ABSTRACT
Autophagy, an evolutionally conserved cellular degradation process, plays critical roles in plant development and stress response. Despite the wealth of information on the vital role of autophagy in responses to environmental stresses, little is known about the regulation of autophagy. In this study, we demonstrated that spermidine (Spd), a kind of polyamine, was involved in the regulation of salt tolerance through activating the expression of ATG (autophagy-related) genes and the formation of autophagosomes in cucumber under salt stress. Furthermore, NADPH oxidase-derived apoplastic H2O2-mediated Spd-induced salt tolerance and autophagy. Exogenous Spd significantly increased the tolerance to salt stress and inhibited the accumulation and ubiquitination of insoluble proteins. Foliar application of Spd promoted the transcript levels of ATG genes and autophagosomes formation. Besides, Spd induced the expression of RBOH (respiratory burst oxidase homolog), and the accumulation of H2O2 both in leaves and roots. However, either pretreatment with dimethylthiourea (DMTU, an H2O2 scavenger) or diphenyleneiodonium chloride (DPI, an inhibitor of NADPH oxidase) reduced Spd-induced accumulation of apoplastic H2O2. Importantly, Spd-induced salt tolerance and autophagy were compromised when plants were pretreated with DMTU or DPI. Furthermore, the silencing of ATG4 and ATG7 reduced Spd-induced salt tolerance and autophagosomes formation. Taken together, these results revealed that RBOH-dependent H2O2 mediated the Spd-induced autophagy and salt tolerance in cucumber.
Abbreviations: Asat: light-saturated rate of CO2 assimilation; ATG: autophagy-related; DCF-DA: 2, 7-dichlorofluorescein diacetate; DMTU: dimethylthiourea; DPI: diphenyleneiodonium chloride; DW: dry weight; EL: electrolyte leakage; FW: fresh weight; Fv/Fm: the maximum quantum yield of photosystem II; GFP: green fluorescent protein; MDC: monodansylcadaverine; PDS: phytoene desaturase; PE: phosphatidylethanolamine; PLD: phospholipase D; RBOH: respiratory burst oxidase homolog; ROS: reactive oxygen species; SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis; SIN1: salt induced NAC1; Spd: spermidine; TOR: target of rapamycin; VIGS: virus-induced gene silencing.
KEYWORDS: Autophagy, cucumber, hydrogen peroxide, salt stress, spermidine
Introduction
Cucumber is widely cultivated all over the world, but during the process of cultivation, it often has to endure different environmental stresses, such as high temperature, chilling, salt, and drought stress. More than 800 million hectares of land worldwide suffer from salt stress [1,2]. Salt stress-induced injuries are more severe to cucumber due to its hypersensitivity to salt stress. Under salt stress, plants often absorb excessive salt ions, which result in ionic and osmotic stress and thereby inhibiting plant growth, development, and yield [3–5]. Therefore, it is necessary to understand the mechanism of salt tolerance and develop strategies to overcome the damage caused by salt stress.
Over the decades, plant growth regulators have gradually emerged as one of the key foci of people’s attention because of their important roles in plant growth, development and stress resistance. Spermidine (Spd), a kind of polyamine ubiquitous in plants, is used as a plant growth regulator and plays critical roles in plant responses to salt stress [6,7]. Salt stress significantly increases the activity of enzymes related to Spd biosynthesis and the content of Spd [8]. Exogenous application of Spd or endogenous manipulation of Spd through overexpression of SAMDC (S-adenosylmethionine decarboxylase), a key rate-limiting enzyme in Spd biosynthesis, alleviates the negative effects of salt stress by inhibiting chlorophyll degradation, maintaining the integrity of chloroplast structural, enhancing the photosynthetic rate and increasing the capacity of the enzymatic and non-enzymatic antioxidant system to mitigate oxidative damage [9–12]. Similarly, proteomics analysis revealed the abundance of proteins in photosynthesis, carbon metabolism, amino acid metabolism, signal transduction, and reactive oxygen species (ROS) homeostasis in Spd- and NaCl-treated plants is significantly greater than that in NaCl-treated plants [13,14]. These results indicate that Spd mainly improves the photosynthetic efficiency and antioxidant capacity to enhance plant salt tolerance. Recently, Spd has been shown to induce H2O2 for activating the antioxidant system [15], while the role of Spd-induced H2O2 in other signal pathways remains elusive.
Autophagy is an evolutionarily conserved self-degradation process in eukaryotes. Briefly, intracellular denatured or aggregated proteins and damaged organelles are engulfed by double-membrane phagophores that subsequently mature into autophagosomes; the autophagosome outer membrane fuses with the tonoplast to form single-membrane autophagic body, which is recycled in the vacuole by hydrolase in plants [16,17]. This process is performed by ATG (autophagy-related) genes [16,17]. Numerous studies have revealed that autophagy is vital for responses to abiotic and biotic stresses in plants [18–20]. Knockout or silencing ATG genes results in increased sensitivity to high temperatures attributed to the accumulation of insoluble proteins [21,22]. The expression of ATG genes and autophagosome formation can be induced by salt stress. In addition, salt sensitivity is significantly increased in ATG18a-silenced Arabidopsis, while ectopic overexpression of ATG3 from apple (Malus domestica) in Arabidopsis enhances its tolerance to salt stress [23,24]. These results suggest that autophagy plays a vital role in plant response to salt stress, but the regulation mechanisms of autophagy remain fragmentary. Studies revealing the mechanisms underlying autophagy regulation mainly focus on the TOR (target of rapamycin) kinase. It is well known that TOR negatively regulates autophagy in plants [25]. Moreover, autophagy is transcriptionally regulated by many transcription factors under environmental stresses. For instance, HSFA1A (class A heat shock factor 1A) binds to the promoters of ATG10 and ATG18f to activate their transcription under drought stress [18]. BZR1 (Brassinazole-resistant 1), the key positive regulator in the phytohormone brassinosteroid pathway, induces the transcription of ATG genes and formation of autophagosomes, thereby mediating responses to nitrogen starvation in tomato plants [20]. Furthermore, ROS also mediate the induction of autophagy [19,26]. Plants deficient in RBOH (respiratory burst oxidase homolog) are sensitive to submergence and compromised in hypoxia-induced autophagy [27], indicating that RBOH-dependent H2O2 is essential for triggering autophagy. Interestingly, autophagy is crucial for Spd-induced suppression of necrosis and enhanced longevity in animals [28,29]. However, little information is available concerning the Spd-induced regulation of autophagy in plants under adverse conditions. Here, we report that Spd plays a positive role in salt stress in cucumber plants through the induction of ATG genes and the formation of autophagosomes. In addition, RBOH-dependent H2O2 acts as a signal in Spd-induced autophagy to increase salt tolerance in cucumber plants.
Results
Exogenous Spd application increased salt tolerance in cucumber
To directly analyze the role of Spd in salt tolerance, we firstly compared the tolerance of cucumber plants to salt stress in the presence or absence of Spd. As shown in Figure 1A, there was no significant difference with or without the application of Spd in the absence of NaCl. Salt stress significantly inhibited plant growth, while the application of exogenous Spd attenuated the inhibition by salt stress (Figure 1A–C). The fresh weight (FW) and dry weight (DW) of the salt-stressed plants were significantly lower than that of the Spd-treated plants after 7 d of salt stress (Figure 1B,C). Furthermore, the electrolyte leakage (EL) value, an indicator of cell membrane damage, of the salt-stressed plants was two-fold of the Spd-treated plants after 7 d of salt stress (Figure 1D). The maximum quantum yield of photosystem II (Fv/Fm) that reflects the maximum light energy conversion efficiency of photosystem II was not significantly different in the presence or absence of Spd under normal growth conditions (Figure 1E). In contrast, the value of Fv/Fm in NaCl and NaCl+Spd decreased by 12.38% and 1.16%, respectively, under salt stress at 7 d in comparison to respective control plants (Figure 1E). Although salt stress dramatically reduced the light-saturated rate of CO2 assimilation (Asat) both in NaCl and NaCl+Spd plants, the value of Asat in NaCl+Spd was still higher than that in NaCl plants (Figure 1F). These results suggested that Spd had a positive effect on plant growth under salt stress.
Figure 1.
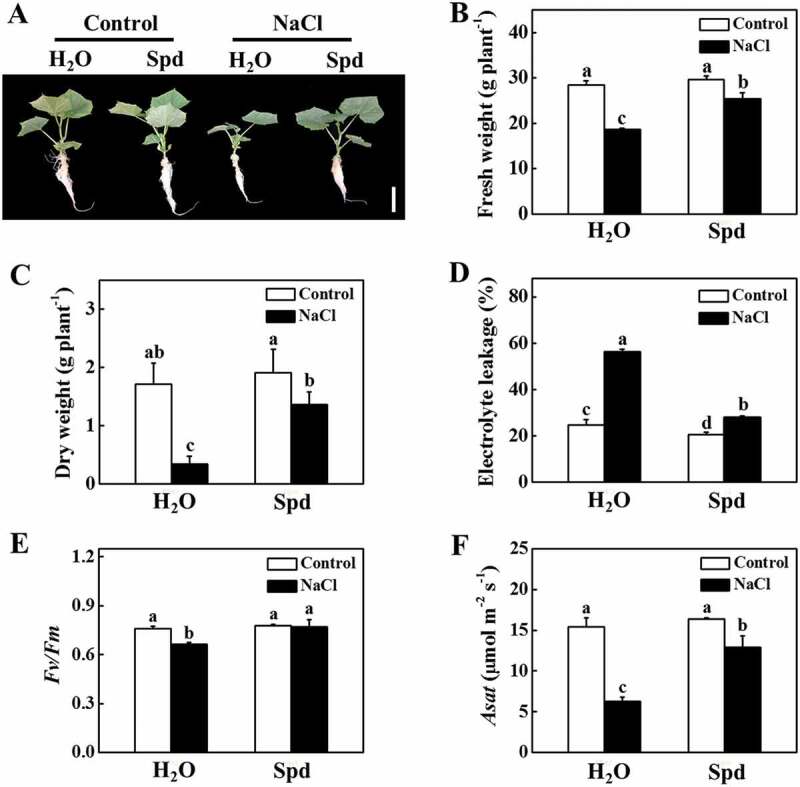
Functional analysis of spermidine (Spd) in response to salt stress in cucumber plants. (A) Increased cucumber salt tolerance by exogenous Spd. Bars: 5 cm. (B) Fresh weight (FW). (C) Dry weight (DW). (D) Electrolyte leakage (EL). (E) The maximum quantum yield of photosystem II (Fv/Fm). (F) The light-saturated rate of CO2 assimilation (Asat). Cucumber plants were exposed to 75 mM NaCl with or without 1 mM Spd treatment for 7 d, and the phenotype, FW, DW, EL, Fv/Fm, and Asat were measured. All data are presented as the means of 3 biological replicates (± SE). Means with the same letter did not significantly differ at P < 0.05, according to Tukey’s test. Three independent experiments were performed with similar results
Association of salt tolerance and the protein-ubiquitin conjugates in cucumber plants under salt stress
It has been demonstrated that abiotic stress causes the accumulation of misfolded and denatured proteins, which aggregate and are recognized by ubiquitin for degradation [30]. Due to the critical role of Spd in plant tolerance to abiotic stresses, we examined whether there was a decreased accumulation of ubiquitinated salt-induced protein aggregates in plants. As shown in Figure 2A, similar levels of insoluble protein were observed in the absence of NaCl in all of the plants. However, there was a distinct increase in the levels of insoluble protein aggregates under salt stress conditions (Figure 2A). The level of insoluble protein aggregates in NaCl+Spd plants was significantly lower than that in NaCl plants after 7 d of salt stress (Figure 2A). To test whether these insoluble proteins were ubiquitinated, we extracted the same amount of total proteins to separate to the soluble and insoluble proteins from plants at 7 d of salt stress (Fig. S1). The protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the anti-ubiquitin monoclonal antibody was used for ubiquitination analysis. As shown in Figure 2B, the levels of ubiquitinated proteins in soluble components were not significantly different between control and salt-stressed plants. While the levels of ubiquitinated proteins in insoluble components in Spd-treated plants were lower than the control plants under normal growth conditions (Figure 2B,C). Salt stress drastically induced the accumulation of ubiquitinated proteins in the insoluble proteins both in NaCl and NaCl+Spd plants, but the level of ubiquitinated proteins in NaCl plants was higher than that in NaCl+Spd plants (Figure 2B,C).
Figure 2.
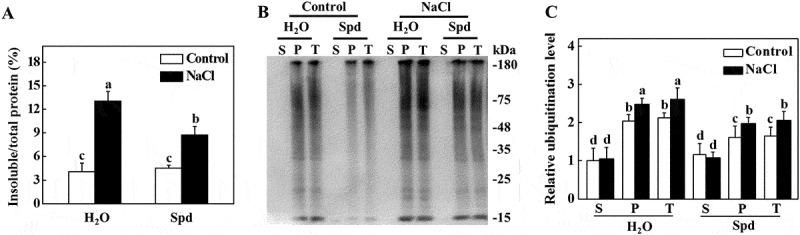
Effects of spermidine (Spd) on the accumulation and ubiquitination of insoluble proteins in the roots of cucumber under salt stress. (A) Increase of insoluble proteins in the roots of cucumber under salt stress. Root tissues from cucumber collected after 7 d of salt stress for the preparation of total, soluble, and insoluble proteins. Total proteins in the starting homogenates and insoluble proteins in the last pellet were determined for the calculation of the percentages of insoluble proteins to total proteins. (B) Ubiquitination of insoluble protein aggregates in cucumber plants collected at 7 d under salt stress. Proteins from the starting homogenates (T), first supernatant fractions (S), and last pellet fractions (P) were subjected to SDS-PAGE and probed with an anti-ubiquitin monoclonal antibody. (C) Relative ubiquitination level normalized to the level of the S of control plants in (B) (n = 3). The level of ubiquitination in (B) was quantified using ImageJ software to calculate the ubiquitination level relative to the S of control plants, which was set to 1. The data are presented as the means of 3 biological replicates (± SE). Means with the same letter did not significantly differ at P < 0.05, according to Tukey’s test. Three independent experiments were performed with similar results
Spd induced the expression of ATG genes and formation of autophagosomes under salt stress
To investigate whether autophagy is involved in Spd-induced salt tolerance, we first examined the expression of 14 ATG genes identified from the Cucumber Genomics Network (http://cucurbitgenomics.org/organism/20) in response to salt stress. As shown in Fig. S2, the transcript levels of ATG genes were slightly increased with exogenous Spd application at 24 h after the treatment. The ATG gene expression was highly elevated at 24 h under salt stress with or without Spd, but the expression levels were more elevated in Spd-treated plants (Fig. S2). To further analyze the effect of Spd on salt-induced autophagy, we used monodansylcadaverine (MDC), an autofluorescent dye, as a probe to detect autophagosomes in plants [31]. Low numbers of punctate fluorescent signals were observed in plants in the absence of NaCl (Figure 3A,B). However, salt stress significantly induced the accumulation of MDC-labeled puncta (Figure 3A,B). The numbers of punctate fluorescence signals in NaCl plants were significantly lower than that in NaCl+Spd plants (Figure 3A,B).
Figure 3.
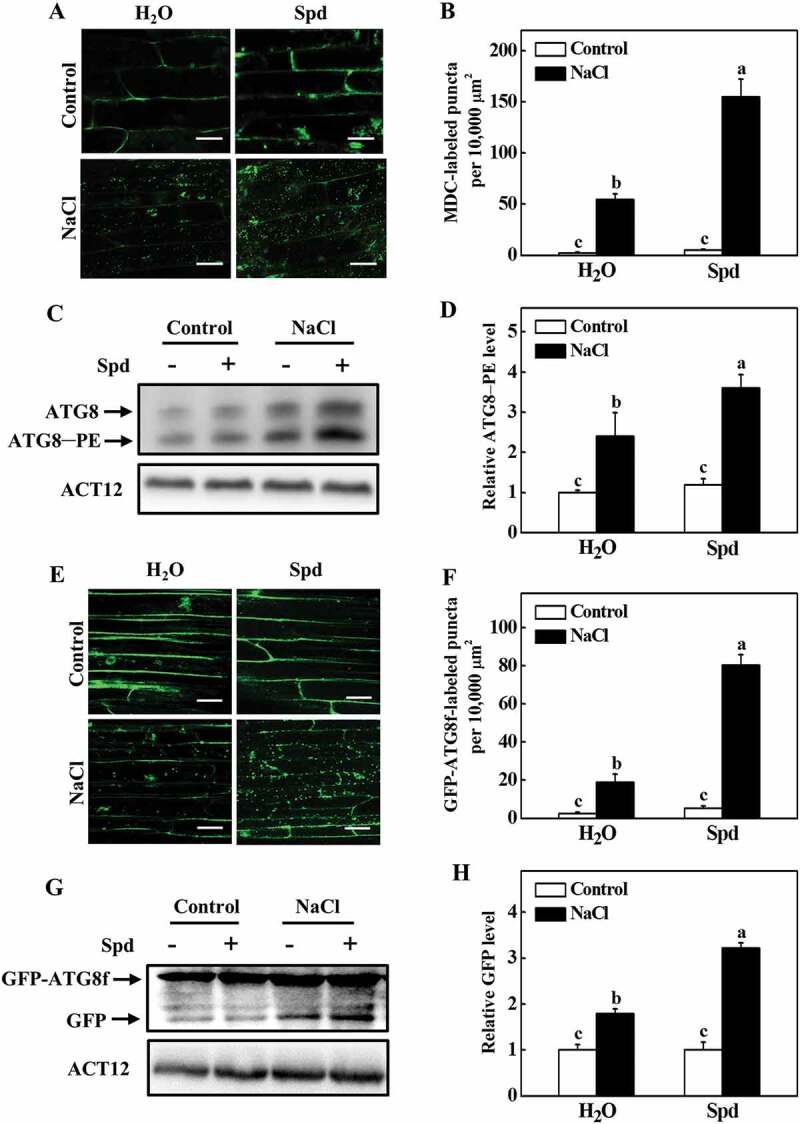
Effects of spermidine (Spd) on the accumulation of autophagosomes in cucumber root under salt stress. (A) MDC-labeled puncta in cucumber roots. Cucumber plants were exposed to 75 mM NaCl stress with or without 1 mM Spd, and the roots were MDC-stained and visualized at 24 h by fluorescence confocal microscopy. MDC-labeled structures are shown as green signals. Bars: 20 μm. (B) Numbers of punctate MDC fluorescence spots per 10,000 μm2 in (A). (C) ATG8 protein levels in the roots of cucumber. ATG8–PE and ATG8 are the lipidated and nonlipidated forms of ATG8, respectively. ACT12 was used as a loading control for the immunoblotting analysis. (D) Relative ATG8–PE levels in (C) (n = 3). (E) GFP fluorescence in the cells of GFP-ATG8f transgenic hair roots under salt stress at 24 h. Bars: 20 μm. (F) Numbers of GFP-ATG8f-labeled puncta per 10,000 μm2 in (E). (G) Immunoblotting analysis showing the processing of GFP-ATG8f under salt stress. Hair roots were collected at 24 h after the treatment, and anti-GFP antibodies were used for immunoblotting. The GFP-ATG8f fusion and free GFP form are indicated on the left. ACT12 was used as a loading control for the immunoblotting analysis. (H) Relative free GFP levels in (G) (n = 3). Means with the same letter did not significantly differ at P < 0.05, according to Tukey’s test. Three independent experiments were performed with similar results
To confirm these results, we used immunoblotting to analyze the formation of ATG8–phosphatidylethanolamine (PE) conjugates to detect autophagosomes by an anti-ATG8 antibody [32]. We first used Arabidopsis as the positive control to confirm whether the ATG8 antibody could specifically recognize the cucumber proteins. The proteins of Arabidopsis and cucumber were separated by SDS-PAGE with urea, and the immunoblotting analysis was performed with the anti-ATG8 antibody. The faster and slower migrating protein species were detected both in Arabidopsis and cucumber (Fig. S3A). To determine the lipidation status of the ATG8, protein crude extracts from salt-stressed plants were used to collect the soluble and membrane fractions and then assayed these by immunoblotting analysis with or without PLD (phospholipase D) treatment. ATG8 of the salt-stressed plants showed a doublet of faster and slower migrating protein species and the faster migrating species were sensitive to PLD (Fig. S3), indicating that they are lipidated ATG8. Under normal conditions, we could slightly detect the ATG8–PE band in cucumber plants (Figure 3C). Faster migration of the ATG8–PE bands was observed in plants after 24 h of salt stress (Figure 3C). By contrast, the band of ATG8–PE was more abundant in Spd-treated plants after salt stress (Figure 3C,D).
To further confirm these results, we used green fluorescent protein (GFP)-tagged ATG8f (GFP-ATG8f) as a marker to examine the formation of autophagosomes under salt stress [31,33]. Cucumber seedlings expressing GFP-ATG8f in roots induced by Agrobacterium rhizogenes K599 were exposed to salt stress with or without application of Spd, and then the GFP fluorescence was detected by confocal microscopy at 24 h. A few numbers of spherical or punctate structures labeled by GFP-ATG8f was observed in all of the plants under control conditions (Figure 3E). The numbers of GFP-ATG8f-labeled structures significantly increased both in NaCl and NaCl+Spd plants but were more prominent in NaCl+Spd plants (Figure 3E,F). When plants suffer from environmental stresses, autophagosomes are formed by lipidation of ATG8 protein with PE and transported into vacuoles by fusion with vacuolar membranes to form autophagic bodies, during which GFP-ATG8 is degraded and released free relatively stable GFP. Therefore, the accumulation level of free GFP reflects the activity of autophagy [34]. As shown in Figure 3G, salt stress enhanced the degradation of GFP-ATG8f, as observed by the accumulation of free GFP. In contrast, the abundance of free GFP in NaCl+Spd plants was more profound than that in NaCl plants (Figure 3G,H). Taken together, these results suggested that Spd was involved in both salt-induced expressions of ATG genes and formation of autophagosomes.
Spd induced the transcription of RBOH and accumulation of H2O2
Apoplastic H2O2 can act as a molecular signal to trigger autophagy, which plays a critical role in response to salt stress [19,23]. Therefore, we examined the impacts of Spd on the transcript of RBOH, encoding NADPH oxidase and production of apoplastic H2O2, and H2O2 accumulation in cucumber plants. The transcript levels of RBOH increased both in the leaves and roots of plants with Spd treatment (Fig. S4A and S4B). The accumulation of H2O2 was measured by a spectrophotometry-based method and 2, 7-dichlorofluorescein diacetate (DCF-DA) staining. The content of H2O2 in the leaves and roots of salt-treated plants was induced (Figure 4A,B). Nevertheless, salt plus Spd (NaCl+Spd)-induced elevation of H2O2 content was more profound compared with only salt (NaCl)-stressed plants, especially at 12 h (Figure 4A,B). DCF-DA staining showed that the salt-induced fluorescence signal was greatly increased in stems and roots of NaCl- and NaCl+Spd-treated plants, while the effect was more profound in NaCl+Spd-treated plants (Figure 4C–F), which was in agreement with the results of H2O2 content. Interestingly, subcellular localization analysis revealed that H2O2 predominantly accumulated in the apoplast of the root cells (Fig. S4C). Taken together, these results showed that Spd induced the expression of RBOH and the accumulation of H2O2 in the leaves and roots of cucumber under salt stress.
Figure 4.
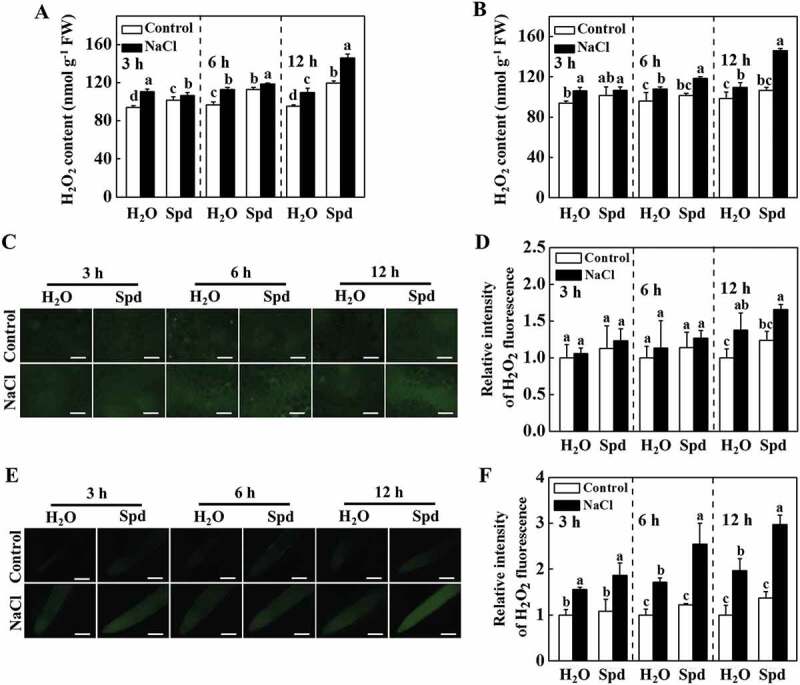
Effects of spermidine (Spd) on the accumulation of H2O2 in cucumber under salt stress. (A) H2O2 content in leaves. (B) H2O2 content in roots. (C) DCF-DA staining of H2O2 accumulation in stems. Bars: 300 μm. (D) Relative intensity of H2O2 fluorescence in (C). (E) DCF-DA staining of H2O2 accumulation in roots. Bars: 300 μm. (F) Relative intensity of H2O2 fluorescence in (E). Cucumber leaves, stems and roots were collected at 3 h, 6 h, and 12 h under 75 mM NaCl with or without 1 mM Spd treatment, and the accumulation of H2O2 was measured. All data are presented as the means of 3 biological replicates (± SE). Means with the same letter did not significantly differ at P < 0.05, according to Tukey’s test. Three independent experiments were performed with similar results. FW, fresh weight
Involvement of apoplastic H2O2 in the Spd-induced salt tolerance and autophagosomes formation
In order to evaluate the role of H2O2 in salt tolerance, we first detected the effects of foliar treatment with different concentrations of H2O2 on plant salt tolerance. Foliar application of a low concentration of H2O2 (1 mM) slightly increased salt tolerance, while the treatment with a moderate concentration of H2O2 (10 mM) markedly enhanced salt tolerance accompanied by higher levels of FW, DW, Fv/Fm, and Asat (Fig. S5A-D). By contrast, the Asat of plants showed no significant changes when treated with a high concentration of H2O2 (20 mM) compared with the only salt-treated plants (Fig. S5D). The foliar application of 10 mM H2O2 increased the number of MDC-labeled puncta and GFP-ATG8f-labeled structures in the present or absence of NaCl, but these effects were more obvious under salt stress (Fig. S5E-H). Thus, plants treated with the appropriate concentration of H2O2 could significantly attenuate salt-induced damage and trigger the formation of autophagosomes.
To investigate whether apoplastic H2O2 accumulation was involved in Spd-induced salt tolerance, we analyzed the effects of dimethylthiourea (DMTU; an H2O2 scavenger) and diphenyleneiodonium chloride (DPI; an inhibitor of NADPH oxidase) on Spd-mediated salt tolerance, the expression of ATG genes and the formation of autophagosomes. Significantly, both Spd and H2O2 effectively attenuated salt stress-induced the increase of EL and the decline of FW, DW, Fv/Fm, and Asat, but these effects were compromised with the pretreatment with either DMTU or DPI (Figure 5, S6A and S6B). Similarly, foliar application of Spd induced the accumulation of H2O2 in leaves, stems, and roots as reflected by the measurement of H2O2 content, DCF-DA, and CeCl3 staining under salt stress, and the effects were mostly inhibited by pretreatment with DMTU or DPI (Figure 6 and S6C). These results suggested that Spd-induced salt tolerance and H2O2 accumulation depended on NADPH oxidase.
Figure 5.
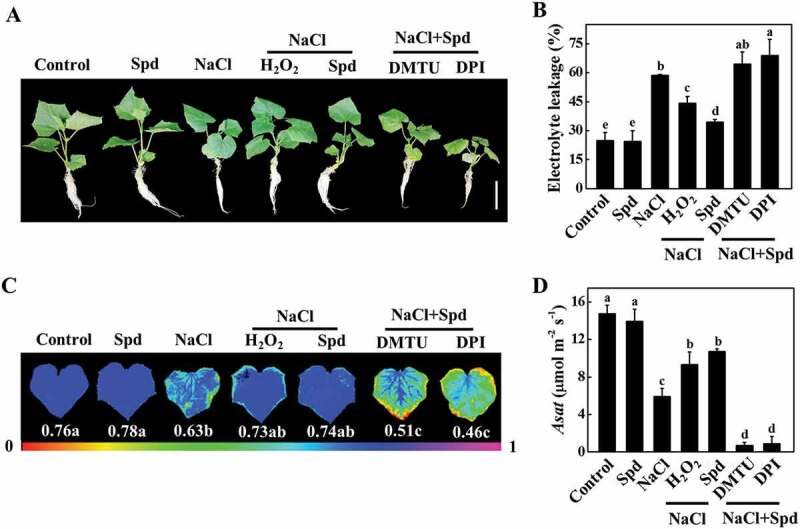
Functional analysis of spermidine (Spd)-induced H2O2 in response to salt stress in cucumber. (A) Decreased cucumber salt tolerance by the treatment with H2O2 scavenge or inhibitor. Bars: 5 cm. (B) Electrolyte leakage (EL). (C) The maximum quantum yield of photosystem II (Fv/Fm). (D) The light-saturated rate of CO2 assimilation (Asat). Cucumber plants were pretreated with 5 mM DMTU or 0.1 mM DPI for 3 h, then exposed to 75 mM NaCl with 1 mM Spd or 10 mM H2O2 for 7 d, and the phenotype, EL, Fv/Fm, and Asat were measured. Results are presented as the means of 3 biological replicates (± SE). Means with the same letter did not significantly differ at P < 0.05, according to Tukey’s test. Three independent experiments were performed with similar results
Figure 6.
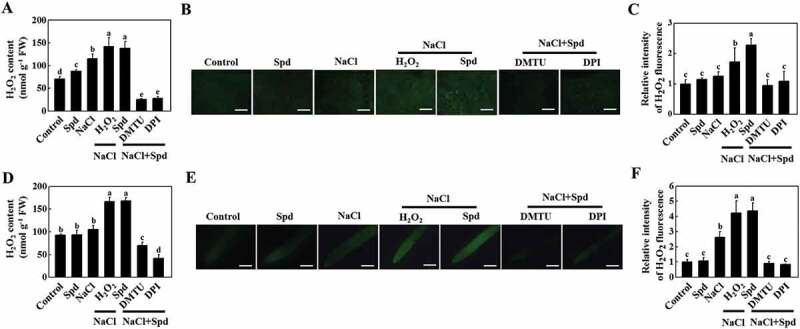
NADPH oxidase-mediated spermidine (Spd)-induced accumulation of H2O2 in cucumber. (A) H2O2 content in leaves. (B) DCF-DA staining of H2O2 accumulation in stems. Bars: 300 μm. (C) Relative intensity of H2O2 fluorescence in (B). (D) H2O2 content in roots. (E) DCF-DA staining of H2O2 accumulation in roots. Bars: 300 μm. (F) Relative intensity of H2O2 fluorescence in (E). Cucumber plants were pretreated with 5 mM DMTU or 0.1 mM DPI for 3 h, then exposed to 75 mM NaCl with 1 mM Spd or 10 mM H2O2. After the treatment for 12 h, the leaves, stems, and roots were collected for the detection of H2O2 content. Results are presented as the means of 3 biological replicates (± SE). Means with the same letter did not significantly differ at P < 0.05, according to Tukey’s test. Three independent experiments were performed with similar results
To verify the role of apoplastic H2O2 in Spd-induced autophagy, we detected the expression of ATG genes and the formation of autophagosomes of plants foliar application of DMTU or DPI combined with Spd at 24 h under salt stress. As shown in Fig. S7, the expression of ATG genes was induced by salt and Spd, but these effects were completely prevented by pretreatment with DMTU or DPI. Either foliar application of H2O2 or Spd drastically increased the number of MDC-stained and GFP-ATG8f-labeled punctate fluorescence signals under salt stress compared with the control plants (Figure 7A–C). However, the number of punctate fluorescence signals of MDC staining and GFP-ATG8f was sharply decreased in the plants treated with Spd+DMTU or Spd+DPI in the presence of NaCl compared with the NaCl treatment (Figure 7A–C). Similarly, the immunoblotting analysis showed that Spd significantly induced the accumulation of ATG8–PE and free GFP in the presence of NaCl, but completely blocked in the plants that were pretreated with DMTU or DPI (Figure 7D–G). Therefore, RBOH-derived H2O2 was involved in Spd-induced autophagy.
Figure 7.
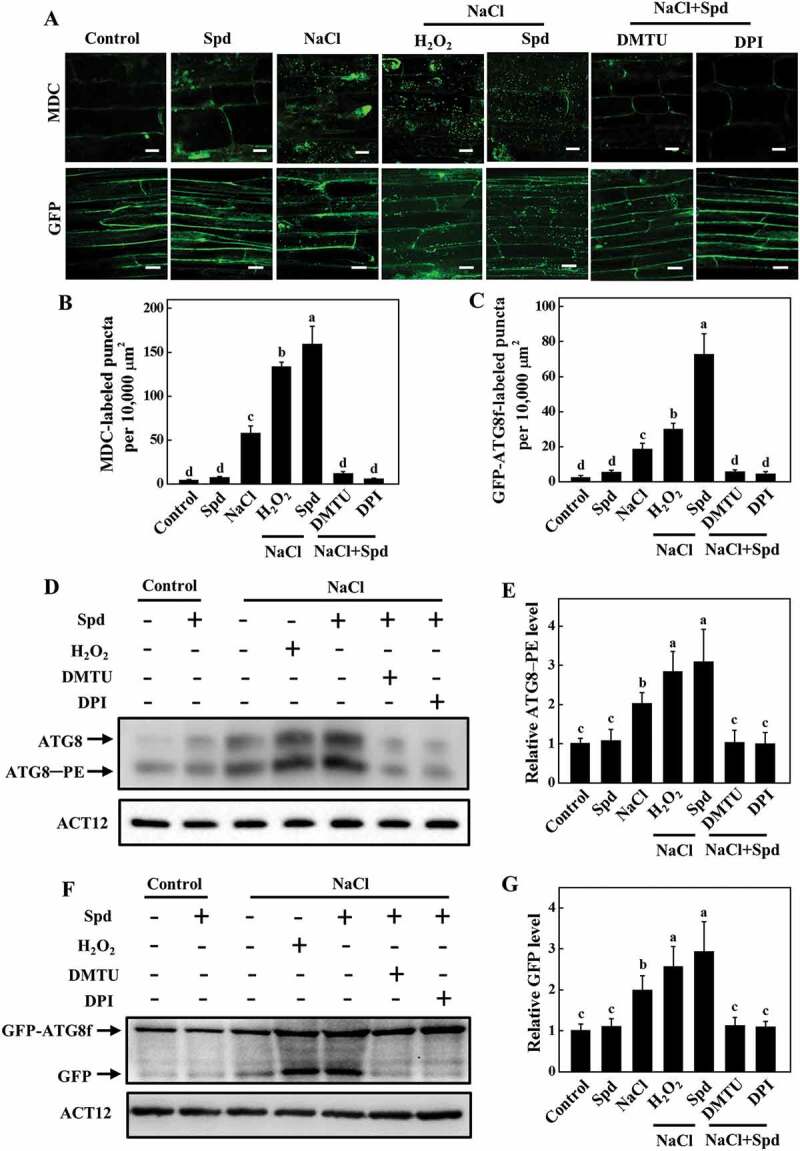
Involvement of H2O2 in spermidine (Spd)-induced accumulation of autophagosomes in the roots of cucumber under salt stress. (A) Visualization of the accumulation of MDC- (upper) and GFP-ATG8f-labeled puncta (lower) in cucumber roots. Cucumber plants were pretreated with 5 mM DMTU or 0.1 mM DPI for 3 h, then exposed to 75 mM NaCl with 1 mM Spd or 10 mM H2O2. After salt treatment for 24 h, roots were MDC-stained or directly visualized at 24 h by fluorescence confocal microscopy. Bars: 20 μm. (B) The number of MDC-labeled puncta per 10,000 μm2 in (A). (C) Numbers of GFP-ATG8f-labeled puncta per 10,000 μm2 in (A). (D) ATG8 protein levels in the roots of cucumber. ATG8–PE and ATG8 are the lipidated and nonlipidated forms of ATG8, respectively. ACT12 was used as a loading control for the immunoblotting analysis. (E) Relative ATG8–PE levels in (D). (F) Immunoblotting analysis showing the processing of GFP-ATG8f under salt stress. Hair roots were collected at 24 h after treatment and anti-GFP antibodies were used for immunoblotting. The GFP-ATG8f fusion and free GFP form are indicated on the left. ACT12 was used as a loading control for the immunoblotting analysis. (G) Relative free GFP levels in (F). Means with the same letter did not significantly differ at P < 0.05, according to Tukey’s test. Three independent experiments were performed with similar results
Functional analysis of autophagy in Spd-induced salt tolerance
To investigate the role of autophagy in Spd-induced salt tolerance, ATG4 and ATG7 genes were silenced using a cucumber green mottle mosaic virus vector (pV190), which has been successfully used for virus-induced gene silencing (VIGS) in cucurbit plants including cucumber [35]. Silencing of PDS (phytoene desaturase), a key gene in the carotenoid biosynthesis pathway, showed the photobleaching phenotypes in cucumber, and the expression of ATG4 and ATG7 decreased by approximately 60 to 70% in their own silenced plants in comparison to pV190 control plants (Fig. S8A and S8B). Although the growth of pV190, ATG4- and ATG7-silenced plants was significantly inhibited under salt stress, the FW and DW of pV190 plants were still higher than that of the ATG4- and ATG7-silenced plants (Fig. S8C-E). Furthermore, we observed a lower number of MDC-labeled puncta in all of the plants in the absence of NaCl (Figure 8A,B). The number of punctate fluorescent signals significantly increased in pV190 plants under salt stress, and the application of Spd further promoted the accumulation of punctate fluorescent signals (Figure 8A,B). However, the number of punctate fluorescent signals in ATG4- and ATG7-silenced plants dramatically lower than that in pV190 plants under salt stress with or without Spd treatment (Figure 8A,B). Similarly, the abundance of ATG8–PE in ATG4- and ATG7-silenced plants was significantly lower than that observed in pV190 plants under salt stress (Figure 8C,D). Therefore, autophagy played a critical role in Spd-induced salt tolerance and the formation of autophagosomes.
Figure 8.
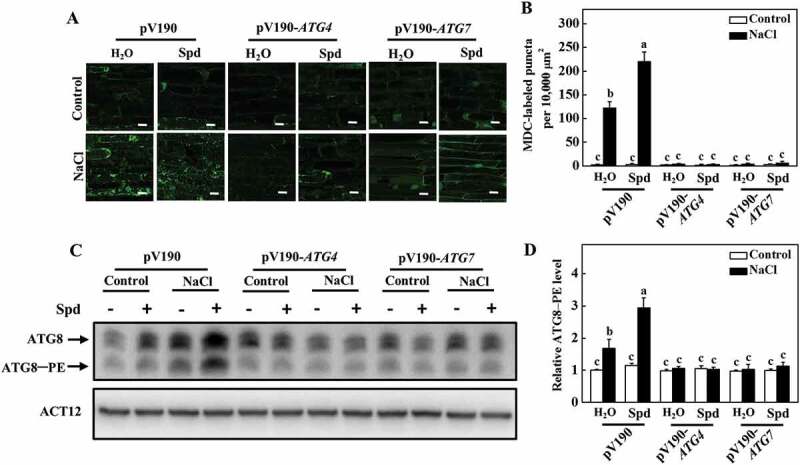
Effects of ATG4 and ATG7 on spermidine (Spd)-induced the formation of autophagosomes. (A) MDC-labeled puncta in the roots of pV190, pV190-ATG4, and pV190-ATG7 plants. Cucumber plants were exposed to 75 mM NaCl stress with or without 1 mM Spd, and the roots were MDC-stained and visualized at 24 h by fluorescence confocal microscopy. MDC-labeled structures are shown as green signals. Bars: 20 μm. (B) The number of MDC-labeled puncta per 10,000 μm2 in (A). (C) ATG8 protein levels in the roots of pV190, pV190-ATG4, and pV190-ATG7 plants. ATG8–PE and ATG8 are the lipidated and nonlipidated forms of ATG8, respectively. ACT12 was used as a loading control for the immunoblotting analysis. (D) Relative ATG8–PE levels in (C). Means with the same letter did not significantly differ at P < 0.05, according to Tukey’s test. Three independent experiments were performed with similar results
Discussion
Salt stress has an adverse effect on plant growth and development, which eventually minimizes crop yield [36,37]. Numerous studies have revealed that Spd, an important plant growth regulator, is involved in response to salt stress in plants [38,39]. Furthermore, autophagy also plays a vital role in plant response to abiotic stresses [19,40]. However, the relationship between Spd and autophagy in plants is unknown. In this study, we demonstrated that Spd increased salt stress tolerance by upregulating ATG genes and the formation of autophagosomes. In addition, RBOH-mediated H2O2 was involved in Spd-induced autophagy and salt tolerance. These results shed new light on our understanding of the action mechanism of Spd in improving plant stress tolerance.
Under normal growth conditions, the content of Spd in plants was lower, but salt stress significantly promoted the accumulation of Spd in cucumber and other plants [8,12]. Since Spd is intrinsically present in plants and is essential for plants’ adaptation to adverse conditions, it has been widely used as an environmentally friendly plant growth regulator to alleviate various abiotic stress [6,41]. Salt stress significantly inhibits plant growth, while exogenous Spd promotes the expression of genes relating to antioxidant enzymes, increases the activity of antioxidant enzymes, maintains the dynamic balance of ROS, and improves the tolerance to salt in plants [9,42–44]. Furthermore, the application of Spd mitigates salt-induced decline of photosynthetic rate through inhibiting chlorophyll degradation, elevating non-photochemical energy loss, and xanthophyll cycle [45,46]. In agreement with previous studies, we found that salt stress dramatically inhibited plant growth, while the treatment with Spd significantly ameliorated the adverse effect of salt stress by maintaining higher levels of FW, DW, Fv/Fm, and Asat (Figure 1). These results suggested that Spd might enhance salt tolerance of plants by improving their photosynthetic capacity and antioxidant capacity.
Peptide synthesis and degradation of proteins regulate virtually all aspects of plant growth and development [47]. Abiotic stresses, such as salt, can cause damage to plant proteins, resulting in protein denaturation and aggregation [18,21]. A large number of reports indicate that stress-caused denatured proteins cannot be restored to their original state, and thus they are recognized, ubiquitinated, and recycled [48,49]. Ubiquitination is an enzymatic protein degradation process that degrades denatured and damaged proteins and involves three consecutive catalysis steps mediated by E1, E2, and E3 enzymes [50,51]. Here, we discovered a remarkable increase in insoluble protein content in salt-stressed plants compared to control plants (Figure 2A). Furthermore, the level of ubiquitination of the insoluble proteins in NaCl-treated plants significantly increased in comparison to the control plants (Figure 2B,C). However, it dramatically decreased in the plants treated with Spd (Figure 2B,C), indicating that Spd alleviated the salt-induced ubiquitinated protein aggregates and accumulation.
The ubiquitin-proteasome system and autophagy are the two major intracellular protein quality control pathways that are responsible for cellular proteostasis by ensuring the timely degradation of misfolded or damaged proteins [52]. However, the ubiquitin-proteasome system fails to degrade the polymeric aggregated proteins due to its narrow proteasome entrance channel [18,53]. To maintain protein homeostasis, these protein aggregates are immediately redirected to autophagy for degradation. Numerous studies have demonstrated that autophagy is a major pathway for the degradation of protein aggregates [18,53]. Here, we found that salt stress increased the expression of ATG genes (Fig. S2) and the formation of autophagosomes in cucumber plants (Figure 3). In addition, the decline of accumulation of ubiquitinated insoluble proteins in Spd-induced salt tolerance was associated with the higher activity of autophagy. These results suggest that the induction of autophagy is crucial for Spd-induced salt tolerance in cucumber plants. Interestingly, feeding Spd in animals, such as humans, mice, nematodes, and flies, triggers autophagy by inhibition of several acetyltransferases, resulting in reducing the occurrence of disease and extended lifespan [54,55]. These results indicate that Spd could induce autophagy both in plants and animals.
Many developmental and physiological processes are mediated by H2O2, a critical signaling molecule with relatively high stability and diffusion capacity through the membranes to different cellular compartments [56,57]. Numerous studies have suggested that H2O2 plays an important role in acclimation-induced salt tolerance. A recent report showed that a NAC transcription factor, GmSIN1 (salt induced NAC1), is rapidly induced by salt stress in soybean, and root growth and salt tolerance are enhanced in GmSIN1-overexpressing plants, while silencing GmSIN1 attenuates the tolerance to salt stress in soybean plants [58]. Further analysis demonstrated that GmSIN1 directly regulates the expression of GmNCED3s and GmRBOHBs, promoting the accumulation of abscisic acid and H2O2, which further induce the expression of GmSIN1 and amplify the initial salt stress signals [58]. Strikingly, RBOHs-dependent H2O2 mediates salt tolerance by improving the accumulation of proline, Na+/K+ homeostasis, and early stomatal closure [59–62]. Furthermore, Spd-induced H2O2 acts as a molecular signal that increases the activity of antioxidant enzymes and reduces salt stress-induced damage, leading to improved plant growth and enhanced tolerance to salt in cucumber [15]. In the present study, treatment with Spd resulted in the elevated expression of RBOH and the accumulation of H2O2, while inhibition of RBOH-dependent H2O2 signaling completely abolished Spd-mediated salt tolerance (Figures 4 and 5). Taken together, the RBOH-dependent production of H2O2 was critical for the Spd-induced tolerance to salt. On the contrary, a large number of studies revealed that both the exogenous application of Spd and endogenous Spd manipulation by overexpression of the genes related to Spd biosynthesis could enhance the activity of the antioxidant enzyme to remove the excessive H2O2, thereby improving salt tolerance [42,63]. This implies that Spd might have a dual role in regulating the homeostasis of H2O2 under salt stress.
Under abiotic stresses, normal physiological metabolism of plants is impaired and a large number of denatured proteins are accumulated [18,20,21]. It is well known that autophagy is essential for the degradation of denatured proteins and stress tolerance [20,64,65]. Interestingly, it is to be noted that H2O2 acts as the signal molecule that mediates the induction of autophagy in plants as well as in other organisms [66]. For instance, under nutrient starvation and salt stress, the induction of autophagy could be blocked by NADPH oxidase inhibitors [23]. The production of ROS by NADPH oxidases is necessary for the activation of autophagy and subsequent plant tolerance to submergence [27]. Furthermore, abscisic acid could contribute to the establishment of autophagy by the activation of RBOH [67]. In animal cells, some reagents, such as ABT-737, tanshinone I, and ketamine, induce autophagy via ROS-dependent pathway [68–70]. In this study, Spd treatment induced the upregulation of ATG genes and autophagosomes formation, which were accompanied by the increase of H2O2 (Figures 6, 7 and S7). However, pretreatment with DMTU or DPI prevented the accumulation of H2O2 and compromised the Spd-induced expression of ATG genes and the formation of autophagosomes under salt stress in cucumber plants (Figures 6, 7 and S7). In addition, the silencing of ATG4 and ATG7 compromised Spd-induced salt tolerance and autophagosomes formation (Figure 8 and S8C-E). Thus, H2O2 likely acts as a secondary messenger that mediates Spd-induced autophagy to enhance salt tolerance in cucumber.
In conclusion, we have provided a comprehensive analysis of Spd-induced autophagy and salt tolerance mechanism in cucumber (Figure 9). When cucumber plants were treated with exogenous Spd, the expression of RBOH was induced, which promoted the accumulation of H2O2 both in the leaves and roots. Then, H2O2 acted as a signal that stimulated the expression of ATG genes and the formation of autophagosomes, which engulfed salt stress-damaged proteins or protein aggregates for further reuse, resulting in increased plant tolerance to salt stress. To the best of our knowledge, this is the first study that systematically illustrated the mechanism of Spd-induced regulation of autophagy under salt stress in cucumber plants.
Figure 9.
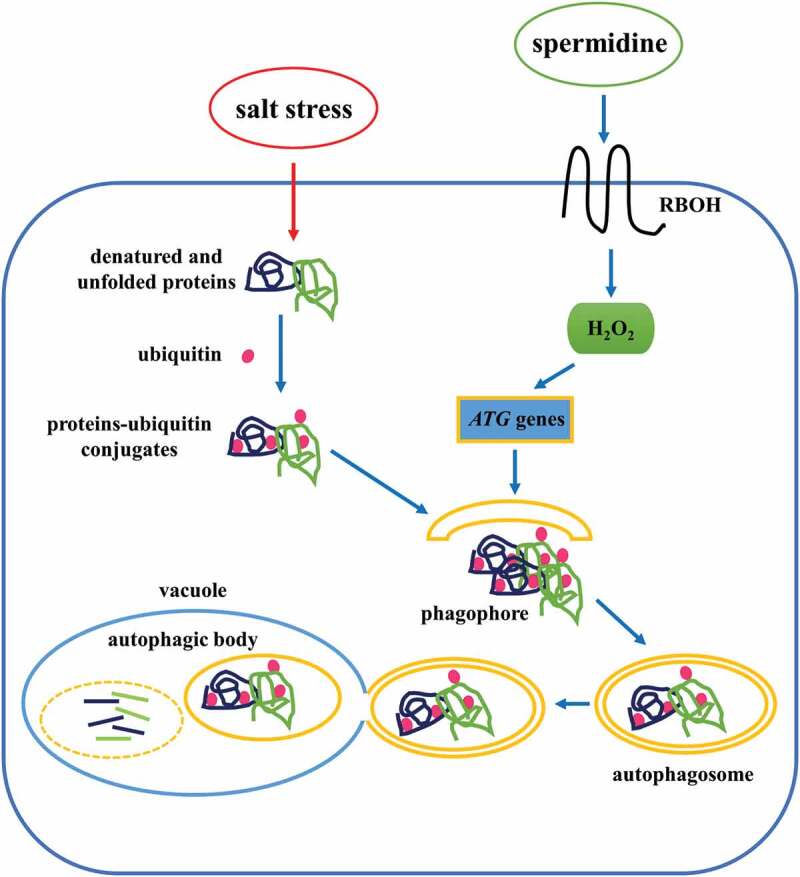
A proposed model for the induction of autophagy by spermidine (Spd) in cucumber under salt stress. Application of Spd induced the expression of RBOH to accumulate H2O2, which acted as a signal to activate autophagy. The increase in autophagy resulted in promoting the degradation of insoluble ubiquitinated protein aggregates to enhance salt tolerance
Materials and Methods
Plant materials and experimental design
Cucumber (Cucumis sativus L. cv. Jinchun No. 4) seeds were purchased from Nanjing Jinfeng Seedling Co., LTD (HGL0024) and used in all experiments. Seeds were germinated and grown in the dark for 24 h at 28 ± 1°C. The germinated seeds were then sown in plastic trays (41 × 41 × 5 cm) covered with a mixture of peat (Jiangsu Xingnong Substrate Technology Co., LTD, A-011) and vermiculite (Jiangsu Xingnong Substrate Technology Co., LTD, A-012) (2:1, v:v). The growth conditions were as follows: a 14/10 (light/dark) photoperiod, 25/18°C (day/night), 600 µmol m−2 s−1 photosynthetic photon flux density, and 75–80% relative humidity.
When the second leaves were fully expanded, the cucumber seedlings were transferred into plastic tanks filled with half-strength Hoagland solution (Shanghai YuanMu Biological Technology Co., LTD, R18965), with 12 plants per tank. For salt stress, 75 mM NaCl was added to the nutrient solution when the third leaves of cucumber were fully expanded. For the treatment with Spd, cucumber plants were sprayed with 1 mM Spd (MP Biomedicals, 152068) every 2 d, and the control plants were sprayed with an equal amount of distilled H2O. To investigate the function of H2O2 in salt stress, different concentrations (1, 10, and 20 mM) of H2O2 (Sinopharm Chemical Reagent, 10011218) were sprayed on the leaves every 2 d. To test the role of H2O2 in Spd-induced salt tolerance, cucumber leaves were pretreated with 0.1 mM DPI (Sigma-Aldrich, D2926) or 5 mM DMTU (Sigma-Aldrich, D188700) for 3 h, and then the plants were sprayed with 1 mM Spd or 10 mM H2O2. The nutrient solution was replaced every 3 d and all experiments were performed with three replicates. Leaf, stem, and root samples were harvested at 3 h, 6 h, and 12 h after salt treatment to measure H2O2 content. The autophagic activity was detected at 24 h after the treatment.
Generation of cucumber with transgenic roots
For generating transgenic cucumber roots overexpressing ATG8f, the full-length coding sequences of GFP and tomato ATG8f were amplified with specific primers (Table S1) by overlapping PCR and ligated to plant transformation vector pFGC1008 (donated by Dr. Jie Zhou of Zhejiang University). The resulting plasmid was transformed into Agrobacterium rhizogenes strains K599 (TOLOBIO, CC96315). Transgenic cucumber hair roots were obtained as previously described [71]. The hypocotyls of 3–5 d old aseptic cucumber seedlings were infected with Agrobacterium rhizogenes, and then grown in the tissue culture bottles filled with 1/2 Murashige & Skoog solid medium (Qingdao Hope Bio-Technology Co., LTD, HB8469-6). When the hair roots developed well, the original roots were cut to obtain the plants overexpressing GFP-ATG8f and transferred them into the nutrient solution for further treatment.
Determination of growth parameters, photosynthetic rate, chlorophyll fluorescence, and electrolyte leakage
For measuring the FW and DW of cucumber plants, the plants at 7 d of salt stress were washed with distilled water and dried with absorbent paper. The FW was measured by electronic balance (Sartorius, Goettingen, Germany). Then, the plants were enclosed in the envelopes and placed in an oven (Shanghai Yiheng Scientific Instrument Co., LTD, China) at 105°C for 30 min. Afterward, the oven temperature was adjusted to 75°C to obtain a constant DW.
After salt stress for 7 d, the Asat was measured with the portable photosynthesis system (LI-6400; Li-COR, Lincoln, NE, USA), maintaining the CO2 concentration at 380 μmol mol−1 and photosynthetic photon flux density at 1000 μmol m−2 s−1.
Cucumber plants were dark-adapted for 30 min to measure the Fv/Fm with the portable fluorometer (PAM-2100, Walz, Effeltrich, Germany) as previously described [72]. The value of EL was measured after salt treatment for 7 d, according to previously described [73].
Total RNA extraction and gene expression analysis
Total RNA was isolated from cucumber roots using RNAsimple Total RNA Kit (Tiangen, DP419) according to the manufacturer’s instruction. Total RNA (1 μg) was used to reverse transcribed to cDNA using the HiScript II Q RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme, R223-01). The quantitative real-time PCR (qPCR) assays were performed using the ChamQ SYBR qPCR Master Mix (Vazyme, Q311-02) in the StepOnePlus™ Real-Time PCR System (Applied Biosystems, USA). The PCR conditions consisted of denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 58°C for 10 s, and extension at 72°C for 20 s. The cucumber actin gene was used as an internal control. Gene-specific primers were designed according to cDNA sequences, as described in Table S2. Relative gene expression was calculated as described by Livak and Schmittgen [74].
Protein extraction and immunoblotting
For protein extraction, cucumber roots or Arabidopsis leaves were ground in liquid nitrogen and homogenized in extraction buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.2% Triton X-100 [Solarbio, T8200], 1 mM phenylmethylsulphonyl fluoride [Solarbio, P8340], 10 mM dithiothreitol). After the homogenates filtering through a 300-μm and 100-μm nylon mesh (Dongguan Jufeng Mesh Products Co., LTD, ZH-NL10), the filtered homogenates were used to measure the total protein concentrations. The same amount of total proteins was centrifuged at 2200 g for 5 min to collect the supernatant, which was used to measure the soluble proteins. The pellet fractions were resuspended with the extraction buffer and subjected to the low-speed centrifugation. This process was repeated five times, and the last pellets were resuspended to detect the insoluble proteins. The protein concentrations of the total, soluble, and insoluble proteins were detected using Bio-Rad Protein Assay Kit II (5000002).
For detection of the lipidation of ATG8, the proteins were extracted and the membrane fractions were collected by centrifugation at 100000 g for 1 h as previously described [75]. The pellet was solubilized in extraction buffer with 0.5% of Triton X-100, and clarified at 16000 g for 10 min to obtain the solubilized membrane fraction. The resulting membrane fraction was incubated for 1 h at 37°C with or without Streptomyces chromofuscus PLD (Sigma-Aldrich, P0065), and stopped by the addition of SDS-PAGE sample buffer.
For immunoblotting analysis of the ubiquitinated, GFP, and actin proteins, the denatured proteins were separated with 12% SDS-PAGE. For the detection of ATG8, the denatured proteins were separated using 13.5% SDS-PAGE with 6 M urea (Guangzhou Jinhuada Chemical Reagent Co., LTD, 1.20601.010). After the proteins were separated using SDS-PAGE, they were transferred to a polyvinylidene fluoride membrane (Millipore, IPVH00010). The membranes were blocked in TBS buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween 20 [Solarbio, T8220]) for 1 h at room temperature, and then incubated for 1 h in TBS buffer containing a mouse anti-ubiquitin monoclonal antibody (Sigma-Aldrich, U0508), rabbit anti-ATG8 polyclonal antibody (Agrisera, AS142769), mouse anti-GFP monoclonal antibody (Cell Signaling Technology, 2955), or rabbit anti-actin polyclonal antibody (Abcam, ab197345). Subsequently, the membranes were incubated for 1 h in TBS buffer with a goat anti-mouse HRP-linked antibody (Millipore, AP124P) or goat anti-rabbit HRP-linked antibody (Cell Signaling Technology, 7074), and the complexes on the blot were visualized using the SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, 34580). The band intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Quantitative assay and histochemical detection of H2O2
The content of H2O2 was measured as previously described [76]. The histochemical staining of H2O2 by CeCl3 (Macklin, C804404) staining was performed as described previously [77]. H2O2 production in tissues was detected using DCF-DA (Sigma-Aldrich, 35845) as previously described [62]. Signal intensities of fluorescence in the images were quantified by ImageJ software. Values were corrected for background. Data were presented as the means of fluorescence intensity relative to the control plants.
Autophagosomes detection
To visualize the accumulation of MDC-labeled puncta, cucumber roots were excised and then immediately vacuum infiltrated with 100 μM MDC (Sigma-Aldrich, 30432) for 30 min, and washed three times with 50 mM phosphate buffer (Solarbio, P1020). The fluorescent signals were monitored under an LSM 800 confocal microscope (Zeiss, Germany), excited by a wavelength of 405 nm and detected at 400 to 580 nm. Transgenic cucumber roots overexpressing GFP-ATG8f were harvested, and autophagosomes were visualized by the LSM 800 confocal microscope excited by a wavelength of 488 nm and detected at 493 to 558 nm.
VIGS constructs and Agrobacterium-mediated virus infection
pV190 VIGS vector, constructed by the lab of Dr. Qinsheng Gu (Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences), was used for the silencing of ATG4, ATG7, and PDS genes in this study. For silencing of these genes, 300-bp fragments of these genes were obtained by PCR amplification with the gene-specific primers (Table S3) and ligated into the pV190 vector using the ClonExpress II One Step Cloning Kit (Vazyme, C112). The resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101 (TOLOBIO, CC96304). When the cotyledons were fully expanded, the A. tumefaciens-mediated virus infection was performed as described by Liu et al [35]. The expression of ATG4 and ATG7 in the roots of VIGS plants was detected using qPCR, and the plants that showed about 30 to 40% transcript levels of control plants were used for salt treatment.
Statistical analysis
At least 3 independent replicates were used for each determination. Data were statistically analyzed by analysis of variance (ANOVA), and the significance of treatment differences was analyzed with Tukey’s test at P < 0.05.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jie Zhou (Zhejiang University) for donating the plant transformation vector. We thank Dr. Yuehua Ma (College of Horticulture, Nanjing Agricultural University) for assistance in the detection of autophagosomes using LSM 800 confocal microscope.
Funding Statement
This work was supported by the National Key Research and Development Program of China (2018YFD1000800), the National Natural Science Foundation of China (31801902), the China Postdoctoral Science Foundation (2019M661867 and 2019T120433), the Fundamental Research Funds for the Central Universities (KJQN201928) and the China Agriculture Research System (CARS-23-B12).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Munns R, Tester M.. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. [DOI] [PubMed] [Google Scholar]
- [2].Yang YQ, Guo Y. Unraveling salt stress signaling in plants. J Int Plant Biol. 2018;60:796–804. [DOI] [PubMed] [Google Scholar]
- [3].Tang XL, Mu XM, Shao HB, et al. Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Cri Rev Biotechnol. 2015;35:425–437. [DOI] [PubMed] [Google Scholar]
- [4].Gupta B, Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics. 2014;2014:701596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhu M, Zhou M, Shabala L, et al. Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance. Plant Cell Environ. 2017;40:1009–1020. [DOI] [PubMed] [Google Scholar]
- [6].Chen D, Shao Q, Yin L, et al. Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front Plant Sci. 2019;9:1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yi Z, Li S, Liang Y, et al. Effects of exogenous spermidine and elevated CO2 on physiological and biochemical changes in tomato plants under iso-osmotic salt stress. J Plant Growth Regul. 2018;37:1222–1234. [Google Scholar]
- [8].Duan JJ, Li J, Guo SR, et al. Exogenous spermidine affects polyamine metabolism in salinity-stressed Cucumis sativus roots and enhances short-term salinity tolerance. J Plant Physiol. 2008;165:1620–1635. [DOI] [PubMed] [Google Scholar]
- [9].Li J, Hu L, Zhang L, et al. Exogenous spermidine is enhancing tomato tolerance to salinity-alkalinity stress by regulating chloroplast antioxidant system and chlorophyll metabolism. BMC Plant Biol. 2015;15:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nahar K, Hasanuzzaman M, Rahman A, et al. Polyamines confer salt tolerance in mung bean (Vigna radiata L.) by reducing sodium uptake, improving nutrient homeostasis, antioxidant defense, and methylglyoxal detoxification systems. Front Plant Sci. 2016;7:1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lou Y, Guan R, Sun M, et al. Spermidine application alleviates salinity damage to antioxidant enzyme activity and gene expression in alfalfa. Ecotoxicology. 2018;27:1323–1330. [DOI] [PubMed] [Google Scholar]
- [12].Gong B, Wan XF, Wei M, et al. Overexpression of S-adenosylmethionine synthetase 1 enhances tomato callus tolerance to alkali stress through polyamine and hydrogen peroxide cross-linked networks. Plant Cell Tiss Org. 2016;124:377–391. [Google Scholar]
- [13].Sang T, Shan X, Li B, et al. Comparative proteomic analysis reveals the positive effect of exogenous spermidine on photosynthesis and salinity tolerance in cucumber seedlings. Plant Cell Rep. 2016;35:1769–1782. [DOI] [PubMed] [Google Scholar]
- [14].Du J, Guo S, Sun J, et al. Proteomic and physiological analyses reveal the role of exogenous spermidine on cucumber roots in response to Ca(NO3)2 stress. Plant Mol Biol. 2018;97:1–21. [DOI] [PubMed] [Google Scholar]
- [15].Wu J, Shu S, Li C, et al. Spermidine-mediated hydrogen peroxide signaling enhances the antioxidant capacity of salt-stressed cucumber roots. Plant Physiol Biochem. 2018;128:152–162. [DOI] [PubMed] [Google Scholar]
- [16].Michaeli S, Galili G, Genschik P, et al. Autophagy in plants - what’s new on the menu? Trends Plant Sci. 2016;21:134–144. [DOI] [PubMed] [Google Scholar]
- [17].Avin-Wittenberg T, Baluska F, Bozhkov PV, et al. Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J Exp Bot. 2018;69:1335–1353. [DOI] [PubMed] [Google Scholar]
- [18].Wang Y, Cai SY, Yin LL, et al. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy. 2015;11:2033–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Avin-Wittenberg T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019;42:1045–1053. [DOI] [PubMed] [Google Scholar]
- [20].Wang Y, Cao JJ, Wang KX, et al. BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 2019;179:671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhou J, Wang J, Cheng Y, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoSGenet. 2013;9:e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhou J, Wang J, Yu JQ, et al. Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci. 2014;5:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu YM, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–963. [DOI] [PubMed] [Google Scholar]
- [24].Wang P, Sun X, Jia X, et al. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017;256:53–64. [DOI] [PubMed] [Google Scholar]
- [25].Liu YM, Bassham DC. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE. 2010;5:e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li LL, Tan J, Miao YY, et al. ROS and autophagy: interactions and molecular regulatory mechanisms. Cell Mol Neurobiol. 2015;35:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen L, Liao B, Qi H, et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy. 2015;11:2233–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Madeo F, Eisenberg T, Buttner S, et al. Spermidine a novel autophagy inducer and longevity elixir. Autophagy. 2010;6:160–162. [DOI] [PubMed] [Google Scholar]
- [29].Eisenberg T, Knauer H, Schauer A, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. [DOI] [PubMed] [Google Scholar]
- [30].Guerra DD, Callis J. Ubiquitin on the move: the ubiquitin modification system plays diverse roles in the regulation of endoplasmic reticulum- and plasma membrane-localized proteins. Plant Physiol. 2012;160:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang Y, Yu BJ, Zhao JP, et al. Autophagy contributes to leaf starch degradation. Plant Cell. 2013;25:1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kwon SI, Cho HJ, Kim SR, et al. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 2013;161:1722–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xiao S, Gao W, Chen QF, et al. Overexpression of Arabidopsis Acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 2010;22:1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li FQ, Chung T, Vierstra RD. Autophagy-related11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014;26:788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu M, Liang ZL, Aranda MA, et al. A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods. 2020;16:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang Y, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018;217:523–539. [DOI] [PubMed] [Google Scholar]
- [37].Morton MJL, Awlia M, Al-Tamimi N, et al. Salt stress under the scalpel- dissecting the genetics of salt tolerance. Plant J. 2019;97:148–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Seifi HS, Shelp BJ. Spermine differentially refines plant defense responses against biotic and abiotic stresses. Front Plant Sci. 2019;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shu S, Chen L, Lu W, et al. Effects of exogenous spermidine on photosynthetic capacity and expression of Calvin cycle genes in salt-stressed cucumber seedlings. J Plant Res. 2014;127:763–773. [DOI] [PubMed] [Google Scholar]
- [40].Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. [DOI] [PubMed] [Google Scholar]
- [41].Wang Y, Gong XW, Liu WK, et al. Gibberellin mediates spermidine-induced salt tolerance and the expression of GT-3b in cucumber. Plant Physiol Biochem. 2020;152:147–156. [DOI] [PubMed] [Google Scholar]
- [42].Li S, Jin H, Zhang Q. The effect of exogenous spermidine concentration on polyamine metabolism and salt tolerance in zoysiagrass (Zoysia japonica Steud) subjected to short-term salinity stress. Front Plant Sci. 2016;7:1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang N, Shi X, Guan Z, et al. Treatment with spermidine protects chrysanthemum seedlings against salinity stress damage. Plant Physiol Biochem. 2016;105:260–270. [DOI] [PubMed] [Google Scholar]
- [44].ElSayed AI, Rafudeen MS, El-hamahmy MAM, et al. Enhancing antioxidant systems by exogenous spermine and spermidine in wheat (Triticum aestivum) seedlings exposed to salt stress. Funct Plant Biol. 2018;45:745–759. [DOI] [PubMed] [Google Scholar]
- [45].Sheng S, Yuan LY, Guo SR, et al. Effects of exogenous spermidine on photosynthesis, xanthophyll cycle and endogenous polyamines in cucumber seedlings exposed to salinity. Afr J Biotechnol. 2012;11:6064–6074. [Google Scholar]
- [46].Khoshbakht D, Asghari MR, Haghighi M. Effects of foliar applications of nitric oxide and spermidine on chlorophyll fluorescence, photosynthesis and antioxidant enzyme activities of citrus seedlings under salinity stress. Photosynthetica. 2018;56:1313–1325. [Google Scholar]
- [47].Chen LY, Hellmann H. Plant E3 ligases: flexible enzymes in a sessile world. Mol Plant. 2013;6:1388–1404. [DOI] [PubMed] [Google Scholar]
- [48].Liu LJ, Cui F, Li QL, et al. The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 2011;21:957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhou J, Zhang Y, Qi JX, et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014;10:e1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Stone SL. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front Plant Sci. 2014;5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Trujillo M. News from the PUB: plant U-box type E3 ubiquitin ligases. J Exp Bot. 2018;69:371–384. [DOI] [PubMed] [Google Scholar]
- [52].Zientara-Rytter K, Subramani S. The roles of ubiquitin-binding protein shuttles in the degradative fate of ubiquitinated proteins in the ubiquitin-proteasome system and autophagy. Cells. 2019;8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li WM, Nie TJ, Xu HD, et al. Chaperone-mediated autophagy: advances from bench to bedside. Neurobiol Dis. 2019;122:41–48. [DOI] [PubMed] [Google Scholar]
- [54].Madeo F, Bauer MA, Carmona-Gutierrez D, et al. Spermidine: a physiological autophagy inducer acting as an anti-aging vitamin in humans? Autophagy. 2019;15:165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Eisenberg T, Abdellatif M, Schroeder S, et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016;22:1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].He H, Van Breusegem F, Mhamdi A. Redox-dependent control of nuclear transcription in plants. J Exp Bot. 2018;69:3359–3372. [DOI] [PubMed] [Google Scholar]
- [57].Waszczak C, Carmody M, Kangasjarvi J. Reactive oxygen species in plant signaling. Annu Rev Plant Biol. 2018;69:209–236. [DOI] [PubMed] [Google Scholar]
- [58].Li S, Wang N, Ji D, et al. A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell. 2019;31:2107–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ma LY, Zhang H, Sun LR, et al. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J Exp Bot. 2012;63:305–317. [DOI] [PubMed] [Google Scholar]
- [60].Ben Rejeb K, Lefebvre-De Vos D, Le Disquet I, et al. Hydrogen peroxide produced by NADPH oxidases increases proline accumulation during salt or mannitol stress in Arabidopsis thaliana. New Phytol. 2015;208:1138–1148. [DOI] [PubMed] [Google Scholar]
- [61].Niu M, Huang Y, Sun S, et al. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J Exp Bot. 2018;69:3465–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yi CY, Yao KQ, Cai SY, et al. High atmospheric carbon dioxide-dependent alleviation of salt stress is linked to respiratory burst oxidase 1 (RBOH1)-dependent H2O2 production in tomato (Solanum lycopersicum). J Exp Bot. 2015;66:7391–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ji M, Wang K, Wang L, et al. Overexpression of a S-adenosylmethionine decarboxylase from sugar beet M14 increased Araidopsis salt tolerance. Int J Mol Sci. 2019;20:1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xiang Y, Contento AL, Bassham D. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy. 2007;3:257–258. [DOI] [PubMed] [Google Scholar]
- [65].Xiong Y, Contento AL, Nguyen PQ, et al. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Perez-Perez ME, Lemaire SD, Crespo JL. Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 2012;160:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Signorelli S, Tarkowski LP, Van den Ende W, et al. Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 2019;24:413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yao XX, Li XN, Zhang D, et al. B-cell lymphoma 2 inhibitor ABT-737 induces Beclin1-and reactive oxygen species-dependent autophagy in Adriamycin-resistant human hepatocellular carcinoma cells. Tumor Biol. 2017;39:695965. [DOI] [PubMed] [Google Scholar]
- [69].Shan ZF, Wei LQ, Yu SQ, et al. Ketamine induces reactive oxygen species and enhances autophagy in SV-HUC-1 human uroepithelial cells. J Cell Physiol. 2019;234:2778–2787. [DOI] [PubMed] [Google Scholar]
- [70].Jian SG, Liang C, Lian MX, et al. Tanshinone I induces apoptosis and protective autophagy in human glioblastoma cells via a reactive oxygen species-dependent pathway. Int J Mol Med. 2020;45:983–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Meng D, Yang Q, Dong BY, et al. Development of an efficient root transgenic system for pigeon pea and its application to other important economically plants. Plant Biotechnol J. 2019;17:1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lu CM, Qiu NW, Wang BS, et al. Salinity treatment shows no effects on photosystem II photochemistry, but increases the resistance of photosystem II to heat stress in halophyte Suaeda salsa. J Exp Bot. 2003;54:851–860. [DOI] [PubMed] [Google Scholar]
- [73].Cao WH, Liu J, He XJ, et al. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007;143:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- [75].Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009;149:220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Brennan T, Frenkel C. Involvement of hydrogen-peroxide in regulation of senescence in pear. Plant Physiol. 1977;59:411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bestwick CS, Brown IR, Bennett MHR, et al. Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell. 1997;9:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


