ABSTRACT
Telomeres protect chromosome ends from nucleolytic degradation, uncontrolled recombination by DNA repair enzymes and checkpoint signaling, and they provide mechanisms for their maintenance by semiconservative DNA replication, telomerase and homologous recombination. The telomeric long noncoding RNA TERRA is transcribed from a large number of chromosome ends. TERRA has been implicated in modulating telomeric chromatin structure and checkpoint signaling, and in telomere maintenance by homology directed repair, and telomerase – when telomeres are damaged or very short. Recent work indicates that TERRA association with telomeres involves the formation of DNA:RNA hybrid structures that can be formed post transcription by the RAD51 DNA recombinase, which in turn may trigger homologous recombination between telomeric repeats and telomere elongation. In this review, we describe the mechanisms of TERRA recruitment to telomeres, R-loop formation and its regulation by shelterin proteins. We discuss the consequences of R-loop formation, with regard to telomere maintenance by DNA recombination and how this may impinge on telomere replication while counteracting telomere shortening in normal cells and in ALT cancer cells, which maintain telomeres in the absence of telomerase.
KEYWORDS: Telomeres, TERRA, R-loops, RAD51, shelterin proteins, homologous recombination
Introduction
Telomeres correspond to the physical ends of eukaryotic chromosomes. They consist of short tandem DNA repeats that are generally rich in guanine and thymine bases. In vertebrates, telomeres consist of 5ʹ-TTAGGG-3ʹ repeats in the DNA strand containing the 3ʹ end. The overall length of human telomeres varies between roughly 3,000–15,000 bp. Telomeres have a 3ʹ overhang of roughly 50–200 nucleotides [1,2] and are associated with several hundred proteins [3,4,5]. Most abundant and best characterized is the shelterin protein complex comprising up to six different polypeptides [6]. TRF1 (telomeric repeat-binding factor 1) and TRF2 bind as homodimers the double stranded part of telomeres. POT1 (protection of telomeres 1) binds specifically to the single-stranded 5ʹ-TTAGGG-3ʹ repeats which are present at the 3ʹ end of telomeres [7]. Alternatively, POT1 is thought to bind to single-stranded 5ʹ-TTAGGG-3ʹ repeats present as displaced strand internally, when telomeres adopt the T-loop configuration (Figure 1). In T-loops, the telomeric 3ʹ overhang is tucked into the double-stranded part of the telomere, base pairing with the complementary 5ʹ-CCCTAA-3ʹ repeats [8]. POT1 can be physically linked to TRF1 and TRF2 through protein interactions involving the shelterin components TPP1 and TIN2 (TRF1-interacting nuclear factor 2). TPP1 also enhances the affinity of POT1 for telomeric DNA, and the interaction of POT1 with the other shelterins is required for telomere association of POT1, possibly by increasing its local concentration [9]. Rap1 (repressor activator protein 1) is recruited to vertebrate telomeres via TRF2.
Figure 1.
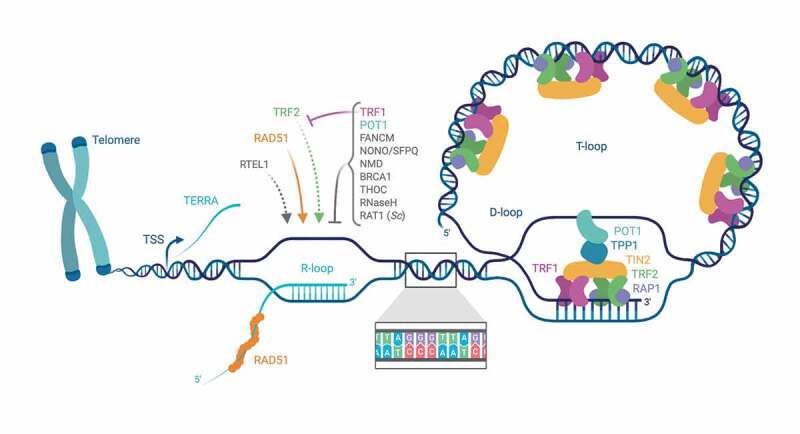
Cartoon of the nucleoprotein structure of chromosome ends; telomeric R-loops and its regulators. Among the numerous proteins associating with telomeres, the shelterin protein complex components are the most abundant, comprising the dsDNA-binding proteins TRF1 and TRF2, the ssDNA-binding protein POT1, TIN2, TPP1 and Rap1. Telomeres have a 3ʹ overhang, which can invade the dsDNA region, forming a D-loop which is termed T-loop. Telomeres are transcribed into TERRA. TERRA transcription starts at subtelomeric regions, extending toward chromosome ends. The vast majority of TERRA molecules is not polyadenylated and largely colocalizes with telomeres. TERRA binding to telomeres can occur through direct base-pairing with telomeric DNA, forming R-loop structures, leaving a displaced G-rich DNA strand. Factors which are suspected to regulate TERRA association with telomeres are indicated. TERRA association and R-loop formation at telomeres depend on the RAD51 DNA recombinase, which binds TERRA and catalyzes TERRA R-loop formation in vitro. Also, the helicase RTEL1 was proposed to stimulate TERRA association with chromosome ends via R-loops. The shelterin TRF2 can also bind TERRA and stimulate R-loop formation in vitro – a process counteracted in vivo by TRF1. Loss of POT1 was found to result in a striking accumulation of telomeric R-loops, suggesting a role in preventing the accumulation of such structures. Proteins involved in the NMD RNA surveillance pathway, including UPF1, UPF2 and SMG1, were also shown to prevent TERRA association with telomeres. The DNA recombination factor BRCA1 was as well shown to modulate TERRA binding to telomeres, preventing R-loop-associated telomeric DNA damage. The ATPase/translocase FANCM resolves RNA:DNA hybrids in vitro and counteracts telomeric R-loop accumulation in ALT cells. The THO multi-subunit complex is present at human and budding yeast telomeres, and was found to prevent R-loop accumulation and telomere shortening in budding yeast. RNase H enzymes – which remove RNA-DNA hybrids through the endonucleolytic cleavage of the engaged RNA moiety – were found to regulate TERRA R-loops, with a prominent role in ALT cells. In S. cerevisiae, Rat1 nuclease, as well as RNase H2, are preferentially recruited to long telomeres in S phase, preventing TERRA accumulation and formation of R-loops prone to pose an obstacle to telomeric replication. The Illustration was created with BioRender.com
A large number of additional telomeric proteins that are less abundant at chromosome ends have been identified through studies of telomere maintenance by telomerase, genetic and protein interaction screens and upon purification of crosslinked telomeric chromatin, followed by mass spectrometry analysis [3,4,5]. Most of these proteins are not telomere specific and many of them bind to telomeres only under certain conditions. Notably, a significant number of proteins identified at telomeres are linked to RNA metabolism, which can be rationalized by the fact that telomeres are transcribed into the telomeric repeat containing RNA (TERRA) [10,11].
TERRA has been detected in a large number of eukaryotes including humans and other vertebrates [10,11], yeast [12,13,14], plants [15] and protozoa [16]. Thus, telomere transcription occurs despite the fact that telomeric chromatin has heterochromatic characteristics [17,18]. At human telomeres, histone H3 is frequently trimethylated at Lys9 and Lys27, and nucleosomes are narrowly spaced [18] due to the lack of the linker histone H1[3]. The discovery of telomere transcription was also counterintuitive at the time, as the telomere position effect – which reflects the variegated repression of genes experimentally placed next to telomeres – suggested a general absence of transcription at telomeres [19,20]. TERRA transcription proceeds from promoters residing in the subtelomeric regions into the telomeric tract [10,21–23]. Thus, human TERRA starts with sequences stemming from different subtelomeres and it ends with numerous 5ʹ-UUAGGG-3ʹ repeats [24]. In Trypanosoma brucei, telomere transcription occurs mainly at the telomere which contains the active VSG gene [25]. In other eukaryotes where this has been characterized, telomere transcription is common to several or all chromosome ends. The assignment of TERRA molecules to individual chromosome ends is ambiguous in many species including humans, as subtelomeres are rich in repetitive sequences which have not been well annotated. Interestingly, the regulation of TERRA expression at individual chromosome ends may differ from one to another. In Saccharomyces cerevisiae for example, the major double strand telomere-binding protein Rap1p regulates TERRA transcription and degradation [26]. At subtelomeres containing the so-called Y’ repeat elements, Rap1p recruits Rif1 and Rif2 to downregulate TERRA transcription. At subtelomeres containing X repeat elements, Rap1-mediated TERRA repression involves the Sir2/3/4 histone deacetylase and Rif1/2 complexes. Also in humans, TERRA transcription has been detected at a large number of chromosome ends. At some telomeres, TERRA promoters harbor CpG islands which are negatively regulated by DNA methyltransferases that can modify these sequences [22,23,27]. A second class of TERRA promoters lacks CpG islands and is insensitive to DNA methylation [23]. In mouse cells, TERRA expression has also been detected at several chromosome ends [28]. In addition, a more abundant noncoding 5ʹ-UUAGGG-3ʹ repeat containing RNA termed PAR-TERRA which is involved in the pairing of homologous sex chromosomes has been described. The PAR-TERRA primary structure has not been elucidated so far and its transcription has not been demonstrated to proceed into the terminal telomeric repeats, as seen for canonical TERRA transcripts [29]. CHIRT-seq experiments indicate binding of PAR-TERRA throughout the genome as well as at chromosome ends, suggesting functions that are particular to this RNA. We do not cover PAR-TERRA in this review.
Telomeres are dynamic structures. They change their composition during the cell cycle and upon shortening and damage, to mediate DNA damage checkpoint signaling and repair. Telomere shortening is a common phenomenon in human somatic cells, as differentiated cells do not express the catalytic subunit of telomerase hTERT [30]. Short telomeres elicit ATM- and ATR-dependent checkpoint signaling to induce cellular senescence which represses the growth of precancerous lesions that have lost normal growth control [31]. At short telomeres, T-loops are thought to unfold due to low levels of TRF2, which enables ATM activation. Low levels of POT1 allow binding of RPA to single-stranded telomeric DNA, leading to ATR-ATRIP recruitment and checkpoint activation. TERRA is another major player which triggers local remodeling events at individual chromosome ends that may have suffered from damage or severe shortening. TERRA expression is enhanced at telomeres from which TRF2 has been depleted [21] and its expression increases from telomeres when they get shorter in budding yeast [32] and in human cells [33]. Thus, through its accumulation at short or damaged telomeres through increased expression and recruitment, TERRA can act as a recruitment platform for DNA repair enzymes at telomeres that need their attention.
TERRA R-loop formation and regulation at chromosome ends
TERRA is transcribed at human and yeast telomeres by RNA polymerase II. The 5ʹ-UUAGGG-3ʹ tract of human TERRA is heterogeneous in length. For most TERRA molecules the length varies between 100–400 nucleotides [24], though longer TERRA transcripts approaching the ends of chromosomes can also be detected [27,33]. More than 90% of human TERRA is not polyadenylated, frequently terminating with the sequence 5ʹ-UUAGG-3ʹ[24]. The mechanisms of 3ʹend formation are not well understood. The 3ʹends of the non-polyadenylated fraction might simply occur due to termination of transcription within telomeric chromatin that might impede efficient read-through. The poly(A) tail of the polyadenylated fraction of TERRA is generated by the canonical poly(A) polymerase, at least in budding yeast [12]. If polyadenylation of TERRA is preceded by RNA cleavage as for mRNAs is not known.
Analyses by fluorescence in situ hybridization and cellular fractionation studies indicate that TERRA is enriched at human telomeres [10]. In addition, approximately half of TERRA is not tightly associated with chromatin. Notably, all poly(A)+ TERRA was detected in a nucleoplasmic fraction and not on chromatin [24] suggesting that its functions do not involve physical interactions with telomeres. In contrast, non-polyadenylated TERRA colocalizes to a large extent with telomeres. In principle, it may be retained at telomeres through interactions with telomere binding proteins or through base-pairing with telomeric DNA, forming so-called R-loop structures in which the TERRA:telomeric DNA RNA/DNA helix causes displacement of the telomeric strand containing 5ʹ-TTAGGG-3ʹ DNA repeats (Figure 1). Several proteins have been identified to regulate TERRA association with telomeres. Using a reporter system in which TERRA was expressed ectopically with an RNA tag (PP7 stem loops) that is recognized by the bacteriophage PP7 coat protein fused to GFP, we could demonstrate that TERRA associates with telomeres post transcription, through the formation of R-loop structures [34] (Figure 2). This association is dependent on TERRA’s 5ʹ-UUAGGG-3ʹ repeats suggesting that the R-loops form within the telomeric repeats. However, the length and exact position of the R-loop structures has not been determined. Telomere association and R-loop formation depend on the RAD51 DNA recombinase [34] since its depletion reduces TERRA foci at telomeres (Figure 2). Interestingly, while endogenous TERRA is rare in S phase, transgenic TERRA expressed from plasmids with a heterologous promoter loses this control and efficiently associates with telomeres in S phase nuclei that were identified through pulse labeling with EdU (Figure 2). This association is also reduced upon RAD51 depletion indicating the involvement of RAD51. The presence of transgenic TERRA at telomeres during replication can explain its interference with telomere replication (see below). Also of note, though RAD51 is an essential protein, its substantial depletion by siRNA only slightly increased the number cells in G2/M and it did not substantially affect the fraction of cells in S phase during which endogenous TERRA levels are low (Figure 3).
Figure 2.
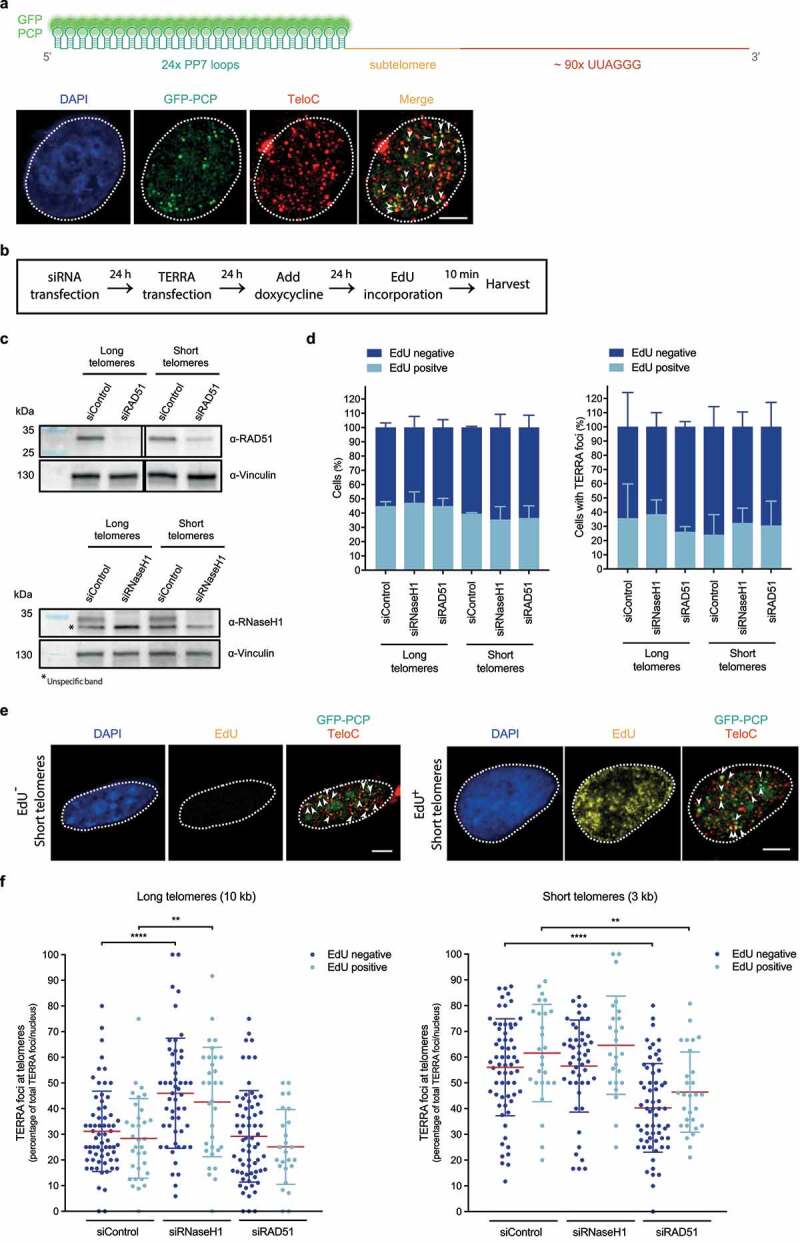
Transgenic TERRA associates with telomeres in and outside S phase. a) Top: Depiction of transiently expressed chimeric TERRA, comprising twenty-four Pseudomonas aeruginosa phage 7 (PP7) stem-loops – recognized by GFP-tagged dimerized PP7 Coat Protein (PCP) –, and a subtelomere-derived sequence, followed by UUAGGG tandem repeats. Illustration created with BioRender.com. Bottom: Immunofluorescence of GFP (green) was employed to analyze co-localization of transiently expressed PP7-fused 15q-TERRA transcripts with telomeres (red) identified by Fluorescence in situ hybridization (FISH) (as described in ref 34). Representative images are shown and were acquired with a Leica SP8 confocal microscope. White dashed line outlines the nuclear region and was determined based on DAPI-staining. White arrowheads indicate co-localization of GFP-PCP with telomeric FISH signals. Scale bar indicates 5 μm. b) HeLa cells with long (10 kilobase average) or short (3 kb average) telomeres were transfected with siRNA pools to down-regulate RAD51 or RNase H1 mRNA levels. PP7-15q-TERRA-coding constructs were transfected and their expression was induced with doxycycline for 24 hours. Cells were then pulse-labeled with 10 μM of 5-ethynyl-2ʹ-deoxyuridine (EdU) (Invitrogen) for 10 min and harvested. c) Western blotting was used to evaluate knockdown efficiency of RAD51 (top) or RNase H1 (bottom). Vinculin is shown as a loading control. Representative blots of three biologically-independent experiments are shown. d) Percentage of EdU-positive cells in the total population of cells (left) or cells displaying TERRA foci (right), across indicated conditions. After EdU pulse-labeling, cells were fixed in 4% paraformaldehyde for 10 min at room temperature. Anti-GFP immunofluorescence was performed as described in ref 34. After fixation of bound primary and secondary antibodies, cells were permeabilized with a detergent solution (0.1% Triton X-100, 0.02% SDS in 1x PBS) for 5 min, followed by pre-blocking with 2% bovine serum albumin (BSA) in 1x PBS for 30 min. Cells were then incubated with a click-it reaction (4 mM copper sulfate, 100 mM sodium ascorbate and 4 μM Alexa Fluor 488 Azide (Invitrogen) in 1x PBS) for 30 min in a humidity-chamber, followed by three 1x PBS washes, permeabilization with a detergent solution (indicated above) for 3 min and 4% paraformaldehyde fixation for 5 min. FISH staining was then carried out following the procedure described in ref 34. At least 460 total cells and 74 cells displaying TERRA foci were analyzed per condition, across three independent biological replicates. Data are means ± s.d. e) Representative images of EdU-negative (left) and EdU-positive (right) HeLa cells with short telomeres, obtained as described in d. Representative images were acquired with a Leica SP8 confocal microscope. White dashed line outlines the nuclear region and was determined based on DAPI-staining. EdU signal is shown, as well as GFP-PCP and TeloC merged signals. White arrowheads indicate co-localization of GFP-PCP with telomeric FISH signals. Scale bars indicate 5 μm. f) The percentage of PP7-15q-TERRA foci colocalizing with telomeric FISH signals per nucleus was assessed by GFP immunofluorescence, EdU Click-it and telomeric FISH as described in d. At least 74 cells were analyzed per condition, across three independent biological replicates. Data are means ± s.d. One-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test was used, comparing all conditions with non-targeting siRNA (siControl): **P < 0.01; ****P < 0.0001. All statistical analysis was performed using GraphPad Prism. All images were processed and analyzed with Image J
Figure 3.
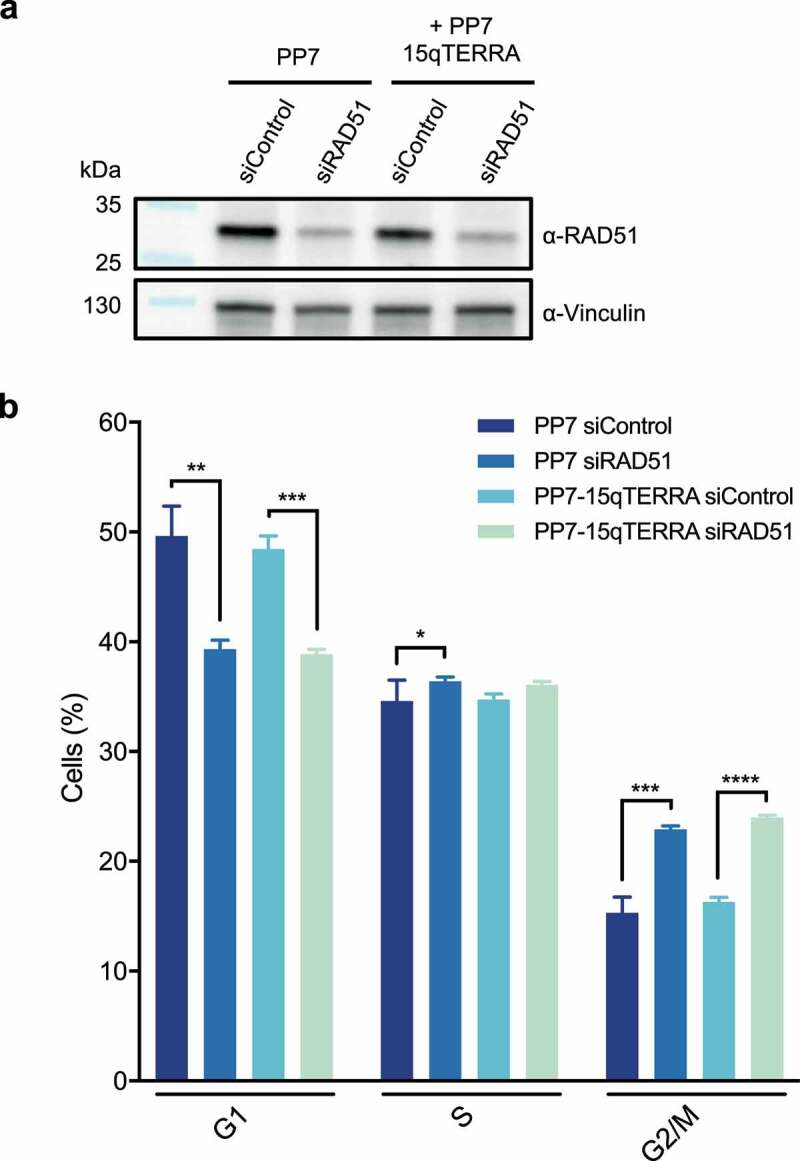
The fraction of cells in S phase is not strongly impacted by siRNA-mediated depletion of RAD51, with or without PP7-15q-TERRA inducible expression. a) Western blotting was used to evaluate siRNA-mediated knockdown efficiency of RAD51, with and without PP7-15q-TERRA inducible expression. Vinculin is shown as a loading control. Representative blots of three biologically-independent experiments are shown. b) Distribution of cells of indicated conditions in G1, S or G2/M cell cycle phases. To evaluate cell cycle distribution of cells with depleted RAD51, DNA content was assessed by flow cytometry analysis of fixed DAPI-stained cells. Briefly, 1 million cells/condition was harvested by trypsinization. Cells were washed twice with cold 1x PBS and resuspended in 1 ml cold 70% ethanol while vortexing. After 30 min, ethanol was aspirated and fixed cells were resuspended in 1x PBS containing 0.2 μg/ml RNase, DNase-free (Merck) and incubated at 37°C for 15 min. 1x PBS containing 2 μg/ml DAPI (BioChemica) was then added to stain DNA. Samples were processed with a BD LSR Fortessa. 20.000 events were acquired per condition, per biological replicate. Data are means ± s.d. of three independent biological replicates. Two-tailed unpaired t-tests were used to calculate P-values: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Flow cytometry data were analyzed with FlowJo and statistical analysis was performed using GraphPad Prism
Since RAD51 promotes homology search and strand invasion of recombining DNA molecules, the data suggested that RAD51 may also home TERRA through an analogous mechanism. In support of this model, RAD51 is bound to TERRA in cellular extracts and it binds the 5ʹ-UUAGGG-3ʹ repeats of TERRA in vitro with high affinity. Furthermore, RAD51 catalyzes strand invasion of TERRA into plasmid DNA in vitro and it is required for the formation of telomeric R-loops by endogenous TERRA [34]. It will be important to determine how RAD51 distinguishes TERRA from the bulk of other nuclear RNAs it may not act upon. The 5ʹ-UUAGGG-3ʹ repeats of TERRA provide a unique signature for this RNA. Also, these G-rich repeats can form G-quadruplex structures which might be recognized by RAD51. Alternatively, other TERRA-binding proteins might facilitate a preferential association of RAD51 with TERRA. BRCA2 facilitates RAD51 binding to single-stranded DNA during double strand break repair by homologous recombination. BRCA2 depletion reduced TERRA association with telomeres, but it also diminished the presence of RAD51 in the nucleus. Thus, it remains uncertain if BRCA2 promotes RAD51 binding to TERRA. Notably, these results are strikingly different from what has been seen for highly transcribed genes, which may retain R-loops from transcription and for which BRCA2 was reported to diminish R-loops through a collaboration with the TREX-2 mRNP biogenesis and export complex [35]. Interestingly, BRCA1 – which among others also promotes RAD51-mediated homologous recombination for DNA repair – is a second DNA recombination factor that was recently shown to physically interact with TERRA as well as TERRA R-loops [36]. In contrast to RAD51, however, BRCA1 counteracts telomeric R-loops [36]. The underlying mechanism remains uncertain, but it was proposed to involve BRCA1 interactions with XRN2, which is the ortholog of yeast Rat1. Rat1 is a 5ʹ-3ʹ RNA exonuclease which is involved in transcription termination, among others. Rat1 degrades TERRA in S. cerevisiae [12]. To what extent other recombination factors may regulate TERRA at telomeres remains to be investigated. The recent findings already hint toward important differences between RNA and DNA mediated homology search. However, if TERRA strand invasion of telomeric DNA initiates at TERRA 3ʹends this mechanism could explain why poly(A)+ TERRA is not retained on chromatin.
While RAD51 is likely to catalyze strand invasion of TERRA into telomeric DNA, several telomere-associated proteins have also been identified to regulate strand invasion and telomere retention (Figure 1). Among the shelterin components, TRF1, TRF2 and POT1 play critical roles. The N-terminal basic domain of TRF2 can bind TERRA and stimulate R-loop formation in vitro [37]. This activity is prevented in vitro and in vivo by TRF1 through its N-terminal acidic domain. Whether TRF2 promotes R-loops under physiological conditions is not known, though TRF2 depletion per se does not decrease R-loop formation at telomeres [34,37]. POT1 binds specifically to the single-stranded G-rich telomeric DNA but not to the corresponding 5ʹ-UUAGGG-3ʹ repeats in TERRA [38]. POT1 deletion in human cells causes rapidly dramatic telomere elongation by RAD51-mediated homologous recombination [39]. At the same time the telomeric R-loops are increased. It is unclear if TERRA R-loops are formed as an immediate consequence of POT1-loss and if they are a prerequisite for telomere recombination. Perhaps more likely, POT1-loss liberates the single-stranded telomeric DNA for RPA binding which subsequently, with assistance of BRCA2, becomes replaced by RAD51. The increased local concentration of RAD51 may facilitate the binding of TERRA by RAD51 in a succeeding step, which will trigger strand invasion and R-loop formation. The DNA strand that is displaced by the TERRA R-loop may recruit additional RPA and RAD51 in a feedforward loop to first promote RPA-dependent DNA checkpoint signaling and subsequently replacement by RAD51 to mediate homologous recombination (Figure 4).
Figure 4.
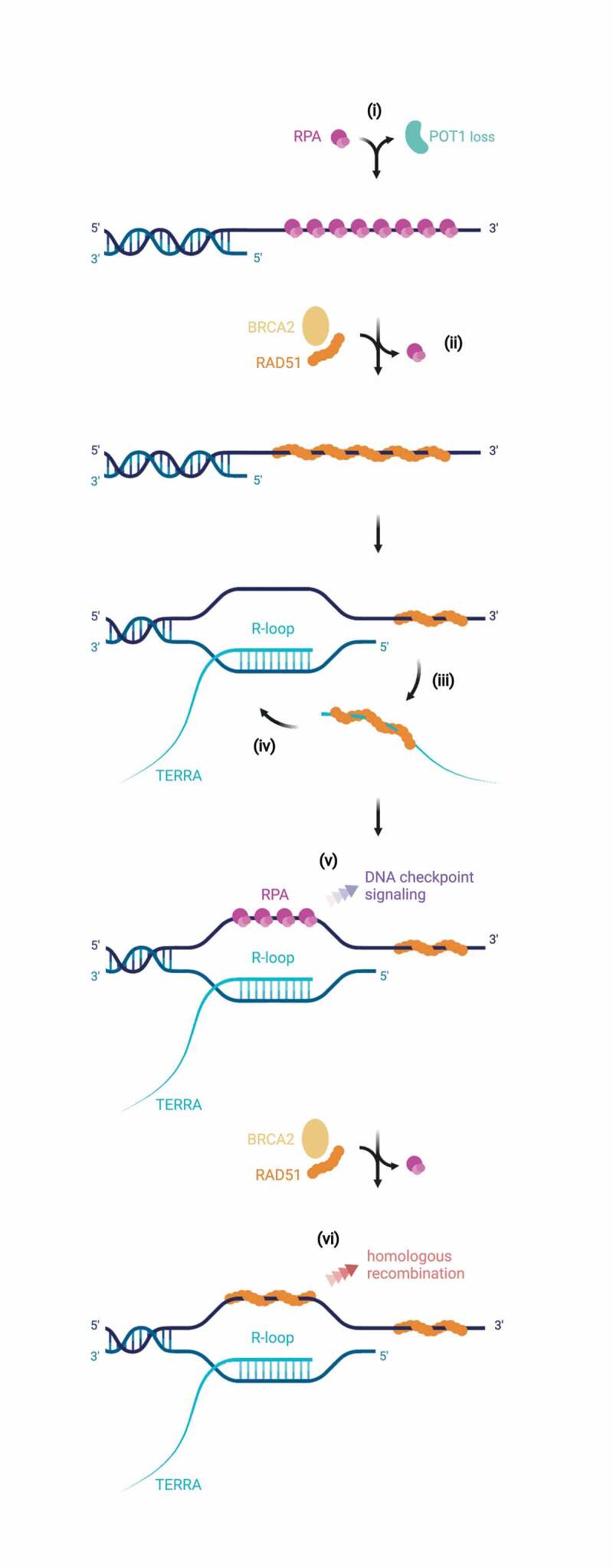
Possible R-loop- and RAD51-mediated telomere elongation mechanism upon loss of POT1. Loss of the ssDNA-binding shelterin POT1 at human telomeres leaves exposed the telomeric 3ʹ overhang – which can be bound by RPA (i). With assistance of BRCA2, RAD51 may displace and replace RPA (ii). This increased concentration of RAD51 at telomeres may facilitate binding of RAD51 to TERRA (iii), which subsequently can trigger strand invasion and R-loop formation (iv). Upon R-loop formation, the displaced single-stranded DNA strand can be bound by RPA – serving as a key platform for ATR activation (v). Assisted by BRCA2, RAD51 may then substitute RPA, mediating homologous recombination at the telomere (vi) eventually resulting in HDR-mediated telomere elongation (observed as a striking consequence of conditional deletion of POT1 in human cells). Illustration created with BioRender.com
The FANCM protein, whose mutation has been associated with Fanconi Anemia, is an ATPase associated with DNA branch migration. Its roles at telomeres have been characterized in U2OS cells [40,41], which are ALT cells (for alternative lengthening of telomeres) that use DNA recombination to maintain telomeric DNA repeats. Strikingly, FANCM depletion caused an increase in R-loops both at telomeres and elsewhere in the genome. In addition, FANCM can resolve telomeric R-loops in vitro using its RNA:DNA helicase activity suggesting that it directly participates in R-loop resolution [40]. NONO/SFPQ heterodimers, which are involved in various aspects of RNA metabolism in the nucleus, have also been implicated in suppressing telomeric R-loops [42]. Their depletion increased R-loop signals partially colocalizing with telomeres in nuclei of U2OS cells. Most recently, the RTEL1 helicase has been implicated in TERRA regulation [43]. RTEL1 has well-established crucial roles during telomere replication resolving T-loops in S phase as well as telomeric G-quadruplex structures [44]. The new work shows that RTEL1 also binds in vitro TERRA 5ʹ-UUAGGG-3ʹ repeats when adopting a G-quadruplex structure [43]. Strikingly, RTEL1 deletion caused strong increase in TERRA levels, while TERRA association with telomeres was significantly diminished. It was proposed that RTEL1 facilitates TERRA association with chromosome ends through a stimulatory effect on telomeric R-loops, which in turn could prevent additional transcription from the R-loop containing telomeres [43]. This proposed mode of action deviates from the documented roles of RTEL1 at G-quadruplex forming DNA sequences elsewhere in the genome where RTEL1 dismantles R-loops [45]. Therefore, it will be interesting to further dissect the mechanism and also test if and to what extent the S phase-specific telomere recruitment of RTEL1 by dephosphorylated TRF2 may contribute [46]. Certainly, also other models could be at play. For example, if RTEL1 affected TERRA 3ʹ end formation preventing its polyadenylation, absence of RTEL1 and TERRA polyadenylation might also stabilize this RNA and trigger its dissociation from chromosome ends [24].
Proteins involved in nonsense mediated RNA decay (NMD) of mRNAs containing premature stop codons also regulate TERRA stability and R-loop formation. These proteins, though most abundant in the cytoplasm, are also present in nuclei and can be detected at telomeres by chromatin immunoprecipitation, suggesting direct roles at telomeres [10]. Depletion of the NMD proteins UPF1, UPF2, SMG1 and SMG6 increased TERRA foci at telomeres, while not markedly impacting on total TERRA levels. This suggested that these proteins displace TERRA from telomeres, though they might also degrade this RNA locally at chromosome ends. Consistent with such models, depletion of UPF1, UPF2 and SMG1 also increased association of transgenic TERRA with telomeres [34]. Live cell imaging of TERRA should allow to discriminate if NMD proteins may influence the dynamics of TERRA interaction with chromosome ends or if they are involved in its local destruction at the telomere.
Regulation of TERRA during the cell cycle and with telomere length
Telomeres change their composition during S phase of the cell cycle to mediate their replication by the semiconservative DNA replication machinery and end maintenance by telomerase. Also, TERRA levels are regulated during the cell cycle. In budding yeast, TERRA levels and telomeric R-loops increase from G1 to S phase, but they drop during S phase, reaching lower levels by the time of semiconservative DNA replication of telomeres in late S phase – possibly to prevent interference with the replisome [32]. The low levels of TERRA in late S phase are a consequence of the recruitment of Rat1 nuclease to telomeres at this stage. In human cells, TERRA levels also decrease during S phase, reaching lowest levels as cells proceed from late S to G2 [24]. This cell cycle control is lost in cells carrying defects in the chromatin remodeling enzyme ATRX or the DNA methyl transferase DNMT3b (see below). However, the exact mechanisms of regulation at the transcriptional and posttranscriptional level remain to be elucidated.
TERRA has been suspected to regulate telomerase activity, and in vitro TERRA is a potent inhibitor of the human telomerase enzyme as it tightly binds to the RNA template [47]. In vivo in budding yeast, however, positive roles of TERRA for telomerase have been proposed. TERRA colocalizes with telomerase in early S phase, which then has been suggested to guide telomerase to specifically short telomeres promoting their preferential elongation [48]. Interestingly, TERRA molecules from short telomeres appeared to reassociate with the short telomeres from which they originated, which could be explained by a homology-driven search mechanism that we describe in this review for human TERRA. In fission yeast, TERRA has also been proposed to be a positive regulator of telomerase, but a positive effect of TERRA overexpression on telomere length was only observed upon treatment of cells with histone deacetylase inhibitors [49]. Finally, roles of TERRA have been postulated in human cells for promoting POT1 association with the G-rich telomeric strand after DNA replication [50]. hnRNPA1 is a major TERRA-binding protein, binding its 5ʹ-UUAGGG-3ʹ repeats [51], while also having strong affinity for single-stranded 5ʹ-TTAGGG-3ʹ DNA repeats [52]. As TERRA levels decline in late S phase, hnRNPA1 is liberated from TERRA – now binding the single-stranded telomeric DNA. Thus, hnRNPA1 may displace RPA from the G-rich telomeric DNA strand, which is the major single-strand DNA-binding protein associating with all single-stranded DNA for replication. As TERRA reaccumulates after S phase, hnRNPA1 may reassociate with TERRA and liberate the single-stranded G-rich telomeric strand for POT1 binding. More detailed knowledge on the regulation of TERRA expression and its interaction with telomerase, hnRNPA1 and other protein partners during the cell cycle may be required to clarify these issues.
TERRA association with chromosome ends also increases as telomeres get shorter. In budding yeast, this is due to increased transcription [48] and the absence of Rat1 nuclease and RNase H2 (a member of the Ribonuclease H family of enzymes that specifically degrade the RNA moiety in DNA:RNA hybrid structures) at short telomeres [32]. Conversely, at longer telomeres, the telomeric Rif2 recruits efficiently RNase H2 and Rat1. In human cells, telomere transcription increases at short telomeres due to a decreased presence of trimethylated H3K9, which recruits HP1 [33]. In addition, ectopically expressed TERRA is recruited to short telomeres more efficiently by the RAD51 recombinase [34]. The underlying mechanism for this has not yet been uncovered in human cells. However, RNase H1 might be involved, as its depletion increased telomere association of ectopic TERRA strongly at long but not at short telomeres [34] (Figure 2).
Consequences of R-loop formation: replication interference and stimulation of HDR
The formation of TERRA-mediated telomeric R-loops appears to be beneficial when telomeres become damaged or when they are very short. In such cases, telomeres activate DNA damage checkpoint signaling to induce cell cycle arrest or cellular senescence, they may be fused to one another by DNA repair enzymes if they completely lose capping function and become mistaken as DNA double strand breaks [31]. Alternatively, short telomeres are healed and re-elongated by telomerase or by homology directed repair. For the latter, single-stranded G-rich telomeric DNA may invade the telomeric DNA of adjacent chromosomes which is followed by elongation of the invading 3ʹ end by DNA polymerase δ [53,54]. This is followed by fill-in synthesis of the lagging strand, leading to telomere extension by conservative synthesis of telomeric DNA. The mechanism is referred to as break-induced replication. Recombination mediated telomere synthesis is prevalent in ALT cancer cells that lack telomerase. However, recombination mediated telomere synthesis, though partially repressed, is also at play in normal or telomerase positive mammalian cells. This notion is supported by the finding that Brca2 deletion or Rad51 depletion in mouse embryonic fibroblasts leads to telomere damage and shortening [55] and telomere-internal double strand breaks can be repaired by homologous recombination [56]. In budding yeast, deletion of telomerase causes continuous telomere shortening. Intriguingly, TERRA R-loop formation at short telomeres promotes homology directed repair-dependent re-elongation, preventing premature onset of cellular senescence [32]. This repair of short telomeres is active in cells that have not yet engaged the ALT pathway, demonstrating the importance of homology directed repair for telomere homeostasis in normal cells.
The first evidence that TERRA R-loops can promote homology directed repair and telomere elongation came from studies in human ALT cancer cells [57]. Coinciding with telomere recombination, TERRA expression and R-loop formation are increased in ALT cells. Telomeric R-loops cause replication stress at chromosome ends, which is a hallmark of ALT telomeres. On the other hand, inhibition of TERRA transcription was recently shown to decrease marks of DNA replication stress at telomeres, while telomere maintenance by ALT activity was impaired [58]. Interestingly, it appears that TERRA R-loops must achieve optimal levels to stimulate homology directed repair, while not completely destroying telomere integrity by overly severe replication stress [57]. RNase H1 and FANCM, which associate with ALT telomeres, play critical roles in keeping telomeric R-loops at tolerable levels [40,41,59]. FANCM depletion is selectively toxic in ALT cells. In its absence DNA replication stress at telomeres increases to unbearable levels. This stress is caused through telomeric R-loops, as RNase H1 overexpression suppressed the telomere replication stress of FANCM depleted ALT cells [40,59]. Of note, ALT cells lose the cell cycle control of TERRA expression due to mutations in chromatin remodeling protein ATRX [60]. Thus, TERRA remains high in S phase and G2 – consistent with its effects on telomere replication and elongation. Non-repressed TERRA in S phase may also, through its binding of the telomerase RNA template, reinforce telomerase repression in ALT cells.
While break-induced replication of telomeres can proceed through Rad51-dependent and independent mechanisms in yeast [61], the different synthesis pathways which operate in human ALT cells have not been fully dissected. Several studies documented roles of RAD51 in ALT cells. Depletion of RAD51 decreased the formation of ALT-associated promyelocytic leukemia bodies (APBs) in ALT cells in which telomeres gather for ALT DNA synthesis [62]. Also, chemical inhibition of RAD51 in ALT cells interfered with telomere maintenance [63] and RAD51 depletion with the frequency of telomere extension events [64]. In addition, human RAD51 facilitates long-range telomere movement and telomere clustering in ALT cells [65]. On the other hand, based on RAD51-depletion experiments, other studies described RAD51-independent mechanisms for break-induced telomere synthesis in ALT cells. A major mechanism involves RAD52, while a second ill-defined mechanism can be observed in the absence of RAD52 [54,66]. Remarkably, RAD52 can bind RNA:DNA duplexes [67,68] as well as single and double stranded DNA and thus may associate with TERRA R-loops to mediate a PCNA-RFC-Polδ-dependent pathway for conservative DNA synthesis that extends telomere length. Overall, the published work demonstrates that distinct mechanisms contribute to telomere elongation in human ALT cells. In our view, however, it may be premature to exclude crucial roles of RAD51 in a subset of the mechanisms, as this protein is required for cell viability and essential functions may have been retained in the knockdown studies.
At least three mechanisms could be envisioned of how TERRA and TERRA R-loops promote telomere recombination and elongation (Figure 5). First, the telomeric R-loops will lead to displacement of the telomeric G-rich strand which may facilitate loading of the RAD51 recombinase and other recombination proteins (Figure 5(a)). Second, R-loops can lead to replication stress and DNA double strand breaks, which can then be repaired by homology directed repair (Figure 5(b)). Third, the increased levels of TERRA at chromosome ends through R-loops in ALT cells may not only require RAD51 – as is the case in telomerase-positive cells – but in turn also increase the local concentration of this TERRA binding recombinase to sustain DNA recombination (Figure 5(c)).
Figure 5.
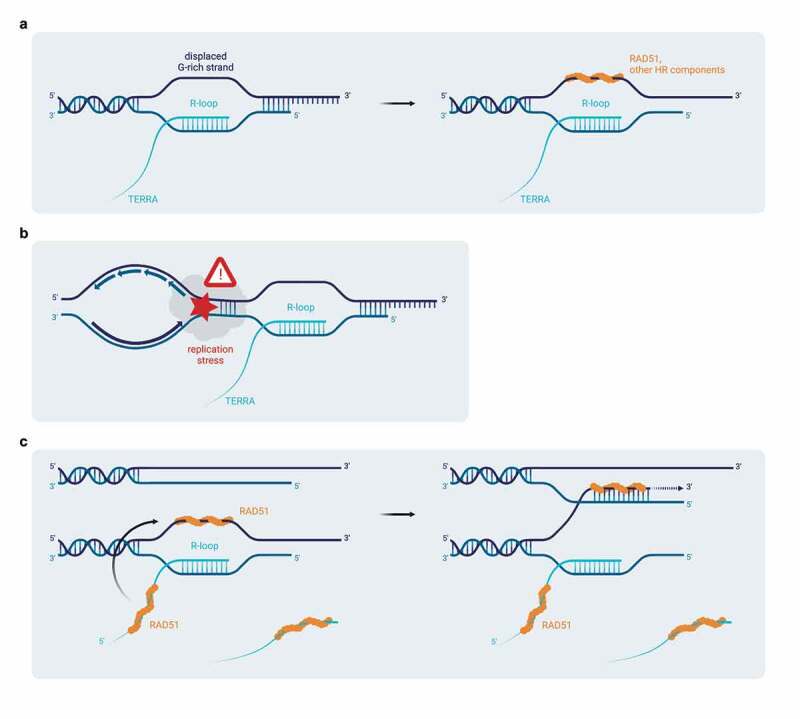
Possible mechanisms for TERRA R-loop-mediated stimulation of homologous recombination at telomeres. a) Upon R-loop formation, the displaced single-stranded DNA strand may facilitate the loading of the RAD51 recombinase and other recombination factors at telomeres. b) Telomeric R-loops pose a structural barrier that hinders progression of the replication machinery. Replication fork stalling may culminate in the formation of DNA double-strand breaks, which can then be repaired by homology-directed repair. c) RAD51-mediated stimulation of TERRA R-loops and association of RAD51 with the displaced DNA strand – exposed as a consequence of R-loop formation – may lead to local increased concentration of this recombinase and other recombination factors at telomeres, which in recombination-prone ALT cells may sustain telomeric homologous recombination and telomere elongation. Illustration created with BioRender.com
R-loops had been originally considered as toxic by-products of transcription which generate obstacles for DNA and RNA polymerases, inducing DNA damage upon replication [69,70]. Also at telomeres, R-loops do interfere with the semiconservative DNA replication machinery. The THO complex, which has been implicated in removing nascent RNA from chromatin, is present at telomeres in human cells and in budding yeast [4,71]. Deletion of THO components in S. cerevisiae increases telomeric R-loops, leading to replication stress at telomeres and telomere shortening [71]. As discussed above, RNase H enzymes also counteract R-loops. In their absence the accumulation of telomeric R-loops leads to telomere loss and accelerated senescence in recombination-deficient budding yeast, supporting the notion that R-loops interfere with telomere replication [72]. Finally, depletion of NMD factors in human cells led to increased TERRA at telomeres and frequent loss of telomeric DNA [10]. Specifically, UPF1 depletion leads to inefficient replication of telomeres synthesized by leading strand synthesis, suggesting a role of this helicase in removing TERRA from leading strand telomeres for their replication [73].
Hypomorphic mutations in the de novo DNA methyltransferase DNMT3b cause ICF (Immunodeficiency, Centromeric instability and Facial anomalies) syndrome type I [27]. Subtelomeric DNA sequences are hypomethylated in ICF type I syndrome cells, correlating with strongly increased TERRA levels. These cells also display increased telomeric R-loops throughout the cell cycle, telomere damage, accelerated telomere shortening and premature replicative senescence, all being consistent with the interference of TERRA R-loops with telomere replication [74]. However, a direct demonstration of causative roles of DNMT3b deficiency and subtelomeric DNA demethylation in the short telomere phenotype has been cumbersome. The DNA methylation status at subtelomeres cannot be rescued in the cellular ICF models with ectopic DNMT3b, indicative of a persistent epigenetic memory [75].
Arguably, the most direct evidence for TERRA interfering with telomere replication comes from experiments in which TERRA was expressed ectopically from plasmids [34]. As discussed above, transgenic TERRA associates with telomeres post transcription through R-loops that are formed in a RAD51-dependent manner. Interference with telomere replication in human cells gives rise to so-called telomere fragility, in which the telomeric FISH signal on metaphase chromosomes is smeared or double [76]. Notably, expression of transgenic TERRA caused a significant increase of telomere fragility. This fragility was suppressed by RNase H1 overexpression or RAD51 depletion, demonstrating that it is a direct consequence of the physical presence of TERRA as R-loop at chromosome ends, even in the absence of increased RNA polymerase II and transcription at the telomere [34].
Conclusions
The finding that TERRA can associate with telomeres post transcription through the formation of R-loops was unexpected. Generally, it had been assumed that R-loops are formed during transcription mostly at the 5ʹ and 3ʹ ends of GC-skewed regions. However, in addition to TERRA, several papers reported on the formation of R-loops post transcription in trans. The long noncoding RNA APOLO associates with multiple loci in the genome of Arabidopsis thaliana post transcription through R-loops modulating gene expression through decoy of polycomb repressive complex 1 components [77]. The mechanism of R-loop formation by APOLO has remained unknown. In S. cerevisiae, it has been reported that Rad51p mediates hybridization of transcripts to homologous chromosomal loci distinct from their site of synthesis [78], though another paper disputed this conclusion [79]. Finally, R-loop formation is well documented for the bacterial CRISPR-Cas9 DNA endonuclease [80], in which Cas9 bends the DNA to allow guide RNA infiltration into the double helix. The mechanism of TERRA R-loop formation post transcription depends on the RAD51 recombinase which binds TERRA, and which can catalyze this reaction in vitro. It will be important to dissect the detailed mechanisms in the future. For instance, it is unclear how TERRA and telomere binding proteins may influence this reaction to achieve substrate specificity for TERRA and preferential invasion at short telomeres. It will also be important to determine if other factors of the recombination machinery which are required for strand invasion of recombining DNA molecules may participate in the TERRA strand invasion reaction. Finally, it will important to determine if and to what extent telomeric R-loops arise during transcription in cis and if RAD51 also plays a role in their establishment.
R-loops cause DNA damage and genome instability, presumably through their interference with DNA polymerases during replication. In addition, several papers reported also on beneficial roles of R-loops, for example for the repair of DNA double breaks [67,68,81]. The work on TERRA suggests that this RNA preferentially associates with short or damaged telomeres, where it may coordinate the DNA damage response, as well as repair processes. As discussed above, excellent evidence has been provided that TERRA promotes telomere elongation by homologous recombination of short telomeres [32,57]. But does the mechanism also involve induction of replication stress and fork collapse, as a prerequisite to induce HDR? Replication stress is prevalent at telomeres in ALT, supporting the notion that these cells indeed must manage a labile balance between telomere loss by fork collapse and elongation by HDR [40,57].
Acknowledgments
We thank Eftychia Kyriacou (EPFL) and Claus Azzalin (iMM, Lisboa) for comments on the manuscript. We also acknowledge support from the Histology, BioImaging and Optics Platform and Flow Cytometry Research Core Facilities at the School of Life Sciences of EPFL.
Funding Statement
The laboratory was supported by the Swiss National Science Foundation (SNFS grant 310030_184718), the SNFS-funded National Center of Competence in Research (NCCR) RNA and disease network (grant 182880) and the European Union’s Horizon 2020 research and innovation programme under grant agreement 812829.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Makarov VL, Hirose Y, Langmore JP.. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88(5):657–666. [DOI] [PubMed] [Google Scholar]
- [2].Chai W, Shay JW, Wright WE.. Human telomeres maintain their overhang length at senescence. Mol Cell Biol. 2005;25(6):2158–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Déjardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136(1):175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Grolimund L, Aeby E, Hamelin R, et al. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat Commun. 2013;4(1):2848. [DOI] [PubMed] [Google Scholar]
- [5].Bartocci C, Diedrich JK, Ouzounov I, et al. Isolation of chromatin from dysfunctional telomeres reveals an important role for Ring1b in NHEJ-mediated chromosome fusions. Cell Rep. 2014;7(4):1320–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].de Lange T. Shelterin-Mediated Telomere Protection. Annu Rev Genet. 2018;52(1):223–247. [DOI] [PubMed] [Google Scholar]
- [7].Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292(5519):1171–1175. [DOI] [PubMed] [Google Scholar]
- [8].Doksani Y, Wu JY, de Lange T, et al. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Takai KK, Kibe T, Donigian JR, et al. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell. 2011;44(4):647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Azzalin CM, Reichenbach P, Khoriauli L, et al. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. [DOI] [PubMed] [Google Scholar]
- [11].Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10(2):228–236. [DOI] [PubMed] [Google Scholar]
- [12].Luke B, Panza A, Redon S, et al. The Rat1p 5' to 3' exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32(4):465–477. [DOI] [PubMed] [Google Scholar]
- [13].Bah A, Wischnewski H, Shchepachev V, et al. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012;40(7):2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Greenwood J, Cooper JP. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 2012;40(7):2956–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vrbsky J, Akimcheva S, Watson JM, et al. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6(6):e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nanavaty V, Sandhu R, Jehi SE, et al. Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids. Nucleic Acids Res. 2017;45(10):5785–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tardat M, Déjardin J. Telomere chromatin establishment and its maintenance during mammalian development. Chromosoma. 2018;127(1):3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tommerup H, Dousmanis A, de Lange T. Unusual chromatin in human telomeres. Mol Cell Biol. 1994;14:5777–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gottschling DE, Aparicio OM, Billington BL, et al. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. [DOI] [PubMed] [Google Scholar]
- [20].Baur JA, Zou Y, Shay JW, et al. Telomere position effect in human cells. Science. 2001;292(5524):2075–2077. [DOI] [PubMed] [Google Scholar]
- [21].Porro A, Feuerhahn S, Delafontaine J, et al. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun. 2014;5(1):5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nergadze SG, Farnung BO, Wischnewski H, et al. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15(12):2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Feretzaki M, Renck Nunes P, Lingner J. Expression and differential regulation of human TERRA at several chromosome ends. RNA. 2019;25(11):1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Porro A, Feuerhahn S, Reichenbach P, et al. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol. 2010;30(20):4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Saha A, Gaurav AK, Pandya UM, et al. Tb TRF suppresses the TERRA level and regulates the cell cycle-dependent TERRA foci number with a TERRA binding activity in its C-terminal Myb domain. Nucleic Acids Res. 2021;49(10):5637–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Iglesias N, Redon S, Pfeiffer V, et al. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011;12(6):587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yehezkel S, Segev Y, Viegas-Péquignot E, et al. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17(18):2776–2789. [DOI] [PubMed] [Google Scholar]
- [28].Viceconte N, Loriot A, Lona Abreu P, et al. PAR-TERRA is the main contributor to telomeric repeat-containing RNA transcripts in normal and cancer mouse cells. RNA. 2021;27(1):106–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chu H-P, Froberg JE, Kesner B, et al. Lee JT. PAR-TERRA directs homologous sex chromosome pairing. Nat Struct Mol Biol. 2017;24(8):620–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. [DOI] [PubMed] [Google Scholar]
- [31].Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Graf M, Bonetti D, Lockhart A, et al. Telomere length determines TERRA and R-loop regulation through the cell cycle. Cell. 2017;170(1):72–85.e14. [DOI] [PubMed] [Google Scholar]
- [33].Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat Struct Mol Biol. 2012;19(9):106–121. [DOI] [PubMed] [Google Scholar]
- [34].Feretzaki M, Pospisilova M, Valador Fernandes R, et al. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature. 2020;587(7833):303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bhatia V, Barroso SI, García-Rubio ML, et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511(7509):362–365. [DOI] [PubMed] [Google Scholar]
- [36].Vohhodina J, Goehring LJ, Liu B, et al. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nat Commun. 2021;12(1):3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee YW, Arora R, Wischnewski H, et al. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat Struct Mol Biol. 2018;25(2):147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci U S A. 2010;107(2):651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Glousker G, Briod A-S, Quadroni M, et al. Human shelterin protein POT 1 prevents severe telomere instability induced by homology-directed DNA repair. EMBO J. 2020;39(23):e104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Silva B, Pentz R, Figueira AM, et al. FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat Commun. 2019;10(1):2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lu R, O’Rourke JJ, Sobinoff AP, et al. The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nat Commun. 2019;10(1):2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Petti E, Buemi V, Zappone A, et al. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat Commun. 2019;10(1):1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ghisays F, Garzia A, Wang H, et al. RTEL1 influences the abundance and localization of TERRA RNA. Nat Commun. 2021;12(1):3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vannier J-B, Pavicic-Kaltenbrunner V, Petalcorin MIR, et al. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149(4):795–806. [DOI] [PubMed] [Google Scholar]
- [45].Wu BR, Vogel I, Ö Ö, et al. RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. Nat Struct Mol Biol. 2020;27:424–437. [DOI] [PubMed] [Google Scholar]
- [46].Sarek G, Kotsantis P, Ruis P, et al. CDK phosphorylation of TRF2 controls t-loop dynamics during the cell cycle. Nature. 2019;575(7783):523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38(17):5797–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cusanelli E, Romero CAP, Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell. 2013;51(6):780–791. [DOI] [PubMed] [Google Scholar]
- [49].Moravec M, Wischnewski H, Bah A, et al. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 2016;17(7):999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Flynn RL, Centore RC, O’Sullivan RJ, et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471(7339):532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Redon S, Zemp I, Lingner J. A three-state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res. 2013;41(19):9117–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ding J, Hayashi MK, Zhang Y, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13(9):1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lydeard JR, Jain S, Yamaguchi M, et al. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448(7155):820–823. [DOI] [PubMed] [Google Scholar]
- [54].Dilley RL, Verma P, Cho NW, et al. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. 2016;539(7627):54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Badie S, Escandell JM, Bouwman P, et al. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol. 2010;17:1461–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Doksani Y, de Lange T. Telomere-internal double-strand breaks are repaired by homologous recombination and PARP1/Lig3-dependent end-joining. Cell Rep. 2016;17(6):1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Arora R, Lee Y, Wischnewski H, et al. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun. 2014;5(1):5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Silva B, Arora R, Bione S, et al. TERRA transcription destabilizes telomere integrity to initiate break-induced replication in human ALT cells. Nat Commun. 2021;12(1):3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pan X, Chen Y, Biju B, et al. FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops. Sci Rep. 2019;9(1):19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Flynn RL, Cox KE, Jeitany M, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347(6219):273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol. 2001;21(5):1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].O’Sullivan RJ, Arnoult N, Lackner DH, et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol. 2014;21(2):167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang T, Zhang Z, Shengzhao G, et al. Strand break-induced replication fork collapse leads to C-circles, C-overhangs and telomeric recombination. PLoS Genet. 2019;15(2):e1007925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sobinoff AP, Allen JA, Neumann AA, et al. BLM and SLX4 play opposing roles in recombination‐dependent replication at human telomeres. EMBO J. 2017;36:2907–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Cho NW, Dilley RL, Lampson MA, et al. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell. 2014;159(1):108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang J-M, Yadav T, Ouyang J, et al. Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep. 2019;26(4):955–968.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yasuhara T, Kato R, Hagiwara Y, et al. Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell. 2018;175(2):558–570.e11. [DOI] [PubMed] [Google Scholar]
- [68].Tan J, Duan M, Yadav T, et al. An R-loop-initiated CSB-RAD52-POLD3 pathway suppresses ROS-induced telomeric DNA breaks. Nucleic Acids Res. 2020;48:1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].García-Muse T, Aguilera A. R loops: from physiological to pathological roles. Cell. 2019;179(3):604–618. [DOI] [PubMed] [Google Scholar]
- [70].Marnef A, Legube G. R-loops as Janus-faced modulators of DNA repair. Nat Cell Biol. 2021;23(4):305–313. [DOI] [PubMed] [Google Scholar]
- [71].Pfeiffer V, Crittin J, Grolimund L, et al. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013;32(21):2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Balk B, Maicher A, Dees M, et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20(10):1199–1205. [DOI] [PubMed] [Google Scholar]
- [73].Chawla R, Redon S, Raftopoulou C, et al. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J. 2011;30(19):4047–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sagie S, Toubiana S, Hartono SR, et al. Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nat Commun. 2017;8(1):14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Toubiana S, Gagliardi M, Papa M, et al. Persistent epigenetic memory impedes rescue of the telomeric phenotype in human ICF iPSCs following DNMT3B correction. Elife. 2019;8:e47859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sfeir A, Kosiyatrakul ST, Hockemeyer D, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ariel F, Lucero L, Christ A, et al. R-loop mediated trans action of the APOLO long noncoding RNA. Mol Cell. 2020;77(5):1055–1065.e4. [DOI] [PubMed] [Google Scholar]
- [78].Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA–DNA hybrids and associated chromosome instability. Elife. 2013;2:e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lafuente-Barquero J, García-Rubio ML, Martin-Alonso MS, et al. Harmful DNA:RNA hybrids are formed in cis and in a Rad51-independent manner. Elife. 2020;9:e56674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Pessina F, Giavazzi F, Yin Y, et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol. 2019;21(10):1286–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]


