Abstract
The Rpb6 subunit of RNA polymerase II is one of the five subunits common to three forms of eukaryotic RNA polymerase. Deletion and truncation analyses of the rpb6 gene in the fission yeast Schizosaccharomyces pombe indicated that Rpb6, consisting of 142 amino acid residues, is an essential protein for cell viability, and the essential region is located in the C-terminal half between residues 61 and 139. After random mutagenesis, a total of 14 temperature-sensitive mutants were isolated, each carrying a single (or double in three cases and triple in one) mutation. Four mutants each carrying a single mutation in the essential region were sensitive to 6-azauracil (6AU), which inhibits transcription elongation by depleting the intracellular pool of GTP and UTP. Both 6AU sensitivity and temperature-sensitive phenotypes of these rpb6 mutants were suppressed by overexpression of TFIIS, a transcription elongation factor. In agreement with the genetic studies, the mutant RNA polymerases containing the mutant Rpb6 subunits showed reduced affinity for TFIIS, as measured by a pull-down assay of TFIIS-RNA polymerase II complexes using a fusion form of TFIIS with glutathione S-transferase. Moreover, the direct interaction between TFIIS and RNA polymerase II was competed by the addition of Rpb6. Taken together, the results lead us to propose that Rpb6 plays a role in the interaction between RNA polymerase II and the transcription elongation factor TFIIS.
The RNA polymerase II of Schizosaccharomyces pombe consists of 12 subunits (35), corresponding to RPB1 to RPB12 of the Saccharomyces cerevisiae RNA polymerase II (44, 45). Two large subunits, Rpb1 and Rpb2, are the homologues of the β′ and β subunits of prokaryotic RNA polymerase, while the two small subunits, Rpb3 and Rpb11, have limited sequence homologies with the N-terminal assembly domain of the bacterial α subunit. These four subunits, Rpb1, Rpb2, Rpb3, and Rpb11, together are considered to form the enzyme core which corresponds to the bacterial core enzyme, with the subunit structure α2ββ′ (19, 35). In the case of RNA polymerase formation in Escherichia coli, subunit assembly proceeds sequentially in the order 2α→α2→α2β→α2ββ′ (core enzyme)→α2ββ′ς (holoenzyme) (16). The assembly core of S. pombe was identified as an Rpb2-Rpb3-Rpb11 ternary complex that corresponds to the α2β complex (19). Little is known, however, about the functions of the other eight subunits, among which five, Rpb5, Rpb6, Rpb8, Rpb10, and Rpb12, are common to all three forms of eukaryotic RNA polymerase (17, 44, 47).
Previously we analyzed the subunit-subunit contact network of S. pombe RNA polymerase II, using far-Western blotting, chemical cross-linking, glutathione S-transferase (GST) pull-down assays, and yeast two-hybrid screening (14, 26, 48). All of the small subunits were found to bind the two large subunits, Rpb1 or Rpb2, but direct interaction between small subunits was indicated for only a few combinations. In particular, Rpb6 was found to make contact with three small subunits, Rpb5, Rpb7, and Rpb8, as well as two large subunits, Rpb1 and Rpb2. The essential role of Rpb6 in the formation of functional RNA polymerase II has also been supported by the findings that (i) S. cerevisiae RPB6 is an essential gene for cell growth (25, 46); (ii) an RPB6 mutation of S. cerevisiae can suppress a temperature-sensitive mutation of RPB1 (5); (iii) Rpo26 (identical to RPB6) of S. cerevisiae plays a role in the assembly of both RNA polymerases I and II (28); and (iv) an S. cerevisiae mutant RNA polymerase I lacking the ABC23 subunit (identical to RPB6) is virtually inactive in RNA synthesis in vitro but regains activity upon the addition of RPB6 (21). The Rpb6 homologues exist in not only eukaryotic RNA polymerases but also archaeal (20) and some viral (23) RNA polymerases. The sequence of Rpb6 family proteins is highly conserved among these RNA polymerases (34). Together, these observations suggest that Rpb6 plays an essential role(s) in the assembly and/or functions of RNA polymerases I, II, and III.
To gain further insight into the structure-function relationship of Rpb6, we examined the minimum essential segment of S. pombe Rpb6 by making a set of N- and C-terminal deletion mutants. Further, we isolated a number of temperature-sensitive S. pombe mutants, each carrying a single mutation in the rpb6 gene, by replacement of the chromosomal rpb6 gene by the PCR-mutagenized rpb6 genes. The results indicate that the C-terminal half is essential for cell viability, but mutations conferring the temperature-sensitive phenotype clustered along the entire sequence of Rpb6, presumably reflecting the involvement of Rpb6 in contact with multiple subunits. Some of the rpb6 mutations in the essential region were found to be suppressed by overexpression of TFIIS, a transcription elongation factor, suggesting direct protein-protein contact between Rpb6 and TFIIS. Some biochemical studies support the notion that one of the targets of TFIIS function is the Rpb6 subunit.
MATERIALS AND METHODS
S. pombe strains and media.
The S. pombe strains used were JY741 (h− ura4-D18 leu1 ade6-M216) and JY742 (h+ ura4-D18 leu1 ade6-M210). The diploid strain used for disruption of the rpb6 gene was made by mating these two strains. Cells were grown in medium YY, SD, or MM (3).
Construction of an S. pombe mutant lacking the rpb6 gene.
Plasmid pRpb6::ura4, used for construction of the S. pombe rpb6 disruptant, was prepared as follows. The ura4 coding sequence was PCR amplified and inserted into pBluescript at a BamHI site; a DNA fragment of about 1 kbp including the rpb6 5′-flanking sequence between −1032 and −12 was isolated from pETrpb6NH (14) and inserted between EcoRI and PstI sites; a fragment of about 1 kbp including the rpb6 3′-flanking sequence between +648 and +1637 was inserted between NotI and SacI sites. The smaller EcoRI-SacI fragment including the rpb6 5′-flanking sequence, the ura4 coding sequence, and the rpb6 3′-flanking sequence was transformed into S. pombe carrying pREP81-Rpb6, which expressed the intact Rpb6 only in the absence of thiamine. Transformation was carried out by the electroporation method (13, 15). Ura+ transformants were selected, and the integration of ura4 at the rpb6 locus on the chromosome was confirmed by PCR to yield the S. pombe rpb6::ura4 disruptant.
Complementation assay of the rpb6 disruptant.
For complementation assay of the rpb6 disruptant, a set of expression plasmids for the entire or partial sequence of rpb6 was constructed. The rpb6 sequences amplified by PCR using Pfu DNA polymerase (Takara) and pETrpb6NH (14) as the template were inserted between NdeI and BamHI sites of pAI-ARS vector (Table 1). pRpb6WT contained the intact full-sized rpb6, while pRpb6NTM and pRpb6CTM series plasmids expressed N- and C-terminal deletion mutant Rpb6 proteins, respectively (Table 1). After transformation into the rpb6 disruptant, an S. pombe rpb6::ura4 strain harboring plasmid pREP81-Rpb6, Leu+ transformants were selected on plates lacking leucine. To test the function of deletion mutant Rpb6 proteins, we examined the viability of transformants after suppressing the expression of intact Rpb6 protein derived from the plasmid pREP81-Rpb6 by the addition of thiamine.
TABLE 1.
Plasmids used in this study
| Plasmid | Construction |
|---|---|
| pRpb6::ura4 | pBluescript containing rpb6 5′-flanking sequence, ura4 coding sequence, and rpb6 3′-flanking sequence |
| pRpb6::Rpb6NH8 | pBluescript containing rpb6 5′-flanking sequence, His8-rpb6 coding sequence, and rpb6 3′-flanking sequence |
| pREP81-Rpb6 | pREP81 containing intact S. pombe rpb6 coding sequence |
| pREP41-TFIIS | pREP41 containing S. pombe TFIIS coding sequence |
| pAI-ARS | pBluescript containing ura4 at HindIII site, ARS1 at EcoRI site, and rpb6 5′- and 3′-flanking sequences between EcoRI and SacI sites |
| pRpb6WT | pAI-ARS containing intact rpb6 coding sequence |
| pRpb6NTM (−40) | pAI-ARS containing N-terminal 40-residue deletion rpb6 |
| pRpb6NTM (−50) | pAI-ARS containing N-terminal 50-residue deletion rpb6 |
| pRpb6NTM (−60) | pAI-ARS containing N-terminal 60-residue deletion rpb6 |
| pRpb6NTM (−70) | pAI-ARS containing N-terminal 70-residue deletion rpb6 |
| pRpb6CTM (−3) | pAI-ARS containing C-terminal 3-residue deletion rpb6 |
| pRpb6CTM (−6) | pAI-ARS containing C-terminal 6-residue deletion rpb6 |
| pRpb6CTM (−60/−3) | pAI-ARS containing N-terminal 60- and C-terminal 3-residue deletion rpb6 |
| pGEX2T-SpIIS | pGEX2T containing the S. pombe TFIIS gene |
| pET21b-Rpb6CH | pET21b containing S. pombe rpb6 cDNA |
Construction of S. pombe 6NH producing His8-tagged Rpb6.
Plasmid pRpb6::Rpb6NH8, used as the PCR template for generation of the recombinant gene coding for Rpb6 fused to an octahistidine (His8) tag at the N terminus, was prepared as follows. A DNA segment containing the entire coding sequence of rpb6 except for the initiation codon and the 3′-flanking sequence to +1164 was PCR amplified using genomic DNA as the template and inserted into pBluescript KS(+) between BamHI and SacI sites; an rpb6 5′-flanking sequence between −302 and −12 was inserted at the 5′ terminus of the rpb6 coding sequence between EcoRI and BamHI; then a sequence coding for His8 including the initiation codon ATG was inserted at the BamHI site. PCR amplification was carried out with the resulting plasmid Rpb6::Rpb6NH as the template and a pair of primers with the sequences 5′-AAGAATTCAAAGTAATAGTAACAAATAGAC-3′ and 5′-AAGAGCTCATTATACCTTGTAAATTTCGC-3′. PCR products were transformed into S. pombe rpb6::ura4 to yield S. pombe 6NH carrying the recombinant rpb6 gene for production of Rpb6 with an H8 tag at the N terminus.
Construction of S. pombe rpb6 mutants.
Mutagenesis of rpb6 was performed by PCR using Taq DNA polymerase and pRpb6::pRpb6NH template and in the presence of 0.25 mM MnCl2 to reduce the fidelity of DNA synthesis (12, 43, 49). The PCR-amplified DNA including the coding sequence for His-tagged Rpb6 was transformed into S. pombe rpb6::ura4 by the electroporation method. The transformed cells were screened for viable colonies on SD plates lacking leucine but containing 5-fluoro-orotic acid, 20 mM thiamine, and 0.2 mg of phloxin B per ml. After incubation at 30°C for 4 days, the temperature was raised to 36°C, and temperature-sensitive colonies were selected by phloxin B color selection.
Expression plasmids for TFIIS.
cDNA for TFIIS was isolated from an S. pombe cDNA library by using a PCR-amplified TFIIS probe based on the S. pombe PPR2 sequence (45). The S. pombe TFIIS expression plasmid (pREP41-TFIIS) was constructed by inserting the PCR-amplified TFIIS coding sequence into vector pREP41 between NdeI and BamHI sites. The E. coli expression plasmid (pGEX2T-SpIIS) for the GST-TFIIS fusion was constructed by inserting the TFIIS-coding sequence into pGEX2T at the BamHI site.
Purification of RNA polymerase II.
Wild-type (6NH) and mutant S. pombe strains were grown in YE medium supplemented with 75 ml of adenine, uracil, and leucine per liter. Cells (20 g) were disrupted in an extraction buffer (50 mM Tris-HCl [pH 7.6 at 4°C], 0.5 M NaCl, 1 mM phenylmethanesulfonyl fluoride [PMSF], 10% glycerol) with a bead beater. After centrifugation at 15,000 rpm for 39 min, the supernatant was loaded onto a Ni2+-nitrilotriacetic acid agarose column (0.5-ml bed volume). After washing with extraction buffer containing 0.5% NP-40, proteins were eluted with extraction buffer containing 200 mM imidazole. The eluted proteins were dialyzed against buffer A (50 mM Tris-HCl [pH 7.8], 1 mM dithiothreitol [DTT], 0.1 mM EDTA, 20% glycerol) and loaded onto a DEAE-Sephadex A25 column (1-ml bed volume). Proteins were eluted with 7.5 ml of a linear gradient of ammonium sulfate from 50 to 500 mM. The RNA polymerase II was eluted at about 250 mM ammonium sulfate.
Nonspecific transcription assay.
Promoter-independent denatured DNA-directed RNA synthesis was carried out essentially as described by Azuma et al. (8). In brief, the reaction mixture contained 50 mM Tris-HCl (pH 7.8 at 37°C); 2 mM MnCl2; 0.5 mM DTT; 50 mM ammonium sulfate; 0.5 mM each ATP, GTP, and CTP; 7 μM UTP; 0.2 μCi of [3H]UTP (Amersham); 2 μg of heat-denatured calf thymus DNA; 50 μg of α-amanitin per ml; and RNA polymerase II. RNA synthesis was carried out at 37°C for 20 min.
Purification of TFIIS.
E. coli DH5 containing the expression plasmid for GST-TFIIS or GST was grown in Luria-Bertani medium. Expression of the recombinant proteins was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were disrupted in a lysis buffer (50 mM Tris-HCl [pH 7.8], 100 mM NaCl, 1 mM DTT, 5% glycerol, 1 mM PMSF, 0.1% NP-40, 0.3 mg of lysozyme per ml). Crude extract was mixed with glutathione-Sepharose 6B beads (Amersham Pharmacia), and the bead-bound proteins were eluted with an elution buffer (50 mM Tris-HCl [pH 7.8], 100 mM NaCl, 1 mM DTT, 5% glycerol, 1 mM PMSF, 0.1% NP-40, 5 mM glutathione). For use in transcription assay, the GST-TFIIS fusion protein was cleaved by thrombin.
GST pull-down assay.
Affinity beads were prepared by mixing purified GST or GST-TFIIS proteins at a protein concentration of 2 mg/ml with glutathione-Sepharose 4B beads (Amersham Pharmacia). Crude extracts of wild-type and mutant S. pombe were prepared essentially as described by Azuma et al. (8), with the slight modification that the ammonium sulfate precipitates were dialyzed against a pull-down buffer (50 mM Tris-HCl [pH 7.8], 10% glycerol, 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 0.1% Triton X-100). The samples containing approximately 50 mg in 100 ml were mixed with 10 ml of the affinity beads; after incubation at 4°C for 60 min, the beads were harvested by centrifugation and washed three times with 0.5 ml each of pull-down buffer with or without 150 mM NaCl.
RESULTS
Deletion mapping of Rpb6.
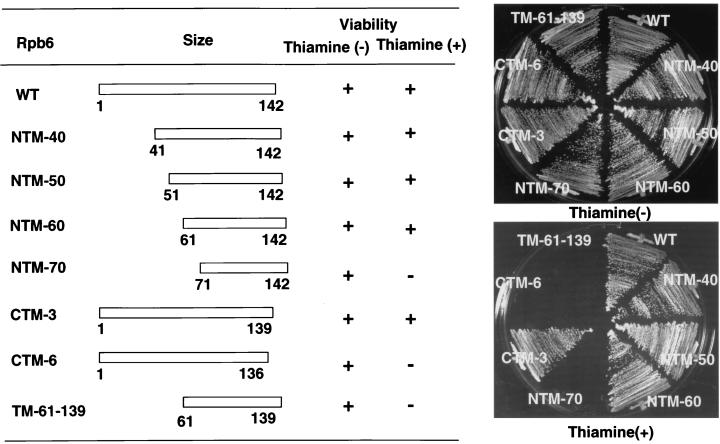
To set up the screening system for rpb6 mutations, we constructed an S. pombe rpb6::ura4 mutant devoid of the rpb6 gene on the chromosome with Rpb6 supplied by an expression plasmid. Since the rpb6 gene on the plasmid is under the control of the nmt1 promoter, Rpb6 was expected to be synthesized only in the absence of thiamine addition (24). The mutant S. pombe thus constructed was unable to grow in the presence of thiamine. Thus, we concluded that rpb6 is an essential gene for S. pombe growth. Into this rpb6 disruptant, we introduced a set of compatible plasmids expressing various degrees of both N- and C-terminal deletion mutants of Rpb6 and tested the in vivo function of truncated Rpb6 proteins in the absence of intact Rpb6 expression. As summarized in Fig. 1, the N-terminal deletion to residue 61 did not affect function as measured by cell viability, while the C-terminal deletion of six amino acid residues made Rpb6 inactive. The results indicate that the region essential for Rpb6 function is located within the C-terminal half of Rpb6 between residues 61 and 139. The sequence of this region is highly conserved among Rpb6 homologues from seven organisms so far sequenced (see Fig. 3). Based on the deletion mapping of S. pombe Rpb6 from both termini, we constructed a minimum fragment consisting of residues 61 to 139, lacking both N- and C-terminal dispensable regions. This minimum fragment was, however, unable to support cell growth.
FIG. 1.
Functional analysis of truncated mutants of Rpb6. Expression plasmids for N- and C-terminal deletion mutants of rpb6 were constructed using vector pAI-ARS and transformed into S. pombe SpRpb6::ura4 containing pREP81-Rpb6 (Table 2). In the absence of thiamine, the intact Rpb6 is expressed from pREP81-Rpb6 (agar plate, upper panel); its expression is repressed by the addition of thiamine (agar plate, lower panel). The functional integrity of truncated Rpb6 mutants was tested in the absence of intact Rpb6.
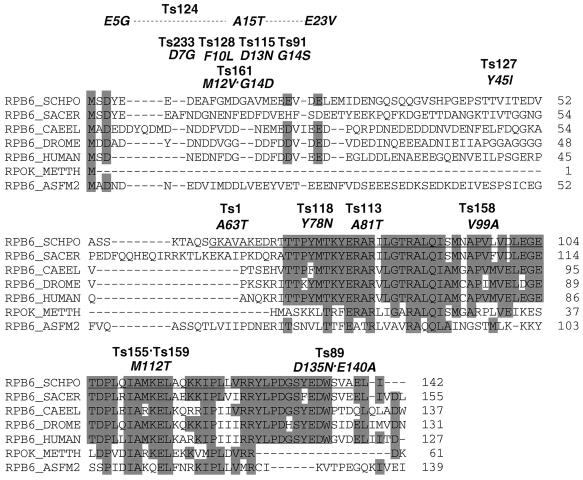
FIG. 3.
Sequence of the rpb6 gene from the temperature-sensitive rpb6 mutants. The rpb6 gene was PCR amplified from total DNA of the 14 temperature-sensitive rpb6 mutants isolated in this study and sequenced. The positions of mutations are shown for the S. pombe (SCHP0) rpb6 gene together with those of all the known rpb6 homologues (SACER, S. cerevisiae; CAEEL, Caenorhabditis elegans; DROME, Drosophila melanogaster; HUMAN, Homo sapiens; METTH, Methanobacterium thermoautotrophicum; ASFM2, African swine fever virus gene 2). The sequences conserved among the rpb6 homologues of these seven species are shaded.
Isolation of Rpb6 mutants.
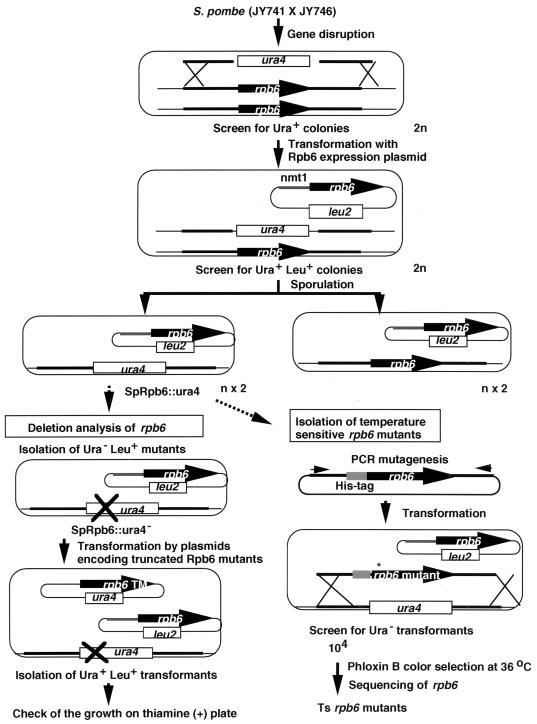
To isolate S. pombe mutants carrying an amino acid substitution in the rpb6 gene, the rpb6::ura4 gene in the rpb6 disruptant was replaced by PCR-mutagenized rpb6 by homologous recombination, and Ura− recombinants were isolated on a 5-fluoroorotic acid-containing plate (see Fig. 2 for an outline and Materials and Methods for experimental details). For quick isolation of mutant RNA polymerases, a His8 tag sequence was added at the N terminus of rpb6. Starting from 104 Ura− colonies, we have so far isolated 14 independent temperature-sensitive mutants, which cannot grow at 36°C, each carrying a single (or multiple in a few cases) mutation in the rpb6 gene.
FIG. 2.
Genetic manipulations of the S. pombe rpb6 gene. The rpb6 disruptant was constructed by homologous recombination after transformation of plasmid pRpb6::ura4 into S. pombe JY741 (ura4 leu1 ade6) and screening for Ura+ transformants. The haploid containing the ura4+ allele was used for the functional analysis of rpb6 deletion mutants and the generation of temperature-sensitive rpb6 mutants. Details are described in Materials and Methods.
The entire Rpb6-coding region was PCR amplified from all 14 temperature-sensitive mutants, and the complete sequences were determined for all PCR products. Eight mutants carried a single (or triple for one mutant) mutation in the N-terminal region between residues 5 and 23 (Table 2). In particular, mutations were clustered in a narrow region from residues 10 to 14 (Fig. 3). This was unexpected because the N-terminal region is dispensable for cell growth (Fig. 1) and because none of the S. cerevisiae RPB6 temperature-sensitive mutants carried mutations in the N-terminal dispensable region (28).
TABLE 2.
Fission yeast rpb6 mutants
| Designation | rpb6 mutation(s) | Rpb6 change | Growth at 36°C | 6AU sensitivity |
|---|---|---|---|---|
| 6NH (wild type) | None | None | Normal | Resistant |
| Ts1 | G198A, T288A | A63T | Tsa | Sensitive |
| Ts89 | G403A, A419C | D135N, E140A | Ts (leaky) | Resistant |
| Ts91 | G40T, G132A, A348G | G14S | Ts (leaky) | Resistant |
| Ts113 | G241A | A81T | Ts (leaky) | Resistant |
| Ts115 | G37T | D13N | Ts (leaky) | Resistant |
| Ts118 | T232A | Y78N | Ts (leaky) | Resistant |
| Ts124 | A14G, G43A, A68T | E5G, A15T, E23V | Ts (leaky) | Resistant |
| Ts127 | C134T | Y45I | Ts | Resistant |
| Ts128 | T30A | F10L | Ts (leaky) | Resistant |
| Ts155 | T335C, A402G | M112T | Ts | Sensitive |
| Ts158 | T296C | V99A | Ts | Sensitive |
| Ts159 | T335C | M112T | Ts | Sensitive |
| Ts161 | A34G, G41A, T120C | M12V, G14D | Ts (leaky) | Resistant |
| Ts233 | A20G | D7G | Ts (leaky) | Resistant |
Ts, temperature sensitive.
Six mutants carried a single (or in one case double) mutation in the C-terminal essential region downstream from residue 61. Mutations in the most conserved region of Rpb6 crucial for functions must have rendered S. pombe lethal as in the case of S. cerevisiae (28).
Growth characteristics of the Rpb6 mutants.
Growth of all temperature-sensitive mutants was monitored on a rich medium plate after up-shift from the permissive (30°C) to the nonpermissive (36°C) temperature. Five mutants, Ts1 (A63T), Ts127 (Y45I), Ts155 (M112T), Ts158 (V99A), and Ts159 (M112T), stopped growing after 5 days, while others continued to grow, albeit at reduced rates. Detailed analysis was then carried out for seven mutants, Ts1 (A63T), Ts158 (V99A), and Ts159 (M112T) from the first group and Ts89 (D135N and E139A), Ts113 (A81T), Ts118 (Y78N), and Ts127 (T45I) from the second group; these mutants carried a single mutation (or double in the case of Ts89) in the essential region except for Ts127, which had a Thr45Ile mutation in the nonessential region.
Growth of these seven mutants and of the parental strain 6NH was monitored in a liquid minimal medium containing adenine, leucine, and uracil after temperature up-shift from 30 to 36°C. All three group I mutants (Ts1, Ts158, and Ts159) and one group II mutant (Ts127) stopped growing after the temperature up-shift, but the other three leaky mutants continued to grow at reduced rates (data not shown).
Growth of the Rpb6 mutants was also examined on a plate containing 6AU, which inhibits IMP dehydrogenase and leads to limitations in GTP and UTP pools (11, 21). After 5 days at the permissive temperature (30°C), the growth of three mutants, Ts1, Ts158, and Ts159, was significantly reduced (these mutants are hereafter classified as group I mutants), suggesting that Rpb6 plays a role in the catalytic activity of RNA synthesis. However, the other four group II mutants grew as fast as the wild type even in the presence of 6AU (Table 2).
Functional interaction in vivo of Rpb6 with TFIIS.
The S. cerevisiae mutants lacking the PPR2 gene encoding the elongation factor TFIIS (or SII) are sensitive to 6AU (27) because of the elongation arrest of RNA chains due to limitation in nucleotide pools (11). Likewise, some RPB1 (subunit 1) and RPB2 (subunit 2) mutants of S. cerevisiae are sensitive to 6AU (4, 22), suggesting that these RNA polymerase II mutants are defective at the step of RNA chain elongation. One possibility raised by this consideration is that Rpb6 is involved in transcription elongation.
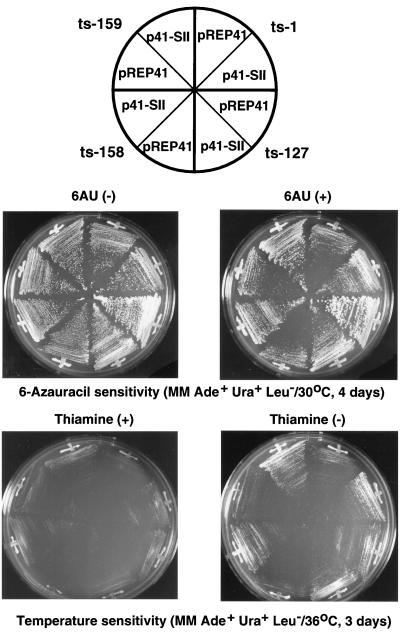
We then tried to suppress the 6AU-sensitive phenotype of three group I rpb6 mutants, Ts1, Ts158, and Ts159, by high-level expression of the S. pombe ppr2 gene. As shown in Fig. 4, the 6AU sensitivity of the three group I mutants was suppressed in the presence of multicopy plasmid pREP41-TFIIS (or p41-SII) encoding TFIIS (Table 1). Growth of the 6AU-insensitive mutant Ts127 was, however, not affected by overexpression of TFIIS. The 6AU sensitivity of Ts127 was as low as that of the wild type (data not shown). The temperature-sensitive phenotype of the three group I rpb6 mutants was also suppressed by introducing multiple copies of the TFIIS expression plasmid. Thus, we concluded that both 6AU sensitivity and the temperature-sensitive phenotype were conferred by the same mutation. Since suppression of the mutant phenotypes by TFIIS is allele specific, one possibility is that TFIIS directly interacts with Rpb6; if this is the case, the TFIIS contact site is located between residues 63 and 112 within the C-terminal essential region of Rpb6.
FIG. 4.
Suppression of sensitivities of rpb6 mutants to 6AU and high temperature by multiple copies of the TFIIS expression plasmid. Three 6AU-sensitive and temperature-sensitive rpb6 mutants (Ts1, Ts158, and Ts159) and one 6AU-insensitive temperature-sensitive mutant (Ts127) were transformed into S. pombe carrying either TFIIS expression plasmid pREP41-TFIIS or control plasmid pREP41. The transformants were grown on plates with or without 6AU (upper panels) and in the presence or absence of thiamine (lower panels). Note that the 6AU sensitivity of Ts127 was as low as that of wild-type S. pombe strain 6NH.
Interaction in vitro of RNA polymerase II with TFIIS.
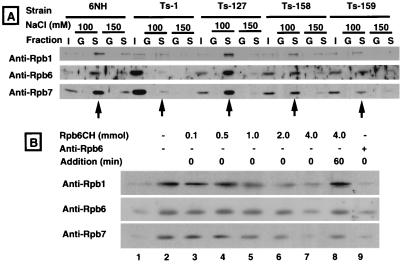
To set up the assay system for direct protein-protein interaction between RNA polymerase II and TFIIS, we expressed a GST fusion form of TFIIS in E. coli and purified the recombinant TFIIS to apparent homogeneity by glutathione-Sepharose column chromatography. The purified GST-TFIIS was mixed with partially purified RNA polymerase II from the wild-type S. pombe and some Rpb6 temperature-sensitive mutants. GST-TFIIS complexes formed were isolated by using glutathione-Sepharose beads. The recovery of RNA polymerase II in the unbound and bead-bound fractions was measured by Western blot analysis using anti-Rpb1, anti-Rpb6, and anti-Rpb7 antibodies. The assay system was used to examine possible influence of the Rpb6 mutations on TFIIS-RNA polymerase II interaction. As shown in Fig. 5A, the RNA polymerase II of wild-type 6NH and Ts127 mutant S. pombe was recovered in the GST-TFIIS fraction, while the yield of RNA polymerase II in the complex fraction was significantly reduced for the three mutants, Ts1, Ts158, and Ts159 (compare lanes I [input] and S [GST-TFIIS complex]; 10% volumes of the input samples were analyzed in lanes I).
FIG. 5.
GST pull-down assay of the mutant RNA polymerase II. (A) Crude extracts of four rpb6 S. pombe mutants (Ts1, Ts127, Ts158, and Ts159) and the wild-type parent 6NH were mixed in vitro with either GST or GST-TFIIS fusion protein. Complexes formed in the presence of 100 or 150 mM NaCl were isolated by using glutathione-Sepharose beads, separated by SDS-PAGE on a 12% gel, and analyzed by Western blotting using anti-Rpb1, anti-Rpb6, and anti-Rpb7 antibodies. Lanes I, input crude extracts (1/10 of the total volume analyzed); lanes G, glutathione bead-bound fractions from GST-cell extract mixtures; S, glutathione bead-bound fractions from GST-TFIIS-cell extract mixtures. Arrows indicate the TFIIS-bound RNA polymerase II subunits at 100 mM NaCl (note that the amounts in lanes G and S are 10 times more than those in lanes I). (B) Competition assay of TFIIS complex formation. Mixtures of an S. pombe cell extract and the purified GST-TFIIS were incubated for 120 min in the presence of increasing amounts of the purified Rpb6CH protein (lanes 3 to 7). The GST-TFIIS-RNA polymerase II complexes were isolated using glutathione-Sepharose beads and subjected to SDS-PAGE (12.5% gel) followed by Western blot analysis using anti-Rpb1, anti-Rpb6, and anti-Rpb7 antibodies. Lane 8, Rpb6 was added at 60 min after the formation of the GST-TFIIS-RNA polymerase complex; lane 9, the cell extract was treated for 30 min with anti-Rpb6 antibody prior to the complex formation assay. Immunostaining was performed as described previously (14), using ECL Western blot detection reagents (Amersham Pharmacia).
If the observed interaction between RNA polymerase II and TFIIS was attributed to the direct contact between Rpb6 and TFIIS, complex formation must be hindered by the addition of Rpb6 protein. To test this possibility, increasing amounts of the purified recombinant Rpb6 were added to the complex formation assay. As shown in Fig. 5B, the addition of free Rpb6 interfered with formation of the RNA polymerase II-TFIIS complex, as detected by immunostaining using anti-Rpb1, anti-Rpb6, and anti-Rpb7 antibodies. After formation of the RNA polymerase II-TFIIS complex, the exogenous addition of excess Rpb6 did not reduce the level of complex (Fig. 5B, lane 8), suggesting the tight binding of TFIIS to the RNA polymerase II. Results of the competition of TFIIS-RNA polymerase II interaction by Rpb6 strongly suggest that one target of TFIIS contact on RNA polymerase II is located on Rpb6.
Transcription stimulation in vitro by TFIIS.
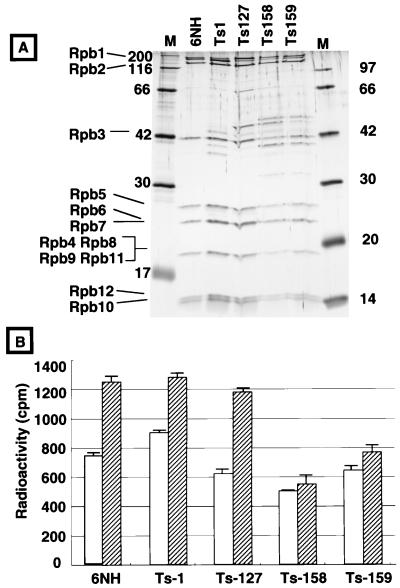
To test the functional interaction in vitro between the RNA polymerase and TFIIS, we purified the RNA polymerase II from wild-type S. pombe and some rpb6 mutants by Ni2+-agarose affinity chromatography (Fig. 6A shows sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] patterns) and measured the activity of promoter-independent nonspecific transcription for the RNA polymerases in the presence and absence of TFIIS (Fig. 6B). The activities of the wild-type (6NH) and Ts127 mutant RNA polymerases were activated more than 1.7- and 1.9-fold, respectively, by the addition of TFIIS, but the activation level was significantly reduced for the mutant RNA polymerases with mutations in the essential region of Rpb6. The stimulation levels were less than 1.3-, 1.1-, and 1.2-fold for Ts1, Ts158, and Ts159 RNA polymerases, respectively. This preliminary assay supports the conclusion that Rpb6 is one target of TFIIS binding on the RNA polymerase II and the contact site on Rpb6 is located within the C-terminal essential region.
FIG. 6.
Transcription stimulation by TFIIS of wild-type and mutant RNA polymerase II. (A) RNA polymerase II was partially purified from the wild type (6NH) and four mutants (Ts1, Ts127, Ts158, and Ts159). Protein composition was analyzed by SDS-PAGE, and the gel was stained with silver. (B) Nonspecific RNA synthesis was carried out in the presence (shaded bars) or absence (open bars) of TFIIS, using the partially purified RNA polymerase II. The reaction mixtures and reaction conditions were as described in Materials and Methods. Standard errors of two independent assays are shown by bars.
DISCUSSION
RNA synthesis in vitro by the RNA polymerases I and II from S. cerevisiae is inhibited by the addition of anti-RPB6 antibodies (9, 36). The S. cerevisiae RNA polymerase I lacking ABC23 (RPB6) is defective in basal transcription activity (21). Mutant studies herein described support the concept that Rpb6 is an essential subunit for the function of S. pombe RNA polymerase II. In support of the essential role of Rpb6 in the functions of all three RNA polymerases, Rpb6 homologues exist in wide varieties of the RNA polymerase from eukaryotes, archaea, and some DNA viruses (20, 23, 41). RPB6 of S. cerevisiae is functionally interchangeable with the corresponding subunits from human and fission yeast proteins (25, 40, 41).
Deletion analysis indicated that the essential region for Rpb6 function is located in the C-terminal half. The functional map of S. pombe Rpb6 is in good agreement with that of S. cerevisiae RPB6 (28). The dispensable nature of the N-terminal proximal region to residue 43 has been observed for the S. cerevisiae RPB6 subunit consisting of 155 amino acid residues (28). In agreement with these findings, the sequence conservation of Rpb6 family proteins is higher for the C-terminal region (Fig. 3). Seven of the 14 Rpb6 temperature-sensitive mutants isolated in this study were, however, found to carry mutations in the N-terminal half. In particular, mutations are clustered within a narrow region between residues 10 and 20. Since this region is not present in the Rpb6 homologue of archaea and is not conserved among eukaryotes (Fig. 3), the N-terminal protruding tail may have a nonessential but unique regulatory or control function for the Rpb6 structure, specific for S. pombe.
One novel finding in our mutant studies is the functional interaction of Rpb6 with transcription elongation factor TFIIS (or SII). TFIIS, originally isolated as a stimulation factor involved in transcription elongation by the RNA polymerase II (38), is present throughout eukaryotes, archaea (26), and a group of DNA viruses (2, 10, 33). During transcription elongation, TFIIS induces the cleavage of nascent RNA at the pause or arrest sites and thereby enhances transcription elongation (6, 31, 32). As in the case of bacterial GreA and GreB proteins, TFIIS directly binds to the RNA polymerase and stimulates its RNA synthesis activity by cutting off nascent RNA chains at 3′ ends (18, 37, 42). The TFIIS of S. cerevisiae is composed of three domains, I, II, and III; the nuclear magnetic resonance structures have been solved for the C-terminal proximal domains II and III (29). Domains II and III are known to be essential for interaction with the RNA polymerase II (1, 7, 39). S. cerevisiae mutants carrying mutations in the PPR2 gene for TFIIS have been isolated; these mutants show high-level sensitivity to 6AU that inhibits IMP dehydrogenase and ultimately results in limitation in GTP and UTP pools (11). Previous genetic studies of S. cerevisiae RNA polymerase II indicated functional interactions of TFIIS with two large subunits, RPB1 (4) and RPB2 (22, 30). In agreement with this prediction, a mutant RPB1 which showed decreased binding affinity to TFIIS was isolated (48). Here we showed that Rpb6 is also involved in the functional interaction between the RNA polymerase II and TFIIS. Several lines of evidence support the concept of direct contact between Rpb6 and TFIIS: (i) the mutations affecting 6AU sensitivity are allele specific (Fig. 4); (ii) the RNA polymerase II-TFIIS interaction is competitively inhibited by the exogenous addition of Rpb6 (Fig. 5); and (iii) the transcription enhancement by TFIIS is interfered with by the addition of anti-Rpb6 but not antibodies against other subunits (36).
The contact sites of TFIIS on both of the two largest subunits, RPB1 and RPB2, of S. cerevisiae RNA polymerase II have been suggested based on the observations that some mutations in the genes coding for the two largest subunits confer increased sensitivity to 6AU (4, 22), but the direct interaction of mutant RNA polymerases with TFIIS has not been examined for these RPB1 and RPB2 mutants. If TFIIS makes direct contact with the two largest subunits, one possible mechanism is that TFIIS interacts with the RNA polymerase II at the boundary formed among Rpb1, Rpb2, and Rpb6. In fact, Rpb6 of S. pombe interacts with both Rpb1 and Rpb2 (14). More direct assays of protein-protein contacts are required to define the exact contact target of TFIIS among three candidate subunits of RNA polymerase II and also to exclude the possibility that the effect of rpb6 mutations is indirect due to general structural changes in mutant RNA polymerase II.
ACKNOWLEDGMENTS
This work was supported by grants-in-aid from the Ministry of Education, Science and Culture of Japan and by Core Research for Evolutional Science and Technology of Japan Science and Technology Corporation.
REFERENCES
- 1.Agarwal K, Baek K, Jeon C, Miyamoto K, Ueno A, Yoon H. Stimulation of transcript elongation requires both the zinc finger and RNA polymerase II binding domains of human TFIIS. Biochemistry. 1991;30:7842–7851. doi: 10.1021/bi00245a026. [DOI] [PubMed] [Google Scholar]
- 2.Ahn B-Y, Gershon P D, Jones E V, Moss R. Identification of rpo30, a vaccinia virus RNA polymerase gene with structural similarity to a eucaryotic transcription elongation factor. Mol Cell Biol. 1990;10:5433–5441. doi: 10.1128/mcb.10.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- 4.Archambault J, Lacroute F, Ruet A, Friesen J D. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archambault J, Schappert K T, Friesen J D. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol Cell Biol. 1990;10:6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awrey D E, Weilbaecher R G, Hemming S A, Orlicky S M, Kane C M, Edwards A M. Transcription elongation through DNA arrest site. J Biol Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 7.Awrey D E, Shimasaki N, Koth C, Weilbaecher R, Olmsted V, Kazanis S, Shan X, Arellano J, Arrowsmith C H, Kane C M, Edwards A M. Yeast transcript elongation factor (TFIIS), structure and function. J Biol Chem. 1998;273:22595–22605. doi: 10.1074/jbc.273.35.22595. [DOI] [PubMed] [Google Scholar]
- 8.Azuma Y, Yamagishi M, Ueshima R, Ishihama A. Subunits of the Schizosaccharomyces pombe RNA polymerase II: enzyme purification and structure of the subunit 3 gene. Nucleic Acids Res. 1993;21:3749–3754. doi: 10.1093/nar/21.16.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beant B, Huet J, Sentenac A, Fromageot P. Analysis of yeast RNA polymerases with subunit-specific antibodies. J Biol Chem. 1983;258:11968–11973. [PubMed] [Google Scholar]
- 10.Dixon L K, Twigg S R K, Baylis S A, Vydelingum S, Bristow C, Hammond J M, Smit G L. Nucleotide sequence of a 55 kbp region from the right end of the genome of a pathogenic African swine fever virus isolate. J Gen Virol. 1994;75:1655–1684. doi: 10.1099/0022-1317-75-7-1655. [DOI] [PubMed] [Google Scholar]
- 11.Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 12.Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polyphosphate chain reaction. Arch Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 13.Grimm C, Kohli J. Observations on integrative transformation in Saccharomyces pombe. Mol Gen Genet. 1988;215:87–93. doi: 10.1007/BF00331308. [DOI] [PubMed] [Google Scholar]
- 14.Ishiguro A, Kimura M, Yasui K, Iwata A, Ueda S, Ishihama A. Two large subunits of the fission yeast RNA polymerase II provide platforms for the assembly of small subunits. J Mol Biol. 1998;279:703–712. doi: 10.1006/jmbi.1998.1823. [DOI] [PubMed] [Google Scholar]
- 15.Ishiguro J, Kobayashi W. A practical method for fission yeast transformation by electroporation. Jpn J Genet. 1995;70:1–6. doi: 10.1266/jjg.70.1. [DOI] [PubMed] [Google Scholar]
- 16.Ishihama A. Subunit assembly of Escherichia coli RNA polymerase. Adv Biophys. 1981;14:1–35. [PubMed] [Google Scholar]
- 17.Ishihama A, Kimura M, Mitsuzawa H. Subunits of yeast RNA polymerases in structure and function. Curr Opin Microbiol. 1998;1:190–196. doi: 10.1016/s1369-5274(98)80010-6. [DOI] [PubMed] [Google Scholar]
- 18.Izban M G, Luse D S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′-5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 19.Kimura M, Ishiguro A, Ishihama A. RNA polymerase II subunits 2, 3 and 11 form a core subassembly with DNA binding activity. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 20.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanzendörfer M, Smid A, Klinger C, Schults P, Sentenac A, Carles C, Riva M. A shared subunit belongs to the eukaryotic core RNA polymerase. Genes Dev. 1997;11:1037–1047. doi: 10.1101/gad.11.8.1037. [DOI] [PubMed] [Google Scholar]
- 22.Lennon J C, III, Wind M, Saunders L, Hock M B, Reins D. Mutations in RNA polymerase II and elongation factor IIS severely reduce mRNA levels in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5771–5779. doi: 10.1128/mcb.18.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Z, Kutish G F, Sussman M D, Rock D L. An African swine fever virus gene with a similarity to eukaryotic RNA polymerase subunit 6. Nucleic Acids Res. 1993;21:2940. doi: 10.1093/nar/21.12.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maundrell K. nmt1 of fission yeast. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 25.McKune K, Woychik N A. Functional substitution of an essential yeast RNA polymerase subunit by a highly conserved mammalian counterpart. Mol Cell Biol. 1994;14:4155–4159. doi: 10.1128/mcb.14.6.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyao T, Honda A, Qu Z, Ishihama A. Mapping of Rpb3 and Rpb5 contact sites on two large subunits, Rpb1 and Rpb2, of the fission yeast RNA polymerase II. Mol Gen Genet. 1998;259:123–129. doi: 10.1007/s004380050796. [DOI] [PubMed] [Google Scholar]
- 27.Nakanishi T, Nakano A, Nomura K, Sekimizu K, Natori S. Structure-function relationship of yeast SII in terms of stimulation of RNA polymerase II, arrest relief, and suppression of 6-azauracil sensitivity. J Biol Chem. 1995;270:8991–8997. doi: 10.1074/jbc.270.15.8991. [DOI] [PubMed] [Google Scholar]
- 28.Nouraini S, Archambault J, Friesen J D. Rpo26p, a subunit common to yeast RNA polymerases, is essential for the assembly of RNA polymerases I and II and for stability of the largest subunits of these enzymes. Mol Cell Biol. 1996;16:5985–5996. doi: 10.1128/mcb.16.11.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olmsted V K, Awrey D E, Koth C, Shan X, Morin P E, Kazanis S, Edwards A M, Arrowsmith C H. Yeast transcript elongation factor (TFIIS), structure and function. J Biol Chem. 1998;273:22589–22594. doi: 10.1074/jbc.273.35.22589. [DOI] [PubMed] [Google Scholar]
- 30.Powell W, Reines D. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reines D, Chamberlin M J, Kane C K. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J Biol Chem. 1989;264:10799–10809. [PubMed] [Google Scholar]
- 32.Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigez J M, Salas M L, Viñuela E. Genes homologous to ubiquitin-conjugating proteins and eukaryotic transcription factor SII in African swine fever virus. Virology. 1992;186:40–52. doi: 10.1016/0042-6822(92)90059-x. [DOI] [PubMed] [Google Scholar]
- 34.Sakurai H, Ishihama A. Gene organization and protein sequence of the small subunits of Schizosaccharomyces pombe RNA polymerase II. Gene. 1997;196:165–174. doi: 10.1016/s0378-1119(97)00222-9. [DOI] [PubMed] [Google Scholar]
- 35.Sakurai H, Mitsuzawa H, Kimura M, Ishihama A. Rpb4 subunit of the fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes. Mol Cell Biol. 1999;19:7511–7518. doi: 10.1128/mcb.19.11.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawadogo M, Huet J, Fromageot P. Similar binding site for P37 factor on yeast RNA polymerases A and B. Biochem Biophys Res Commun. 1980;96:258–264. doi: 10.1016/0006-291x(80)91208-5. [DOI] [PubMed] [Google Scholar]
- 37.Sawadogo M, Sentenac A, Fromageot P. Interaction of a new polypeptide with yeast RNA polymerase B. J Biol Chem. 1980;255:12–15. [PubMed] [Google Scholar]
- 38.Sekimizu K, Nakanishi Y, Mizuno D, Natori S. Purification and preparation of antibody to RNA polymerase II stimulatory factors from Ehrlich ascites tumor cells. Biochemistry. 1989;18:1582–1587. doi: 10.1021/bi00575a031. [DOI] [PubMed] [Google Scholar]
- 39.Shimoaraiso M, Nakanishi T, Kubo T, Natori S. Identification of the region in yeast S-II that defines species specificity in its interaction with RNA polymerase II. J Biol Chem. 1997;272:26550–26554. doi: 10.1074/jbc.272.42.26550. [DOI] [PubMed] [Google Scholar]
- 40.Shpakovski G V. The fission yeast Schizosaccharomyces pombe rpb6 gene encodes the common phosphorylated subunit of RNA polymerase and complements a mutation in the corresponding gene of Saccharomyces cerevisiae. Gene. 1994;147:67–69. doi: 10.1016/0378-1119(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 41.Shpakovski G V, Acker I, Wintzerith M, Lacroix J-F, Thuriaux P, Vigneron M. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sopta M, Carthew R W, Greenblatt L. Isolation of three proteins that bind to mammalian RNA polymerase II. J Biol Chem. 1985;260:10353–10360. [PubMed] [Google Scholar]
- 43.Svetlov V, Cooper T G. Efficient PCR-based random mutagenesis of subgenic (100 bp) DNA fragments. Yeast. 1998;14:89–91. doi: 10.1002/(SICI)1097-0061(19980115)14:1<89::AID-YEA194>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 44.Thuriaux P, Sentenac A. Yeast nuclear RNA polymerases. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. II. Gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1–48. [Google Scholar]
- 45.Williams A A, Kane C M. Isolation and characterization of the Schizosaccharomyces pombe gene encoding transcript elongation factor TFIIS. Yeast. 1996;12:227–236. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C227::AID-YEA905%3E3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 46.Woychik N A, Liao S, Kolodziej P A, Young R A. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 1990;4:313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- 47.Woychik N A, Young R A. Exploring RNA polymerase II structure and function. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 227–242. [Google Scholar]
- 48.Wu J, Awrey D E, Edwards A M, Archambault J, Friesen J D. In vitro characterization of mutant yeast RNA polymerase II with reduced binding for elongation factor TFIIS. Proc Natl Acad Sci USA. 1996;93:11552–11557. doi: 10.1073/pnas.93.21.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasui K, Ishiguro A, Ishihama A. Location of subunit-subunit contact sites on RNA polymerase II from the fission yeast Schizosaccharomyces pombe. Biochemistry. 1998;37:5542–5548. doi: 10.1021/bi972939b. [DOI] [PubMed] [Google Scholar]
- 50.Zou C, Fujita N, Igarashi K, Ishihama A. Mapping the Escherichia coli RNA polymerase contact site I for cAMP receptor protein. Mol Microbiol. 1992;6:2599–2605. doi: 10.1111/j.1365-2958.1992.tb01437.x. [DOI] [PubMed] [Google Scholar]