ABSTRACT
Streptococcus suis is one of the important emerging zoonotic pathogens. Serotype 2 is most prevalent in patients worldwide. In the present study, we first isolated one S. suis serotype 7 strain GX69 from the blood culture of a patient with septicemia complicated with pneumonia in China. In order to deepen the understanding of S. suis serotype 7 population characteristics, we investigated the phylogenetic structure, genomic features, and virulence of S. suis serotype 7 population, including 35 strains and 79 genomes. Significant diversities were revealed in S. suis serotype 7 population, which were clustered into 22 sequence types (STs), five minimum core genome (MCG) groups, and six lineages. Lineages 1, 3a, and 6 were mainly constituted by genomes from Asia. Genomes of Lineages 2, 3b, and 5a were mainly from Northern America. Most of genomes from Europe (41/48) were clustered into Lineage 5b. In addition to strain GX69, 13 of 21 S. suis serotype 7 representative strains were classified as virulent strains using the C57BL/6 mouse model. Virulence-associated genes preferentially present in highly pathogenic S. suis serotype 2 strains were not suitable as virulence indicators for S. suis serotype 7 strains. Integrative mobilizable elements were widespread and may play a critical role in disseminating antibiotic resistance genes of S. suis serotype 7 strains. Our study confirmed S. suis serotype 7 is a non-negligible pathotype and deepened the understanding of the population structure of S. suis serotype 7, which provided valuable information for the improved surveillance of this serotype.
KEYWORDS: Streptococcus suis serotype 7, zoonotic pathogens, phylogeny, integrative mobilizable elements, virulence
Introduction
Streptococcus suis is an important emerging zoonotic pathogen responsible, among other infections, for septicemia, meningitis, endocarditis, and arthritis in humans [1]. To date, serotyping is an important routine diagnostic procedure and is widely used for subtyping S. suis strains. Among 29 confirmed serotypes (1–19, 21, 23–25, 27–31, and 1/2) and 28 novel cps types [2–5], serotype 2 is most frequently isolated from clinical cases in swine and humans worldwide [1,6]. Two outbreaks featured by high rates of streptococcal toxic-shock-like syndrome (STSLS) were caused by S. suis serotype 2 sequence type (ST)7 strains in China [7,8]. Recently, the prevalence of serotype 14 has also increased among sporadic human cases in China [9]. Serotype 9 has become the most prevalent serotype in diseased pigs in some European counties [1,10], and one human case of serotype 9 infection was reported [11]. Serotypes 4, 5, 16, 21, 24, and 31 have also been reported in human infections [1,12]. Serotype 7 is an important serotype frequently isolated from diseased pigs in European countries, North America, and Thailand [13–18], and it was also related to severe herd problems of meningitis and arthritis in nursery and grower pigs [14].
In the present study, an S. suis serotype 7 strain was isolated for the first time from the blood culture of a patient with septicemia complicated with pneumonia in China, suggesting that some serotype 7 strains may possess zoonotic potential. Except for limited epidemiologic studies [19] and in vitro survival assay in swine blood [20], little information is available for the phylogeny, evolution, and pathogenicity of S. suis serotype 7 population. The present study included 35 strains and 79 genomes of strains from 1999 to 2019 originating from nine countries to represent the S. suis serotype 7 population. The phylogenetic relationship, dissemination mechanisms of antibiotic resistance (AR) genes, variation of cps arrangements, and virulence were investigated to elucidate the population structure, genomic features, evolution, and pathogenicity of S. suis serotype 7.
Materials and methods
Case description
On 22 July 2016, a 71-year-old female patient with a history of hypertension was admitted to the First People’s Hospital of Yulin in Yulin city because of repeating fever and chill (highest body temperature of 39.5°C), cough, and abdominal pain for five days. A computerized tomography scan image indicated inflammation of both lungs. The serum level of high-sensitivity C-reactive protein and total counts of white blood cells were 112.77 mg/L and 5.68 × 109/L, respectively. The neutrophil percentage was 72.7%. The patient’s blood pressure was 103/63 mm Hg. Meropenem, piperacillin/tazobactam, and levofloxacin were given as antibiotic therapy. The patient recovered and was discharged ten days later. A strain (named GX69) was isolated from the blood culture of the patient. The strain was confirmed as S. suis by amplifying S. suis-specific recN gene [21]. GX69 was first identified as serotype 7 by the agglutination test using the serum purchased from Statens Serum Institute, Copenhagen, Denmark, and further confirmed with a molecular serotyping method [22].
Bacterial strains, genomes, and sequencing
For comparison purposes, 35 strains and 79 genomes were used in this study (Table 1). Twenty-seven of them were from China (24 of them were sequenced in the present study), 23 from the United States of American (USA), 22 from the United Kingdom (UK), 16 from Canada (sequenced in the present study), 13 from Spain (sequenced in the present study), 9 from the Netherlands (3 of them were sequenced in the present study), 2 from France (sequenced in the present study), 1 from Germany (sequenced in the present study) and Denmark each. Genomes of unspecified origin were from Genbank database. All genomes were re-confirmed to belong to S. suis by analysing their full length of 16s rRNA sequences [23] and recN gene specific to S. suis [24]. In addition, these genomes harboured S. suis serotype 7 specific wzy gene. They were isolated from 1999 to 2019.
Table 1.
The information of strains and genomes used in the study.
| Lineage | Name of Strain | MCG | Serotype | Sequence Type | cps subtype | Accession number | Host | Isolation source | Location | Year | AR genes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage1 | 93.01Ba | 1 | 7 | 1609 | 7-II | SAMN17982935 | Diseased pig | Heart | Spain | 2001 | tet(W) |
| YS12 | 1 | 7 | 17 | 7-Ib | SAMN02469508 | Healthy pig | Nasopharynx swab | CN | 2012 | tet(O) | |
| WUSS415a | 1 | 7 | 1611 | 7–1 | SAMN17982954 | Healthy pig | Tonsil | CN | 2017.12 | ant(6)-Ia, erm(B), tet(M) | |
| WUSS417a | 1 | 7 | 1611 | 7–1 | SAMN17982955 | Healthy pig | Tonsil | CN | 2017.12 | ant(6)-Ia, erm(B), tet(M) | |
| Lineage2 | 2145959a | 2 | 7 | 1613 | 7-1b | SAMN18117666 | Diseased pig | Brain | Canada | 2018 | tet(O), erm(B) |
| 2255955a | 2 | 7 | 89 | 7-5a | SAMN18117668 | Diseased pig | Brain | Canada | 2019 | tet(O), erm(B) | |
| 2120811a | 2 | 7 | 89 | 7-Ia | SAMN18117667 | Diseased pig | Liver | Canada | 2018 | tet(O), erm(B) | |
| 2108284a | 2 | 7 | 89 | 7-Ib | SAMN18117669 | Diseased pig | Joint | Canada | 2018 | tet(O), erm(B) | |
| 128.01Ba | 2 | 7 | 24 | 7-II | SAMN17982937 | Diseased pig | Brain | Spain | 2001 | tet(O),aph(3’)-IIIa,ant(6)-Ia,sat-4,erm(B) | |
| 255Ba | 2 | 7 | 24 | 7-II | SAMN17982942 | Diseased pig | Lung | Spain | 1999 | tet(O),aph(3’)-IIIa,ant(6)-Ia,sat-4,erm(B) | |
| 173Ba | 2 | 7 | 24 | 7-II | SAMN17982941 | Diseased pig | Brain | Spain | 1999 | tet(O),aph(3’)-IIIa,ant(6)-Ia,sat-4,erm(B) | |
| 126.01Ba | 2 | 7 | 24 | 7-II | SAMN17982936 | Diseased pig | Brain | Spain | 2001 | tet(O),aph(3’)-IIIa,ant(6)-Ia,sat-4,erm(B) | |
| Lineage3a | Ssuis120 | 3 | 7 | 373 | 7-Ib | SRR9123103 | Diseased pig | Meninges | USA | 2016 | tet(O) |
| Ssuis95 | 3 | 7 | 373 | 7-Ia | SRR9123148 | Diseased pig | Joint | USA | 2016 | tet(O) | |
| 2018WUSS020a | 3 | 7 | 373 | 7-Ib | SAMN17982957 | Healthy pig | Tonsil | CN | 2018.09 | tet(O) | |
| 2018WUSS025a | 3 | 7 | 373 | 7-Ia | SAMN17982958 | Healthy pig | Tonsil | CN | 2018.09 | tet(O) | |
| 2018WUSS017a | 3 | 7 | 373 | 7-Ia | SAMN17982956 | Healthy pig | Tonsil | CN | 2018.09 | tet(O) | |
| WUSS401a | 3 | 7 | 373 | 7-Ia | SAMN17982952 | Healthy pig | Tonsil | CN | 2017.12 | tet(O), erm(B) | |
| 2019WUSS018a | 3 | 7 | 373 | 7-Ib | SAMN17982962 | Healthy pig | Tonsil | CN | 2019.11 | tet(O), erm(B) | |
| 2019WUSS020a | 3 | 7 | 373 | 7-Ib | SAMN17982964 | Healthy pig | Nasopharynx swab | CN | 2019.11 | tet(O), erm(B) | |
| 2019WUSS019a | 3 | 7 | 373 | 7-Ib | SAMN17982963 | Healthy pig | Tonsil | CN | 2019.11 | tet(O), erm(B) | |
| 2019WUSS017a | 3 | 7 | 373 | 7-Ib | SAMN17982961 | Healthy pig | Tonsil | CN | 2019.11 | tet(O), erm(B) | |
| GX69a | 3 | 7 | 373 | 7-Ia | SAMN18029937 | Patient | Blood | CN | 2016 | tet(O), erm(B) | |
| WUSS255a | 3 | 7 | 373 | 7-Ib | SAMN17982946 | Healthy pig | Nasopharynx swab | CN | 2017.10 | tet(O), erm(B) | |
| WUSS382a | 3 | 7 | 373 | 7-Ia | SAMN17982951 | Healthy pig | Tonsil | CN | 2017.12 | tet(O), erm(B) | |
| WUSS318a | 3 | 7 | 373 | 7-Ia | SAMN17982949 | Healthy pig | Tonsil | CN | 2017.12 | tet(O), erm(B) | |
| WUSS316a | 3 | 7 | 373 | 7-Ia | SAMN17982948 | Healthy pig | Tonsil | CN | 2017.12 | tet(O), erm(B) | |
| Lineage3b | Ssuis359 | 3 | 7 | 373 | 7-Ia | SAMN11854340 | Diseased pig | Lung | USA | 2017 | tet(O), erm(B) |
| Ssuis93 | 3 | 7 | 94 | 7-Ia | SRR9123171 | Diseased pig | Brain | USA | 2016 | tet(O) | |
| 2156696a | 3 | 7 | 94 | 7-Ia | SAMN18117674 | Diseased pig | brain | Canada | 2018 | – | |
| Ssuis51 | 3 | 7 | 980 | 7-III | SRR9123229 | Diseased pig | Lung | USA | 2015 | tet(O), erm(B) | |
| Ssuis118 | 3 | 7 | 979 | 7-Ia | SRR9123101 | Diseased pig | Brain | USA | 2016 | tet(O), erm(B) | |
| Ssuis98 | 3 | 7 | 94 | 7-Ia | SRR9123145 | Diseased pig | Brain | USA | 2015 | erm(B), tet(O) | |
| Ssuis39 | 3 | 7 | 94 | 7-III | SRR9123191 | Diseased pig | Brain | USA | 2015 | tet(O), erm(B) | |
| Ssuis40 | 3 | 7 | 94 | 7-Ia | SRR9123184 | Diseased pig | Meninges | USA | 2015 | tet(O), erm(B) | |
| 2130772a | 3 | 7 | 839 | 7-Ia | SAMN18117675 | Diseased pig | Brain | Canada | 2018 | tet(O), erm(B) | |
| Ssuis45 | 3 | 7 | 94 | 7-Ia | SRR9123062 | Diseased pig | Brain | USA | 2015 | – | |
| Ssuis109 | 3 | 7 | 94 | 7-Ia | SRR9123073 | Diseased pig | Brain | USA | 2015 | erm(B) | |
| Ssuis303 | 3 | 7 | 94 | 7-III | SRR9123157 | Diseased pig | Joint | USA | 2016 | – | |
| Ssuis41 | 3 | 7 | 94 | 7-Ia | SRR9123183 | Diseased pig | Brain | USA | 2015 | erm(B) | |
| Ssuis46 | 3 | 7 | 94 | 7-III | SRR9123063 | Diseased pig | Lung | USA | 2014 | tet(O) | |
| Ssuis324 | 3 | 7 | 94 | 7-III | SRR9123265 | Diseased pig | Brain | USA | 2017 | – | |
| Ssuis108 | 3 | 7 | 94 | 7-Ia | SRR9123068 | Diseased pig | Brain | USA | 2015 | erm(B) | |
| Lineage4 | WUSS004a | 4 | 7 | 225 | 7-Ib | SAMN17982943 | Diseased pig | / | CN | 2016 | tet(O), erm(B) |
| WUSS029a | 4 | 7 | 225 | 7-Ib | SAMN17982945 | Healthy pig | / | CN | unknown | tet(O), erm(B) | |
| Ssuis8 | 4 | 7 | 225 | 7-Ib | SRR9123237 | Diseased pig | Brain | USA | 2014 | aph(3’)-IIIa,ant(6)-Ia, sat-4, erm(B), tet(O) | |
| Ssuis11 | 4 | 7 | 225 | 7-Ib | SRR9123252 | Diseased pig | Brain | USA | 2014 | ant(6)-Ia(2),ant(9)-Ia, aph(3’)-IIIa, sat-4, erm(B), tet(O) | |
| 2234142a | 4 | 7 | 1614 | 7-Ib | SAMN18117670 | Diseased pig | Brain | Canada | 2019 | erm(B) | |
| WUSS013a | 4 | 7 | 225 | 7-Ib | SAMN17982944 | Diseased pig | / | CN | unknown | tet(O), erm(B) | |
| Ssuis100 | 4 | 7 | 225 | 7-Ib | SRR9123151 | Diseased pig | Meninges | USA | 2015 | tet(O) | |
| 2225102a | 4 | 7 | 32 | 7-Ib | SAMN18117671 | Diseased pig | Heart | Canada | 2019 | tet(O), erm(B) | |
| YS63a | 4 | 7 | 32 | 7-Ia | SAMN20087851 | Healthy pig | Nasopharynx swab | CN | 2012 | tet(O),erm(B) | |
| YS66 | 4 | 7 | 32 | 7-Ia | SAMN02469560 | Healthy pig | Nasopharynx swab | CN | 2012 | tet(O),erm(B) | |
| 2211488a | 4 | 7 | 34 | 7-Ia | SAMN18117672 | Diseased pig | Heart | Canada | 2019 | tet(O), erm(B) | |
| S12R | 4 | 7 | 907 | 7-Ia | SAMEA3233991 | Diseased pig | Lung | UK | 2010 | – | |
| SS1022 | 4 | 7 | 907 | 7-Ia | SAMEA1316674 | Pig | / | UK | 2014 | – | |
| 2135990a | 4 | 7 | 32 | 7-Ib | SAMN18117673 | Diseased pig | Brain | Canada | 2018 | tet(O), erm(B) | |
| Lineage5a | 2139811a | 4 | 7 | 1610 | 7-Ia | SAMN18117676 | Diseased pig | Brain | Canada | 2018 | tet(O), erm(B) |
| 2288194a | 4 | 7 | 1610 | 7-Ia | SAMN18117677 | Diseased pig | Brain | Canada | 2019 | tet(O), erm(B) | |
| 2175452a | 4 | 7 | 971 | 7-Ia | SAMN18117678 | Diseased pig | Abdominal liquid | Canada | 2019 | tet(O), erm(B) | |
| 2274226a | 4 | 7 | 971 | 7-Ia | SAMN18117679 | Diseased pig | Brain | Canada | 2019 | tet(O), erm(B) | |
| Ssuis265 | 4 | 7 | 971 | 7-Ia | SRR9123206 | Healthy pig | Tonsil | USA | 2016 | tet(O), erm(B) | |
| Lineage5b | 2207481a | 4 | 7 | 971 | 7-Ia | SAMN18117680 | Diseased pig | Lung | Canada | 2019 | tet(O), erm(B) |
| GD-0031 | 4 | 7 | 29 | 7-Ia | SAMEA3595225 | Diseased pig | CSF | Netherlands | 2002 | erm(B),tet(O) | |
| GD-0067 | 4 | 7 | 29 | 7-Ia | SAMEA3595239 | Diseased pig | CSF | Netherlands | 2004 | erm(B),tet(O) | |
| GD-0061 | 4 | 7 | 854 | 7-Ia | SAMEA3595236 | Diseased pig | CSF | Netherlands | 2004 | dfrF,ant(6)-Ia, tet(O), cat-TC | |
| 2138579a | 4 | 7 | 29 | 7-Ib | SAMN18117681 | Diseased pig | Brain | Germany | 2018 | tet(O), erm(B) | |
| 2180644a | 4 | 7 | 29 | 7-Ia | SAMN18117682 | Diseased pig | Brain | Spain | 2019 | tet(O) | |
| WUSS366a | 4 | 7 | 29 | 7-Ib | SAMN17982950 | Healthy pig | Tonsil | CN | 2017.12 | – | |
| WUSS302a | 4 | 7 | 29 | 7-Ia | SAMN17982947 | Healthy pig | Tonsil | CN | 2017.11 | tet(O), erm(B) | |
| S11O | 4 | 7 | 29 | 7-Ia | SAMEA3233988 | Diseased pig | Lung | UK | 2010 | lnu(B), aph(3’)-IIIa, lsaC,ant(9)-Ia | |
| SS1018 | 4 | 7 | 29 | 7-Ia | SAMEA1316697 | Pig | / | UK | 2014 | aph(3’)-IIIa, lnu(B), lsaC,ant(9)-Ia | |
| Ssuis77 | 4 | 7 | 973 | 7-Ib | SRR9123095 | Pathogenic | Liver | USA | 2014 | erm(B) | |
| 2243014a | 4 | 7 | 29 | 7-Ia | SAMN18117684 | Diseased pig | Brain | Canada | 2019 | tet(O), erm(B) | |
| 2114366a | 4 | 7 | 29 | 7-Ib | SAMN18117683 | Diseased pig | Joint | France | 2018 | aph(3’)-IIIa,lnu(B),lsaC,ant(9)-Ia | |
| S10W | 4 | 7 | 29 | 7-Ia | SAMEA3233977 | Diseased pig | Lung | UK | 2010 | tet(W),erm(B) | |
| Ssuis139 | 4 | 7 | 29 | 7-Ia | SRR9123173 | Diseased pig | Joint | USA | 2016 | tet(O), erm(B) | |
| Ssuis136 | 4 | 7 | 29 | 7-Ib | SRR9123182 | Diseased pig | Meninges | USA | 2016 | tet(O), erm(B) | |
| SS1000 | 4 | 7 | 29 | 7-Ia | SAMEA1316594 | Pig | / | UK | 2014 | tet(W),erm(B) | |
| SS1051 | 4 | 7 | 29 | 7-Ib | SAMEA1316581 | Pig | / | UK | 2014 | erm(B),tet(O) | |
| S15R | 4 | 7 | 29 | 7-Ia | SAMEA3234014 | Diseased pig | Brain | UK | 2010 | erm(B),tet(O) | |
| 2148719a | 4 | 7 | 29 | 7-Ia | SAMN18117685 | Diseased pig | Spleen | Netherlands | 2018 | tet(O), erm(B) | |
| 149Ba | 4 | 7 | 29 | 7-Ia | SAMN17982938 | Diseased pig | Heart | Spain | 1999 | – | |
| LS4E | 4 | 7 | 29 | 7-II | SAMEA3233917 | Healthy pig | Tonsil | UK | 2011 | dfrF,tet(M) | |
| GD0094 | 4 | 7 | 29 | 7-Ib | SAMEA3595252 | Diseased pig | CSF | Netherlands | 2006 | – | |
| 12V457 | 4 | 7 | 29 | 7-Ib | SAMEA3595206 | Diseased pig | CSF | Netherlands | 2006 | erm(B),tet(O) | |
| LS6Z | 4 | 7 | 29 | 7-II | SAMEA3233938 | pig | / | UK | 2011 | dfrF,tet(M) | |
| 150Ba | 4 | 7 | 29 | 7-Ia | SAMN17982939 | Diseased pig | Lymphatic gland | Spain | 1999 | – | |
| D9 | 4 | 7 | 29 | 7-Ib | SAMN02603321 | Pig | / | CN | / | erm(B),tet(O) | |
| LSS83 | 4 | 7 | 29 | 7-II | SAMEA1316699 | Pig | / | UK | 2014 | dfrF,tet(M) | |
| 2114361a | 4 | 7 | 29 | 7-Ia | SAMN18117687 | Diseased pig | Brain | Spain | 2018 | tet(O), erm(B) | |
| 2245605a | 4 | 7 | 29 | 7-Ia | SAMN18117686 | Diseased pig | Joint | Spain | 2019 | tet(O), erm(B) | |
| 2245604a | 4 | 7 | 29 | 7-Ia | SAMN18117688 | Diseased pig | Spleen | Netherlands | 2019 | tet(O), erm(B) | |
| S10D | 4 | 7 | 29 | 7-II | SAMEA3233974 | Diseased pig | Brain | UK | 2010 | tet(M) | |
| LOLA-SS008 | 4 | 7 | 29 | 7-II | SAMEA1316648 | Pig | / | UK | 2014 | dfrF,tet(M) | |
| 2260249a | 4 | 7 | 29 | 7-II | SAMN18117689 | Diseased pig | Joint | Netherlands | 2019 | tet(O), erm(B) | |
| LS5F | 4 | 7 | 29 | 7-II | SAMEA3233926 | Healthy pig | Tonsil | UK | 2011 | dfrF,tet(M) | |
| GD-0070 | 4 | 7 | 29 | 7-Ib | SAMEA3595240 | Diseased pig | CSF | Netherlands | 2005 | erm(B),tet(O) | |
| LSS85 | 4 | 7 | 29 | 7-II | SAMEA1316680 | Pig | / | UK | 2014 | dfrF,tet(M) | |
| LL-W | 4 | 7 | 29 | 7-II | SAMEA3233877 | Diseased pig | / | UK | 2010 | dfrF,tet(M) | |
| LS3D | 4 | 7 | 29 | 7-II | SAMEA3233910 | Healthy pig | Tonsil | UK | 2011 | dfrF,tet(M) | |
| SS1007 | 4 | 7 | 29 | 7-II | SAMEA1316618 | Pig | / | UK | 2014 | tet(M) | |
| LSS84 | 4 | 7 | 29 | 7-II | SAMEA1316689 | Pig | / | UK | 2014 | dfrF,tet(M) | |
| 2138581a | 4 | 7 | 29 | 7-Ia | SAMN18117690 | Diseased pig | Spleen | France | 2018 | tet(O), erm(B) | |
| 8074 | 4 | 7 | 29 | 7-Ia | SAMN02469536 | Diseased pig | / | Danmark | 1980' | – | |
| 2138556a | 4 | 7 | 29 | 7-Ia | SAMN18117691 | Diseased pig | Brain | Spain | 2018 | – | |
| LSS56 | 4 | 7 | 29 | 7-II | SAMEA1316549 | Pig | / | UK | 2014 | dfrF,tet(M) | |
| 2270437a | 4 | 7 | 29 | 7-II | SAMN18117692 | Diseased pig | Brain | Spain | 2019 | tet(O), erm(B) | |
| 151Ba | 4 | 7 | 29 | 7-Ia | SAMN17982940 | Diseased pig | Heart | Spain | 1999 | – | |
| LOLA-SS009 | 4 | 7 | 29 | 7-II | SAMEA1316611 | Pig | / | UK | 2014 | tet(M) | |
| LL-X | 4 | 7 | 29 | 7-II | SAMEA3233878 | Diseased pig | Brain | UK | 2010 | tet(M) | |
| WUSS413a | 4 | 7 | 29 | 7-Ib | SAMN17982953 | Healthy pig | Tonsil | CN | 2017.12 | tet(O), erm(B) | |
| Lineage6 | 2018WUSS100a | 7–2 | 7 | 1612 | 7-III | SAMN17982959 | Healthy pig | Tonsil | CN | 2019.01 | ant(6)-Ia, aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(M) |
| 2018WUSS101a | 7–2 | 7 | 1612 | 7-III | SUB9103358 | Healthy pig | Tonsil | CN | 2019.01 | ant(6)-Ia, aac(6′)-Ie-aph(2″)-Ia, erm(B), tet(M) |
genomes were sequenced in the present study. Non-indicated genomes from NCBI.
/: not available; –: None of AR genes
In the present study, the complete genome of strain GX69 was sequenced using PacBio Sequel platform and Illumina NovaSeq PE150, whereas the draft genomes were sequenced using Illumina NovaSeq PE150. Sequencing libraries were generated using the methods described previously [25]. The valid reads filtered low-quality reads were assembled into contigs and scaffolds with SOAPdenovo (release 1.04). Genes were predicted by using Glimmer 3.02, and gene orthologs were determined by using GO (Gene ontology) V20171011, KEGG (Kyoto Encyclopedia of Gene and Genmomes) V20181107, and COG (Clusters of Othologous Database) V20171127.
Bioinformatics analysis
MLST and MCG typing
The multilocus sequence type (MLST) and the minimum core genome (MCG) group of the genomes were determined by using PubMLST (https://pubmlst.org/bigsdb?db=pubmlst_ssuis_seqdef&page=sequenceQuery), and a method previously described [26], respectively.
Phylogenetic analysis
Single-nucleotide polymorphisms (SNPs) were detected using Bowtie 2, and MUMmer v3.23 for sequencing reads and complete genomes, respectively, and the genome sequence of SC84 (accession No. FM252031) [27] was used as a reference. The mutational SNP sites were selected based on the method described in a previous study [26], and then the phylogenetic tree was constructed using the maximum likelihood method by FastTree v2.1.10. Streptococcus pneumoniae ATCC 700669 (accession No. NC_011900) was used as an outgroup to root the tree. The tree was presented using FigTree v1.4.0.
Detection of S. suis virulence-associated genes, AR genes and AR genes associated with mobile genetic elements (MGEs)
Distributions of virulence-associated genes and regions of difference (RDs) preferentially present in highly pathogenic S. suis serotype 2 strains were investigated among S. suis serotype 7 genomes, consisting of genes mrp, sly, epf, sao, nadR, NisR, NisK, SalR, SalK, revS, ofs, RD6, RD12, RD14, RD21, RD29, RD40, RD53, and RD60 [28,29]. Genes having a global match region at <80% of the amino-acid sequence with an identity of <80% were determined to be absent.
AR genes were analysed by searching Comprehensive Antibiotic Resistance database (CARD) and Antibiotic Resistance genes database (ARDB). A resistance gene was only regarded as a homologue in tested strains if it showed at least 80% identity in amino-acid sequence across 80% of the length of the protein [30]. The prophages and ICEs were predicted by PHAST (http://phast.wishartlab.com/) and ICEberg (https://db-mml.sjtu.edu.cn/ICEberg/), respectively. For the identification of integrative and conjugative elements (ICEs), signature proteins including integrase, relaxase, and VirB4 were typed using the database from a previous study [31]. Search strategies and the definitions of integrative mobilizable elements (IMEs) and cis-IMEs (CIMEs) were carried out according to the methods previously described [31,32].
Analysis of cps gene cluster
Each cps gene cluster was extracted from the genomes and compared with that of the serotype 7 reference strain 8074 (GenBank accession No. BR001004.1). The homology groups (HGs) of cps genes were assigned according to the nomenclature described in a previous study [33]. The sequence comparison of cps gene cluster was performed using blastN programme in BLAST with an e-value cutoff of e-10 and was visualized using an in-house Perl script (https://github.com/dupengcheng/BlastViewer).
Infection experiments
The virulence of strain GX69 from the patient and 21 additional representative strains based on the distribution in the phylogenetic tree were tested. For comparison, the highly pathogenic and well-characterized S. suis serotype 2 reference strain P1/7 (ST1) [34] was included [35,36]. C57BL/6 mice (6 weeks old, female) were injected intraperitoneally with 5 × 107 CFU of S. suis strain in 1 mL PBS or 1 mL PBS only as a control group. The infection dose of each strain was confirmed by plating the serial dilutions of the suspension onto the Todd–Hewitt broth (THB, Oxoid Ltd, London, UK) agar before and after the infection. Each infected group contained ten mice, and the mock-infected group contained five mice. The mortality was recorded per six hours within 24 h post-infection and per 12 h from 24 h to 96 h post-infection. The experiment was performed independently at least twice for each strain. The mortality of each infected group was calculated via the Kaplan–Meier method. S. suis serotype 7 strains initiating lethal infection with a mortality ≥80% at 96 h post-infection were classified as virulent strains.
Investigation of antimicrobial susceptibility profiles
To determine whether the AR genes in genomes conferred the predicted resistance to the corresponding bacteria, we used the MIC-test strip (Liofilchem, Roseto degli Abruzzi, Italy) to assess the antimicrobial susceptibility of strains carrying AR genes. The following antibiotics were tested: clindamycin (0.016–56 μg/mL), erythromycin (0.016–256 μg/mL), azithromycin (0.016–256 μg/mL), tetracycline (0.016–256 μg/mL), gentamicin (0.016–256 μg/mL), kanamycin (0.016–256 μg/mL), and streptomycin (0.064–1024 μg/mL). For tetracycline, azithromycin, erythromycin, and clindamycin, breakpoints were used as recommended by the Clinical and Laboratory Standard Institute (CLSI) guidelines 2019 (M100-S29) for Streptococcus spp. Viridans group. No breakpoint values of streptomycin, kanamycin, and gentamicin were available for Streptococcus. Their breakpoints were taken from a previous study [37].
Statistics
The survival curves of different infected groups were compared using Gehan–Breslow–Wilcoxon test. For the test, a p-value < .05 was considered to be significant.
Nucleotide sequence accession numbers
The sequences of the genomes sequenced in the study were deposited in the GenBank under accession numbers listed in Table 1.
Ethical approval
This study and the application of the animal experiments with code 2020-024 were reviewed and approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention.
Results
MLST and MCG typing
Among 114 genomes, 22 different STs were identified, revealing high heterogeneity of S. suis serotype 7 population. ST29 (n = 47) was most prevalent, followed by ST373 (n = 16), ST94 (n = 12), ST225 (n = 6), ST24 (n = 4), ST32 (n = 4), ST971 (n = 4), ST89 (n = 3), ST907 (n = 2), ST1610 (n = 2), ST1611 (n = 2), and ST1612 (n = 2). The remaining ST17, ST34, ST839, ST854, ST973, ST979, ST980, ST1609, ST1613, and ST1614 only contained one strain each. The strain GX69 from the patient was ST373 which was prevalent in China, whereas ST29 and ST94 were predominant in Europe and North America, respectively (Table 1).
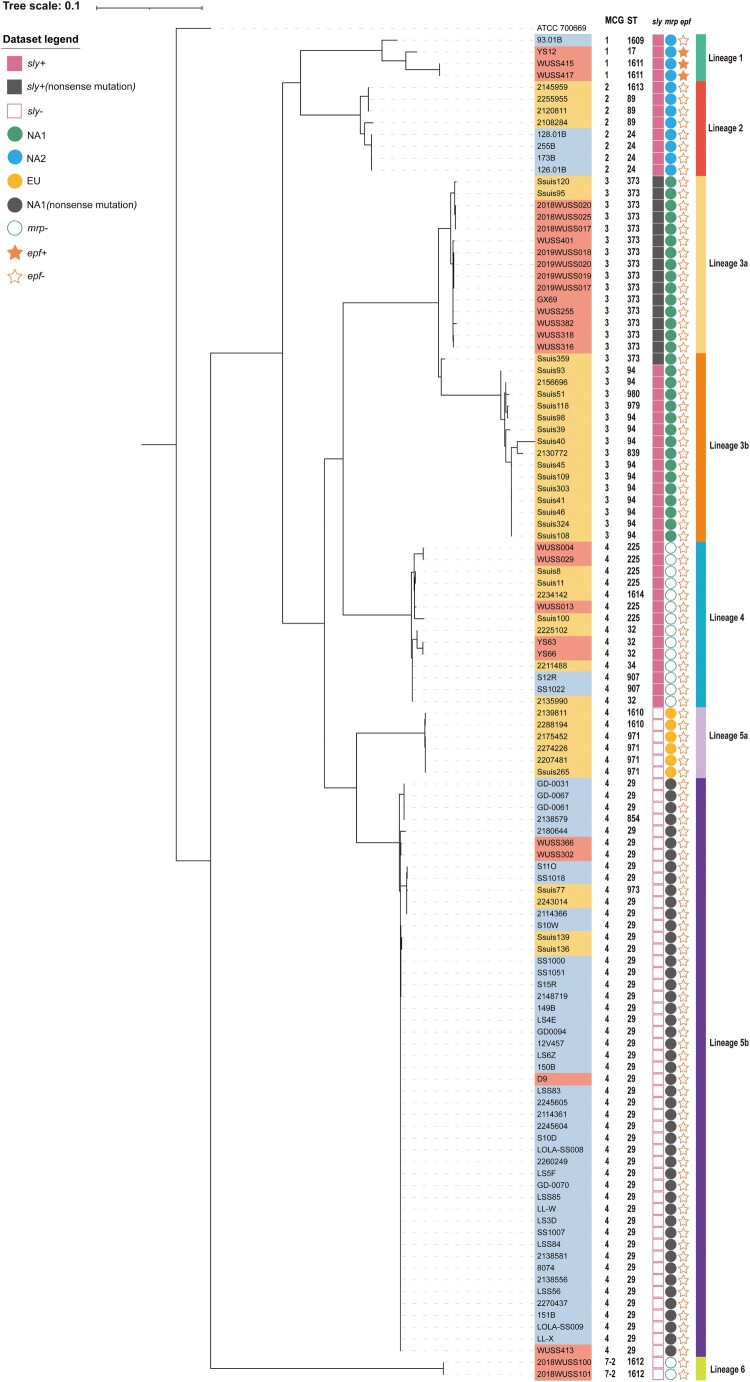
The 114 genomes were clustered into five MCG groups, including MCG groups 1, 2, 3, 4, and 7–2. MCG group 4 was predominant and included ST29 strains. It is noteworthy that genomes of MCG group 4 were composed of 10 STs and 69 genomes widely distributed in all nine countries. Five STs and 31 genomes were classified into MCG group 3, including the strain GX69 from the patient. Eight and four genomes were classified into MCG groups 2 and 1, respectively. Both of them contained 3 STs. Two ST1612 genomes were classified into MCG group 7–2 (Figure 1).
Figure 1.
A maximum-likelihood phylogenetic tree of 114 S. suis serotype 7 genomes. The phylogenetic tree was constructed based on mutational SNPs differences across the whole core genome. The S. pneumoniae ATCC 700669 was used as an outgroup to root the tree. The strains were coloured based on the isolation regions, grey for Europe, orange-yellow for North America, and orange-red for China. The scale is given as the number of substitutions per variable site.
Based on the distribution of mutational SNPs in core genomes, 114 genomes were clustered into six lineages. Each MCG group consisted of one lineage, except for MCG group 4. Both Lineages 4 and 5 were composed of MCG group 4, whereas contained 14 and 55 genomes, respectively. Lineage 3 was divided into Lineages 3a and 3b. Interestingly, genomes of Lineage 3a and Lineage 3b were mainly from China and USA, respectively. Compared with Lineage 3a, composed of ST373 genomes, Lineage 3b was mainly composed of ST94 genomes. Lineages 5a and 5b were mainly composed of ST971 and ST29 genomes, respectively (Figure 1).
The difference in virulence among S. suis serotype 7 strains.
In order to evaluate the virulence level of S. suis serotype 7 population, we compared the survival level of C57BL/6 mice infected with S. suis highly pathogenic serotype 2 strain P1/7, strain GX69, and additional 21 serotype 7 representative strains. Most mice infected with S. suis serotype 7 strains showed obvious septic signs during the infection, such as rough hair coat, swollen eyes, weakness, and shivering. The apparent diversity in the survival curves of mice infected with S. suis serotype 7 strains were observed. A significant difference was observed in survival curves between mice infected with strains P1/7 and GX69 (p = .0002), which attributed to the differences in mortality at the early phase of the infection. Mice infected with P1/7 had a 20% survival rate at 12 h post-infection, while mice infected with strain GX69 had a 65% survival rate at the same time point. Notably, the survival levels of mice infected with strain GX69 dramatically decreased after 12 h post-infection. Its survival rate decreased to 10% at 24 h post-infection and was identical to that of strain P1/7 (Table 2, Figure S1A). Thus, strain GX69 possessed the capacity to initiate lethal infection in C57BL/6 mice and was classified as a virulent strain.
- Among additional 21 serotype 7 representative strains, the mortalities of mice infected with eight S. suis serotype 7 strains at 96 h post-infection were less than 50%. These strains were classified as lowly virulent strains. Interestingly, half of them were isolated from diseased pigs.
- None of the mice infected with strains 128.01B, 173B, WUSS316, WUSS302, and 8074 died within the infection period (Table 2), even though strains 128.01B, 173B, and 8074 were isolated from diseased pigs.
- The survival mice infected with strains 126.01B (p < .0001), WUSS382 (p < .0001), and 2018WUSS100 (p < .0001) were significantly higher than that of mice infected with strain GX69 (Table 2). The strain 126.01B was isolated from diseased pig.
- The survival levels of mice infected with remaining 13 S. suis serotype 7 representative strains were significantly higher than that of mice infected with strains P1/7. However, the mortalities of mice infected with these strains at 96 h post-infection reached or exceeded 80% (Table 2). These strains were classified as virulent strains. It is noteworthy that seven of them were isolated from healthy pigs.
- The survival curves of mice infected with YS63 (p < .0001), WUSS013 (p < .0001), and WUSS029 (p = .0021) were significantly different from that of mice infected strain GX69, since mice infected with three strain mainly died after 24 h post-infection (Table 2 and Figure S1B). Two strains YS63 and WUSS029 were isolated from healthy pigs.
- The survival curves of mice infected with 10 strains YS12 (p = .2817), WUSS415 (p = .7557), 93.01B (p = .0592), 2018WUSS020 (p = .2705), 2019WUSS020 (p = .8214), WUSS004 (p = .0838), WUSS413 (p = .2015), 149B (p = .7383), 150B (p = .3338), and 151B (p = .9834) were similar to that of mice infected with strain GX69 (Table 2 and Figure S1C). Therefore, they were classified as virulent strains. Among them, strains YS12, WUSS415, 2018WUSS020, 2019WUSS020, and WUSS413 were isolated from healthy pigs.
Table 2.
The value of mortality and statistical comparison in the survival assay.
| Lineage | Strains | The mean mortality of each infected group at different post-infection time pointsa | Virulence levelb | p valuec | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6h | 12h | 18 h | 24h | 36h | 48 h | 60h | 72h | 96 h | Compare to P1/7 infected group | Compare to GX69 infected group | Compare to control group | |||
| lineage1 | YS12 | 0 ± 0 | 50 ± 7 | 75 ± 4 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | V | .0009 | .2817 | <.0001 |
| WUSS415 | 0 ± 0 | 10 ± 7 | 85 ± 4 | 90 ± 7 | 90 ± 7 | 90 ± 7 | 90 ± 7 | 90 ± 7 | 90 ± 7 | V | <.0001 | .7557 | <.0001 | |
| 93.01B | 0 ± 0 | 0 ± 0 | 45 ± 32 | 90 ± 0 | 90 ± 0 | 95 ± 4 | 95 ± 4 | 95 ± 4 | 100 ± 0 | V | <.0001 | .0592 | <.0001 | |
| lineage2 | 126.01B | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 10 ± 0 | 20 ± 0 | L | <.0001 | <.0001 | .1468 |
| 128.01B | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | L | <.0001 | <.0001 | >.9999 | |
| 173B | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | L | <.0001 | <.0001 | >.9999 | |
| lineage3a | GX69 | 0 ± 0 | 35 ± 14 | 65 ± 7 | 90 ± 4 | 90 ± 7 | 90 ± 7 | 90 ± 7 | 90 ± 7 | 90 ± 7 | V | .0002 | / | <.0001 |
| WUSS316 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | L | <.0001 | <.0001 | >.9999 | |
| WUSS382 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 20 ± 0 | 30 ± 7 | 35 ± 11 | 40 ± 7 | 40 ± 7 | L | <.0001 | <.0001 | .0262 | |
| 2018WUSS020 | 0 ± 0 | 23 ± 3 | 57 ± 7 | 70 ± 5 | 83 ± 3 | 83 ± 3 | 83 ± 3 | 83 ± 3 | 83 ± 3 | V | <.0001 | .2705 | .0001 | |
| 2019WUSS020 | 0 ± 0 | 30 ± 7 | 70 ± 7 | 80 ± 0 | 80 ± 0 | 80 ± 0 | 80 ± 0 | 80 ± 0 | 80 ± 0 | V | .0001 | .8214 | .0002 | |
| lineage4 | YS63 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 45 ± 11 | 65 ± 11 | 90 ± 7 | 90 ± 7 | 90 ± 7 | 90 ± 7 | V | <.0001 | <.0001 | <.0001 |
| WUSS004 | 0 ± 0 | 23 ± 11 | 37 ± 12 | 70 ± 8 | 90 ± 5 | 93 ± 5 | 93 ± 5 | 93 ± 5 | 93 ± 5 | V | <.0001 | .0838 | <.0001 | |
| WUSS013 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 10 ± 7 | 60 ± 21 | 85 ± 4 | 100 ± 0 | V | <.0001 | <.0001 | <.0001 | |
| WUSS029 | 0 ± 0 | 10 ± 7 | 15 ± 4 | 45 ± 18 | 75 ± 11 | 95 ± 4 | 95 ± 4 | 95 ± 4 | 95 ± 4 | V | <.0001 | .0021 | <.0001 | |
| Lineage5b | WUSS302 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | L | <.0001 | <.0001 | >.9999 |
| WUSS413 | 0 ± 0 | 20 ± 5 | 47 ± 7 | 83 ± 10 | 97 ± 3 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | V | <.0001 | .2015 | <.0001 | |
| 149B | 0 ± 0 | 37 ± 14 | 50 ± 17 | 93 ± 3 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | V | <.0001 | .7383 | <.0001 | |
| 150B | 0 ± 0 | 0 ± 0 | 75 ± 3 | 90 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | V | <.0001 | .3338 | <.0001 | |
| 8074 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | L | <.0001 | <.0001 | >.9999 | |
| 151B | 0 ± 0 | 33 ± 12 | 60 ± 14 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | V | <.0001 | .9834 | <.0001 | |
| lineage6 | 2018WUSS100 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 15 ± 9 | 30 ± 6 | 40 ± 6 | 40 ± 6 | 40 ± 6 | 45 ± 3 | L | <.0001 | <.0001 | .0168 |
| P1/7 | 60 ± 7 | 80 ± 0 | 90 ± 0 | 90 ± 0 | 90 ± 0 | 90 ± 0 | 90 ± 0 | 90 ± 0 | 90 ± 0 | H | / | .0002 | <.0001 | |
| Control | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | / | <.0001 | <.0001 | / | |
The mortality represented as mean ± SED (calculated via the Kaplan–Meier method) at different post-infection time points was present.
H indicates highly virulent, V indicates virulent, and L indicates lowly virulent.
The survival curves of different infected groups were compared using Gehan–Breslow–Wilcoxon test.
Interestingly, all three tested strains from Lineage 1 were classified as virulent strains, while all strains from Lineages 2 and 6 belonged to lowly virulent strains. On the contrary, Lineages 3a, 4, and 5b contained both virulent and lowly virulent strains.
Distribution of putative S. suis virulence-related genes
Only three genomes (YS12, WUSS415, and WUSS417) of Lineage 1 were positive for epf. Sixteen genomes from Lineages 4 and 6 were mrp gene negative. Most of the genomes (98/114, 86.0%) contained putative full-length mrp gene copies. Based on the variation in the central portion of the gene, mrp was grouped into three subtypes EU, NA1, and NA2 [38]. Subtype NA2 (n = 12) was only present in genomes of Lineages 1 and 2. Subtype EU (n = 6) was only found in genomes of Lineage 5a. All genomes of Lineage 3 and 5b harboured subtype NA1(n = 31). Compared with those of Lineage 3, mrp gene of Lineage 5b (n = 49) did not contain an intact open reading frame because of a frameshift mutation in 2.1 kb, which may result in the truncated MRP expression. sly gene was only present in genomes of Lineages 1, 2, 3, and 4. A premature stop codon was present in sly gene of ST373 genomes of Lineage 3 (Figure 1).
Genes nadR, NisR, NisK, SalR, and SalK were absent from all serotype 7 genomes. Genes revS and ofs were only present in genomes of Lineage 1. sao gene was widely distributed in serotype 7 genomes, except for genomes of Lineage 6. RD6 was present in strains WUSS415, WUSS417, and YS12, while the remaining RDs tested in the study were absent from all serotype 7 genomes.
The distribution of AR genes in S. suis serotype 7 genomes
Thirteen genomes did not harbour any AR genes. Totally, 216 AR genes were present in the remaining 101 genomes. The AR genes belonged to six categories tetracycline, macrolides/lincosamides/streptogramin (MLS), lincosamide, aminoglycosides, trimethoprim, and chloramphenicol (Table 1).
The tetracyclines resistance genes
Ninety-three genomes carried tetracycline-resistant genes. Three types of tetracycline-resistant genes were found among them, consisting of tet(O), tet(M), and tet(W). tet(O) was the prevalent tetracycline-resistant gene and was present in 72 genomes. Eighteen genomes carried tet(M) gene, mainly from Lineage 5b (14/18). tet(W) gene was present in three genomes.
The MLS and lincosamide resistance genes
Three types of genes were found, consisting of genes erm(B), lsaC, and lnuB. The MLS resistance gene erm(B), encoding rRNA adenine N-6-methyltransferase, was prevalent and present in 72 genomes. The lincosamides-streptogramin A resistance gene lsaC was found in three genomes of Lineage 6, which simultaneously harboured lincosamide resistance gene lnuB.
The aminoglycosides resistance genes
Fifteen genomes carried aminoglycosides resistance genes, including streptomycin resistance gene ant6ia encoding aminoglycoside O-nucleotidylyltransferase (n = 13), kanamycin resistance gene aph(3′)-IIIa encoding aminoglycoside O-phosphotransferase (n = 9), spectinomycin resistance gene ant9ia encoding aminoglycoside 3′-phosphotransferase (n = 4), and gentamicin and kanamycin resistance gene aac(6′)-Ie-aph(2″)-Ia encoding aminoglycoside acetyltransferase (n = 2).
The trimethoprim and chloramphenicol resistance genes
Eleven genomes harboured trimethoprim resistance gene dfrF encoding dihydrofolate reductase. It is noteworthy that all genomes carried dfrF gene were from Lineage 6. One genome of Lineage 6 harboured chloramphenicol resistance gene cat-TC encoding chloramphenicol acetyltransferase.
Antimicrobial susceptibility profiles of available strains
To investigate whether AR genes conferred resistance to host strains, we tested the antimicrobial susceptibility of available strains, including 25 Chinese and five Spanish strains, carrying genes responsible for resistance to tetracycline, erythromycin, clindamycin, streptomycin, kanamycin, or gentamycin. Thirty strains harboured tetracycline resistance genes were all resistant to tetracycline, with a MIC value between 12 and 128 μg/mL. Concomitant resistance to erythromycin and clindamycin was found in all strains (n = 25) carrying erm(B) gene due to the overlapping ribosomal binding sites of the two antibiotics. MIC values for both antibiotics were between 128 and >256 μg/mL. A high level of kanamycin (MICs > 256 μg/mL) resistance was found in four strains carrying aph3-IIIa gene. The MIC values of both gentamycin and kanamycin were >256 μg/mL in two strains carrying aac(6′)-Ie-aph(2″)-Ia gene. The MIC values of streptomycin were >1024 μg/mL in nine strains carrying the ant6ia gene (Table S1). The data confirmed that these AR genes conferred corresponding antibiotic resistance phenotypes to their host.
AR genes associated with MGEs
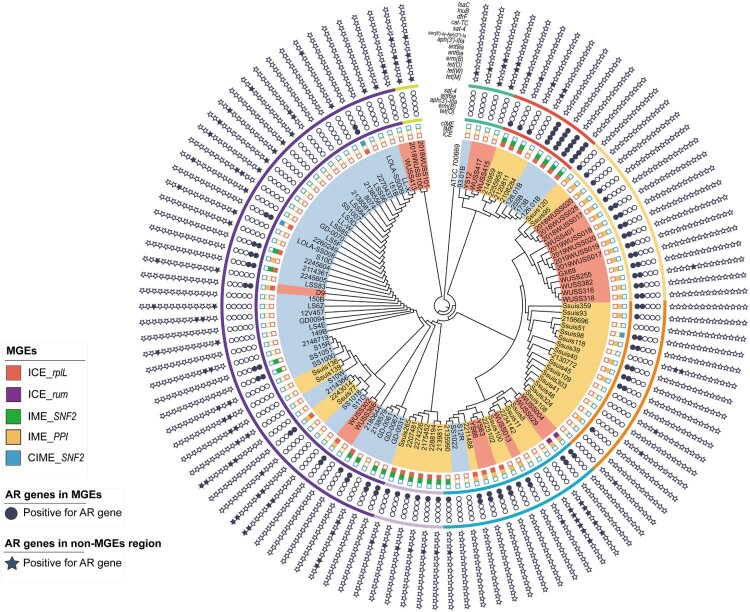
To investigate the mechanism to disseminate AR genes, the MGEs harbouring AR genes in S. suis serotype 7 genomes were predicted. Among 114 genomes, 27 ICEs, 56 IMEs, and three CIMEs (absent of the integrase and relaxase genes) with a complete sequence were detected. These ICEs were distributed in Lineages 2, 4, and 5, whereas IMEs were distributed in Lineages 2, 3, 4, and 5. Totally, 111 of 216 AR genes were present in these MGEs (Figure 2).
Figure 2.
The distribution of AR genes in MGEs and non-MGEs regions of S. suis serotype 7 genomes. The inner circle is the distribution of ICEs, IMEs, and CIMEs in S. suis serotype 7 genomes. Each panel of boxes filled with different colors represents ICEs, IMEs, and CIMEs integrated into the different locus of corresponding genomes, and hollow boxes represent the absence of ICEs, IMEs, or CIMEs in corresponding genomes. The middle circle showed the AR genes carried by ICEs, IMEs, or CIMEs. Each filled circle represents the corresponding AR gene present on corresponding ICEs, IMEs, or CIMEs. The outer circle showed the AR genes located into non-MGEs regions in S. suis serotype 7 genomes. Each filled star represents the corresponding AR gene present on non-MGEs regions in corresponding genomes.
Twenty-five ICEs were inserted into rplL locus. All of them harboured a 15-bp att sequence 5′-TTATTTAAGAGTAAC-3′ in the flanking region. ICESsuWUSS029 and ICESsuWUSS004 were integrated into downstream of rum gene. Both of them harboured the 14-bp att sequences 5’-CACGTGGAGTGCGT-3′ and 5′-CATGTTGAAGTTGT-3′ in the 5′ and 3′ flanking regions, respectively. All ICEs were classified as Tn5252 group and harboured intact signature proteins VirB4, integrase, and canonical relaxase of the MobP family. Fifty-six AR genes resistant to tetracycline, MLS, and aminoglycosides were present in these ICEs (Figure 2).
The genes SNF2 and PPI encoding a putative adenine-specific DNA methylase and a putative peptidylprolyl isomerase, respectively, are two specific insertion hot spots for integrating IMEs or CIMEs [32]. In the present study, 34 and 22 IMEs were integrated into the PPI and SNF2 genes, respectively. All three CIMEs were integrated into SNF2 gene (Figure 2). All IMEs or CIMEs harboured an 11-bp inverted repeat 5′-TTTTGCGGACA-3′ in their flanking region. Interestingly, 25 IMEs and two CIMEs were integrated into ICEs and all AR genes in the ICEs were carried by these integrated IMEs and CIMEs. The remaining 31 IMEs and one CIMEs were located in non-ICE regions. Thirty-two tet(O) and 23 ermB genes were present in these IMEs and CIMEs. The integrases of all IMEs were identical and belonged to serine integrase type 3, regardless of their integration site. Meanwhile, the relaxases of the IMEs belonged to the MobV superfamily. Based on their integrase and relaxase types, all IMEs belonged to IME_Class_6. It is noteworthy that all AR genes responsible for resistance to aminoglycosides, trimethoprim, and chloramphenicol were not present in the above MGEs.
Differences of cps gene clusters among strains
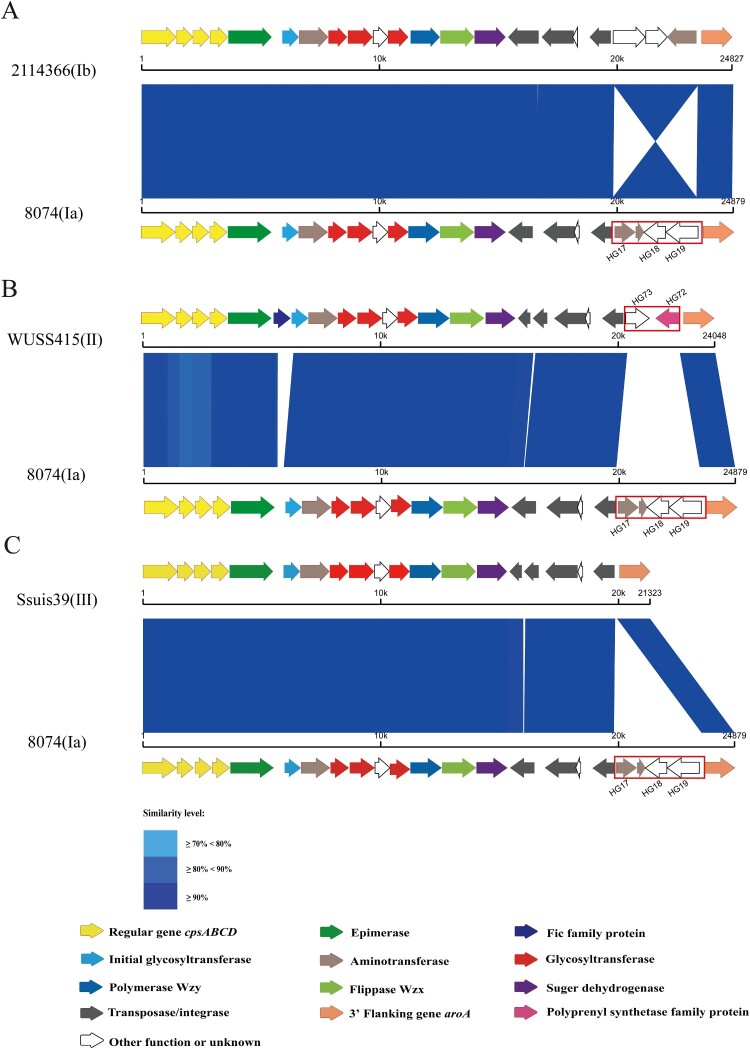
The cps gene clusters of 114 genomes were located between the orfZ-orfX region and the aroA gene, which belonged to pattern I-a [33]. Based on the variable presence of HG17 (Aspartate aminotransferase), HG18 (Tetratricopeptide repeat protein) and HG19 (Hypothetical protein) of the serotype 7 reference strain 8074 (GenBank accession No. BR001004.1), four subtypes of cps gene clusters were found among 114 genomes (Figure 3).
Figure 3.
The schematic comparison of the cps gene cluster subtype Ia to that of Ib (A), II (B), and III (C). Each colored arrow represents the gene whose predicted function is shown in the blow panel. HG17, HG18, HG19, HG72, and HG73 genes are indicated. The aroA gene is located on the 3′ side of each locus. Regions of over 70% identity were marked by blue shading.
HG17, HG18, and HG19 were present in cps gene clusters of 84 genomes that belonged to cps7-I. The arrangement of HG17, HG18, and HG19 in cps gene clusters of 54 genomes similar to that of the serotype 7 reference strain 8074 was nominated as cps7-Ia, while the arrangement of them inverted in cps gene clusters of 30 genomes was nominated as cps7-Ib. cps7-Ib was mainly present in genomes from Lineage 5b (n = 11), Lineage 4 (n = 9) and Lineage 3a (n = 7).
Replacement of HG17, HG18, and HG19 by HG72 and HG73 was found in cps gene clusters of 23 genomes which were nominated as cps7-II. These genomes were distributed in Lineage 5b (n = 16), Lineage 2 (n = 4), and Lineage 1 (n = 3), from European countries and China.
HG17, HG18, and HG19 were absent in seven cps gene clusters which were nominated as cps 7-III. Five of them were from the USA and distributed in Lineage 5b, while two were from China and consisted of Lineage 6.
Discussion
In the present study, we first reported a S. suis serotype 7 strain (GX69) isolated from a patient with septicemia complicated with pneumonia. The strain GX69 was ST373 and belonged to MCG group 3, whereas ST1 and ST7 are predominant in S. suis strains from patients [39], mainly clustered into MCG group 1 [26]. The genotype of S. suis classical virulence markers in strain GX69 was mrpNA1 sly+ epf –, whereas the prevalent genotype of corresponding virulence markers in human strains was mrpEU sly+ epf+ or mrpNA2 sly+ epf+[9,40]. It is noteworthy that a premature stop codon was present in sly gene of strain GX69 and may result in the truncated SLY expression. To evaluate the virulence of strain GX69, the survival level was compared with that of highly pathogenic S. suis serotype 2 strain P1/7. Significant differences at the early phase of infection and the similarity at the middle phase of infection were observed between the two strains in the mouse infection model. Our result confirmed that GX69 was a virulent strain and possessed the capacity to initiate lethal infection, even though virulence-associated genes and RDs preferentially present in highly pathogenic S. suis serotype 2 strains were almost absent in strain GX69. We proposed that S. suis serotype 7 may be considered as a potential zoonotic pathotype, and further investigation of S. suis serotype 7 population is needed to improve the prevention and control strategies.
In the present study, S. suis serotype 7 population composed of 35 strains and 79 genomes of strains from 1999 to 2019 in nine countries was investigated. Twenty-two STs and five MCG groups were identified among S. suis serotype 7 genomes clustered into six lineages based on the distribution of mutational SNPs in the core genomes. Interestingly, since most predominant ST29, ST373, and ST94 were prevalent in respective regions, it suggests that the evolution of S. suis serotype 7 population was relevant to the geographical distribution. The evolutionary affinity between ST373 and ST94 was revealed in that they belonged to MCG group 3 and were clustered into Lineage 3. It is noteworthy that significant heterogeneity was observed within ST373 strains, which were clustered into three clades. Similar heterogeneity was previously reported in phylogenetic analysis of ST1[40], ST7 [9], and ST25 [41].
Based on the results of the survival assay using the C57BL/6 mouse model, the strain GX69 and over 60% additional representative strains tested (13/21) were classified as virulent strains. Among 13 virulent strains, the survival curves of ten S. suis serotype 7 representative strains were similar to that of strain GX69. In a recent study, 82.6% S. suis serotype 7 strains from North America were pathogenic based on the clinical information and site of isolation [20]. In the present study, seven strains isolated from healthy pigs were classified as virulent strains, and two of them belong to ST373, the same ST of strain GX69. Therefore, we proposed that the public health threat of S. suis serotype 7, especially those virulent ST373 strains, should not be ignored. Coincidentally, healthy pigs were reported to be a reservoir of strains with high virulence potential in humans [39,42]. Moreover, four strains isolated from diseased pigs were classified as lowly virulent. A correlation between the virulence level of strains and their origin (diseased or healthy pigs) could not be observed in the present study. A similar result was also reported in our previous study [43]. It should be noted that the presence of clinical signs in pigs may also depend on co-infection with some viral and bacterial pathogens [42]. Thus, the public health significance of strains may not be accurately evaluated only based on the clinical information of their host.
In the present study, three classical virulence markers mrp, sly, and epf were not critical virulence indicators of the S. suis serotype 7 strains. However, a significant correlation of genotypes and variations of three genes and their distribution in lineages was observed, suggesting that these genes correlate with the evolution of S. suis serotype 7 population rather than virulence. Most virulence-associated genes preferentially present in highly pathogenic S. suis serotype 2 strains were absent from all serotype 7 genomes. Previous studies also reported that these virulence markers studied in S. suis serotype 2 strains were not suitable as virulence markers for S. suis non-serotype 2 strains [3,12,44]. Thus, S. suis serotype 7 virulent strains may utilize a different pathogenesis strategy. Because of the high diversity of virulence levels within S. suis serotype 7 population, further studies are necessary to identify reliable virulence indicators of S. suis serotype 7 strains. Using multiple animal models to accurately pathotype ST373 strains combined with comparative genomic analysis of ST373 strains with different virulence levels may be feasible.
Six categories of AR genes are present in S. suis serotype 7 genomes. The predominant categories were tetracycline and MLS resistance genes. High rates of resistance to tetracycline, macrolide, lincosamide, and erythromycin have been reported in both human and pig isolates of S. suis in the last 20 years [45–49]. Tetracycline, lincosamide, and macrolide are used extensively for therapy and metaphylaxis in the swine industry [32,50,51], contributing to the emergence and spread of associated resistance. The most prevalent tetracycline resistance gene was tet(O). This is different from what was previously reported for serotype 2 strains, which have been shown to mainly carry tet(M) and tet(W) [45,48].
Previous studies have shown that MGEs play a significant role in the horizontal transfer of AR genes in S. suis [31,32]. Twenty-seven ICEs carrying AR genes were found in S. suis serotype 7 genomes, although intact prophages carrying AR genes were not detected. Conversely, AR genes in S. suis serotype 31 population were majorly present in prophages [12]. In the present study, two types of DNA cargo of IMEs and CIMEs with AR genes were integrated into genes SNF2 or PPI of all ICEs. Similar IMEs were also integrated into SNF2 gene of ICESsuZJ20091101-1(KX077882.1), ICESsuLP081102 (KX077885.1), ICESsuJH1301 (KX077887.1) [31], and ICESsD9 [52]. A similar CIME was also inserted into the same integration site of ICESsuBSB6 [53]. Likely, the exchange, acquisition, and deletion of the IME/CIME module may contribute to the evolution of ICEs. In the present study, all AR genes in ICEs were carried by these IMEs and CIMEs. Moreover, IMEs and CIMEs carrying AR genes were also present in non-ICE regions of additional 32 S. suis serotype 7 genomes.
IMEs were reported to be more widespread than ICEs in S. suis [32]. In this work, IMEs were also found to be highly prevalent in S. suis serotype 7 genomes. Over 50% AR genes identified in serotype 7 genomes were present in IMEs. IMEs mainly carried tetracycline, erythromycin, and lincosamide resistance genes. We propose that IMEs may play a critical role in the horizontal transfer of these AR genes in S. suis serotype 7. Interestingly, the proportion of genome carrying IMEs was higher in Lineages 2, 3a, 4, and 5a. Our data indicated that the transmission patterns of AR genes might be related to the evolution of serotype 7 population. CIMEs are decayed IMEs, which are cis-mobilizable elements without integration and relaxase genes but with attL and attR sites. CIMEs carrying tet(O) and ermB genes were found in two ICEs and non-ICE region of one additional genome. Further study is needed to investigate the role of CIMEs in the transmission of AR genes.
Finally, different organizations of cps loci were observed among S. suis serotype 7 population. These differences can be attributed to the variable presence of HG17, HG18, and HG19. The function of HG17 was related to aminotransferase, while the functions of both HG18 and HG19 were ATP-binding proteins. HG17, HG18, and HG19 existed widely in cps gene clusters of S. suis serotypes 4, 5, 17, 18, 19, and 23 reference strains. It is noteworthy that HG17, HG18, and HG19 were also inverted in cps gene clusters of S. suis serotypes 17 and 23 reference strains. Among 23 cps gene clusters of S. suis serotype 7 genomes, HG17, HG18 and HG19 were replaced by HG72 and HG73. The functions of HG72 and HG73 were related to carboxyvinyltransferase and unknown, respectively. HG72 and HG73 also existed in cps gene clusters of S. suis serotypes 11 and 30 reference strains. Based on the agglutination test results, the capsular antigenic phenotype was not affected by the variable presence of HG17, HG18, and HG19. Therefore, these HGs may not be involved in the forming of serotype 7-specific epitopes. The subtype cps7-I was most predominant among S. suis serotype 7 population and dispersed in different lineages and geographical regions. On the other hand, subtype cps7-II was majorly present in strains from Europe (such as UK and Spain), while it was absent in strains from North America. Different cps subtypes may enhance the fitness of corresponding host strains in specific environments.
In conclusion, our data confirmed S. suis serotype 7 is a non-negligible pathotype and deepened the understanding of S. suis serotype 7 population. Geographically dependent characteristics were revealed in the evolution of S. suis serotype 7 population. Our study provided valuable information for the improved surveillance of S. suis serotype 7 strains. Further studies are needed to identify the virulence indicators to predict the public health significance of S. suis serotype 7 strains.
Supplementary Material
Acknowledgements
HZ and ZW designed the project; HZ drafted the manuscript; ZW, MG and AV reviewed the manuscript. PL, and JW carried out the experiments and generated the data; PL, AE, WM, MG, AV, PD, ML, and HZ analysed the data. All authors have read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Science and Technology Major Project from the Ministry of Health of the People's Republic of China [grant number 2017ZX10303405-002] and the National Natural Science Foundation of China [grant number 81572044].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Goyette-Desjardins G, Auger JP, Xu J, et al. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microb Infect. 2014 Jun;3(6):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bojarska A, Janas K, Pejsak Z, et al. Diversity of serotypes and new cps loci variants among Streptococcus suis isolates from pigs in Poland and Belarus. Vet Microbiol. 2020;240:108534. [DOI] [PubMed] [Google Scholar]
- 3.Huang J, Liu X, Chen H, et al. Identification of six novel capsular polysaccharide loci (NCL) from Streptococcus suis multidrug resistant non-typeable strains and the pathogenic characteristic of strains carrying new NCLs. Transbound Emerg Dis. 2019;66(2):995–1003. [DOI] [PubMed] [Google Scholar]
- 4.Zheng H, Qiu X, Roy D, et al. Genotyping and investigating capsular polysaccharide synthesis gene loci of non-serotypeable Streptococcus suis isolated from diseased pigs in Canada. Vet Res. 2017;48(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu X, Bai X, Lan R, et al. Novel capsular polysaccharide loci and new diagnostic tools for high-throughput capsular gene typing in Streptococcus suis. Appl Environ Microbiol. 2016;82(24):7102–7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huong VT, Ha N, Huy NT, et al. Epidemiology, clinical manifestations, and outcomes of Streptococcus suis infection in humans. Emerg Infect Dis. 2014;20(7):1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, Jing H, Chen Z, et al. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis. 2006;12(6):914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye C, Zhu X, Jing H, et al. Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg Infect Dis. 2006;12(8):1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M, Du P, Wang J, et al. Genomic Epidemiology of Streptococcus suis sequence type 7 sporadic infections in the Guangxi Zhuang autonomous region of China. Pathogens. 2019;8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultsz C, Jansen E, Keijzers W, et al. Differences in the population structure of invasive Streptococcus suis strains isolated from pigs and from humans in The Netherlands. PLoS One. 2012;7(5):e33854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerdsin A, Hatrongjit R, Gottschalk M, et al. Emergence of Streptococcus suis serotype 9 infection in humans. J Microbiol Immunol Infect. 2017;50(4):545–546. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Sun J, Bian C, et al. The population structure, antimicrobial resistance, and pathogenicity of Streptococcus suis cps31. Vet Microbiol. 2021;259:109149. [DOI] [PubMed] [Google Scholar]
- 13.Prufer TL, Rohde J, Verspohl J, et al. Molecular typing of Streptococcus suis strains isolated from diseased and healthy pigs between 1996-2016. PLoS One. 2019;14(1):e0210801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unterweger C, Baums CG, Hocher M, et al. [Clinical situation, diagnosis and prevention of a Streptococcus suis serotype 7 problem on a farm]. Berl Munch Tierarztl Wochenschr. 2014 May-Jun;127(5–6):194–201. [PubMed] [Google Scholar]
- 15.Nutravong T, Angkititrakul S, Jiwakanon N, et al. Identification of major Streptococcus suis serotypes 2, 7, 8 and 9 isolated from pigs and humans in upper northeastern Thailand. Southeast Asian J Trop Med Public Health. 2014 Sep;45(5):1173–1181. [PubMed] [Google Scholar]
- 16.Tian Y, Aarestrup FM, Lu CP.. Characterization of Streptococcus suis serotype 7 isolates from diseased pigs in Denmark. Vet Microbiol. 2004;103(1–2):55–62. [DOI] [PubMed] [Google Scholar]
- 17.Tarradas C, Perea A, Vela AI, et al. Distribution of serotypes of Streptococcus suis isolated from diseased pigs in Spain. Vet Rec. 2004;154(21):665–666. [DOI] [PubMed] [Google Scholar]
- 18.MacLennan M, Foster G, Dick K, et al. Streptococcus suis serotypes 7, 8 and 14 from diseased pigs in Scotland. Vet Rec. 1996;139(17):423–424. [DOI] [PubMed] [Google Scholar]
- 19.Rieckmann K, Seydel A, Szewczyk K, et al. Streptococcus suis cps7: an emerging virulent sequence type (ST29) shows a distinct, IgM-determined pattern of bacterial survival in blood of piglets during the early adaptive immune response after weaning. Vet Res. 2018;49(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrada AA, Gottschalk M, Rossow S, et al. Serotype and genotype (Multilocus sequence type) of Streptococcus suis isolates from the United States serve as predictors of pathotype. J Clin Microbiol. 2019;57(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishida S, Tien le HT, Osawa R, et al. Development of an appropriate PCR system for the reclassification of Streptococcus suis. J Microbiol Meth. 2014;107:66–70. [DOI] [PubMed] [Google Scholar]
- 22.Bai X, Liu Z, Ji S, et al. Simultaneous detection of 33 Streptococcus suis serotypes using the luminex xTAG(R) assay. J Microbiol Meth. 2015;117:95–99. [DOI] [PubMed] [Google Scholar]
- 23.Chatellier S, Harel J, Zhang Y, et al. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int J Syst Bacteriol. 1998;48(Pt 2):581–589. [DOI] [PubMed] [Google Scholar]
- 24.Tien LHT, Nishibori T, Nishitani Y, et al. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet Microbiol. 2013;162(2-4):842–849. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Yi X, Liang P, et al. Investigation of the genomic and pathogenic features of the potentially zoonotic Streptococcus parasuis. Pathogens. 2021 Jul 2;10(7):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Zhang W, Zheng H, et al. Minimum core genome sequence typing of bacterial pathogens: a unified approach for clinical and public health microbiology. J Clin Microbiol. 2013;51(8):2582–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye C, Zheng H, Zhang J, et al. Clinical, experimental, and genomic differences between intermediately pathogenic, highly pathogenic, and epidemic Streptococcus suis. J Infect Dis. 2009;199(1):97–107. [DOI] [PubMed] [Google Scholar]
- 28.Zheng H, Du P, Qiu X, et al. Genomic comparisons of Streptococcus suis serotype 9 strains recovered from diseased pigs in Spain and Canada. Vet Res. 2018;49(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng X, Zheng H, Lan R, et al. Identification of genes and genomic islands correlated with high pathogenicity in Streptococcus suis using whole genome tiling microarrays. PLoS One. 2011;6(3):e17987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Yang X, Qin J, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nature Commun. 2013;4:2151. [DOI] [PubMed] [Google Scholar]
- 31.Huang J, Ma J, Shang K, et al. Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: A probable mobile genetic elements reservoir for other streptococci. Front Cell Infect Microbiol. 2016;6(118):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Libante V, Nombre Y, Coluzzi C, et al. Chromosomal conjugative and mobilizable elements in Streptococcus suis: major actors in the spreading of antimicrobial resistance and bacteriocin synthesis genes. Pathogens. 2020;9(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okura M, Takamatsu D, Maruyama F, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl Environ Microbiol. 2013;79(8):2796–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holden MT, Hauser H, Sanders M, et al. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One. 2009;4(7):e6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lachance C, Gottschalk M, Gerber PP, et al. Exacerbated type II interferon response drives hypervirulence and toxic shock by an emergent epidemic strain of Streptococcus suis. Infect Immun. 2013;81(6):1928–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lachance C, Segura M, Gerber PP, et al. Toll-like receptor 2-independent host innate immune response against an epidemic strain of Streptococcus suis that causes a toxic shock-like syndrome in humans. PLoS One. 2013;8(5):e65031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marie J, Morvan H, Berthelot-Herault F, et al. Antimicrobial susceptibility of Streptococcus suis isolated from swine in France and from humans in different countries between 1996 and 2000. J Antimicrob Chemother. 2002;50(2):201–209. [DOI] [PubMed] [Google Scholar]
- 38.Fittipaldi N, Fuller TE, Teel JF, et al. Serotype distribution and production of muramidase-released protein, extracellular factor and suilysin by field strains of Streptococcus suis isolated in the United States. Vet Microbiol. 2009;139(3-4):310–317. [DOI] [PubMed] [Google Scholar]
- 39.Dong X, Chao Y, Zhou Y, et al. The global emergence of a novel Streptococcus suis clade associated with human infections. EMBO Mol Med. 2021;13(7):e13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callejo R, Zheng H, Du P, et al. Streptococcus suis serotype 2 strains isolated in Argentina (South America) are different from those recovered in North America and present a higher risk for humans. JMM Case Rep. 2016;3(5):e005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Athey TB, Teatero S, Takamatsu D, et al. Population structure and antimicrobial resistance profiles of Streptococcus suis serotype 2 sequence type 25 strains. PLoS One. 2016;11(3):e0150908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obradovic MR, Segura M, Segales J, et al. Review of the speculative role of co-infections in Streptococcus suis-associated diseases in pigs. Vet Res. 2021;52(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng H, Lan R, Zheng X, et al. Comparative genomic hybridization identifies virulence differences in Streptococcus suis. PLoS One. 2014;9(2):e87866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong W, Zhu Y, Ma Y, et al. Multilocus sequence typing and virulence genotyping of Streptococcus suis serotype 9 isolates revealed high genetic and virulence diversity. FEMS Microbiol Lett. 2017;364(22):1–8. [DOI] [PubMed] [Google Scholar]
- 45.Palmieri C, Varaldo PE, Facinelli B.. Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front Microbiol. 2011;2:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Princivalli MS, Palmieri C, Magi G, et al. Genetic diversity of Streptococcus suis clinical isolates from pigs and humans in Italy (2003-2007). Euro Surveill. 2009;14(33):1–7. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C, Ning Y, Zhang Z, et al. In vitro antimicrobial susceptibility of Streptococcus suis strains isolated from clinically healthy sows in China. Vet Microbiol. 2008;131(3-4):386–392. [DOI] [PubMed] [Google Scholar]
- 48.Ye C, Bai X, Zhang J, et al. Spread of Streptococcus suis sequence type 7, China. Emerg Infect Dis. 2008;14(5):787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wisselink HJ, Veldman KT, Van den Eede C, et al. Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licensed in veterinary medicine. Vet Microbiol. 2006;113(1–2):73–82. [DOI] [PubMed] [Google Scholar]
- 50.Xu Z, Xie J, Peters BM, et al. Longitudinal surveillance on antibiogram of important gram-positive pathogens in southern China, 2001 to 2015. Microb Pathog. 2017;103:80–86. [DOI] [PubMed] [Google Scholar]
- 51.Seitz M, Valentin-Weigand P, Willenborg J.. Use of antibiotics and antimicrobial resistance in veterinary medicine as exemplified by the swine pathogen Streptococcus suis. Curr Top Microbiol Immunol. 2016;398:103–121. [DOI] [PubMed] [Google Scholar]
- 52.Huang K, Song Y, Zhang Q, et al. Characterisation of a novel integrative and conjugative element ICESsD9 carrying erm(B) and tet(O) resistance determinants in Streptococcus suis, and the distribution of ICESsD9-like elements in clinical isolates. J Global Antimicrob Resis. 2016;7:13–18. [DOI] [PubMed] [Google Scholar]
- 53.Huang J, Chen L, Li D, et al. Emergence of a vanG-carrying and multidrug resistant ICE in zoonotic pathogen Streptococccus suis. Vet Microbiol. 2018;222:109–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.