (cSO2), defined as the fraction of time for which cSO2 was +/− 10 cSO2 from the mean value during the NIRS measurement (VAR-cSO2). We observed that fluctuations in cSO2 were significantly elevated during the early maturation phase (4 weeks post-delivery) in infants with severe white matter injuries (t test; p = 0.025; Figure 1E).
Furthermore, we evaluated the predictive potential of aEEG and NIRS signatures at each time point for neuroimaging outcome. We found that although mean cSO2 had no predictive value for detecting brain injury, VAR-cSO2 had some predictive power 4 weeks post-delivery (area under the curve [AUC] = 0.70). The level of BS was most predictive 2 weeks post-delivery (AUC = 0.80) and by combining BS and VAR-cSO2 (BS/100 × VAR-cSO2), predictive potential was at AUC = 0.79 2 weeks post-delivery and increased to AUC = 0.74 2 and 4 weeks post-delivery (Figures 1F and 1G).
Therefore, we conclude that neuroimaging outcome is preceded by a delayed onset of electrocortical maturation. This transient suppression of neurophysiological development and increased instability in cranial oxygen saturation at approximately 2 to 4 weeks post-delivery is predictive for PWMI at term-equivalent age and may have an important role in its pathology.
Neonates with brain injury display a pro-inflammatory polarized T cell response
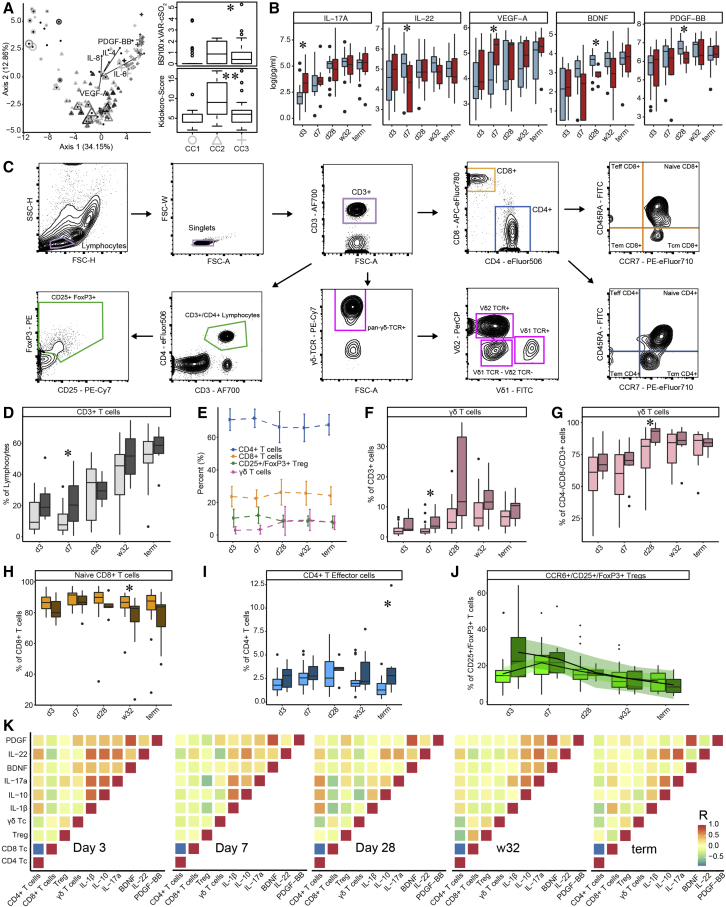
Although T cells are an important link between the gut microbiome and the brain (Benakis et al., 2016) their ontogenesis during early life, and their association with perinatal brain damage in extremely premature infants, remains poorly understood. To investigate temporal changes in T cell dynamics in relation to perinatal brain damage, we investigated T cell subsets via fluorescence-activated cell sorting (FACS) and their corresponding cytokine response via multiplex immunoassays in blood samples from 40 extremely premature infants at 5 time points. Out of these, four died, 26 displayed age-adequate MRIs or only mild brain injuries, and ten infants were diagnosed with severe pathological brain injuries. Principal coordinates analysis (PCoA) of cytokine response revealed three distinct clusters of samples: one early-life cluster (Cyto-Cluster 1; mean days post-delivery: 17) and two clusters from later time points (Cyto-Cluster 2 and 3; mean days post-delivery: 30 and 35, respectively) (Figure 2A). Cyto-Cluster 2 was enriched in samples from infants suffering brain damage (38% in Cyto-Cluster 2 versus 10% in Cyto-Cluster 3). Strikingly, Kidokoro scores at term-equivalent age, as well as BS combined with VAR-cSO2 (BS/100 × VAR-cSO2) were higher in Cyto-Cluster 2 as compared with Cyto-Cluster 3 (Wilcoxon; p = 0.035 and 0.0088, respectively). Levels of interleukins 4, 6, and 8 (IL-4, IL-6, and IL-8), vascular endothelial growth factor-A (VEGF-A), and platelet-derived growth factor-BB (PDGF-BB) were significantly associated with the observed variance of the PCoA, with VEGF-A driving the formation of Cyto-Cluster 2 and the other cytokines being associated with Cyto-Cluster 3. Furthermore, we built a correlation network of secreted cytokines and chemokines to reveal connectivity within their expression. Most notably, we found that networks produced with samples from infants without severe brain injuries displayed a more central role of PDGF-BB as compared with infants with severe brain injuries, whose networks were rather centered around interleukin 17a (IL-17a) (Figures S2A–S2C). Also, by employing topographical data analysis of cytokine levels, we observed a divergence in the developmental trajectory in premature infants 1 month after birth, which converges at term age (Figure S2D).
Figure 2.
T cell ontogenesis in extremely premature infants with and without severe brain injury
(A) Principal coordinates analysis (PCoA)—biplot of sequestered cytokine and chemokine composition. Silhouette scoring identified 3 main cyto-Clusters (as indicated by different symbols). The shade of the symbols (gray to black) is determined by days post-delivery—the older the infant the darker the symbol. The size of the symbols is determined by the Kidokoro Score as assessed at term-equivalent age via cMRI—the higher the score the larger the symbol. Significantly correlated (p < 0.05) cytokines/chemokines are plotted as arrows. In addition, box plots next to PCoA show the range of combined values for burst suppression and variance of cranial oxygenation (BS/100 × VAR-cSO2), as well as the range of Kidokoro Scores in the respective cyto-clusters (cyto-cluster 1, CC1; cyto-cluster 2, CC2; cyto-cluster 3, CC3).
(B) Blood cytokine/chemokine concentrations in infants with (red) and without (blue) severe BI (3 days post-delivery, d3; 7 days post-delivery, d7; 28 days post-delivery, d28; 32 weeks of gestational age, w32; term-equivalent age = term).
(C) Representative images illustrating the gating strategy for FACS analysis. Differently colored gates mark gating for respective cell populations. Magenta, untargeted; blue, T helper cells; orange, cytotoxic T cells; green, T regulatory cells; pink, γδ T cells. For box plots in (D) and (F–J), darker shade represents data from infants with, and lighter shade from infants without, severe BI.
(D) Proportion of CD3+ cells during hospitalization.
(E) Relative distribution of T regulatory (green), T helper (blue), γδ T (pink), and cytotoxic T cells (orange).
(F–I) Box plots showing the expression of T cell receptors relative to the given parental population throughout hospitalization.
(J) Box plots showing the expression of CCR6+/T regulatory cells with the addition of smoothed lines resulting from locally estimated scatterplot smoothing (LOESS) to indicate trends of development.
(K) Spearman correlation analysis to reveal associations between T cell populations and selected sequestered cytokines/chemokines at respective time points (p < 0.001). Red color indicates positive correlation, blue color indicates negative correlation. Box plots show group median and interquartile range. Asterisks represent p values: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
When comparing cytokine expression between infants with and without brain injury, we observed that peripheral blood T cells in infants with brain injury had elevated production of IL-17a 3 days post-delivery (Wilcoxon; p = 0.048) and VEGF-A 7 days post-delivery (Wilcoxon; p = 0.034), accompanied by reduced interleukin 22 (IL-22) 7 days post-delivery (Wilcoxon; p = 0.044; Figure 2B). Although these differences had disappeared by 28 days post-delivery, samples from infants with brain injury 28 days post-delivery exhibited reduced production of the neuroprotectants brain-derived neurotrophic factor (BDNF) (Wilcoxon; p = 0.012) and PDGF-BB (Wilcoxon; p = 0.026) (Figure 2B). These data suggest that a pro-inflammatory T cell polarization via IL-17a early after birth underlies latter aberrant diversification, during which a dysregulated VEGF-A response is key for the pathogenesis of brain injury. Additional cytokines and chemokines that were quantified but did not show significant differences between infants with and without brain injuries are shown in Figure S2G.
We then profiled T regulatory, T helper, cytotoxic, and γδ T cells from the same samples via FACS (Figures 2D–2J). T helper cells (TH:CD4+/CD3+) were the most abundant CD3+ lymphocytes, followed by cytotoxic T cells (TH:CD8+/CD3+), regulatory T cells (Treg:CD25+FoxP3+/CD3+CD4+), and γδ T cells (γδPAN/CD3+) (Figure 2E). Infants with brain injuries had increased levels of T cells 1 week after birth (Wilcoxon; p = 0.039; Figure 2D), which corresponded with elevated levels of γδ T cells in the CD3+ population (Wilcoxon; p = 0.027; Figure 2F). The majority of γδ T cells exist as double negative (DN; CD8 and CD4) T cells. Shortly after birth, ∼60% of the cells within the overall DN T cell population expressed a PAN-γδ T cell marker. 1 month after birth, DN PAN-γδ T cells increased in relative numbers and were significantly more abundant in infants with brain injuries (Wilcoxon; p = 0.031; Figure 2G). In addition, infants with severe brain injuries had fewer naive CD8+ T cells at 32 weeks corrected GA (Wilcoxon; p = 0.031; Figure 2H) and more effector CD4+ T cells at term-equivalent age (Wilcoxon; p = 0.014; Figure 2I). Although not statistically significant, we observed elevated numbers of migration-stimulating CCR6+ T regulatory cells (Yamazaki et al., 2008) in infants with brain injuries shortly after birth (Figure 2J). Additional T cell receptors that were quantified but did not show significant differences between infants with and without brain injuries are shown in Figure S2H.
Next, we sought to unravel the interplay of VEGF-A, interleukins, and the neuroprotectants PDGF-BB and BDNF with different T cell populations via correlation analysis (Figure 2K). The presented values are extracted from a correlation analysis of all measured cytokines and T cells. T helper cells were negatively associated with cytotoxic T cells at all time points, as well as with γδ T cells at three and 28 days post-delivery and at 32 weeks GA. 3 days post-delivery, VEGF-A was negatively associated with the neuroprotectants PDGF-BB and BDNF. Seven days post-delivery T helper cells were negatively associated with IL-10 and IL-22, while T regulatory cells were negatively associated with IL-1beta and IL-17a. γδ T cells, however, were positively associated with IL-8 and PDGF-BB. At 28 days post-delivery, VEGF-A was associated positively with the increase of γδ T cells, but negatively with T regulatory cells. This positive association disappeared in later time points, whereas the negative association with T regulatory cells persisted. Furthermore, we assessed the influence of cytokine and chemokine secretion levels, as well as Kidokoro scores and myelination scores assessed via cMRI at term age, on the variance of given T cell populations via canonical correlation analysis (CCA). We found that the composition of T regulatory cells, T helper cells, cytotoxic T cells, and γδ T cells across all samples was significantly affected by the expression of IL-17a (p value: 0.004), IL-12P40 (p value: 0.004), PDGF-BB (p value: 0.002), and IL-4 (p value: 0.028), as well as cranial myelination at term age (p value 0.028), with a combined explanatory power of all variables of R2 = 0.26. (Figure S2E).
We conclude that the increase of γδ T cells starting 7 days post-delivery may be involved in the pathogenesis of PWMI. This initial polarization correlates with IL-8, IL-17a, VEGF-A, and PDGF-BB secretion at first. Subsequently, a strong link between VEGF-A and γδ T cells persists, whereas the further secretion of neuroprotectants PDGF-BB and BDNF is suppressed in infants with severe brain damage.
Overgrowth of Klebsiella is associated with brain damage
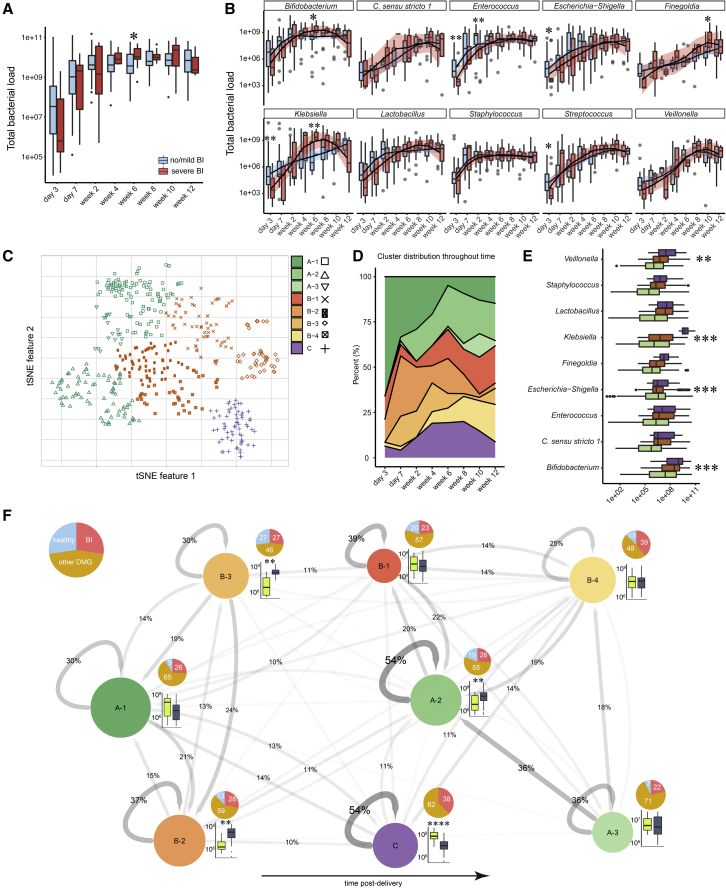
The microbiota is an important instigator of immunological tolerance, as the nature and quantity of microbial antigens determine lymphocyte responses (Sekirov et al., 2010). We employed 16S rRNA gene-targeted qPCR and 16S rRNA gene amplicon sequencing to quantify total bacterial load in stool samples and for quantitative microbiome profiling of community composition. We observed an increase in bacterial load over time that plateaued by 2 weeks post-delivery, reaching a concentration of ∼109 cells per g feces (Figure 3A). 6 weeks post-delivery, patients with brain injuries had slightly higher bacterial loads (Wilcoxon; p = 0.03), but otherwise, colonization dynamics were very similar to patients without brain injury. Overall, the gut microbiota was dominated by 10 abundant and prevalent genera (Bifidobacterium, Escherichia-Shigella, Enterococcus, Lactobacillus, Staphylococcus, Streptococcus, Klebsiella, Clostridium sensu stricto 1, Veillonella, and Finegoldia), which occurred in a minimum of 20% of samples with an abundance of at least 105 cells and together comprised 75% of all 16S rRNA amplicon sequence variants (ASVs; Figure S3A). We observed that strictly anaerobic core bacteria (Clostridia, Finegoldia, and Veillonella) occurred sparsely early post-delivery and increased in abundance in later periods (Figure 3B; Pearson correlation, R = 0.53, 0.62, and 0.48, respectively, p < 0.001; Figure S3C). Strikingly, Klebsiella was on average 1.7-fold more abundant 4 weeks post-delivery (Wilcoxon; p = 0.0049), but less abundant shortly post-delivery in infants with severe brain injury (Wilcoxon; p = 0.0066; Figure 3B). Consistent with this observation, there was a statistically significant association of Klebsiella pneumoniae ASV_c07_vnh_xuf with severe brain injury (edgeR differential abundance test; p = 0.0467). This strain was isolated from stool of a premature infant suffering from severe brain injury, and its genome features several potential virulence factors as well as genes allowing it to colonize in the inflamed gut, including biofilm formation potential, a repertoire of antibiotic resistance genes, iron-scavenging siderophores, and the genes for dissimilatory nitrate reduction to ammonium (Tables S2 and S3). Furthermore, we observed increased Bifidobacterium sp. 6 weeks post-delivery, and Finegoldia sp. shortly before discharge, in infants with severe brain injuries (Wilcoxon; p = 0.0455 and 0.033, respectively). However, infants without severe brain injuries furthermore had elevated levels of Enterococcus, Escherichia-Shigella, and Streptococcus (Wilcoxon; p = 0.0083, 0.028, and 0.045, respectively) 3 days post-delivery, as well as more Enterococcus 2 weeks post-delivery (Wilcoxon; p = 0.0065; Figure 3B).
Figure 3.
The microbiome of extremely premature infants and its diversification corresponding to brain injury
(A) Total bacterial cell counts per gram feces over time.
(B) Absolute abundances of the most abundant bacterial genera in infants with (red) and without (blue) severe BI.
(C) T-distributed stochastic neighbor embedding (tSNE) ordination of all 16S rRNA gene amplicon libraries corrected by absolute abundances and 16S rRNA gene copy number. Hierarchical clustering (Ward’s method) identified 3 main clusters (green, orange, and purple) and 8 sub-clusters (symbols).
(D) Area plot illustrating the percentage distribution of sub-clusters throughout time.
(E) Absolute abundance of bacterial genera in respective main clusters.
(F) Network illustrating the trajectories of microbiome succession in extremely premature infants. Circles correspond to tSNE sub-clusters. Circle size indicates the number of patients found in a sub-cluster for at least one sampling time. Arrows represent transitions between clusters, whereby the thickness and shade of the arrow indicates the probability of the transition. Recursive arrows represent the percentage of consecutive observations within the same cluster. Pie charts show the distribution of patients in respective clusters diagnosed as healthy (blue, lacking any diagnosis listed in Table 1), with severe BI (red), or other pathologies (other DMG, see Table 1; ochre). Sub-clusters are arranged according to the average patient age of samples in the cluster, ordered from youngest (left) to oldest (right). Box plots next to circles show the absolute abundance of Klebsiella (bright green) and Escherichia-Shigella (dark blue) in the associated sub-cluster. Box plots show group median and interquartile range. Smoothed lines result from locally estimated scatterplot smoothing (LOESS) and indicate trends of microbiome development. Asterisks represent p values: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In order to better understand microbiome assembly and dynamics, we used T-distributed stochastic neighbor embedding (tSNE) to identify distinct microbiome states. This analysis revealed three major microbiome clusters that could be further subdivided into eight sub-clusters, as determined by silhouette score cluster optimization (Figures 3C, 3D, and S3D). Overall, the clusters had different bacterial loads (repeated measures ANOVA; p < 0.001), with cluster C having an increased load compared with clusters A and B (Tukey’s post-hoc test; p = 0.0017 and 0.0003, respectively). Cluster A divided into three and cluster B into four sub-clusters, respectively, whereas cluster C did not form sub-clusters. Furthermore, the bacterial genera Veillonella, Klebsiella, Escherichia-Shigella, and Bifidobacterium had differential abundances within the clusters (repeated measures ANOVA; p = 0.002, < 0.001, < 0.001, and 0.001). Additionally, Veillonella was elevated in cluster C as compared with cluster A (Tukey’s post-hoc test; p = 0.0012), Klebsiella was elevated in cluster C as compared with clusters A and B (Tukey’s post-hoc test; both p ≤ 0.001), Escherichia-Shigella was elevated in clusters A over cluster B (Tukey’s post-hoc test; p < 0.001), and Bifidobacterium was elevated in cluster C as compared with clusters A and B (Tukey’s post-hoc test; p = 0.036 and < 0.001, respectively; Figure 3E).
We next used the sub-clusters to examine microbiome dynamics and to determine whether the microbiome undergoes characteristic trajectories which could be indicative of deterministic succession (Figure 3F). We observed a trend in the frequency of clusters over the first 12 weeks of life, with early-life clusters A1 and B2 gradually becoming less prominent concomitant with increases in the prevalence of A2 and B1. At later time points, cluster B4 also became more frequently detected. clusters A2 and C had the highest stability, i.e., the highest probability of the microbiome staying in the same cluster at the subsequent time point. In cluster C, Klebsiella was the dominant genus, whereas in cluster A2 Escherichia-Shigella was more abundant. As no fecal samples of healthy infants were observed in cluster C, we conclude that Klebsiella domination and overgrowth in the gut may be key for driving the assembly of a distinctive and remarkably stable microbiome that is associated with neurological pathologies.
Furthermore, mode of delivery and heavy use of antibiotics have been discussed as potential drivers of aberrant microbial succession (Aguilar-Lopez et al., 2021) Although we observed no difference in time spent on antibiotics during hospitalization between infants with and without severe brain injuries, there was a significant association of severe brain injury with spontaneous delivery (Table 1). Neither the duration of antibiotic administration nor delivery mode, however, were significantly associated with the proportion of samples per patient classified as cluster-C microbiota (chi-square test, p = 0.792 and 0.723, respectively). This suggests that spontaneous birth involves particular risks for pathological brain development but does not imply subsequent development of cluster-C-like microbiota.
Enteral nutrition drives gut fermentation but does not control bacterial load
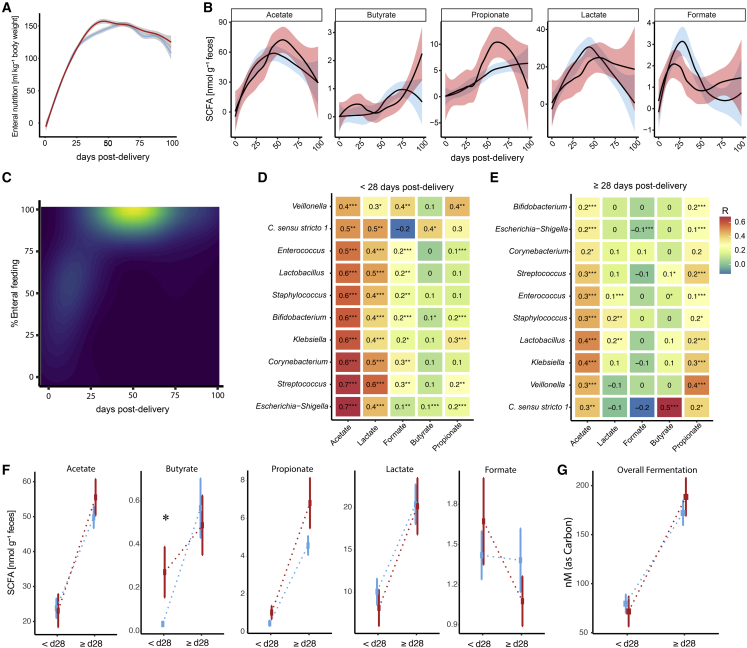
Though the healthy human adult gut is considered to be a largely anoxic environment, the GIT of premature infants has been reported to be partially oxic (Singer et al., 2019) and therefore must transition to anoxia in the course of post-natal development. In the absence of terminal electron acceptors such as oxygen, bacteria must gain energy via substrate-level phosphorylation, and in the gut this is typically accomplished by fermentation of dietary or host-derived compounds into short-chain fatty acids (SCFAs), which can act as neurological (Dalile et al., 2019) and immunological (Parada Venegas et al., 2019) signaling molecules. Although the major source of nutrients for the gut microbiota may well be expected to be derived from enteral nutrition, we observed that bacterial load was uncoupled from the quantity of enteral nutrition, with bacterial load plateauing several weeks before the enteral nutrition amount (Figures 3A and 4A). This may be due to the presence of alternative substrates such as meconium, mucins, or post-natal intestinal cell sloughing. Conversely, growth restricting effects could be introduced by molecules such as bile acids, maternal antibodies, and alternative limiting nutrients such as iron.
Figure 4.
SCFAs in stool of extremely premature infants
(A) Enteral feeding over time for infants with (red) and without severe BI (blue).
(B) Fecal SCFAs in nmol g−1 feces throughout hospitalization of premature infants with (red) and without (blue) severe BI.
(C) Weighted contour plot showing the association of SCFAs with percentage of enteral feeding and days post-delivery. Brighter color indicates higher SCFA concentrations.
(D–F) (D) Heatmap of Pearson correlations of bacterial genus abundance with SCFA levels [nmol] before 28 days post-delivery and (E) at or after 28 days post-delivery (F) SCFA concentrations in infants with brain injuries (red) and infants without (blue) before 28 days post-delivery (< d28) and at or after (> d28).
(G) Differences in overall fermentation measured as nM per Carbon (nM_C) between infants with (red) and without severe BI (blue) before 28 days post-delivery (< d28) and at after (> d28). Squares show group median, error bars show standard deviation. Smoothed lines result from LOESS and indicate trends of microbiome development. Asterisks represent p values: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We did not observe statistically significant differences between infants with and without severe brain injuries concerning the amount of acetate, butyrate, propionate, lactate, and formate measured in stool at specific sampling time points (Figure 4B). Overall, SCFA production peaked at approximately 50 days post-delivery and, unlike bacterial load, appeared to be driven by the amount of enteral nutrition (Figure 4C). Acetate and lactate can be produced by either aerobic or anaerobic metabolism, but as bacteria transition from aerobic to anaerobic metabolism, bacteria oxidize pyruvate to acetyl-CoA and formate, rather than to acetyl-CoA, CO2, and NADH (White, 2000). Interestingly, we observed the highest accumulations of formate 1 month post-delivery. This was followed by a gradual increase in propionate and butyrate, indicating that anaerobic fermentation activity gradually increases over time. This shift to anoxia is mirrored in the increase in the obligate anaerobic taxa (Figures 3B and 4B). Therefore, we pooled samples before and after 28 days post-delivery to reveal characteristics of an oxygen-influenced and anoxic premature infant gut. After day 28 post-delivery, Clostridium sensu strictu 1 correlated positively with butyrate production, and Veillonella correlated with acetate and propionate (Pearson correlation; R = 0.5, 0.3, and 0.4, respectively, p < 0.001; Figure 4E), consistent with its ability to produce acetate and propionate via fermentation (Ng and Hamilton, 1971). However, as we detect small amounts of butyrate and propionate early post-delivery as well, we conclude that although aerobic metabolism might be dominant early after birth, fermentation can exist in parallel, presumably in anaerobic micro-niches. To test if either early or late fermentation processes have implications for brain damage, we evaluated the association of SCFAs with severe brain injury. Therefore, we calculated a mean value per patient within pools of samples before and after 28 days post-delivery (Figures 4F and 4G). We found that at early time points patients with brain damage had significantly elevated levels of butyrate and slightly elevated levels of propionate (Wilcoxon; p = 0.005 and 0.068, respectively), but that there was no significant difference at later time points, suggesting that an early transition to gut anoxia may be associated with brain injury.
Biomarkers of the gut-microbiota-immune-brain axis predict PWMI
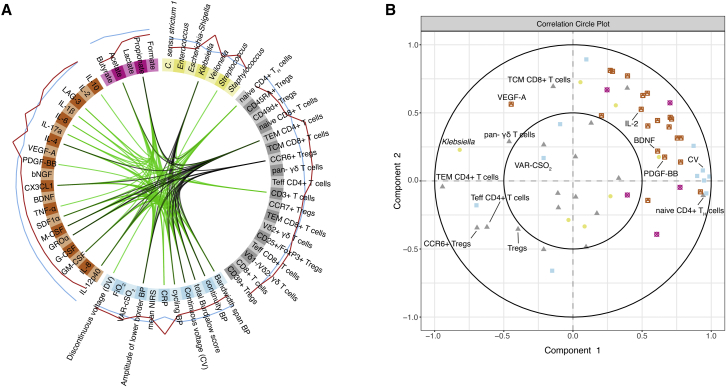
In order to identify robust biomarkers of brain damage in our dataset, we employed an integrative analysis of microbiome, metabolome, clinical, immunological, and neurophysiological data (Singh et al., 2019). The strongest indicators for white matter injury were elevated Klebsiella levels, peripheral blood C-reactive protein (CRP), specific T cell populations (effector memory T cells TH:Tem CD4+/CD4+, CCR6+ T cell receptor and γδ T cells), and increased T cell production of several cytokines and chemokines (IL-2, LAG-3, VEGF-A, IL1-beta, and IL-17a) (Figure 5A). Next, we evaluated correlations across different data types (Figure 5B). We observed an association of Klebsiella levels with γδ T cells, VEGF-A secretion, CD4+ T effector memory cells, and CCR6+ T regulatory cells. Concomitantly, Klebsiella levels are inversely associated with the secretion of many other cytokines/chemokines including the neuroprotectants PDGF-BB and BDNF.
Figure 5.
The gut-microbiota-immune-brain axis of extremely premature infants and biomarkers of brain injury
(A) Data integration analysis for biomarker discovery using Latent variable approaches (Diablo). Integration of microbiome (yellow), metabolite (pink), T cell (gray), cytokine and chemokine secretion (orange), and neurophysiological/clinical (blue) data (n = 84). Lines within the circle represent positive correlations in green, and negative correlations in black (canonical correlation cut-off: +/− 0.65). Lines outside of the circle indicate the predictive potential of corresponding variables for age-adequate or mild (blue) or severe (red) BI.
(B) Correlation circle plot showing associations between variables as measured via canonical correlations. Neighboring of variables indicates correlations between variables.
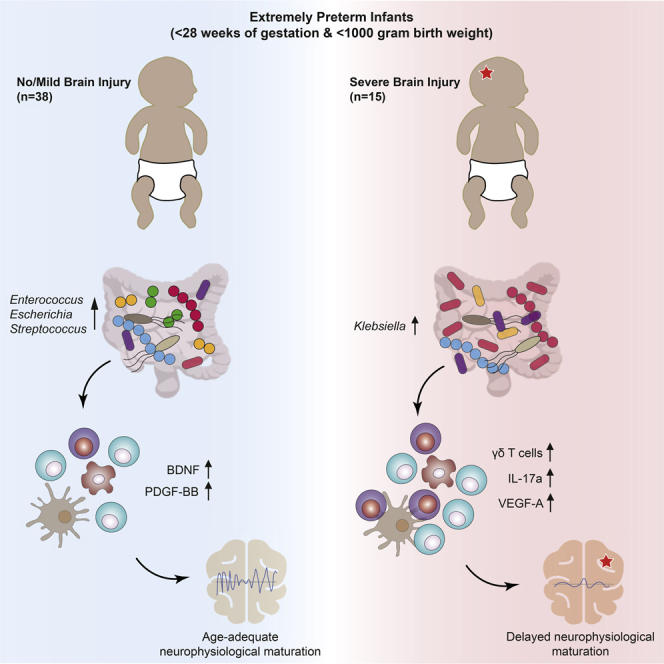
In summary, we have identified several features suggestive of an aberrant gut-microbiota-immune-brain axis development. Infants with severe brain injuries displayed a pro-inflammatory T cell polarization, which was marked by the expansion of γδ T cells in peripheral blood and driven by elevated IL-17a and VEGF-A secretion shortly post-delivery. Subsequently, the onset of neurophysiological maturation was delayed in these infants and was characterized by a suppression of electrocortical potential, an increased instability in cranial oxygen saturation, as well as a lack in secretion of neuroprotectants PDGF-BB and BDNF. Klebsiella overgrowth in the gut was key for driving the assembly of a distinctive and remarkably stable microbiome that associated with severe brain injury. Therefore, we conclude that Klebsiella spp. are involved in the dysregulation of the gut microbiota-immune-brain axis and may exacerbate PWMI.
Discussion
Our study addresses the development of the gut-microbiota-immune-brain axis in extremely premature infants. We observed a characteristic trajectory of extra-uterine brain maturation that can be divided into three major phases: a neurodevelopmental quiescent phase, neurophysiological maturation phase, and a term-equivalent state. Using systems-biology approaches, we identified Klebsiella overgrowth in the gut as highly predictive for brain damage (Figure 5). Microbiome cluster analysis revealed that Klebsiella-dominated gut communities were remarkably stable and observed exclusively in infants with pathologies (Figure 3). This potentially pathological community state is associated with pro-inflammatory and T cell migratory cues during the quiescent phase and an impaired production of neuroprotective agents accompanied by an expansion of γδ T cells during the neurophysiological maturation phase (Figure 2).
1 week post-delivery, we observed elevated levels of CCR6+ T regulatory cells in all infants (Figure 2J) indicating an onset of migratory events in immunological development, which may be triggered in part by the increase in bacterial numbers in the GIT (Figure 3A). Infants with brain injuries had higher levels of CCR6+ T regulatory cells 3 days post-delivery (Figure 2J), as well as elevated peripheral blood levels of the migration stimulant VEGF-A 7 days post-delivery (Figure 2B). VEGF-A plays a key role in the pathogenesis of retinopathy of prematurity (ROP) by promoting abnormal branching of blood vessels emanating from the retina (Kandasamy et al., 2017). Interestingly, all infants with brain injuries were also diagnosed with ROP (Table 1). Infants with ROP but without severe brain damage displayed similar levels of VEGF-A in peripheral blood. However, infants unaffected by ROP or brain injury showed significant reduction of VEGF-A levels 7 days post-delivery as compared with infants with severe brain injury (Wilcoxon; p = 0.0091) and infants with ROP but without severe brain damage (Wilcoxon; p = 0.032). This suggests that increased VEGF-A secretion early post-delivery associated with both ROP and brain injury and that notably severe brain injury includes ROP, but not vice versa.
Furthermore, we observed an increase of γδ T cells in peripheral blood in infants with severe brain injuries at 7 and 28 days post-delivery (Figures 2F and 2G). γδ T cells are important early-life immune cells as they can respond to innate-derived cytokines (Ferreira, 2013; Xu et al., 2020), and gut-resident γδ T cells have been shown to sense microbial colonization and support immunological tolerance and barrier function (Nielsen et al., 2017). During pro-inflammatory conditions, γδ T cells partially disappear from GIT tissue and abandon their intraepithelial support function (Weitkamp et al., 2014). IL-17a is an important signal for γδ T cells to migrate to the CNS, where they aggravate brain injuries (Benakis et al., 2016; Albertsson et al., 2018). Interestingly, in our cohort we observed that infants suffering from brain injury displayed elevated IL-17a levels in peripheral blood shortly after birth (Figure 2E). These data suggest that γδ T cell proliferation and migration is at least partially driven by IL-17a and is involved in the pathogenesis of perinatal brain damage.
The gut microbiota and its metabolic products have significant immunomodulatory potential, including for example being able to drive the increase of IL-17a-producing γδ T cells (Duan et al., 2010). We observed that infants with brain injuries had elevated levels of fecal butyrate, a major microbially produced metabolite, in the first month of life (Figure 4F). Importantly, differences in fecal butyrate or other SCFA levels must not solely arise from altered microbial production but may also result from altered absorption and colonocyte utilization (Litvak et al., 2018). Although butyrate can act positively on IL-10 expression and epithelial barrier function (Zheng et al., 2017), it has also been shown to aggravate GIT barrier dysfunction in patients with active inflammation (Vancamelbeke et al., 2019). As premature infants may have compromised epithelial barrier functions (Hill et al., 2017), these could potentially underlie the elevated fecal butyrate levels in early life, which may antagonize healthy mucosal development.
In premature infants, prescription of broad-spectrum antibiotics (Zwittink et al., 2017) and mode of delivery (Aguilar-Lopez et al., 2021) have been reported to affect gut-microbiota development. Cesarean (C)-sections are generally considered to be the safer delivery method for extremely premature infants, given the increased risk for respiratory distress during spontaneous delivery which can lead to brain injuries (Humberg et al., 2017). This agrees with our observation of heightened incidence for severe brain injury in spontaneously delivered premature infants (Table 1). However, C-sections have been shown to promote the growth of facultative anaerobes such as Enterobacteriaceae (Shao et al., 2019), which in our cohort were predominantly represented by the genera Escherichia-Shigella and Klebsiella. Interestingly, we observed elevated numbers of both genera in infants delivered via C-section shortly post-delivery (Figure 3B), but no association of delivery mode, or duration of antibiotic treatment with the establishment of cluster-C-like microbiota, suggesting that other mechanisms underlie the development of the given pathological community state. Enterobacteriaceae perform mixed-acid fermentation under anaerobic GIT conditions but also thrive by aerobic metabolism when oxygen is available. We observed that infants with severe brain injuries had higher bacterial loads in general, as well as higher numbers of Klebsiella 6 weeks post-delivery (Figures 3A and 3B). Klebsiella–related 16S rRNA gene amplicon sequences, as well as genomic evidence of Klebsiella isolates, indicated that Klebsiella pneumoniae was the dominant species of this genus (Table S2). It has recently been demonstrated that Klebsiella is capable of exploiting pioneer communities in the premature infant gut and establishes during later periods of microbial succession (Rao et al., 2021), though the mechanisms underlying this remain unknown. Klebsiella has previously been associated with sepsis, necrotizing enterocolitis (NEC), and nosocomial infections in neonates (Starzyk-Łuszcz et al., 2017; Wisgrill et al., 2019). Multidrug-resistant and hyper-virulent Klebsiella has emerged globally and is now a major cause of severe infections, particularly in immuno-compromised patients (Lee et al., 2017). Virulent Klebsiella strains can possess pathogenicity factors such as adhesins (Di Martino et al., 1995), type 1 and type 3 pili (Di Martino et al., 2003), siderophores (Holden et al., 2016), a thick capsule, and altered lipopolysaccharides to evade host recognition (Evrard et al., 2010). In addition to potentially producing pro-inflammatory cell components, K. pneumoniae overgrowth may also be responsible for early gut luminal oxygen depletion as well as elevated early-life butyrate levels, either directly via dark fermentation (García-Depraect et al., 2021) or indirectly by facilitating the colonization of obligate anaerobes such as Clostridia (Figure 3B). By doing so, Klebsiella may drive the pro-inflammatory polarization of immune cells and be responsible for the subsequent migration of γδ T cells. It remains unclear, however, whether Klebsiella employs specific modifications that allow host evasion to reach overgrowth, or if certain nutritional sources, e.g., host-derived nitrate, become available under pro-inflammatory conditions (Winter et al., 2013), leading to a subsequent bloom that overwhelms gastrointestinal clearance by mechanisms such as breast-milk-derived immunoglobulins (Gopalakrishna et al., 2019). Future studies should elucidate the mechanisms by which Klebsiella evades clearance from the gut, as well as to reveal the diversity of Klebsiella strains in the premature neonate gut via methods such as shotgun metagenomics.
Furthermore, probiotics are widely discussed as a potentially protective therapy against enteric pathogens. Indeed, prescription of probiotics seems to lower the risk for sepsis and NEC but fails to completely eradicate their occurrence (van Best et al., 2020). Similarly, we find that the daily probiotic administration in our cohort did not completely protect from PWMI. More research concerning the host recognition of probiotic strains during early life, as well as interaction of these with other microbiota in the GIT, is urgently needed for the development of preemptive measures other than broad-spectrum antibiotics.
We conclude that aberrant development of the gut-microbiota-immune-brain axis may play a role in brain injury in extremely premature neonates. Specifically, our data suggest that Klebsiella overgrowth as well as associated alterations in the microbiome may exacerbate brain injury, perhaps by triggering changes in immunological development such as elevated γδ T cell levels. These immunological alterations, combined with the subsequent depletion of neuroprotective agents, may affect neurophysiological development and neurodevelopmental maturation. These findings suggest that novel therapeutic measures targeting the gut-microbiota-immune-brain axis may hold potential for protecting premature infants from PWMI.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 (UCHT-1) | Thermo Fisher | Cat# 56-0038; RRID:AB_10597906 |
| CD4 (RPA-T4) | Thermo Fisher | Cat# 69-0049; RRID:AB_2637466 |
| CD8 (SK1) | Thermo Fisher | Cat# 47-0087; RRID:AB_2016684 |
| CD27 (O323) | Thermo Fisher | Cat# 17-0279;RRID:AB_10671130 |
| CD28 (CD28.6) | Thermo Fisher | Cat# 16-0288-81; RRID:AB_468924 |
| CD45RA (HI100) | Thermo Fisher | Cat# 48-0458; RRID:AB_1272059 |
| γδ-TCR (IMMU510) | Beckman Coulter | Cat# B10247 |
| Vd1-TCR (TS8.2) | Thermo Fisher | Cat# TCR2730; RRID:AB_223624 |
| Vd2-TCR (B6) | BioLegend | Cat# 331410; RRID:AB_1877263 |
| CXCR3 (1C6) | BD Biosciences | Cat# 741005; RRID:AB_2740628 |
| CCR4 (D8SEE) | Thermo Fisher | Cat# 12-1949; RRID:AB_2572591 |
| CXCR5 (MU5UBEE) | Thermo Fisher | Cat# 25-9185; RRID:AB_2573540 |
| CCR10 (1B8) | BD Biosciences | Cat# 564770; RRID:AB_2738942 |
| CCR6 (R6H1) | Thermo Fisher | Cat# 17-1969; RRID:AB_10733388 |
| CD25 (BC96) | Thermo Fisher | Cat# 12-0259-80; RRID:AB_657722 |
| CCR7 (4B12) | Thermo Fisher | Cat# 47-1971; RRID:AB_2573974 |
| CD39 (eBioA1) | BD Biosciences | Cat# 11-0399; RRID:AB_11151149 |
| CD49d (9F10) | BD Biosciences | Cat# 744587; RRID:AB_2742336 |
| Helios (22F6) | BD Biosciences | Cat# 563951; RRID:AB_2738506 |
| CCR5 (3A9) | BD Biosciences | Cat# 741005; RRID:AB_396312 |
| CTLA-4 (BNI3) | BD Biosciences | Cat# 562743; RRID:AB_2737762 |
| Biological samples | ||
| Peripheral blood samples from extremely premature infants | General Hospital of Vienna | N/A |
| Fecal samples from extremely premature infants | General Hospital of Vienna | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Phenol/Chloroform/Isoamylalcohol | Sigma Aldrich | Cat# P3803 |
| Brilliant III Ultra-Fast SYBR Green Master Mix | Agilent Technologies | Cat# 600882 |
| Nuclease free water | Thermo Fisher | Cat# 10977015 |
| Milli-Q water | Merck Millipore | Cat# C205110 |
| AIM V | Thermo Fisher | Cat# 12055091 |
| Phytohemagglutinin-L | Thermo Fisher | Cat# 00-4977-93 |
| VersaLyse Red blood cell lysis solution | Beckman Coulter | Cat# A09777 |
| Fixative Solution IOTest 3 10x concentrate | Beckman Coulter | Cat# A07800 |
| 1% Staining Buffer (PBS w/o Ca2+/Mg2+ + 1% FCS) | Thermo Fisher | Cat# 16140071 |
| Critical commercial assays | ||
| Procartaplex 23 Cytokines | Thermo Fisher | N/A |
| PerFix-nc kit | Beckman Coulter | Cat# 16140071 |
| SequalPrep™ Normalization Plate Kit | Thermo Fisher | Cat# A1051001 |
| Quant-iT PicoGreen dsDNA Assay | Thermo Fisher | Cat# P7589 |
| Deposited data | ||
| 16S rRNA gene amplicon sequencing data | NCBI | PRJNA715072 |
| Klebsiella pneumoniae genome | NCBI | PRJNA715072 |
| Oligonucleotides | ||
| Forward primer rRNA sequencing and qPCR (515F): 5‘-GTG YCA GCM GCC GCG GTA A-3‘ | (Parada et al., 2016) | N/A |
| Reverse primer rRNA sequencing and qPCR (806R): 5‘-GGA CTA CNV GGG TWT CTA AT-3‘ | (Apprill et al., 2015) | N/A |
| Head sequence of forward primer for barcoding (H1): 5‘-GCT ATG CGC GAG CTG C-3‘ | (Herbold et al., 2015) | N/A |
| Head sequence of reverse primer for barcoding (H2): 5‘-TAG CGC ACA CCT GGT A-3‘ | (Pjevac et al., 2021) | N/A |
| Software and algorithms | ||
| R 4.0 | (R Core Team, 2017) | https://www.r-project.org/ |
| FlowJo™ Software | (BD, 2019) | https://flowjo.com/ |
| Cytoscape | (Shannon et al., 2003) | https://doi.org/10.1101/gr.1239303 |
| FASTA/FASTQ demultiplexer | Laros JFJ | github.com/jfjlaros/demultiplex |
| DADA2 | (Callahan et al., 2016) | https://doi.org/10.12688/F1000RESEARCH.8986.1 |
| SILVA database | (Quast et al., 2013) | https://doi.org/10.1093/nar/gks1219 |
| decontam | (Davis et al., 2018) | https://doi.org/10.1186/s40168-018-0605-2 |
| usearch | (Edgar, 2010) | https://doi.org/10.1093/bioinformatics/btq461 |
| CFX Manager Software | Bio-Rad Laboratories | #1845000 |
| R package “Corrplot” | (Wei and Simko, 2021) | https://github.com/taiyun/corrplot |
| R package “qgraph” | (Epskamp et al., 2012) | https://doi.org/10.18637/jss.v048.i04 |
| R package “TDA” | (Fasy et al., 2015) | https://doi.org/hal-01113028 |
| R package “Vegan” | (Oksanen et al., 2013) | https://cran.r-project.org, https://github.com/vegandevs/vegan |
| R package “tSNE” | (Justin Donaldson, 2016) | https://cran.r-project.org/web/packages/tsne/tsne.pdf. |
| R package “ggplot2” | (Wickham, 2011) | https://doi.org/10.1002/wics.147 |
| R package “plotROC” | (Sachs, 2017) | https://doi.org/10.18637/jss.v079.c02 |
| R package “mixomics” | (Singh et al., 2019) | https://doi.org/10.1093/bioinformatics/bty1054 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact David Berry (david.berry@univie.ac.at).
Materials availability
16S rRNA gene amplicon sequencing data and the Klebsiella pneumoniae genome were deposited under the BioProject accession number PRJNA715072.
Experimental model, clinical definitions and subject details
This study was approved by the ethics committee of the Medical University of Vienna (Ethics number 1348/2017). 60 extremely premature infants were enrolled between September 2017 and June 2019 at the Medical University of Vienna as part of the PreMiBraIn study. Inclusion criteria were birth before the 28th week of gestation with less than 1000 g birth weight. Infants with congenital malformations, chromosomal aberrations, maternally transmitted infectious diseases (e.g. HIV, hepatitis A/B/C, etc.) and inborn errors of metabolism were excluded. Clinical parameters of each patient were prospectively monitored by the clinical staff during hospitalization and recorded within the hospitals’ electronic database ICIP (Philips Healthcare Systems). Intrauterine growth restriction was defined as birth weight under the 10th percentile. Culture-proven early-onset sepsis was defined as infection with elevated inflammatory parameters (C-reactive protein; CRP > 0.5 mg dl-1 or IL-6 > 150 pg ml-1) and positive blood culture. Since the detection rate of blood cultures directly after birth was very low in our cohort, we defined the term “clinical-suspected inflammation” for infants with elevated inflammatory markers after birth (CRP > 0.5 mg dl-1 or IL-6 > 150 pg ml-1). Late-onset sepsis (LOS) was defined according to the NEO-KISS protocol for nosocomial infection surveillance for preterm infants (Gastmeier et al., 2004). Clinical LOS as well as LOS with coagulase negative Staphylococcus (CoNS) were defined as an episode with the following characteristics: > 72 hours post-delivery, empiric antibiotic therapy ≥ 5 days, no apparent infection at another body site, and additionally fulfilling any two of the following criteria: temperature > 38°C or < 36.5°C, temperature instability, tachycardia, bradycardia, apnoea, hypotension, hyperglycaemia, metabolic acidosis, prolonged recapillarization time or positive blood infection parameter (CRP > 2 mg dl-1 or IL-6 > 50 pg ml-1). Culture-positive LOS was defined as a clinical infection as described above with the additional growth of a pathogen in the corresponding blood culture. Bronchopulmonary dysplasia (BPD) was defined as supplemental oxygen treatment or oxygen plus respiratory support at 36 weeks postmenstrual age (Higgins et al., 2018). Retinopathy of prematurity (ROP) was diagnosed and staged according to the international consensus guidelines (International Committee for the Classification of Retinopathy of Prematurity, 2005).
Furthermore, every infant received, starting from the first day of life, the probiotic preparation Infloran® (Bifidobacterium bifidum and Lactobacillus acidophilus) until the corrected age of 34 weeks of gestational age (GA). The enteral feeding regimen included the infant’s own pasteurized mother’s milk or pasteurized human donor milk. When 100 ml kg-1 enteral feeding was achieved, the milk was fortified with 4% bovine milk fortifier until term-equivalent age.
Over the course of hospitalization, neurophysiological development was monitored, stool samples were collected, and blood was drawn at intended time points as described in Figure 1A. In total we collected 547 stool samples, 162 blood samples, and obtained and assessed 335 amplitude integrated electroencephalography (aEEG) and 423 near-infrared spectroscopy (NIRS) spectra. Brain injuries were identified and assessed by combined assessment of magnetic resonance imaging (MRI) at term-equivalent age (Kidokoro et al., 2013) and routine ultrasound imaging (Papile et al., 1978; de Vries et al., 1992).
Method details
Stool and blood sample collection
Stool samples from all 60 extremely premature infants were collected from diapers via collection tubes during patient care routines, without the addition of any type of preservation buffer, and stored at -80° until further analysis. Samples were kept on dry ice while aliquoting for the extraction of nucleic acids and analysis of SCFAs respectively. For the isolation of bacteria, additional stool was sampled randomly, and stored in 40% (v:v) glycerol at -80°C. Blood samples were drawn from premature infants at intended time points via arterial catheter or peripheral venous puncture. Sampling of blood was only conducted if combined with clinical routine sampling to minimize stressing the patients. 300 μL blood was sampled into Lithium Heparin S-Monovettes and stored at 4°C for a maximum of 4 hours before further analysis. For each sampling time point, except for “day 3” samples (Figure 1A), we allowed for a +/- 2-day time window for sample collection. “Day 3” samples were collected +1/-2-day time window for sample collection.
Assessment of neurophysiological development
Neurophysiological characteristics from premature infants were assessed at intended time points (Figure 1A). aEEG was used for monitoring of cerebral activity. A total of 5 gold cup electrodes were applied in combination with skin preparation and electrode contact gels. To ensure conduct, a tissue wetted with NaCl 0.9 % solution was placed upon the prepared electrode. The reference electrode was placed either frontal or dorsal of the infants’ head, the remaining were placed on C3, P3, C4 and P4, according to the 10-20 system (Wells, 1928). Electrodes were fixed by either using a bandage wrapped around the infants’ head, or by placing a CPAP hat above. Electrodes were connected to a CFM Olympic Brainz Monitor device (OBM, Natus) and the measurement lasted for at least 3 hours.
For assessment of cerebral oxygenation, NIRS (INVOS, Medtronic) was employed. A cerebral oximetry infant-neonatal sensor was placed on the left side of the forehead with detector placement towards a lateral position below the gold cup electrodes. The probe was connected to a cerebral oximeter and the measurement lasted in parallel to the aEEG measurement, for at least 3 hours. Cranial magnetic resonance imaging (cMRI) images was performed at term-equivalent age (between 36 and 40 weeks of GA), using a standard neonatal MR protocol (Antonov et al., 2017) at 1.5 Tesla.
Analysis of cytokine secretion
25 μL of peripheral whole blood was used for analysis of T cell-specific cytokine production from patients 1-40 after incubation of whole blood with 250 μL AIM V and 20μL phytohemagglutinin-L (PHA-L end concentration: 1.25 μg ml-1) for 72 hours (Movafagh et al., 2011). All incubations were conducted in 96-well plates on 37°C. Afterwards, samples were centrifuged at 1,000 g for 5 minutes and the supernatants were collected and stored at -80°C for subsequent analysis. Cytokines within supernatants were assessed via customized ProcartaPlex immunoassays, following the protocol of each respective assay. Upon completion of each multiplex assay, the amounts of respective cytokines were analyzed via Luminex.
Enumeration and phenotyping of peripheral T cells
50 μL of peripheral whole blood was used for analysis of T helper, cytotoxic T cells, γδ T cells, and T regulatory cells respectively. This analysis was conducted with blood drawn from patients 1-40. The employed monoclonal antibodies are shown in the key resources table. Cellular staining of γδ T, cytotoxic T cells and T helper cells was conducted as described elsewhere (Wistuba-Hamprecht et al., 2014). Staining of T regulatory cells was conducted according to protocol, employing a PerFix-nc kit in combination with Brilliant Violet Staining Buffer for enhanced surface staining, and 100% fetal calf serum (FCS) as a buffer during fixation for subsequent intracellular staining. Stained cells were resuspended in 50 μL staining buffer (1x PBS without Ca2+/Mg2+ + 1% FCS) and analyzed within 4 hours via fluorescence activated cell sorting (FACS).
16S rRNA gene amplicon sequencing and qPCR
547 stool samples of 60 preterm infants were weighed in (20–30 mg) for phenol-chloroform extraction of total nucleic acids (TNA) (Pérez-Brocal et al., 2020), with the inclusion of one bead-beating step, and an additional extraction of one negative control per extraction-batch, consisting of nuclease-free water to allow for the assessment of contamination. TNA were eluted in 40 μl nuclease free water and stored at -20°C until further analysis. Extracted TNA concentration was measured using Quant-iT PicoGreen dsDNA Assay (Thermo Fisher). Extracts served as templates for qPCR amplification of bacterial 16S rRNA genes, as well as for 16S rRNA gene amplicon sequencing.
For the enumeration of gene copy numbers, the primer pair 515F (Parada et al., 2016) and 806R (Apprill et al., 2015), modified with a linker sequence (Herbold et al., 2015), were used for qPCR amplification of 16S rRNA genes using a CFX96 Real-Time System. The concentration of each sample was adjusted to a concentration between 1-20 ng μl-1. PCR assay mixtures consisted of 10 μl Brilliant III Ultra-Fast SYBR Green Master Mix already containing a SureStart Taq DNA polymerase, MgCl2 at a concentration of 2.5 mM, two times 0.8 μl primer solution (400 nM), 7.4 μl sterile nuclease-free water, and 1 μl template DNA solution or adjusted 6.4 μl sterile nuclease-free water and 2 μl template DNA. The PCR amplification program encompassed an initial denaturation step at 95°C for 3 min followed by 40 two-step cycles at 95°C for 10 s and at 55°C for 30 s. In each run a no-template (without TNA) control was included. Genomic DNA of Escherichia coli DH5alpha was used as a standard (highest concentration 25 ng μl-1 following a four-times 1:10 dilution series).
16S rRNA gene amplification and amplicon sequencing was performed at the Joint Microbiome Facility of the Medical University of Vienna and the University of Vienna. The V4 region of the bacterial and archaeal 16S rRNA gene was amplified (30 cycles) from the DNA using the mentioned modified primer pair and barcoded (8 cycles) in a unique dual barcoding setup (UDB-H12). Thereafter, barcoded samples were purified and normalized over a SequalPrep™ Normalization Plate Kit using a Biomek® NXP Span-8 pipetting robot (Beckman Coulter), and pooled and concentrated on columns (Analytik Jena). Indexed sequencing libraries were prepared with the Illumina TruSeq Nano Kit as described previously (Herbold et al., 2015), and sequenced in paired-end mode (600 cycles; V3 chemistry) on an Illumina MiSeq following the manufacturer’s instructions. The workflow systematically included four negative controls (PCR blanks, i.e., PCR-grade water as template) for each 90 samples sequenced. Also, a ZYMObiomics mock community was sequenced and analyzed at the sequencing facility as part of their established quality control routine (Pjevac et al., 2021)
Detection of short chain fatty acids in stool samples
The fecal samples (0.026 ± 0.012 g) were suspended in 1.5 ml milli-Q water by vortexing for 10 minutes at maximum speed. The supernatant after centrifuging samples for 10 minutes at 10,000g, was filtered through a 0.2 μm membrane filter. Filtered samples were 1:2 diluted with milliQ water and analyzed on a 930 Compact IC Flex instrument equipped with an 858 Professional Sample Processor with extended MiPuT (Micro Metrohm intelligent Partial Loop Injection Technique), a Metrohm CO2 Suppressor for inline bicarbonate removal, a Metrosep Organic acids 250/7.8 column, a Metrosep Organic acids Guard/4.6 guard column and an 850 IC conductivity detector (Metrohm, Herisau, Switzerland).
Klebsiella pneumoniae isolation and genome sequencing
Klebsiella pneumoniae was isolated from stool anaerobically on peptone yeast glucose (PYG) agar plates. Colonies were re-streaked multiple times to ensure clonality. After identifying Klebsiella pneumoniae by 16S rRNA gene-targeted PCR, the culture was grown in PYG medium and DNA was isolated via phenol-chloroform extraction as described above. Illumina sequencing libraries were prepared using the NEBNext Ultra II FS DNA Library Prep Kit, and sequencing was performed with an Illumina Miseq (v3 kit, 600 cycles). Sequencing data was quality-trimmed at a phred score of 15 using bbmap (https://sourceforge.net/projects/bbmap/), and assembled with SPAdes 3.11.1 (Bankevich et al., 2012). Contigs shorter than 1000 bp were removed from the assembly, and the quality of the genome draft was assessed with CheckM (Parks et al., 2015) and annotated with RAST.
Quantification and statistical analysis
Processing and analysis of neurological data
Resulting aEEGs were analyzed by visual assessment of Burdjalov total maturational scores (Burdjalov et al., 2003) and percentages of background patterns (Hellstro m-Westas et al., 2006; Klebermass et al., 2011). Routine cranial ultrasound screenings and cMRIs were assessed for brain damage via established scores (Kidokoro et al., 2013), as well as expert radiologist knowledge. Near-infrared spectra (NIRS) were examined individually. Artifacts (e.g., probe fell off forehead) were removed. Variability of cranial oxygen supply was calculated as a percentage of the fraction of time spent with an oxygen supply deviating beyond 10% from the mean of the total measurement. To calculate and visualize receiver operating characteristics (ROC) curves we employed the R package plotROC (Sachs, 2017).
Processing and analysis of immunological data
Ordinations of T cell populations were performed via canonical correlation analysis (CCA) with the cca() function from the R package vegan version 2.5 (Oksanen et al., 2013). The function ordistep() was then used in forward model selection to define the adjusted R2 value for significant cytokines. Subsequently, the respective CCAs were rerun with significant cytokines solely. Missing values for the Multiplex (in vitro Cytokines) and FACS dataset (in vivo T cell ontogenesis) were interpolated using mean value interpolation. Principal Coordinate Analysis (PCoA was conducted based on the abundance-based canberra dissimilarity matrix and visualized with ggplot(). Silhouette Clustering (Oksanen et al., 2012) was employed to identify the optimal number of clusters (Figure S2F). Weighted averages of cytokine and chemokine concentrations were added a posteriori to the ordination plot by computing the covariance of variables with all axes. Afterwards, only positive eigenvalues were selected and the value of covariance was standardized as described in (Legendre and Gallagher, 2001). Furthermore, topographical data analysis (TDA) was employed on cytokine and chemokine data via the TDAmapper R function (https://cran.r-project.org/web/packages/TDAmapper/index.html). TDA was calculated with five intervals, allowing for 70% overlap, and ten clustering bins.
16S rRNA gene amplicon sequence analysis
Amplicon pools were extracted from the raw sequencing data using the FASTQ workflow in BaseSpace (Illumina) with default parameters. Demultiplexing was performed with the python package demultiplex allowing one mismatch for barcodes and two mismatches for linkers and primers. Amplicon sequence variants (ASVs) were inferred using the DADA2 R package version 1.14.1, applying the recommended workflow (Callahan et al., 2016). FASTQ reads were trimmed at 150nt with allowed expected errors of 2. ASV sequences were subsequently merged and classified using SINA version 1.6.0 (Pruesse et al., 2012) and the SILVA database SSU Ref NR 99 release 138 using default parameters (Quast et al., 2013).
Quantification of bacterial load, 16S rRNA gene copy number correction, and analysis
All qPCR reactions were performed in triplicates. All reactions with a standard deviation greater than 25% were repeated. The quality of each qPCR run was assessed via Bio-Rad CFX Manager Software. A standard curve was constructed by plotting the known log-transformed starting quantity against the cycle threshold (Ct–value). The whole E.coli DH5alpha genome was searched via NCBI Reference database and the molecular weight was calculated using an oligonucleotide properties calculator. The molecular weight was needed for the calculation of genome copies μl-1 via Dalton's equation. Multiplication of the genome copies μl-1 with the 16S rRNA gene copies per genome (7) leads to the correct 16S rRNA gene copy numbers μl-1. The standard amount of 16S rRNA gene copy numbers μl-1 of E. coli DH5alpha was 3.7 x 107. This value of the standard was used to calculate back to 16S rRNA gene copy numbers (g-1 of faeces) taking into account the dilution factor for each sample (samples were initially diluted to a starting concentration of < 20ng μl-1).
16S rRNA gene counts were normalized via rarefying to even minimal sampling depth (4,000 reads per sample). Subsequently, a filter was applied to exclude ASVs that occurred in less than three samples overall. Rarefied and filtered counts were multiplied by the qPCR total cell counts, and ASV counts at genus level or above were corrected for the number of 16S rRNA operons (Stoddard et al., 2015). As this method has been shown to be susceptible to errors in quantification of relatively underrepresented ASVs in a sample (Tettamanti Boshier et al., 2020), abundances of rarer taxa should be treated as approximations. Filtered abundance-corrected data was used to conduct a cluster analysis via T-distributed stochastic neighbor embedding (tSNE) of ASVs with 10,000 iterations and a perplexity of 30, employing the R package “Rtsne” (Justin Donaldson, 2016). Silhouette Clustering (Oksanen et al., 2012) was employed to identify the optimal number of microbiome clusters in the dataset (hierarchical clustering; Ward’s method). Furthermore, EdgeR was used to identify count-based differential expression of ASVs between infants with and without brain injuries, using the microbiome analyst platform as described on the developer’s website (www.microbiomeanalyst.ca). To define the extremely premature infant core microbiota, an additional filter was applied to identify ASVs that occurred in a minimum of 20% of samples with an abundance of >105 cells g-1.
Basic statistical analysis and data integration
Basic statistical analysis (Student’s T-test, ANOVA, repeated measures ANOVA, Wilcoxon Test, and chi-square test, Fisher’s exact test) was performed via R version 4.0 and the R package rstatix version 0.7.0. All p-values were adjusted using Bonferroni’s method for Unpaired T Test and Wilcoxon Rank Sum Test, as well as Tukey post-hoc test for ANOVA.
Data Integration Analysis for Biomarker discovery using Latent variable approaches for Omics studies (DIABLO) was used to integrate complementary microbiome, immunological, metabolic, and neurophysiological data. DIABLO analysis was performed using the “Mixomics” R package (Singh et al., 2019). The DIABLO framework builds on Generalized Canonical Correlation Analysis (Tenenhaus and Tenenhaus, 2011) to generalize partial least squares (PLS) for multiple matching data sets. In total we picked the maximum of 80 matching samples to combine our datasets. Only highly prevalent ASVs (occurrence in at least 20% of all samples with an absolute abundance > 105) were included in the analysis. Two components were used (centroid distance and overall error rate) to fine tune the model selection as described on the developer’s website (www.mixomics.org). As a result, we interpreted a correlation value of each feature to its corresponding feature in another dataset (e.g. Klebsiella [microbiome] to effector CD4+ [T cells].
Data visualization
Data was visualized via R version 4.0 and R package ggplot2 version 3.3.3 (Wickham, 2011). Transitions throughout tSNE clusters were modelled via Cytoscape (https://cytoscape.org). For FACS gating the FlowJo™ Software was used.
Additional resources
This study was registered at ClinicalTrials.gov under the number NCT03213275.
Acknowledgments
We thank all families for their participation and all medical staff for their help with sample collection. We thank Clemens Schauberger from the University of South Denmark, Department of Biology, for his computational help and advice. This project was funded by an inter-university cluster project grant between the University of Vienna and the Medical University of Vienna (“PreMiBraIn”), the European Research Council (Starting grant: FunKeyGut 741623), and the Austrian Science Fund (FWF; P27831-B28).
Author contributions
Conceptualization, L.W., D.B., and D.S.; methodology, D.S., M.M., B.W., B.H., P.P., V.G., K.G., K.K.-S., K.D.P., and G.K.; software, D.S., B.H., L.U., and V.G.; investigation, D.S.; writing – original draft, D.S. and D.B.; writing – review & editing, D.S., D.B., L.W., and K.D.P.; resources, P.P., A.B., B.W., D.B., T.V.D.W., and A.S.; supervision, D.B.; and funding acquisition, D.B., L.W., and A.B.
Declaration of interests
The authors declare no competing interests.
Published: September 3, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.08.004.
Contributor Information
David Berry, Email: david.berry@univie.ac.at.
Lukas Wisgrill, Email: lukas.wisgrill@meduniwien.ac.at.
Supplemental information
Data and code availability
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams-Chapman I., Heyne R.J., DeMauro S.B., Duncan A.F., Hintz S.R., Pappas A., Vohr B.R., McDonald S.A., Das A., Newman J.E. Neurodevelopmental impairment Among extremely preterm infants in the neonatal research network. Pediatrics. 2018;141:e20173091. doi: 10.1542/peds.2017-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Lopez M., Dinsmoor A.M., Ho T.T.B., Donovan S.M. A systematic review of the factors influencing microbial colonization of the preterm infant gut. Gut Microbes. 2021;13:1–33. doi: 10.1080/19490976.2021.1884514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsson A.M., Zhang X., Vontell R., Bi D., Bronson R.T., Supramaniam V., Baburamani A.A., Hua S., Nazmi A., Cardell S. γδ T cells contribute to injury in the developing brain. Am. J. Pathol. 2018;188:757–767. doi: 10.1016/j.ajpath.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcon-Giner C., Dalby M.J., Caim S., Ketskemety J., Shaw A., Sim K., Lawson M.A.E., Kiu R., Leclaire C., Chalklen L. Microbiota supplementation with Bifidobacterium and Lactobacillus modifies the preterm infant gut microbiota and metabolome: an observational study. Cell Rep. Med. 2020;1:100077. doi: 10.1016/j.xcrm.2020.100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov N.K., Ruzal-Shapiro C.B., Morel K.D., Millar W.S., Kashyap S., Lauren C.T., Garzon M.C. Feed and wrap MRI technique in infants. Clin. Pediatr. 2017;56:1095–1103. doi: 10.1177/0009922816677806. [DOI] [PubMed] [Google Scholar]
- Apprill A., McNally S., Parsons R., Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015;75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- Holden V.I., Breen P., Houle S., Dozois C.M., Bachman M.A. Klebsiella pneumoniae Siderophores induce inflammation, bacterial dissemination, and HIF-1α stabilization during pneumonia. mBio. 2016;7 doi: 10.1128/mBio.01397-16. e01397–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benakis C., Brea D., Caballero S., Faraco G., Moore J., Murphy M., Sita G., Racchumi G., Ling L., Pamer E.G. Commensal microbiota affects ischemic stroke outcome by regulating intestinal γδ T cells. Nat. Med. 2016;22:516–523. doi: 10.1038/nm.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaz B., Bazin T., Pellissier S. The vagus nerve at the interface of the microbiota-gut-brain axis. Front. Neurosci. 2018;12:49. doi: 10.3389/fnins.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdjalov V.F., Baumgart S., Spitzer A.R. Cerebral function monitoring: a new scoring system for the evaluation of brain maturation in neonates. Pediatrics. 2003;112:855–861. doi: 10.1542/peds.112.4.855. [DOI] [PubMed] [Google Scholar]
- Callahan B.J., Sankaran K., Fukuyama J.A., McMurdie P.J., Holmes S.P. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res. 2016;5:1492. doi: 10.12688/f1000research.8986.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J.F., O’Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., Codagnone M.G., Cussotto S., Fulling C., Golubeva A.V. The microbiota-gut-brain axis. Physiol. Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- Dalile B., Van Oudenhove L., Vervliet B., Verbeke K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019;16:461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries L.S., Eken P., Dubowitz L.M.S. The spectrum of leukomalacia using cranial ultrasound. Behav. Brain Res. 1992;49:1–6. doi: 10.1016/S0166-4328(05)80189-5. [DOI] [PubMed] [Google Scholar]
- Di Martino P., Bertin Y., Girardeau J.P., Livrelli V., Joly B., Darfeuille-Michaud A. Molecular characterization and adhesive properties of CF29K, an adhesin of Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 1995;63:4336–4344. doi: 10.1128/iai.63.11.4336-4344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino P., Cafferini N., Joly B., Darfeuille-Michaud A. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 2003;154:9–16. doi: 10.1016/S0923-2508(02)00004-9. [DOI] [PubMed] [Google Scholar]
- Duan J., Chung H., Troy E., Kasper D.L. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe. 2010;7:140–150. doi: 10.1016/j.chom.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Epskamp S., Cramer A.O.J., Waldorp L.J., Schmittmann V.D., Borsboom D. Qgraph: network visualizations of relationships in psychometric data. J. Stat. Soft. 2012;48 doi: 10.18637/jss.v048.i04. [DOI] [Google Scholar]
- Evrard B., Balestrino D., Dosgilbert A., Bouya-Gachancard J.L., Charbonnel N., Forestier C., Tridon A. Roles of capsule and lipopolysaccharide O antigen in interactions of human monocyte-derived dendritic cells and Klebsiella pneumoniae. Infect. Immun. 2010;78:210–219. doi: 10.1128/IAI.00864-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasy B.T., Kim J., Lecci F., Maria C. Introduction to the R package TDA. 2015. https://hal.inria.fr/hal-01113028/document
- Ferreira L.M.R. Gammadelta T cells: innately adaptive immune cells? Int. Rev. Immunol. 2013;32:223–248. doi: 10.3109/08830185.2013.783831. [DOI] [PubMed] [Google Scholar]
- BD FlowJo™. software version 6.1. 2019. https://flowjo.com/
- García-Depraect O., Muñoz R., Rodríguez E., Rene E.R., León-Becerril E. Microbial ecology of a lactate-driven dark fermentation process producing hydrogen under carbohydrate-limiting conditions. Int. J. Hydr. Energy. 2021;46:11284–11296. doi: 10.1016/j.ijhydene.2020.08.209. [DOI] [Google Scholar]
- Gastmeier P., Geffers C., Schwab F., Fitzner J., Obladen M., Rüden H. Development of a surveillance system for nosocomial infections: the component for neonatal intensive care units in Germany. J. Hosp. Infect. 2004;57:126–131. doi: 10.1016/j.jhin.2003.12.038. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna K.P., Macadangdang B.R., Rogers M.B., Tometich J.T., Firek B.A., Baker R., Ji J., Burr A.H.P., Ma C., Good M. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat. Med. 2019;25:1110–1115. doi: 10.1038/s41591-019-0480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstro m-Westas L., Rosen I., de Vries L.S., Greisen G. Amplitude-integrated EEG classification and interpretation in preterm and term infants. NeoReviews. 2006;7:e76–e87. doi: 10.1542/neo.7-2-e76. [DOI] [Google Scholar]
- Herbold C.W., Pelikan C., Kuzyk O., Hausmann B., Angel R., Berry D., Loy A. A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Front. Microbiol. 2015;6:731. doi: 10.3389/fmicb.2015.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R.D., Jobe A.H., Koso-Thomas M., Bancalari E., Viscardi R.M., Hartert T.V., Ryan R.M., Kallapur S.G., Steinhorn R.H., Konduri G.G. Bronchopulmonary dysplasia: executive summary of a workshop. J. Pediatr. 2018;197:300–308. doi: 10.1016/j.jpeds.2018.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D.R., Huang S., Nagy M.S., Yadagiri V.K., Fields C., Mukherjee D., Bons B., Dedhia P.H., Chin A.M., Tsai Y.-H. Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. eLife. 2017;6:e29132. doi: 10.7554/eLife.29132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humberg A., Härtel C., Paul P., Hanke K., Bossung V., Hartz A., Fasel L., Rausch T.K., Rody A., Herting E. Delivery mode and intraventricular hemorrhage risk in very-low-birth-weight infants: observational data of the German Neonatal Network. Eur. J. Obstet. Gynecol. Reprod. Biol. 2017;212:144–149. doi: 10.1016/j.ejogrb.2017.03.032. [DOI] [PubMed] [Google Scholar]
- International Committee for the Classification of Retinopathy of Prematurity The international classification of retinopathy of prematurity revisited. Arch. Ophthalmol. 2005;123:991–999. doi: 10.1001/archopht.123.7.991. [DOI] [PubMed] [Google Scholar]
- Justin Donaldson M. Package “tsne” Title T-Distributed Stochastic Neighbor Embedding for R (t-SNE), pp. 2–5. 2016. https://cran.r-project.org/web/packages/tsne/tsne.pdf
- Kandasamy Y., Hartley L., Rudd D., Smith R. The association between systemic vascular endothelial growth factor and retinopathy of prematurity in premature infants: a systematic review. Br. J. Ophthalmol. 2017;101:21–24. doi: 10.1136/bjophthalmol-2016-308828. [DOI] [PubMed] [Google Scholar]
- Keunen K., van Elburg R.M., van Bel F., Benders M.J. Impact of nutrition on brain development and its neuroprotective implications following preterm birth. Pediatr. Res. 2015;77:148–155. doi: 10.1038/pr.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro H., Niel J.J., Inder T.E. New MR imaging assessment tool to define brain abnormalities in very preterm infants at term. AJNR Am. J. Neuroradiol. 2013;34:2208–2214. doi: 10.3174/ajnr.A3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebermass K., Olischar M., Waldhoer T., Fuiko R., Pollak A., Weninger M. Amplitude-integrated EEG pattern predicts further outcome in preterm infants. Pediatr. Res. 2011;70:102–108. doi: 10.1203/PDR.0b013e31821ba200. [DOI] [PubMed] [Google Scholar]
- Lee C.R., Lee J.H., Park K.S., Jeon J.H., Kim Y.B., Cha C.J., Jeong B.C., Lee S.H. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front. Cell. Infect. Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P., Gallagher E.D. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- Litvak Y., Byndloss M.X., Bäumler A.J. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362 doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L.G., Walsh B.H., Knutsen C., Neil J.J., Smyser C.D., Rogers C.E., Inder T.E. Brain growth in the NICU: critical periods of tissue-specific expansion. Pediatr. Res. 2018;83:976–981. doi: 10.1038/pr.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S.K., Round J.L., Kasper D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Movafagh A., Heydary H., Mortazavi-Tabatabaei S.A., Azargashb E. The significance application of indigenous phytohemagglutinin (PHA) mitogen on metaphase and cell culture procedure. Iran. J. Pharm. Res. 2011;10:895–903. [PMC free article] [PubMed] [Google Scholar]
- Ng S.K., Hamilton I.R. Lactate metabolism by Veillonella parvula. J. Bacteriol. 1971;105:999–1005. doi: 10.1128/jb.105.3.999-1005.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M.M., Witherden D.A., Havran W.L. γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat. Rev. Immunol. 2017;17:733–745. doi: 10.1038/nri.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño D.F., Zhou Q., Yamaguchi Y., Martin L.Y., Wang S., Fulton W.B., Jia H., Lu P., Prindle T., Zhang F. Cognitive impairments induced by necrotizing enterocolitis can be prevented by inhibiting microglial activation in mouse brain. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aan0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Kindt R., Legendre P., Minchin P.R., O’hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Wagner H., Oksanen M.J. Package ‘vegan’. Community ecology package. version. 2013;2:1–295. [Google Scholar]
- Papile L.A., Burstein J., Burstein R., Koffler H. Incidence and evolution of subependymal and intraventricular hemorrhage: a study of infants with birth weights less than 1,500 gm. J. Pediatr. 1978;92:529–534. doi: 10.1016/S0022-3476(78)80282-0. [DOI] [PubMed] [Google Scholar]
- Parada Venegas D.P., De la Fuente M.K., Landskron G., González M.J., Quera R., Dijkstra G., Harmsen H.J.M., Faber K.N., Hermoso M.A. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada A.E., Needham D.M., Fuhrman J.A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Brocal V., Magne F., Ruiz-Ruiz S., Ponce C.A., Bustamante R., San Martin V., Gutierrez M., Gatti G., Vargas S.L., Moya A. Optimized DNA extraction and purification method for characterization of bacterial and fungal communities in lung tissue samples. Scientific Rep. 2020;10(1):1–15. doi: 10.1038/s41598-020-74137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjevac P., Hausmann B., Schwarz J., Kohl G., Herbold C.W., Loy A., Berry D. An economical and flexible dual barcoding, two-step PCR approach for highly multiplexed amplicon sequencing. Front Microbiol. 2021;12:669776. doi: 10.3389/fmicb.2021.669776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Peplies J., Glöckner F.O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team R: A language and environment for statistical computing. (R Foundation for Statistical Computing) 2017. https://www.R-project.org/
- Rao C., Coyte K.Z., Bainter W., Geha R.S., Martin C.R., Rakoff-Nahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021;591:633–638. doi: 10.1038/s41586-021-03241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs M.C. Plotroc: A tool for plotting ROC curves. J. Stat. Softw. 2017;79 doi: 10.18637/jss.v079.c02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Russell S.L., Antunes L.C., Finlay B.B. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Forster S.C., Tsaliki E., Vervier K., Strang A., Simpson N., Kumar N., Stares M.D., Rodger A., Brocklehurst P. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J.R., Blosser E.G., Zindl C.L., Silberger D.J., Conlan S., Laufer V.A., DiToro D., Deming C., Kumar R., Morrow C.D. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat. Med. 2019;25:1772–1782. doi: 10.1038/s41591-019-0640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Shannon C.P., Gautier B., Rohart F., Vacher M., Tebbutt S.J., Lê Cao K.A. Diablo: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. 2019;35:3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starzyk-Łuszcz K., Zielonka T.M., Jakubik J., Życińska K. Mortality due to nosocomial infection with Klebsiella pneumoniae ESBL<sup/>. Adv. Exp. Med. Biol. 2017;1022:19–26. doi: 10.1007/5584_2017_38. [DOI] [PubMed] [Google Scholar]
- Stoddard S.F., Smith B.J., Hein R., Roller B.R., Schmidt T.M. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklein S., Hilgendorff A., Li M., Förster K., Flemmer A.W., Galiè F., Wunderlich S., Wang D., Stein S., Ehrhardt H. Variable functional connectivity architecture of the preterm human brain: impact of developmental cortical expansion and maturation. Proc. Natl. Acad. Sci. USA. 2020;117:1201–1206. doi: 10.1073/pnas.1907892117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus A., Tenenhaus M. Regularized generalized canonical correlation analysis. Psychometrika. 2011;76:257–284. doi: 10.1007/s11336-011-9206-8. [DOI] [PubMed] [Google Scholar]
- Tettamanti Boshier F.A., Srinivasan S., Lopez A., Hoffman N.G., Proll S., Fredricks D.N., Schiffer J.T. Complementing 16S rRNA gene amplicon sequencing with total bacterial load to infer absolute species concentrations in the vaginal microbiome. mSystems. 2020;5 doi: 10.1128/mSystems.00777-19. e00777–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommiska V., Heinonen K., Lehtonen L., Renlund M., Saarela T., Tammela O., Virtanen M., Fellman V. No improvement in outcome of nationwide extremely low birth weight infant populations between 1996–1997 and 1999–2000. Pediatrics. 2007;119:29–36. doi: 10.1542/peds.2006-1472. [DOI] [PubMed] [Google Scholar]
- van Best N., Trepels-Kottek S., Savelkoul P., Orlikowsky T., Hornef M.W., Penders J. Influence of probiotic supplementation on the developing microbiota in human preterm neonates. Gut Microbes. 2020;12:1–16. doi: 10.1080/19490976.2020.1826747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancamelbeke M., Laeremans T., Vanhove W., Arnauts K., Ramalho A.S., Farré R., Cleynen I., Ferrante M., Vermeire S. Butyrate does not protect against inflammation-induced loss of epithelial barrier function and cytokine production in primary cell monolayers from patients with ulcerative colitis. J. Crohns Colitis. 2019;13:1351–1361. doi: 10.1093/ecco-jcc/jjz064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe J.J. Brain injury in premature infants: a complex amalgam of destructive and developmental disturbances. Lancet Neurol. 2009;8:110–124. doi: 10.1016/S1474-4422(08)70294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner B.B. The contribution of the gut microbiome to neurodevelopment and neuropsychiatric disorders. Pediatr. Res. 2019;85:216–224. doi: 10.1038/s41390-018-0191-9. [DOI] [PubMed] [Google Scholar]
- Wei T., Simko V. R package 'corrplot': visualization of a correlation matrix, (0.90 Version) 2021. https://github.com/taiyun/corrplot
- Weitkamp J.H., Rosen M.J., Zhao Z., Koyama T., Geem D., Denning T.L., Rock M.T., Moore D.J., Halpern M.D., Matta P. Small intestinal intraepithelial TCRγδ+ T lymphocytes are present in the premature intestine but selectively reduced in surgical necrotizing enterocolitis. PLoS One. 2014;9:e99042. doi: 10.1371/journal.pone.0099042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells C.J. Preparation of the patient. Anesthesia & Analgesia. 1928;7:257–262. doi: 10.1213/00000539-192801000-00096. [DOI] [Google Scholar]
- White D. Oxford University Press; 2000. Physiology and Biochemistry of Prokaryotes. [Google Scholar]
- Wickham H. Ggplot2. WIREs. Comp. Stat. 2011;3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- Winter S.E., Winter M.G., Xavier M.N., Thiennimitr P., Poon V., Keestra A.M., Laughlin R.C., Gomez G., Wu J., Lawhon S.D. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisgrill L., Lepuschitz S., Blaschitz M., Rittenschober-Böhm J., Diab-El Schahawi M., Schubert S., Indra A., Berger A. Outbreak of yersiniabactin-producing Klebsiella pneumoniae in a neonatal intensive care unit. Pediatr. Infect. Dis. J. 2019;38:638–642. doi: 10.1097/INF.0000000000002258. [DOI] [PubMed] [Google Scholar]
- Wistuba-Hamprecht K., Pawelec G., Derhovanessian E. OMIP-020: phenotypic characterization of human γδ T-cells by multicolor flow cytometry. Cytometry A. 2014;85:522–524. doi: 10.1002/cyto.a.22470. [DOI] [PubMed] [Google Scholar]
- Woodward L.J., Anderson P.J., Austin N.C., Howard K., Inder T.E. Neonatal MRI to predict neurodevelopmental outcomes in preterm infants. N. Engl. J. Med. 2006;355:685–694. doi: 10.1056/NEJMoa053792. 1018. [DOI] [PubMed] [Google Scholar]
- Xu W., Lau Z.W.X., Fulop T., Larbi A. The aging of γδ T cells. Cells. 2020;9:1–18. doi: 10.3390/cells9051181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Yang X.O., Chung Y., Fukunaga A., Nurieva R., Pappu B., Martin-Orozco N., Kang H.S., Ma L., Panopoulos A.D. CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 2008;181:8391–8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Kelly C.J., Battista K.D., Schaefer R., Lanis J.M., Alexeev E.E., Wang R.X., Onyiah J.C., Kominsky D.J., Colgan S.P. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor–dependent repression of Claudin-2. J. Immunol. 2017;199:2976–2984. doi: 10.4049/jimmunol.1700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwittink R.D., van Zoeren-Grobben D., Martin R., van Lingen R.A., Groot Jebbink L.J., Boeren S., Renes I.B., van Elburg R.M., Belzer C., Knol J. Metaproteomics reveals functional differences in intestinal microbiota development of preterm infants. Mol. Cell. Proteomics. 2017;16:1610–1620. doi: 10.1074/mcp.RA117.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.