ABSTRACT
Huntington disease (HD) manifests a unique macroautophagy/autophagy defect: the presense of cytosolic autophagosomes without substrates or so-called “empty” autophagosomes. It was proposed that mutant HTT (huntingtin; mHTT) disrupts cargo recognition by the selective autophagy receptor SQSTM1/p62 thus leading to the failure of cargo sequestration by phagophores, the precursors to autophagosomes. Here we looked at recent discoveries that liquid-like SQSTM1 droplets can serve as platforms for autophagosome formation, and discussed possible alternative mechanisms for “empty” autophagosome formation in HD inspired by these findings.
KEYWORDS: Autophagosome, huntingtin, Huntington disease, phase transition, SQSTM1/p62 droplet
Introduction
Accumulating evidence suggests that defects in autophagy are common in neurodegenerative diseases (NDs) [1,2]. Among all NDs, defective autophagy in Huntington disease (HD) is quite unique in that not only mutant HTT (huntingtin; mHTT) aggregates are subject to autophagy degradation, but wild-type HTT also serves as a scaffold protein to facilitate selective autophagy [3,4]. In addition, HD cells contain autophagosomes that lack substrates (“empty” autophagosomes), suggesting mHTT itself may interfere with autophagic cargo recognition and sequestration [5]. Both HTT and mHTT interact with SQSTM1/p62 (sequestosome 1) [5,6], a receptor protein for selective autophagy that bridges ubiquitinated cargoes and phagophores [7]. It has also been reported that HTT is required for the interaction of SQSTM1 with phagophores [6]. How mHTT affects autophagic cargo sequestration, however, still remains largely unclear. Here we will discuss a few recent studies on mechanisms by which liquid-like SQSTM1 droplets direct autophagosome formation and possible new insights these discoveries may bring into HD.
SQSTM1 droplets mediate autophagosome formation
The understanding of the mechanisms on how SQSTM1 facilitates autophagy is quickly developing since its first identification as a receptor protein for selective autophagy [7]. It is now known the interaction of SQSTM1 with ubiquitinated cargoes requires its ubiquitin-associated (UBA) domain. The LC3-interacting region (LIR) allows SQSTM1 to interact with phagophores via its binding to MAP1LC3/LC3 (microtubule associated protein 1 light chain 3), a ubiquitin-like protein conjugated to the phagophore membrane after autophagy induction. Besides the interactions with cargoes and phagophores, SQSTM1 can cluster to cytosolic round structures termed SQSTM1 bodies [8]. This clustering of SQSTM1, mediated by its N-terminal PB1 domain, is crucial for SQSTM1 to select cargoes and interact with phagophores [9]. A few recent studies indicated that SQSTM1 bodies are droplets formed via liquid-liquid phase separation (LLPS) [10–12]. The structural and functional properties of SQSTM1 bodies, however, are still not fully understood.
Published early in 2021, two independent studies both discovered that phase separated SQSTM1 droplets can serve as platforms for autophagosome formation [13,14], revealing a new role played by SQSTM1 in selective autophagy. In one study, Kageyama et al found SQSTM1 formed gel-like structures that can be degraded by autophagy in mammalian cells. Very interestingly, the authors also found that autophagosomes emerged in two ways, either enwrapping SQSTM1 droplets or forming right next to them. When the interaction of SQSTM1 with LC3 was challenged by competitive inhibition, the engulfing of SQSTM1 droplets by phagophores was largely reduced and selective autophagy was compromised [13]. Agudo-Canalejo et al also observed that SQSTM1 formed micrometer-sized liquid-like droplets [14]. Differently, they captured LC3-positive punctate structures on the surfaces of SQSTM1 droplets and then focused more on the dynamics of the autophagosome formation process. From a point of view of physics, they considered the selective autophagy of SQSTM1 droplets as the deformation of the droplet via wetting of the phagophore membranes and developed a minimal mathematical model to describe three key factors that affect the droplet-membrane interaction: the droplet-size-dependent surface tension, the instability of the expanding membrane and the asymmetry of the membrane spontaneous curvatures. This model predicted three scenarios: 1) when the surface tension of the droplet is above a critical value, phagophore membranes will expand to achieve complete sequestration of the droplet; 2) when the surface tension is below the critical value and membrane spontaneous curvature favors the droplet side, phagophore membranes will bend and sequester part of the droplet (piecemeal autophagy); 3) when the surface tension is below the critical value and membrane spontaneous curvature disfavors the droplet side, phagophore membranes will bend away from the SQSTM1 droplet and eventually grow to autophagosomes, presumably with cytosolic contents. All the three scenarios were demonstrated in a protein-free in vitro giant unilamellar vesicles (GUVs) system and further confirmed in cultured mammalian cells. More importantly, when the LIR of SQSTM1 was deleted, although adjacent localization of SQSTM1(ΔLIR) droplets and LC3-positive membranes could still be observed, no SQSTM1(ΔLIR) droplets were piecemealed or completely engulfed by phagophores, suggesting that the SQSTM1 LIR-LC3 interaction is required for phagophore membranes to bend toward SQSTM1 droplets.
“Empty” autophagosomes in HD – fail to recognize or fail to engulf?
The existence of “empty” autophagosomes found in HD cells by Martinez–Vicente et al remains an unneglectable concern, especially for strategies aiming at boosting autophagy to reduce mHTT. A thorough understanding of the underlying molecular mechanisms therefore is needed. In their original article where they discovered this phenomenon, Martinez–Vicente et al observed mHTT-SQSTM1 interaction on autophagosomes and proposed that mHTT interfered with SQSTM1-mediated cargo recognition and led to the failure of cargo sequestration by autophagosomes [5]. The later finding that HTT also interacts with SQSTM1 and facilitates its interaction with ubiquitinated selective autophagy cargoes also seemed to fit this hypothesis [6]. However, direct evidence supporting mHTT disrupting SQSTM1-cargo interaction is lacking.
The sequestration of cargoes by selective autophagy requires two events, the recognition of the cargoes and the engulfment of the cargoes by autophagosomes. Intuitively, when “empty” autophagosomes appear, it implies that the formation of these autophagosomes was complete and normal; they just somehow failed to recognize their targets. But is this the only possible scenario? The new findings discussed in the previous section made another scenario also possible: maybe the cargo was recognized, but just could not be properly engulfed?
In the case of HD, given that SQSTM1 droplets can serve as platforms for phagophore membranes to grow and bend, considering the interaction between SQSTM1 and mHTT, we propose a “fail-to-engulf” model, in which there are at least two possible ways mHTT can affect phagophore membranes’ bending and lead to the formation of “empty” autophagosomes on SQSTM1 droplets (Figure 1): 1) mHTT monomers may interact directly with SQSTM1 on the surface of SQSTM1 droplets, therefore affect the spontaneous curvatures of the growing phagophore membranes, make them bend away from the SQSTM1 droplets to complete the formation of intact autophagosomes without cargoes; 2) mHTT oligomers may interfere with SQSTM1 droplet formation, change its liquidity and surface tension, which are crucial for proper bending of phagophore membranes.
Figure 1.
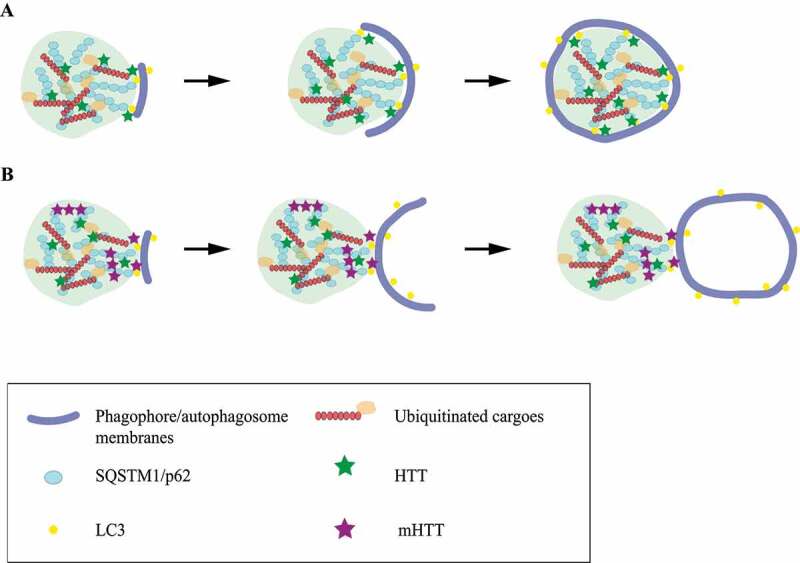
Schematic drawing of the fail-to-engulf model
(A). HTT facilitates an SQSTM1 droplet to properly recognize ubiquitinated cargoes and mediate autophagosome formation, resulting in complete engulfment of the droplet. (B). mHTT monomers interfere with SQSTM1-LC3 interaction on the surface of an SQSTM1 droplet; mHTT oligomers affect the properties of the SQSTM1 droplet; alone or together, mHTT changes the key factors for proper phagophore membrane bending and leads to the formation of an autophagosome without substrates.
Our model might provide a better explanation for some of the previous observations. In isolated autophagosomes, Martinez–Vicente et al found SQSTM1 and HTT were mostly in the lumen in WT cells. In HD cells, however, both SQSTM1 and mHTT were enriched on autophagosomes’ membranes [5]. The fail-to-recognize hypothesis could not explain this lumen-to-membrane change of distribution of SQSTM1 and HTT in autophagosomes. Our fail-to-engulf model, however, will predict SQSTM1 droplets and mHTT to be associated with autophagosome membranes and be absent in autophagosome lumen.
The fail-to-engulf model could also be supported by other experimental results. It has been shown that knockout of HTT leads to decreased SQSTM1-LC3 interaction [6]. mHTT was also reported to impair the interaction of SQSTM1 and LC3 [15]. SQSTM1 bodies co-localized with mHTT aggregates [8], indicating a close interaction between SQSTM1 and mHTT even in the form of protein assemblies of higher molecular complexity. In addition, a number of SQSTM1-binding proteins have been reported to affect the formation, size and fluidity of SQSTM1 droplets [reviewed in [16]]. Furthermore, both HTT and mHTT can undergo phase transition and form assemblies possessing different liquidity features, partially dependent on the length of the polyQ region [reviewed in [17]]. Taken together, it is not unlikely that mHTT alter the physical properties of SQSTM1 droplets and lead to changes of its surface tension.
Discussion
Studies in the past few years have started to establish the involvement of phase transition in diverse aspects of autophagy, including the LLPS of SQSTM1, a selective autophagy receptor protein [18,19]. Latest discoveries revealed yet another role of SQSTM1 droplets in autophagy as platforms of autophagosome formation. Physical modeling predicted an anti-intuitive scenario of phagophore’s growth away from SQSTM1 droplets and this scenario was confirmed both in vitro and in SQSTM1ΔLIR cells. Here we discussed a possible implication of these findings in the pathological intracellular environment of HD and tried to find new explanations for a unique autophagy defect of HD cells: the presence of “empty” autophagosomes. Although current evidence doesn’t go against our model, more studies are needed to further test this model. The key facts to check will be the distribution of SQSTM1 and mHTT and their interaction in HD cells with “empty” autophagosomes. With the accumulation of knowledge on SQSTM1 and mHTT assembly formation, together with the availability of more molecular tools and more advanced microscopies, we believe more details of SQSTM1-mHTT interaction with higher spatial and temporal resolutions could be revealed.
Furthermore, if the “fail-to-engulf” model is correct, it will still be intriguing how the phagophore membranes bending away from the SQSTM1 droplet grow and finally close to form autophagosomes without “accidentally” engulfing any cargoes beside cytosolic contents. One possibility is that the “empty” autophagosomes formed this way is rather small to engulf cargoes like organelles or protein aggregates. The formation of an autophagosome requires the expansion of the initiated phagophore, which needs membrane supplies, followed by a closure step to form a closed vesicle [20,21]. The size and shape of the “empty” autophagosomes formed in HD therefore will be determined at least by membrane spontaneous curvatures and membrane supplies. It is still unclear what membrane sources are used in “empty” autophagosome formation. Nonetheless, time-lapse imaging used by Agudo-Canalejo et al successfully recorded the process of cytosol-sequestrating (but not droplet-sequestrating) autophagosome formation on SQSTM1 droplets in SQSTM1ΔLIR cells [14]. The sizes of these cytosol-sequestrating autophagyosomes are around 0.1–0.3 μm, comparable to the size of most “empty” autophagosomes in HD cells [5]. Applying similar approaches to HD cells will very likely reveal more details of the “empty” autophagosomes formation process. Moreover, autophagy-related proteins ATG2 and ATG9 have been reported to be responsible for lipid transfer to growing phagophores in bulk autophagy [20,21]. Whether they are involved in “empty” autophagosomes formation in HD is also worth studying.
Activating autophagy has appeared to be a promising strategy to lower mHTT [22]. However, autophagic abnormalities such as the “empty” autophagosomes in HD cells raised concerns that cannot be overlooked. Understanding the underlying mechanisms of these abnormalities is a prerequisite to direct further research to target the affected players/steps involved and to avoid attempts that might boost nonfunctional autophagy and even stressing the already compromised autophagosome-lysosome system. If “fail-to-engulf” really is the cause of the “empty” autophagosomes in HD, approaches that can correct the bending of the phagophore membranes on SQSTM1 droplets will be of therapeutic potential. In Kageyama et al’s study, the overexpression of an engineered protein that also contains an LIR could compete with SQSTM1 for LC3 and alter the direction of the bending of phagophore membranes on SQSTM1 droplets, demonstrating that such an approach is indeed possible [13].
Last but not least, both Kageyama et al and Agudo-Canalejo et al found autophagosomes formed next to but not engulfing or piecemealing SQSTM1 droplets when the SQSTM1-LC3 interaction was challenged (either by LIR deletion or competition), and they both proposed these autophagosomes to be involved in bulk autophagy. It will be interesting to test whether these autophagosomes do contain any discernable substrates or do they rather resemble the “empty” autophagosomes in HD cells. If it turns out to be the latter, then “empty” autophagosomes might be a phenomenon not unique to HD.
Acknowledgments
We apologize to colleagues whose works are not cited due to space limitations.
Funding Statement
This work is supported by the National Natural Science Foundation of China (82071432), the Zhejiang Provincial Natural Science Foundation of China (LY20H090019) and the Young Talent grant from the Collaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals at ZJUT.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Menzies FM, Fleming A, Rubinsztein DC.. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015. Jun;16(6):345–357. [DOI] [PubMed] [Google Scholar]
- [2].Giovedì S, Ravanelli MM, Parisi B, et al. Dysfunctional autophagy and endolysosomal system in neurodegenerative diseases: relevance and therapeutic options. Front Cell Neurosci. 2020;14:602116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Martin DD, Ladha S, Ehrnhoefer DE, et al. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 2015. Jan;38(1):26–35. . [DOI] [PubMed] [Google Scholar]
- [4].Ochaba J, Lukacsovich T, Csikos G, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci U S A. 2014. Nov 25;111(47):16889–16894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Martinez-Vicente M, Talloczy Z, Wong E, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010. May;13(5):567–576. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rui YN, Xu Z, Patel B, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015. Mar;17(3):262–275. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lamark T, Svenning S, Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017. Dec 12;61(6):609–624. [DOI] [PubMed] [Google Scholar]
- [8].Bjørkøy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005. Nov 21;171(4):603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wurzer B, Zaffagnini G, Fracchiolla D, et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife. 2015. Sep 28;4:e08941. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sun D, Wu R, Zheng J, et al. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018. Apr;28(4):405–415. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zaffagnini G, Savova A, Danieli A, et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018. Mar 1;37(5). DOI: 10.15252/embj.201798308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sánchez-Martín P, Sou YS, Kageyama S, et al. NBR1-mediated p62-liquid droplets enhance the Keap1-Nrf2 system. EMBO Rep. 2020. Mar 4;21(3):e48902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kageyama S, Gudmundsson SR, Sou YS, et al. p62/SQSTM1-droplet serves as a platform for autophagosome formation and anti-oxidative stress response. Nat Commun. 2021. Jan 4;12(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Agudo-Canalejo J, Schultz SW, Chino H, et al. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature. 2021. Jan 20;591(7848):142–146. [DOI] [PubMed] [Google Scholar]
- [15].Franco-Iborra S, Plaza-Zabala A, Montpeyo M, et al. Mutant HTT (huntingtin) impairs mitophagy in a cellular model of Huntington disease. Autophagy. 2020;24:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Berkamp S, Mostafavi S, Sachse C. Structure and function of p62/SQSTM1 in the emerging framework of phase separation. FEBS J. 2020. Dec 17. DOI: 10.1111/febs.15672. [DOI] [PubMed] [Google Scholar]
- [17].Yang J, Yang X. Phase transition of Huntingtin: factors and pathological relevance. Front Genet. 2020;11:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Noda NN, Wang Z, Zhang H. Liquid-liquid phase separation in autophagy. J Cell Biol. 2020. Aug 3;219(8). DOI: 10.1083/jcb.202004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fujioka Y, Noda NN. Biomolecular condensates in autophagy regulation. Curr Opin Cell Biol. 2021. Jan;11(69):23–29. [DOI] [PubMed] [Google Scholar]
- [20].Melia TJ, Lystad AH, Simonsen A. Autophagosome biogenesis: from membrane growth to closure. J Cell Biol. 2020 Jun 1;219(6):e202002085.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chang C, Jensen LE, Hurley JH. Autophagosome biogenesis comes out of the black box. Nat Cell Biol. 2021. May;23(5):450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Valionyte E, Yang Y, Roberts SL, et al. Lowering mutant Huntingtin levels and toxicity: autophagy-endolysosome pathways in Huntington’s disease. J Mol Biol. 2020. Apr 3;432(8):2673–2691. [DOI] [PubMed] [Google Scholar]


