ABSTRACT
As one of the most severe kinds of neurological damage, spinal cord injury (SCI) contributes to persistent motor dysfunction and involves a large repertoire of gene alterations. The participation of circular RNAs (circRNAs) in neurological recovery following SCI needs to be clarified. In the current work, we attempted to assess the function of hsa_circRNA_0003962/circTYW1 and its underlying mechanism in SCI. By accessing the GEO repository, the expression of circTYW1, microRNA-380 (miR-380), and FGF9 in SCI and sham-operated rats was evaluated. PC12 cells after oxygen-glucose deprivation (OGD) treatment were prepared to mimic the SCI model. circTYW1 and FGF9 were poorly expressed, whereas miR-380 was highly expressed in the spinal cord tissues of SCI rats. circTYW1 promoted neurological recovery in SCI rats and inhibited apoptosis in spinal cord tissues. In PC12 cells exposed to OGD, circTYW1 suppressed PC12 cell apoptosis; however, miR-380 overexpression reversed the protective effect of circTYW1 on PC12 cells. Also, circTYW1 promoted FGF9 expression through competitively binding to miR-380, which activated the ERK1/2 signaling. In summary, our results demonstrated that declines in circTYW1 prevented SCI rats from neurological recovery by regulating the miR-380/FGF9/ERK1/2 axis, which might provide new understanding for SCI treatment.
KEYWORDS: Spinal cord injury, hsa_circRNA_0003962/circTYW1, microRNA-380, fgf9, erk1/2 signaling
1. Introduction
Spinal cord injury (SCI), characterized by damaged sensory and motor functions, disturbs millions of people in the world and normally has life-long consequences [1,2]. With the advance in the comprehension of pathophysiologic mechanisms underlying in the development of SCI, possible treatments with the purpose of ameliorating neural damage have been developed but with modest benefits [3]. Therefore, a better understanding in the molecular mechanisms involved in the pathophysiological processes that ensue after SCI in a more comprehensive manner, particularly those associated with gene alteration, is of paramount importance for developing novel and effective treatment strategies. Circular RNAs (circRNAs) are a family of long noncoding RNAs which have covalently linked ends and have been highlighted to be related to many neurological diseases, such as Parkinson’s disease and schizophrenia [4].
Interestingly, at the time point of 3 days after contusion, 1676 circRNAs were revealed by a circRNA microarray to be differentially expressed in spinal cord in a previous study, suggesting their participation in SCI [5]. In addition, a number of circRNAs are differentially expressed after SCI and play important roles in the proliferation, inflammation, and apoptosis in the central nervous system post-traumatic injury [6]. Moreover, circRNAs are expected to be conserved between species, which will facilitate the clinical transformation for circRNAs in nerve injury [7]. CircRNAs are believed to modulate the transcription of parent genes and drive to form alternatively spliced mRNAs and sponge microRNAs (miRNAs), namely, competing endogenous RNAs (ceRNAs) [8]. For instance, the circRNA_01477/miR-423-5p network has been indicated as a key meditator in the changeable regeneration environment throughout the recovery from SCI [9]. Our circRNA microarray prediction using the Gene Expression Omnibus GSE114426 dataset revealed that circ_0003962, whose host gene is the thirteenth exon of TYW1, was significantly downregulated in spinal cord tissues of 3 rats with SCI. Therefore, circ_0003962/circTYW1 was selected as our target. Overexpression of miR-380-3p suppressed cell proliferation, induced the apoptotic rate, and intensified the toxicity of paraquat in mouse neuroblastoma cells [10]. However, its downstream mechanism remains unclear. Our integrated mRNA microarray and target mRNA prediction using StarBase, TargetScan and miRDB revealed that fibroblast growth factor 9 (FGF9) was the only mRNA that was downregulated in spinal cord tissues of SCI rats and was a target of miR-380. FGF9 is widely expressed in the central nervous system, and FGF9 knockdown induces neuronal apoptosis and inflammation in the cerebellum of mice [11]. The current study was designed to assess the role of circTYW1 in SCI rats and PC12 cells exposed to oxygen-glucose deprivation (OGD). Additionally, we also studied the regulatory mechanism of circTYW1 in neurological recovery after SCI in relation to the miR-380/FGF9 axis, which may help us to understand the mechanism of SCI.
2. Material and Methods
2.1. Bioinformatics analysis
We downloaded SCI-related gene expression microarrays GSE114426, GSE19890 and GSE45006 from the GEO database (Organism: rattus norvegicus). Among them, GSE114426 contained circRNA gene expression microarrays of SCI from three sham-operated rats and three SCI-modeled rats (Rats in the SCI group were subjected to laminectomy plus contusion. Rats were anesthetized at 3 days post-SCI, and a 1 cm long segment of spinal cord, including the injury epicenter, was dissected and collected for the experiment.). GSE19890 contained miRNA gene expression of spinal cord tissues from five sham-operated rats and five spinal cord injured rats (Modeling methods and tissue sample collection was not provided). GSE45006 contained mRNA expression of spinal cord tissues from four SCI rats and four sham-operated rats (Rat thoracic spinal cord (T7) was injured using aneurysm clip impact-compression, and the epicenter area of injured spinal cord was isolated). We used the R Limma package for microarray correction and differential expression analysis, followed by heatmap plotting of differentially expressed genes using the R-pHEATMAP package. We further analyzed the sequence conservation of circTYW1 in Homo sapiens as well as Rattus norvigecus through the circatlas website (https://ngdc.cncb.ac.cn/databasecommons/). Also, the binding miRNAs of circTYW1 were predicted by circBank (http://www.circbank.cn/). StarBase (http://starbase.sysu.edu.cn/), TargetScan (http://www.targetscan.org/vert_72/), and miRDB (http://mirdb.org/) were used to predict the targeting mRNAs of miR-380.
2.2. Animals, SCI procedures and injection
The animals had free access to standard rat chow and water. All experimental procedures were implemented following the guidelines of the Institutional Animal Care and Use Committee of Luoyang Orthopedic Hospital of Henan Province. A total of 30 male Sprague-Dawley rats (6 weeks, 220–250 g) from Animal Experimental Center of Luoyang Orthopedic Hospital of Henan Province were kept in a temperature-controlled (22–23°C), humidity-controlled (55–60%) and light-controlled (12-h light/dark cycle) environment. Thirty rats were divided into 5 groups, of which 6 were recorded as the Sham group and the remaining 24 as the SCI group.
Rats were fixed in a prone position and anesthetized with pentobarbital at 35 mg/kg. A 2–3 cm incision was created in the dorsal midline skin, and vertebral T7-T9 were exposed and stabilized. A laminectomy was conducted at the thoracic level T8. A syringe needle was utilized to stimulate the injury, which was released from a height of 12.5 mm above the surface of the cord. The muscle layers were sutured, followed by disinfection. For the sham-operated rats, an incision was made at the same site, but spinal ligation was not performed. Subsequently, 18 rats with SCI were randomly divided into three groups, and lentivirus (Lv)-negative control (NC), Lv-circTYW1, and Lv-short hairpin (sh) RNA targeting circTYW1 (Lv-shcircTYW1) were injected into the rat spinal cord, respectively.
All lentiviral vectors were from Shanghai Hanbio (Shanghai, China). A laminectomy was carried out at the thoracic level under pentobarbital anesthesia. A polyethylene catheter (PE10, Portex, Smith Medical, Kent, UK) was passed through the T9-12 tail, and the 2-cm free end was exposed to the upper thoracic region. Lentivirus were prepared and titered to 1 × 109 transfection units/mL. Rats in each group were intrathecally injected with 100 μL lentivirus. After 5 d, the SCI procedure was performed, and the motor function of hind limbs was evaluated at 7 d post-surgery.
2.3. Basso, Beattie, and Bresnahan (BBB) scale
The BBB scale was applied to assess locomotor capacity of the hind limbs on the 7th d after spinal cord contusion. BBB scale ranges from 0 to 21, where 0 indicated no movement and 21 indicated normal motor function. Scores were determined by two independent observers who did not perform the experiment, and the scores of the two observers were averaged. After the last behavioral experiment (day 56 after surgery), euthanasia was performed by intraperitoneal injection of 150 mg/kg sodium pentobarbital. Following euthanasia, animal death was confirmed by observing the lack of heartbeat, blink and nerve reflex. Subsequently, spinal cord tissues at T8 from the rats were collected for protein and mRNA assays.
2.4. RNA-fluorescence in situ hybridization (FISH)
Briefly, the tissue sections or PC12 cells were treated with proteinase K (2 μg/mL) for 15 min at room temperature and hybridized with Cy3-labeled antisense RNA overnight at 63°C. The hybridization solution consisted of 50% formamide, 20 mm Tris-HCl (pH = 7.5), 600 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 10% dextran sulfate, 200 μg/mL yeast tRNA and 1× Denhardt solution. After being washed with washing buffer, the sections were incubated with alkaline phosphatase-conjugated anti-DIG antibody (1:5000, ab420; Abcam, Cambridge, UK). Color development was performed in the dark at room temperature in the presence of 4-nitrotetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate (Roche Diagnostics).
2.5. Nissl staining
The 5-µm sections were placed in a mixture of anhydrous ethanol and chloroform (1:1) and allowed to permeate overnight at room temperature. Next, the tissue sections were treated with 100% anhydrous ethanol, 95% ethanol and distilled water. The sections were treated with preheated 0.1% cresyl fast violet at 37°C for 10 min, washed once with triple distilled water, differentiated with 95% ethanol for 5 min, and placed in 100% anhydrous ethanol and xylene. Finally, the tissue sections were fixed for microscopic observation. The average number of motor neurons at five stained sites in the anterior horn of the spinal cord was randomly selected for each group.
Terminal deoxynucleotidyl transferase (TdT)-mediated 2ʹ-Deoxyuridine 5ʹ-Triphosphate (dUTP) nick end labeling (TUNEL)
Apoptosis was measured by TUNEL analysis (Roche Molecular Systems, Inc., Branchburg, NJ, USA) as per the manufacturer’s protocols. Briefly, the spinal cord tissues or PC12 cells were fixed with 4% formaldehyde for 20 min, permeabilized for 10 min at ambient temperature, and finally labeled with TUNEL for 60 min at 37°C in a humidified environment. After staining, the sections were incubated with 6-dimidyl-2-phenylindole solution for 5 min to stain the nuclei. Finally, five regions were randomly selected in each section, and the average number of apoptotic cells per 200 cells was determined.
2.7. Fluoro-Jade C (FJC) staining
The sections (25-μm) were immersed in an 80% ethanol solution containing 1% NaOH for 5 min and in 70% ethanol and distilled water for 2 min. The sections were incubated in 0.06% potassium permanganate solution for 10 min. After water rinsing, the sections were stained with FJC solution (AAT Bioquest, Sunnyvale, CA, USA) for 20 min. The sections were then dried, and covered with fixed medium for further cell counting. The entire process was performed in darkness.
2.8. Immunohistochemistry
Spinal cord sections were collected from sham-operated and SCI rats, embedded in Tissue-Tek OCT and frozen at −80°C until use. The sections (20 μm) were prepared using a cryostat and mounted on Matsunami adhesive-coated slides (Matsunami, Osaka, Japan). The sections were sealed with phosphate buffered saline (PBS) containing 5% bovine serum albumin and 0.1% Triton X-100 for 1 h at room temperature, followed by incubation with primary antibodies against Caspase-3 (Abcam) and phos-ERK1/2 (Thermo Fisher Scientific Inc., Waltham, MA, USA) at 4°C overnight. Immunoreactivity was observed using horseradish peroxidase-coupled secondary antibodies (Thermo Fischer Scientific). Images were captured under a microscope (IX83, Olympus Optical Co., Ltd., Tokyo, Japan).
2.9. RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
A Mini-BEST Universal RNA Extraction Kit (TaKaRa, Kyoto, Japan) was used to extract total RNA from tissues and cells. For circRNA and mRNA, RNA was reversely transcribed to cDNA using the Prime Script RT Master Mix kit (TaKaRa), and qPCR analysis was conducted using SYBR Green Master Mix (TaKaRa) and PCR LightCycler480 (Roche, Diagnostics, Basel, Switzerland). In addition, RNase R (Epicenter Technologies, Madison, WI, USA) was used to confirm the presence of circTYW1 and to eliminate the effects of linear TYW1 RNA. For miRNA, cDNA was synthesized using the PrimeScript™ RT kit (TaKaRa). The expression of miR-380 was detected using TaqMan Universal Master Mix II (Assay ID: 002260; Applied Biosystems, Inc., Foster City, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 (Assay ID: 001973, Applied Biosystems) served as internal controls. The complete sequences of the primers are exhibited in Table 1.
Table 1.
Sequences of oligomers and primers used in the present research
| Targets | Species | Forward (5ʹ-3ʹ) | Reverse (5ʹ-3ʹ) |
| CircTYW1 | Rattus | GATAACTCCTGCTCTCCGAGAAG | CATGCAGCGATGGCTCTCATTC |
| FGF9 | Rattus | CCAGGAAAGACCACAGCCGATT | CCATACAGCTCCCCCTTCTCAT |
| GAPDH | Rattus | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
| miR-380 | Rattus | TGGTTGACCATAGAACATG | GAACATGTCTGCGTATCTC |
| U6 | Rattus | CTCGCTTCGGCAGCACAT | TTTGCGTGTCATCCTTGCG |
Note: circRNAs, circular RNAs; miRNAs, microRNAs; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
2.10. Western blot
On the seventh day following SCI, the rats were anesthetized, and spinal cord tissues were collected. The concentration of protein in each group was 2 μg/μL. Separated and concentrated gels were dispensed. Next, 30 μg protein was added to the corresponding wells. The gels were separated after electrophoresis, and the proteins on the separated gels were transferred to a polyvinylidene fluoride membrane and allowed to stand at room temperature for 2 h with the sealing solution. Subsequently, the membranes were probed at 4°C overnight with the following antibodies against FGF9 (1:1000, Abcam), ERK1/2 (1:1000, Abcam), phos-ERK1/2 (phospho T202 + Y204) (1:1000, Abcam) and GAPDH (1:2000, Cell Signaling Technologies, Beverly, MA, USA). On the second day, the membranes were re-probed with the secondary antibody (1:2000; Abcam) for 2 h at ambient temperature. Finally, protein bands were visualized using an enhanced chemiluminescence solution. The results were analyzed in ImageJ2x software.
2.11. Cell culture and treatment
The PC12 neurons were grown in a 37°C incubator with Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA, USA) plus 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/ mL streptomycin in 5% CO2. Subsequently, the cells were infected using the constructed Lv-negative control (NC), Lv-shcircTYW1 or Lv-circTYW1. At 24 h after infection, the culture medium was replaced with fresh medium for another 48-h culture. Subsequently, PC12 cells stably expressing Lv-circTYW1 were taken, and the constructed miR-380 mimic and the corresponding mimic NC or shRNA targeting FGF9 and the corresponding sh-NC were used to infect PC12 cells, respectively.
For OGD exposure, PC12 cells after infection were stored in glucose-free DMEM (Gibco) and placed in a hypoxic chamber (Ruskinn Technology, Ltd., Cardiff, UK) with 5% N2 and 95% CO2 at 37°C for 2 h. After hypoxia, the cells were incubated in an incubator at 37°C in 5% CO2 for 12 h, and then the hypoxic glucose-free medium was refreshed with normal DMEM. Control cells were cultured under normal conditions for 12 h. The cells were harvested for subsequent experiments.
2.12. Casapse-3/7 activity assay
Relative caspase-3/7 activity was measured using a Caspase-Glo 3/7 kit (catalog number G8090, Promega, Madison, WI, USA). PC12 cell lysate was seeded into 96-well microtiter plates and treated with Caspase-Glo 3/7 analytical mixtures for 30 min. The luminescence was detected using a microplate reader.
2.13. Lactate dehydrogenase (LDH) assay
The cells were seeded into 96-well plates, and the culture supernatant was collected. LDH release levels were then checked using an LDH release assay kit (Beyotime Biotechnology Co., Ltd., Shanghai, China) according to the instructions. The optical density (OD) value at 495 nm was measured using a microplate reader.
2.14. Flow cytometry
The cells were seeded in 6-well plates (3 × 105 cells/well). Then, cell suspension was stained with fluorescein isothiocyanate (FITC)/propidium iodide (PI). Annexin positive/PI negative and Annexin positive/PI positive cells were regarded as apoptotic cells, and apoptosis index refers to the proportion of Annexin positive/PI negative and Annexin positive/PI positive to total cells. A FACScan flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and CellQuest (Becton, Dickinson and Company) software were utilized for analyses.
2.15. Cell counting kit-8 (CCK-8) assay
PC12 cells were incubated with 100 μL medium in 96-well plates with blank wells overnight at 37°C. After 24 h of transfection, hypoxia (2 h) and reoxygenation (22 h) were performed. Subsequently, the cells in each well were reacted with 10 μL CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan) for 1–2 h at 37°C. OD values were detected at 450 nm in each well using a microplate reader.
2.16. Fractionation and export assay
Ribonuclease R (RNase R; Geneseed, Guangzhou, Guangdong, China) was used to characterize circTYW1. Briefly, the extracted RNA was treated with RNase R, and then RT-qPCR was performed to determine the expression of circTYW1 and GAPDH (representative of linear RNA). Cytoplasmic and nuclear RNA from PC12 cells were extracted using the Cytoplasmic and Nuclear RNA Purification Kit (AmyJet Scientific Inc., Wuhan, Hubei, China). The expression of circTYW1, U6 and GAPDH in the cytoplasm and nucleus were then assessed using RT-qPCR. GAPDH and U6 were used as cytoplasmic and nuclear controls, respectively.
2.17. FISH
FISH experiments were performed using Cy5-labeled probes (5ʹ-DIG-ACATCCCCCATGGTCTTCTACTGTCAACT-3ʹ) to detect the expression of circTYW1. After pre-hybridization (1× PBS containing 0.5% Triton X-100), the samples were hybridized in hybridization buffer (1000 mg/mL sheared salmon sperm DNA, 1000 mg/mL yeast transfer RNA, 10 mM dichlorodiphenyltrichloroethane, 4× sodium chloride-sodium citrate buffer, 1× Denhardt’s solution, 10% dextran sulfate and 40% formamide) with specific probes overnight at 60°C and then imaged.
2.18. Dual-luciferase reporter assays
Sequences with predicted binding sites between miR-380 and circ_0003962 or FGF9 3ʹ-untranslated region (3ʹ-UTR) were inserted into the gene vector pmirGLO (Promega Corp., Madison, WI, USA). Next, wild-type for FGF9 (FGF9-wt) or circ_0003962 (circ_0003962-wt) and mutant for FGF9 (FGF9-mt) or circ_0003962 (circ_0003962-mt) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The FGF9-wt and FGF9-mt constructs or circ_0003962-wt and circ_0003962-mt were then co-transfected into PC12 cells with NC mimic or miR-380 mimic. After 24 h of transfection, the cells underwent 2 h of hypoxia and 22 h of reoxygenation were lysed. According to the manufacturer’s instructions for the dual-luciferase assay kit (K801-200, BioVision, Milpitas, CA, USA), a dual-luciferase reporter gene analysis system (Promega) was used. The luciferase activity of the target reporter gene was determined based on the relative light unit (RLU) of firefly luciferase divided by the RLU of renilla luciferase, using renilla luciferase as an internal control.
2.19. Biotin-labeled RNA pull-down assay
The biotinylated miR-380 probe was designed and synthesized by Ribobio (Guangzhou, Guangdong, China). Approximately 1 × 107 PC12 cells were harvested, lysed and sonicated. The miR-380 probe and the Oligo probe were incubated in C-1 magnetic beads (Life Technologies, Carlsbad, CA, USA) at 25°C for 2 h to generate probe-coated beads. The cell lysates were incubated with these probe-coated beads at 4°C overnight. After three washes with wash buffer, the RNA complexes bound to the beads were eluted, and the enrichment levels of circTYW1 or FGF9 in the complexes were detected by RT-PCR or RT-qPCR using the RNeasy Mini kit (Qiagen, Valencia, CA, USA).
2.20. Data analysis
Data were analyzed by GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA, USA) and displayed as mean ± standard deviation (SD). Unpaired t test, one-way or two-way analysis of variance (ANOVA) with Tukey’s post-hoc test were used for statistical analysis depending on the characteristics of the data. All experiments were repeated at least three times, and each experiment was performed in triplicate. The threshold for statistical significance was a p < 0.05.
3. Results
CircRNA_0003962 is poorly expressed in spinal cord tissues of SCI rats and PC12 cells exposed to OGD
We first downloaded a circRNA expression microarray GSE114426 containing spinal cord tissues from three sham-operated rats and three SCI rats from the GEO database. We identified a total of 305 circRNAs with differential expression in SCI-injured rat spinal cord tissues by Limma R package analysis, of which 68 circRNAs were upregulated and 237 circRNAs were downregulated (Figure 1a). Figure 1b and Table 2 shows the top 30 differentially expressed circRNAs. And then, we detected the expression of the top ten differentially expressed circRNAs in the spinal cord tissues of SCI rats. The expression of circ_0003962 was most significantly reduced in SCI rats (Figure 1c). Moreover, the host gene for circ_0003962 is the thirteenth exon of TYW1, and we further found through the circatlas website that hsa-circTYW1 is highly conserved with rno-circTYW1 (Figure 1d). Furthermore, we used OGD to treat PC12 cells to mimic the injury after SCI, and we observed that the expression of circTYW1 in PC12 cells was significantly suppressed after OGD treatment (Figure 1e).
Figure 1.
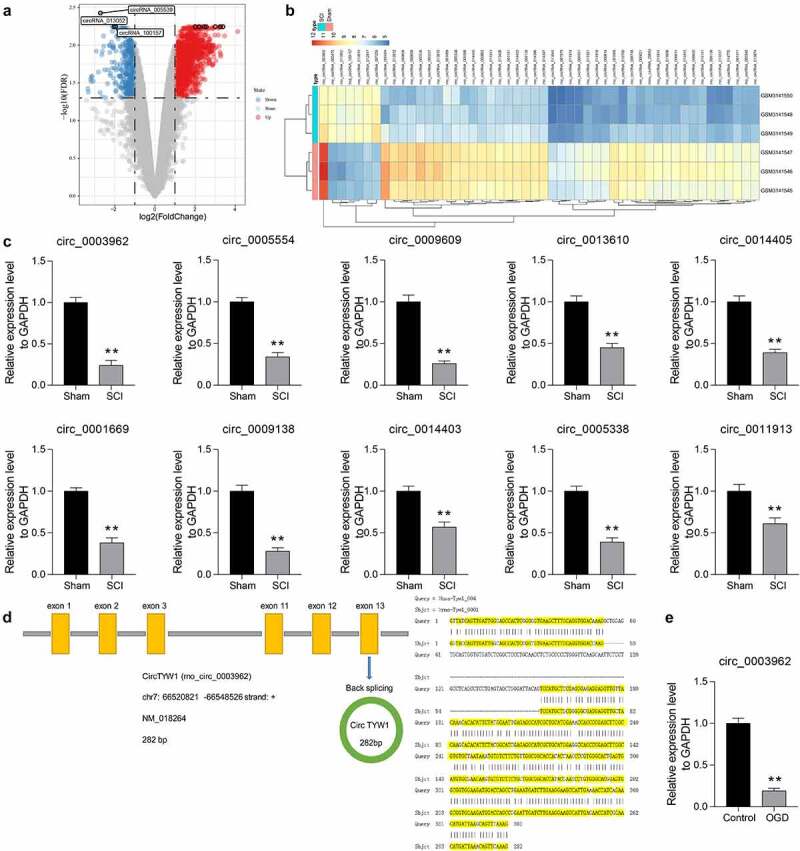
CircRNA_0003962 is downregulated in spinal cord tissues of SCI rats. A-B, circRNA microarray was performed based on 3 spinal cord tissues of SCI and sham-operated rats for volcano plot (a) and heatmap (b) mapping; C, RT-qPCR detection of the top ten differentially expressed circRNAs in spinal cord tissues at T8 from sham-operated and SCI rats on the 56th day after surgery (n = 6); D, The host gene for circ_0003962 and its conservation in Homo sapiens as well as in Rattus norvigecus; E, RT-qPCR detection of circTYW1 expression in OGD-treated PC12 cells. Data were presented as the mean ± SD. Unpaired t test was used for statistical analysis. All experiments were repeated at least three times, and each experiment was performed in triplicate. **p < 0.01 vs sham-operated rats or PC12 cells treated with normal condition
Table 2.
Top 30 differentially expressed genes in GSE114426
| logFC | AveExpr | t | p.Value | adj.p.Val | B | |
| rno_circRNA_005470 | 2.77 | 6.87 | 18.07 | 2.57E-07 | 3.64E-03 | 7.08E+00 |
| rno_circRNA_005537 | −2.94 | 7.99 | −13.54 | 2.00E-06 | 6.36E-03 | 5.53E+00 |
| rno_circRNA_001668 | −3.33 | 7.35 | −13.46 | 2.07E-06 | 6.36E-03 | 5.50E+00 |
| rno_circRNA_014396 | −2.41 | 7.66 | −12.97 | 2.69E-06 | 6.36E-03 | 5.28E+00 |
| rno_circRNA_014397 | −2.49 | 7.72 | −12.73 | 3.06E-06 | 6.36E-03 | 5.17E+00 |
| hsa_circRNA_100157 | 2.03 | 6.88 | 12.24 | 4.04E-06 | 6.36E-03 | 4.94E+00 |
| rno_circRNA_013610 | −3.19 | 7.95 | −12.11 | 4.35E-06 | 6.36E-03 | 4.87E+00 |
| rno_circRNA_005340 | −2.14 | 6.74 | −11.65 | 5.72E-06 | 6.36E-03 | 4.63E+00 |
| rno_circRNA_010919 | 2.17 | 6.91 | 11.49 | 6.29E-06 | 6.36E-03 | 4.55E+00 |
| rno_circRNA_012917 | 2.1 | 7.27 | 11.33 | 6.90E-06 | 6.36E-03 | 4.47E+00 |
| rno_circRNA_010227 | −2.87 | 6.69 | −10.86 | 9.28E-06 | 6.36E-03 | 4.20E+00 |
| rno_circRNA_007979 | −2.21 | 5.9 | −10.81 | 9.60E-06 | 6.36E-03 | 4.17E+00 |
| rno_circRNA_009138 | −2.35 | 7.83 | −10.67 | 1.05E-05 | 6.36E-03 | 4.09E+00 |
| rno_circRNA_001669 | −2.32 | 8.03 | −10.64 | 1.07E-05 | 6.36E-03 | 4.07E+00 |
| rno_circRNA_013612 | −3.21 | 7.78 | −10.63 | 1.07E-05 | 6.36E-03 | 4.07E+00 |
| rno_circRNA_005536 | −3.22 | 8.18 | −10.56 | 1.13E-05 | 6.36E-03 | 4.02E+00 |
| rno_circRNA_011919 | −2.12 | 5.98 | −10.52 | 1.16E-05 | 6.36E-03 | 4.00E+00 |
| rno_circRNA_005338 | −2.2 | 7.92 | −10.51 | 1.16E-05 | 6.36E-03 | 3.99E+00 |
| rno_circRNA_015406 | −2.05 | 7.61 | −10.35 | 1.29E-05 | 6.36E-03 | 3.90E+00 |
| rno_circRNA_001161 | −2.17 | 7.7 | −10.26 | 1.37E-05 | 6.36E-03 | 3.85E+00 |
| rno_circRNA_006620 | −2.25 | 7 | −10.17 | 1.46E-05 | 6.36E-03 | 3.79E+00 |
| rno_circRNA_009608 | −3.17 | 8 | −10.16 | 1.46E-05 | 6.36E-03 | 3.78E+00 |
| rno_circRNA_009136 | −2.61 | 6.62 | −10.15 | 1.47E-05 | 6.36E-03 | 3.78E+00 |
| rno_circRNA_009609 | −3.13 | 7.87 | −10.1 | 1.53E-05 | 6.36E-03 | 3.74E+00 |
| rno_circRNA_013874 | −2.01 | 6.75 | −9.86 | 1.80E-05 | 6.36E-03 | 3.59E+00 |
| rno_circRNA_014770 | −2.69 | 6.68 | −9.83 | 1.84E-05 | 6.36E-03 | 3.57E+00 |
| rno_circRNA_014403 | −2.11 | 7.68 | −9.66 | 2.07E-05 | 6.44E-03 | 3.46E+00 |
| rno_circRNA_003962 | −2.85 | 10.35 | −9.66 | 2.07E-05 | 6.44E-03 | 3.46E+00 |
| rno_circRNA_000331 | −2.38 | 6.16 | −9.66 | 2.08E-05 | 6.44E-03 | 3.46E+00 |
| rno_circRNA_011921 | −2.25 | 6.54 | −9.49 | 2.34E-05 | 6.44E-03 | 3.34E+00 |
Note: FC, foldchange; rno, rattus norvegicus; hsa, homo sapiens.
3.2. Overexpression of circTYW1 promotes neurological recovery in SCI rats
To verify the role of circTYW1 in neurological recovery in SCI rats, we injected circTYW1 overexpression plasmid and shRNA targeting circTYW1 into SCI rats (Figure 2a). We first used RT-qPCR to detect the expression of circTYW1 in rat spinal cord tissues to confirm the successful intervention of circTYW1 (Figure 2b). We subsequently tested the recovery of locomotor function in rats after SCI surgery by BBB scale and found that increasing circTYW1 expression in spinal cord tissues promoted recovery of locomotor function in SCI rats, which was significantly reduced in shcircTYW1-injected rats (Figure 2c). Furthermore, Nissl staining revealed that neuronal activity in rat spinal cord tissues was significantly increased after overexpression of circTYW1 and hampered after downregulation of circTYW1 (Figure 2d). TUNEL and FJC staining showed that knocking-down circTYW1 elevated the proportion of apoptotic cells and injured neurons in rat spinal cord tissues, whereas overexpression of circTYW1 led to opposite experimental results (Figure 2e, f). Furthermore, we used immunohistochemistry to detect the expression of Caspase-3 in rat spinal cord tissues. After overexpression of circTYW1, the staining intensity of Caspase-3 in rat spinal cord tissues was significantly decreased, while in shcircTYW1-injected rats, the staining intensity of Caspase-3 had a significant elevation (Figure 2g).
Figure 2.
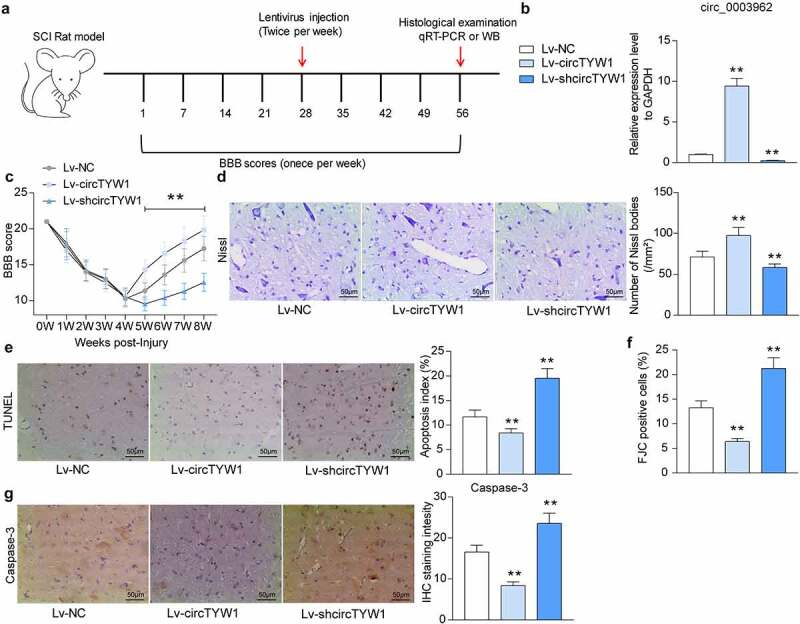
Overexpression of circTYW1 promotes neurological recovery in SCI rats. A, the time course of the experiment; B, expression of circTYW1 in spinal cord tissues of rats by RT-qPCR; C, BBB score assessment of recovery of locomotion in rats after SCI surgery; D, Nissl staining for neuronal activity in rat spinal cord tissues; E, TUNEL staining for the proportion of apoptotic neurons in rat spinal cord tissues; F, FJC staining for the proportion of neurons damaged in rat spinal cord tissues; G, staining intensity of Capsase-3 in rat spinal cord tissues by immunohistochemistry. n = 6. Data were presented as the mean ± SD. One-way ANOVA with Tukey’s post-hoc test was used for statistical analysis. **p < 0.01 vs SCI rats injected with Lv-NC
3.3. circTYW1 inhibits OGD-induced PC12 cell injury
To probe the role of circTYW1 on the protective role of PC12 cell against injury induced by OGD, we overexpressed or knocked-down circTYW1 expression in PC12 cells and determined successful cell infection by RT-qPCR (Figure 3a). Subsequently, we used the Caspase-3/7 kit to detect changes in Caspase 3/7 activity in the cells treated with OGD. Poor expression of circTYW1 remarkably promoted OGD-induced Caspase-3/7 activation, whereas OGD-induced Caspase-3/7 activation was notably reduced in the cells overexpressing circTYW1 (Figure 3b). Furthermore, we assessed the level of LDH release from cells and observed that shcircTYW1 contributed to OGD-induced cytotoxicity, whereas increased expression of circTYW1 in cells significantly decreased LDH release (Figure 3c). Flow cytometry and TUNEL staining detecting changes in apoptosis levels after OGD treatment demonstrated that circTYW1 was neuroprotective (Figure 3d, e). Thereafter, we further assayed cell activity using CCK-8 and found that poor expression of circTYW1 further exacerbated the inhibitory effect of OGD on PC12 cell activity, whereas high expression of circTYW1 attenuated this effect (figure 3f).
Figure 3.
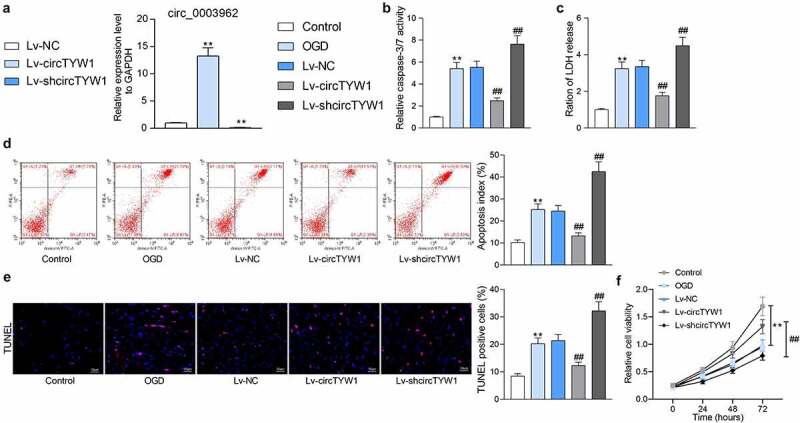
circTYW1 inhibits OGD-induced PC12 cell injury. A, expression of circTYW1 in PC12 cells detected by RT-qPCR. PC12 cells after infection were exposed to OGD or PBS for 12 h. B, Caspase-3/7 kit detection of Caspase-3/7 activity in PC12 cells; C, LDH kit detection of cytotoxicity in PC12 cells; D, flow cytometry detection of the PC12 cell apoptosis; E, TUNEL staining of the PC12 cell apoptosis; F, CCK-8 detection of PC12 cell activity. Data were presented as the mean ± SD. All experiments were repeated at least three times, and each experiment was performed in triplicate. One-way (panels A-E) or two-way ANOVA (panel F) with Tukey’s post-hoc test was used for statistical analysis. **p < 0.01 vs PC12 cells treated with normal condition; ##p < 0.01 vs PC12 cells transfected with Lv-NC and exposed to OGD
3.4. CircTYW1 localizes to the cytoplasm and targets miR-380
Based on the aforementioned experimental results, circTYW1 is displayed to be neuroprotective. To clarify its regulatory mechanism, we first used fractionation and export and FISH assays for the subcellular localization of circTYW1, and noted that circTYW1 was mainly distributed in the cytoplasm (Figure 4a, b). To further analyze the specific cells in which circTYW1 acts, we used RNA-FISH with NeuN immunofluorescence to essentially confirm that circTYW1 was expressed mainly in neurons of spinal cord tissues (Fig S1A).
Figure 4.
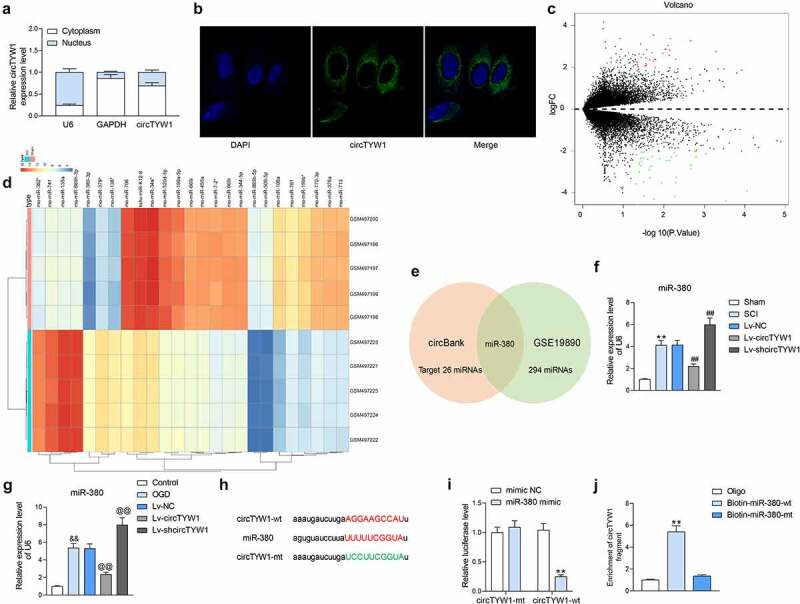
CircTYW1 is localized in the cytoplasm and binds to miR-380. A, subcellular localization of circTYW1 detected by fractionation and export assay; B, subcellular localization of circTYW1 detected by FISH experiments; C-D, GSE19890 microarray (comprised of spinal cord tissues from sham-operated 5 rats and 5 SCI rats) was performed for volcano plot (c) and heatmap (d) mapping; E, Venn diagram showing the number of miRNAs with binding relationship predicted by algorithm circbank and differentially expressed miRNAs in GSE19890 microarray; F, expression of miR-380 in rat spinal cord tissues on the 56th day after surgery by RT-qPCR (n = 6); G, expression of miR-380 in PC12 cells at 12 h after OGD treatment detected by RT-qPCR; H, binding sites of miR-380 to circTYW1; I, dual-luciferase assay determination of the binding relationship between miR-380 and circTYW1; J, biotin-labeled RNA pull down assay detection of the binding relationship between miR-380 and circTYW1. Data were presented as the mean ± SD. All experiments were repeated at least three times, and each experiment was performed in triplicate. One-way (panels F, G and J) or two-way ANOVA (panels A and I) with Tukey’s post-hoc test was used for statistical analysis. **p < 0.01 vs sham-operated rats, mimic NC or Oligo/miR-380-mt treatment; ##p < 0.01 vs SCI rats injected with Lv-NC; && p < 0.01 vs PC12 cells treated with normal condition; @@p < 0.01 vs PC12 cells transfected with Lv-NC and exposed to OGD
We then downloaded a miRNA expression microarray GSE19890 of spinal cord-injured rats from the GEO database. Spinal cord tissues of five sham-operated rats and five spinal cord-injured rats were included in the dataset. A total of 485 differentially expressed miRNAs were screened out (Figure 4c, d), which were intersected with miRNAs that bound to circTYW1 predicted by circbank. miR-380 was found as the only result (Figure 4e). We then examined the miR-380 expression in the spinal cord tissues of rats with SCI and noted that the miR-380 expression was remarkably increased, while further overexpression of circTYW1 significantly suppressed the expression of miR-380 (figure 4f). The consistent results were observed in OGD-treated PC12 cells (Figure 4g).
In order to detect the binding relationship between miR-380 and circTYW1, we designed dual-luciferase and biotin-labeled RNA pull-down experiments. In dual-luciferase experiments, luciferase activity was significantly decreased in 293 T cells transfected with circTYW1-wt and miR-380 mimic (Figure 4h, i). Furthermore, significantly higher levels of circTYW1 enrichment were observed in the complexes pulled down using Biotin-miR-380-wt in PC12 cells than in Oligo and miR-380-mt (Figure 4j). Moreover, we further detected that miR-380 was mainly distributed in the cytoplasm in PC12 cells using RNA-FISH experiments (Fig S1B). The above results indicate that miR-380 can bind to circTYW1.
3.5. miR-380 mimic attenuates the protective effect of circTYW1 on PC12 cells
To further determine the role of miR-380 on neurological recovery following SCI, PC12 cells with circTYW1 knockdown were infected with miR-380 inhibitor, and those overexpressing circTYW1 were infected with miR-380 mimic. RT-qPCR verified the successful intervention (Figure 5a). miR-380 inhibitor significantly attenuated the inhibition of PC12 activity caused by OGD in the presence of shcircTYW1, whereas miR-380 mimic infection in PC12 cells with high expression of circTYW1 had completely opposite results (Figure 5b-f).
Figure 5.
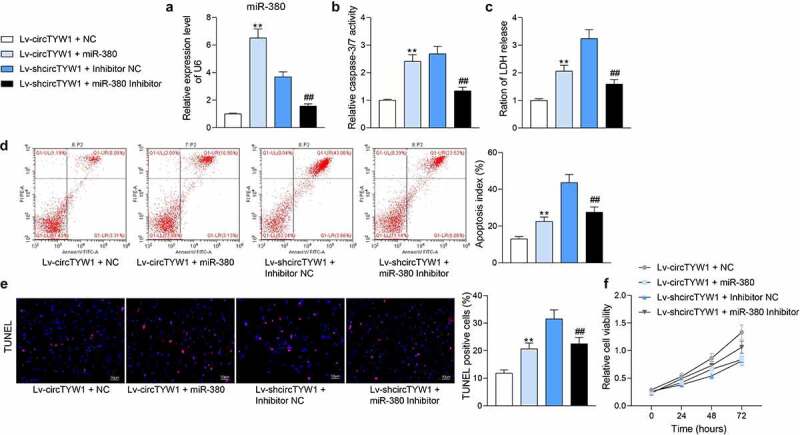
miR-380 mimic attenuates the protective effect of circTYW1 on PC12 cells. A, expression of miR-380 in PC12 cells detected by RT-qPCR; B, Caspase-3/7 kit detection of Caspase-3/7 activity in PC12 cells; C, LDH kit detection of cytotoxicity in PC12 cells; D, flow cytometry detection of the PC12 cell apoptosis; E, TUNEL staining of the PC12 cell apoptosis; F, CCK-8 detection of PC12 cell activity. Data were presented as the mean ± SD. All experiments were repeated at least three times, and each experiment was performed in triplicate. One-way (panels A-E) or two-way ANOVA (panel F) with Tukey’s post-hoc test was used for statistical analysis. **p < 0.01 vs PC12 cells transfected with Lv-circTYW1 + NC and exposed to OGD; ##p < 0.01 vs PC12 cells transfected with Lv-shcircTYW1 + inhibitor NC and exposed to OGD
3.6. FGF9 is bound and negatively regulated by miR-380
We further analyzed the GSE45006 microarray, which contained spinal cord tissues from four SCI rats and four sham-operated rats, and obtained 291 differentially expressed genes (Figure 6a, b). Subsequently, we used StarBase, TargetScan, and miRDB to predict the target mRNAs of miR-380 and cross-sectioned with differentially expressed genes in GSE45006, and we screened out FGF9 (Figure 6c).
Figure 6.
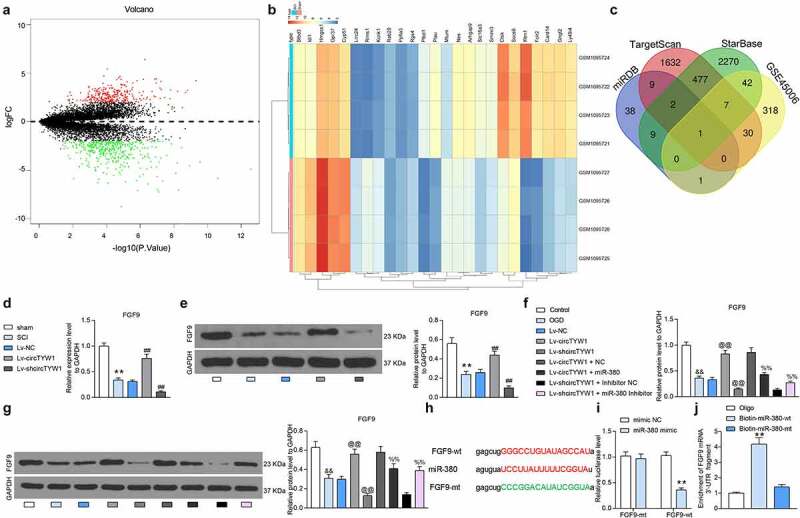
miR-380 targets and regulates FGF9. A-B, mRNA microarray was performed based on 4 spinal cord tissues of SCI and sham-operated rats for volcano plot (a) and heatmap (b) mapping; C, Venn diagram showing the number of miRNAs with binding relationship predicted by algorithms StarBase, TargetScan, miRDB and differentially expressed miRNAs in GSE450006 microarray; D-E, expression of FGF9 mRNA (d) and protein (e) in rat spinal cord tissues by RT-qPCR and western blot (n = 6); F-G, expression of FGF9 mRNA (f) and protein (g) in PC12 cells detected by RT-qPCR and western blot; H, binding sites of miR-380 to FGF9; I, dual-luciferase assay determination of the binding relationship between miR-380 and FGF9; J, biotin-labeled RNA pull-down assay detection of the binding relationship between miR-380 and FGF9. Data were presented as the mean ± SD. All experiments were repeated at least three times, and each experiment was performed in triplicate. One-way (panels D-G and J) or two-way ANOVA (panel I) with Tukey’s post-hoc test was used for statistical analysis. **p < 0.01 vs sham-operated rats, mimic NC or Oligo/miR-380-mt treatment; ##p < 0.01 vs SCI rats injected with Lv-NC; &&p < 0.01 vs PC12 cells treated with normal condition; @@p < 0.01 vs PC12 cells transfected with Lv-NC and exposed to OGD; $$p < 0.01 vs PC12 cells transfected with Lv-circTYW1 + NC and exposed to OGD; %%p < 0.01 vs PC12 cells transfected with Lv-shcircTYW1 + inhibitor NC and exposed to OGD
We then examined the FGF9 expression in spinal cord tissues of the rats with SCI and noted that the FGF9 expression was much lower, while further overexpression of circTYW1 significantly enhanced the expression of FGF9 (Figure 6d, e). In OGD-treated PC12 cells, we found a significant increase in FGF9 expression after overexpression of circTYW1 and a significant decline in FGF9 expression after further overexpression of miR-380 (Figure 6f, g).
In order to detect the binding relationship between miR-380 and FGF9, we designed dual-luciferase and biotin-labeled RNA pull-down experiments. In dual-luciferase experiments, luciferase activity was significantly reduced in 293 T cells transfected with FGF9-wt and miR-380 mimic (Figure 6h, i). Moreover, we found significantly higher levels of FGF9 enrichment in complexes pulled down using Biotin-miR-380-wt in PC12 cells than in Oligo and Biotin-miR-380-mt (Figure 6j). The above results indicate that miR-380 can bind to FGF9.
3.7. The involvement of circTYW1/miR-380/FGF9 axis in regulating the ERK1/2 signaling in SCI
In a study by Chuang et al., it was noted that FGF9 could inhibit MPP-induced oxidative damage in neurons by promoting phosphorylation modification of ERK1/2 [12]. We subsequently detected the activation of ERK1/2 in rat spinal cord tissues by western blot and found that the phosphorylation level of ERK1/2 in rat spinal cord tissues was significantly suppressed after SCI surgery, however, increased expression of circTYW1 resulted in a significant increase in the phosphorylation level of ERK1/2 (Figure 7a). Immunohistochemical staining of phos-ERK1/2 in rat spinal cord tissues showed the consistent experimental results (Figure 7b). The phosphorylation level of ERK1/2 was significantly inhibited after OGD treatment. After further circTYW1 overexpression, there was an increase in phosphorylated ERK1/2. However, after further transfection with miR-380 mimic, there was a significant decline in phosphorylated ERK1/2 in PC12 cells (Figure 7c).
Figure 7.
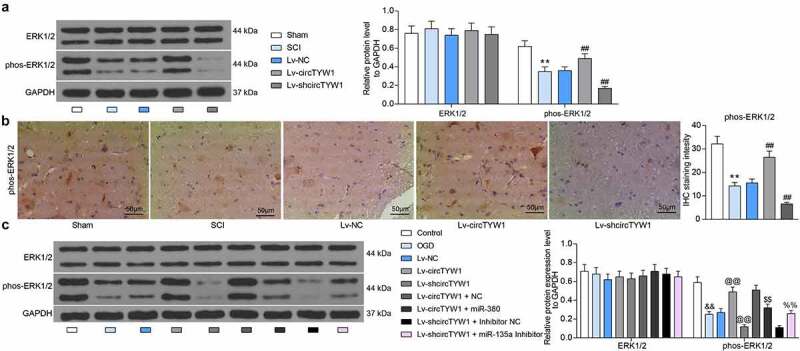
CircTYW1 exerts a neuroprotective role during SCI by regulating the ERK1/2 signaling via the miR-380/FGF9 axis. A, western blot detection of ERK1/2 expression and phosphorylation level in rat spinal cord tissues (n = 6); B, staining intensity of phos-ERK1/2 in rat spinal cord tissues by immunohistochemistry (n = 6); C, western blot detection of ERK1/2 expression and phosphorylation levels in PC12 cells. Data were presented as the mean ± SD. All experiments were repeated at least three times, and each experiment was performed in triplicate. One-way (panel B) or two-way ANOVA (panels A and C) with Tukey’s post-hoc test was used for statistical analysis. **p < 0.01 vs sham-operated rats; ##p < 0.01 vs SCI rats injected with Lv-NC; &&p < 0.01 vs PC12 cells treated with normal condition; @@p < 0.01 vs PC12 cells transfected with Lv-NC and exposed to OGD; $$p < 0.01 vs PC12 cells transfected with Lv-circTYW1 + NC and exposed to OGD; %%p < 0.01 vs PC12 cells transfected with Lv-shcircTYW1 + inhibitor NC and exposed to OGD
3.8. Knockdown of FGF9 impairs the protective effect of circTYW1 on PC12 cells
To further explore the effect of FGF9 on PC12 cells, we further infected PC12 cells overexpressing circTYW1 with shRNA targeting FGF9. Western blot was conducted to verify the transfection efficiency (Figure 8a). Subsequently, we found that the growth of PC12 cells was significantly reduced (Figure 8b) and the proportion of apoptotic cells was significantly increased (Figure 8c, d) after OGD treatment, indicating that knockdown of FGF9 could impair the protective effect of circTYW1 on PC12 cells. In summary, the expression of circTYW1 was significantly reduced in spinal cord-injured rats, which promoted the inhibitory effect of miR-380 on FGF9 and therefore downregulated the activity of the ERK1/2 signaling pathway, affecting the neurological recovery in rats.
Figure 8.
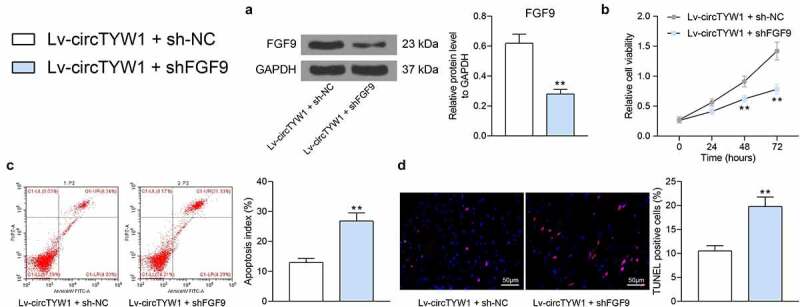
Knockdown of FGF9 impairs the protective effect of circTYW1 on PC12 cells. PC12 cells overexpressing circTYW1 were further infected with shRNA targeting FGF9. A, western blot detection of FGF9 protein expression in PC12 cells; B, CCK-8 detection of PC12 cell activity; C, flow cytometry detection of the PC12 cell apoptosis; D, TUNEL staining of the PC12 cell apoptosis. Data were presented as the mean ± SD. All experiments were repeated at least three times, and each experiment was performed in triplicate. Unpaired t test (panels A, C and D) or two-way ANOVA (panel B) with Tukey’s post-hoc test was used for statistical analysis. **p < 0.01 vs Lv-circTYW1 + sh-NC
4. Discussion
There are around 2 million people suffering from SCI in the world, and the complexity and pathogenic mechanisms following SCI, involving neuronal death and inflammation, complicate the timely treatment and management [13]. CircRNA, a new kind of endogenous noncoding RNAs, is implicated in manifold biological processes, such as acting as miRNA sponges in the process of the assembly of protein complex and regulating the gene transcription activity [14]. The regulatory role for circRNAs has been established through the ceRNA network in the pathogenesis and treatment of SCI [15]. In the current study, the expression of circTYW1, a novel circRNA, was remarkably downregulated in the rats with SCI and PC12 cells exposed to OGD. The findings of the study suggest that circTYW1 promotes neurological recovery following SCI by interfering with the miR-380/FGF9/ERK1/2 axis.
In the present study, poor circRNA_0003962/circTYW1 expression was identified in spinal cord tissues of SCI rats and PC12 cells exposed to OGD. Moreover, enhanced circTYW1 expression potentiated the neurological recovery following SCI in rats, as evidenced by improved motor function and reduced neuronal apoptosis. Identical results were also observed in vitro. Another circRNA circPan3 overexpression attenuated cell apoptosis in cardiomyocytes induced by 2 μM Doxorubicin, as manifested by lowered Caspase-3/7 activities [16]. Besides, the inhibition of circRNA001372 promoted apoptosis and cell damage makers, including LDH, IL-1β, IL-6, IL-17, IL-18, contributing to the declines in cell viability [17]. Under the context of SCI, cicRNA_7079 knockdown enhanced apoptosis in NSC-34 motor neurons [18]. The neuron-protective role of circTYW1 was substantiated in the present study, which has rarely been investigated before. As a consequence, we sought to decipher the downstream biomolecules involved. Intriguingly, circ-HIPK3 was poorly expressed in SCI rats and AGE1.HN and PC12 cells induced by 100 uM CoCl, and overexpression of circ-HIPK3 relieved the neuronal apoptosis through regulating miR-588/DPYSL5 axis in SCI [19]. Additional mechanistical studies in our study revealed that circTYW1 was mainly distributed in the cytoplasm of PC12 cells, indicating its function might be elicited through the ceRNA mechanism. After determining the miR-380 as the target of circTYW1, we performed a rescue experiment in PC12 cells, which corroborated that miR-380 mimic flattened the protective role of circTYW1 against apoptosis in PC12 cells. In line with our data, downregulation of Circ_0001723 induced Caspase-1 protein expression in vitro by upregulating miR-380-3p [20].
Similarly, we combined algorithms StarBase, TargetScan, and miRDB with GSE450006 microarray to screen out FGF9 as a target of miR-380. The targeting relation was then validated through dual-luciferase experiments and biotin-labeled RNA pull-down experiments. Previously, miR-339 has been suggested to inhibit PC12 cell proliferation under the condition of OGD/R, and FGF9 is a direct target of miR-339 and counteracted the effect of miR-339 on the viability and apoptosis of PC12 cells [21]. FGF9 has also been reported to protect neurons from toxicity induced by 1-methyl-4-phenylpyridinium via upregulating heme oxygenase-1 and γ-glutamylcysteine synthetase expression [22]. More specifically, FGF9 ablation led to the impairment of the ERK1/2 signaling in peripheral nerve injury [23]. In consistent, Sumbal et al. observed that FGF9 sustained the activation of the ERK1/2 signaling and promoted proliferation and migration of fibroblasts and mouse Leydig tumor cells [23,24]. In the nervous system, ERK1/2 is important for neuronal differentiation, plasticity and neuronal survival, which may involve transcriptional mediation and/or direct suppression of cell death machinery [25]. Therefore, we postulated that FGF9 is involved in SCI through the modulation of the ERK1/2 signaling. Our following western blot and immunohistochemistry displayed the ERK1/2 signaling deficit in spinal cord tissues of SCI rats and PC12 cells subjected to OGD. Further circTYW1 overexpression expedited the ERK1/2 signaling, while miR-380 upregulation blunted the activation of the ERK1/2 signaling.
In the present study, circTYW1 was downregulated in spinal cord tissues in SCI rats, exogenous circTYW1 evidently promoted the neurological recovery of SCI rats, and circTYW1 sponged miR-380 to regulate the expression of FGF9 and the activation of the ERK1/2 signaling (Figure 9). These experimental results suggest that miR-380 might be a target of circTYW1 in spinal cord tissues. Since miR-380 may have other targets in this microenvironment, it is possible that circTYW1 affects the neurological recovery of SCI rats by regulating other targets. We concluded that circTYW1 promotes neurological recovery following SCI via the miR-380/FGF9/ERK1/2 regulatory axis. Targeting circTYW1 may be a new diagnostic SCI marker and therapeutic target.
Figure 9.
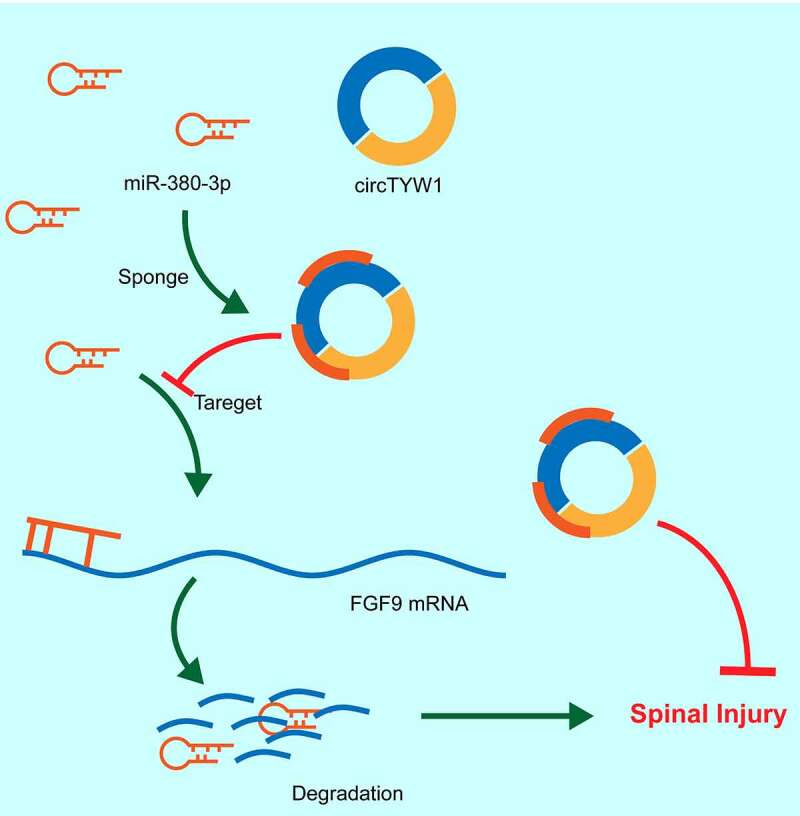
The mechanism diagram. circTYW1 promotes FGF9 expression through competitively binding to miR-380, which in turn activates the ERK1/2 signaling pathway, thus exerting a neuroprotective effect in rats with SCI
Supplementary Material
Acknowledgments
Not applicable.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Courtine G, Sofroniew MV.. Spinal cord repair: advances in biology and technology. Nat Med. 2019;25:898–908. [DOI] [PubMed] [Google Scholar]
- [2].Kjell J, Olson L.. Rat models of spinal cord injury: from pathology to potential therapies. Dis Model Mech. 2016;9:1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Silva NA, Sousa N, Reis RL, et al. From basics to clinical: a comprehensive review on spinal cord injury. Prog Neurobiol. 2014;114:25–57. [DOI] [PubMed] [Google Scholar]
- [4].Floris G, Zhang L, Follesa P, et al. Regulatory Role of Circular RNAs and Neurological Disorders. Mol Neurobiol. 2017;54:5156–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Qin C, Liu CB, Yang DG, et al. Circular RNA Expression Alteration and Bioinformatics Analysis in Rats After Traumatic Spinal Cord Injury. Front Mol Neurosci. 2018;11:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Xu L, Ye X, Zhong J, et al. New Insight of Circular RNAs’ Roles in Central Nervous System Post-Traumatic Injury. Front Neurosci. 2021;15:644239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yao C, Yu B. Role of Long Noncoding RNAs and Circular RNAs in Nerve Regeneration. Front Mol Neurosci. 2019;12:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- [9].Wu R, Mao S, Wang Y, et al. Differential Circular RNA Expression Profiles Following Spinal Cord Injury in Rats: a Temporal and Experimental Analysis. Front Neurosci. 2019;13:1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cai Z, Zheng F, Ding Y, et al . Nrf2-regulated miR-380-3p blocks the translation of Sp3 protein and its mediation of paraquat-induced toxicity in mouse neuroblastoma N2a cells. Toxicol Sci. 2019;171:515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guo M, Chen H, Duan W, et al. FGF9 knockout in GABAergic neurons induces apoptosis and inflammation via the Fas/caspase-3 pathway in the cerebellum of mice. Brain Res Bull. 2020;154:91–101. [DOI] [PubMed] [Google Scholar]
- [12].Chuang JI, Huang JY, Tsai SJ, et al. FGF9-induced changes in cellular redox status and HO-1 upregulation are FGFR-dependent and proceed through both ERK and AKT to induce CREB and Nrf2 activation. Free Radic Biol Med. 2015;89:274–286. [DOI] [PubMed] [Google Scholar]
- [13].Yuan J, Botchway BOA, Zhang Y, et al. Role of Circular Ribonucleic Acids in the Treatment of Traumatic Brain and Spinal Cord Injury. Mol Neurobiol. 2020;57:4296–4304. [DOI] [PubMed] [Google Scholar]
- [14].Qu X, Li Z, Chen J, et al. The emerging roles of circular RNAs in CNS injuries. J Neurosci Res. 2020;98:1485–1497. [DOI] [PubMed] [Google Scholar]
- [15].Peng P, Zhang B, Huang J, et al. Identification of a circRNA-miRNA-mRNA network to explore the effects of circRNAs on pathogenesis and treatment of spinal cord injury. Life Sci. 2020;257:118039. [DOI] [PubMed] [Google Scholar]
- [16].Ji X, Ding W, Xu T, et al . MicroRNA-31-5p attenuates doxorubicin-induced cardiotoxicity via quaking and circular RNA Pan3. J Mol Cell Cardiol. 2020;140:56–67. [DOI] [PubMed] [Google Scholar]
- [17].Wang M, Suo L, Yang S, et al. 001372 Reduces Inflammation in Propofol-Induced Neuroinflammation and Neural Apoptosis through PIK3CA/Akt/NF-kappaB by miRNA-148b-3p. J Invest Surg. 2020;1–11. [DOI] [PubMed] [Google Scholar]
- [18].Yao Y, Wang J, He T, et al. Microarray assay of circular RNAs reveals cicRNA.7079 as a new anti-apoptotic molecule in spinal cord injury in mice. Brain Res Bull. 2020;164:157–171. [DOI] [PubMed] [Google Scholar]
- [19].Zhao J, Qi X, Bai J, et al. A circRNA derived from linear HIPK3 relieves the neuronal cell apoptosis in spinal cord injury via ceRNA pattern. Biochem Biophys Res Commun. 2020;528:359–367. [DOI] [PubMed] [Google Scholar]
- [20].Li X, Lou X, Xu S, et al. Hypoxia inducible factor-1 (HIF-1alpha) reduced inflammation in spinal cord injury via miR-380-3p/ NLRP3 by Circ 0001723. Biol Res. 2020;53:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gao XZ, Ma RH, Zhang ZX. miR-339 Promotes Hypoxia-Induced Neuronal Apoptosis and Impairs Cell Viability by Targeting FGF9/CACNG2 and Mediating MAPK Pathway in Ischemic Stroke. Front Neurol. 2020;11:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang JY, Chuang JI. Fibroblast growth factor 9 upregulates heme oxygenase-1 and gamma-glutamylcysteine synthetase expression to protect neurons from 1-methyl-4-phenylpyridinium toxicity. Free Radic Biol Med. 2010;49:1099–1108. [DOI] [PubMed] [Google Scholar]
- [23].Deng B, Lv W, Duan W, et al. FGF9 modulates Schwann cell myelination in developing nerves and induces a pro-inflammatory environment during injury. J Cell Biochem. 2018;119:8643–8658. [DOI] [PubMed] [Google Scholar]
- [24].Sumbal J, Koledova Z. FGF signaling in mammary gland fibroblasts regulates multiple fibroblast functions and mammary epithelial morphogenesis. Development. 2019. p. 146. [DOI] [PubMed] [Google Scholar]
- [25].Hetman M, Gozdz A. Role of extracellular signal regulated kinases 1 and 2 in neuronal survival. Eur J Biochem. 2004;271:2050–2055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


