ABSTRACT
Pyroptosis is a newly discovered programmed cell death that is associated with tumor progression, prognosis, and treatment response. However, the potential roles of pyroptosis-related genes (PRGs) in the tumor microenvironment (TME) remain unclear. We described the alterations of PRGs in 1109 colorectal cancer (CRC) samples from genetic and transcriptional fields and evaluated their expression patterns from four independent datasets. We identified two distinct molecular subtypes and found that multi-layer PRG alterations were correlated with patient clinicopathological features, prognosis, and TME cell-infiltrating characteristics. Then, a PRG_score for predicting recurrence-free survival (RFS) was constructed and its predictive capability in CRC patients was validated. Consequently, we constructed a highly accurate nomogram for improving the clinical applicability of the PRG_score. A low PRG_score, characterized by increased microsatellite instability-high (MSI-H), mutation burden, and immunity activation, indicated favorable odds of RFS. Moreover, the PRG_score was significantly associated with the cancer stem cell (CSC) index and chemotherapeutic drug sensitivity. Our comprehensive analysis of PRGs in CRC demonstrated their potential roles in the tumor-immune-stromal microenvironment, clinicopathological features, and prognosis. These findings may improve our understanding of PRGs in CRC and pave a new path for the assessment of prognosis and the development of more effective immunotherapy strategies.
KEYWORDS: Pyroptosis, colorectal cancer, tumor microenvironment, microsatellite instability, recurrence-free survival
Introduction
Pyroptosis, a form of inflammatory cell death, is exceptional compared to other types of programmed cell death.1 It is characterized by Gasdermin family-mediated pore formation, followed by cell swelling, lysis, and the release of pro-inflammatory intracellular cytokines including interleukin (IL)-18, IL- 1β, High mobility group box 1 (HMGB1), and adenosine triphosphate (ATP).2 In terms of mechanism, the canonical (caspase-1) and non-canonical inflammasome-induced pyroptosis (caspase-4/5/11) are involved in pyroptosis. Pyroptosis plays a crucial role in the pathogenesis and progression of various cancers, including colorectal cancer (CRC).3,4 CRC is characterized by inherent biological invasiveness and specific radiological and chemical resistance that result in high recurrence rates and progression. Gasdermin D (GSDMD) expression is decreased in CRC cells compared to that in adjacent normal cells, and low GSDMD expression is associated with a worse CRC prognosis. Pyroptosis induced by lipopolysaccharide can inhibit CRC tumorigenesis by promoting GSDMD expression and N-terminal GSDMD membrane translocation to improve chemosensitivity in response to oxaliplatin in CRC cells.5 Knockdown of gasdermin C (GSDMC) can inhibit CRC cell proliferation and tumorigenesis, while GSDMC overexpression promotes cell proliferation, suggesting that GSDMC may be a promising therapeutic target for CRC.6 Yu et al.7 reported that gasdermin E (GSDME) mediated lobaplatin-induced pyroptosis downstream of the ROS/JNK/Bax-mitochondrial apoptosis pathway and caspase-3/-9 activation.
Emerging evidence also indicates crosstalk between pyroptosis and the tumor immune microenvironment.8,9 The tumor microenvironment (TME) has been widely implicated tumor development and progression.10 In addition to tumor cells, the TME also includes fibroblasts, endothelial cells, immune and inflammatory cells, extracellular matrix elements, and diffusible cytokines and chemokines secreted from cancer and stromal cells. The complex crosstalk between tumor cells and nonmalignant cells produces TME that affects cancer development and progression.11 Malignant cells that interact with surrounding cells through the circulatory and lymphatic systems to promote tumor angiogenesis, and induce immune tolerance by releasing cell signaling molecules. The TME can also influence tumor progression, and tumor-infiltrating immune cells (TIICs) within the TME can predict cancer prognosis.12 At present, due to technical limitations, most studies assess only one or two pyroptosis-related genes (PRGs) and cell types, while the anti-tumor effect is characterized by numerous genes interacting in a highly coordinated manner. Hence, a comprehensive understanding of the characteristics of TME cell infiltration mediated by multiple PRGs may provide important insights for understanding the underlying mechanism of CRC tumorigenesis and predicting the response to immunotherapy.
This study comprehensively evaluated the expression profiles of PRGs and obtained a comprehensive overview of the intratumoral immune landscape using two computational algorithms; namely, CIBERSORT and ESTIMATE. First, 1109 patients with CRC were stratified into two discrete subtypes according to PRG expression levels. Patients were then classified into three gene subtypes based on differentially expressed genes (DEGs) identified based on the two pyroptosis subtypes. We further established a scoring system to predict recurrence-free survival (RFS) and characterize the immune landscape of CRC, which accurately predicted patient outcomes and responses to immunotherapy.
Materials and methods
Data sources
Figure S1 shows a map of the process of the present work. Gene expression (fragments per kilobase million, FPKM) and the relevant prognostic and clinicopathological data of CRC were downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) databases. Three GEO CRC cohorts (GSE39582, GSE17536, and GSE38832) and TCGA cohorts were obtained for subsequent analyses. We obtained the raw “CELL” files and performed background adjustment and quantile normalization. The FPKM values of TCGA-colon adenocarcinoma/rectum adenocarcinoma (COAD/READ) were transformed into transcripts per kilobase million (TPM), as previously described, and were believed to be identical to those from microarrays.13,14 Four datasets were combined, and batch effects were eliminated by applying the “Combat” algorithm. We excluded data from patients with no RFS information; thus, a total of 1109 CRC patients were included in the subsequent analyses. Detailed information on these 1109 patients with CRC is presented in Table S1. The clinical variables included age, sex, tumor location, TNM stage, KRAS mutation, BRAF mutation, follow-up time, and survival status.
Consensus clustering analysis of PRGs
Forty-eight PRGs were retrieved from the MSigDB Team (REACTOME_PYROPTOSIS) (http://www.broad.mit.edu/gsea/msigdb/) and previous publications.15 The full details of these genes are shown in Table S2. The R package “ConsensusClusterPlus” was employed for consensus unsupervised clustering analysis to classify patients into distinct molecular subtypes according to PRG expression. This clustering was performed based on the following criteria: First, the cumulative distribution function (CDF) curve increased gradually and smoothly. Second, no groups had a small sample size. Lastly, after clustering, the intra-group correlation increased, while the inter-group correlation decreased. To investigate the differences in PRGs in biological processes, gene set variation analysis (GSVA) was performed with the hallmark gene set (c2.cp.kegg.v7.2) derived from the MSigDB database.
Relationship between molecular subtypes with the clinical features and prognosis of CRC
To examine the clinical value of the two subtypes identified by consensus clustering, we compared the relationships between molecular subtypes, clinicopathological characteristics, and prognosis. The patient characteristics included age, sex, tumor location, TNM stage, KRAS mutation status, and BRAF mutation status. Furthermore, the differences in RFS among different subtypes were assessed using Kaplan–Meier curves generated by the “survival” and “survminer” R packages.
Correlations of molecular subtypes with TME, PD-1, and PD-L1 in CRC
We used the ESTIMATE algorithm to evaluate the immune and stromal scores of each patient. In addition, the fractions of 22 human immune cell subsets of every CRC sample were calculated by the CIBERSORT algorithm.16 Furthermore, the levels of immune cell infiltration in the CRC TME were also determined using a single-sample gene set enrichment analysis (ssGSEA) algorithm.17 We also analyzed the correlations between the two subtypes of PD-1 and PD-L1 expression.
DEG identification and functional annotation
DEGs between the different pyroptosis subtypes were identified using the “limma” package in R with a fold-change of 1.5 and an adjusted p-value of <0.05. To further explore the potential functions of pyroptosis pattern-related DEGs and identify the related gene functions and enriched pathways, functional enrichment analyses were executed on the DEGs using the “clusterprofiler” package in R.
Construction of the pyroptosis-related prognostic PRG_score
The pyroptosis score was calculated to quantify the pyroptosis patterns of the individual tumors. First, the DEGs were subjected to univariate Cox regression analysis to identify those linked to CRC RFS. Second, the patients were classified into different subtype groups (pyroptosis gene subtype A, pyroptosis gene subtype B, and pyroptosis gene subtype C) for deeper analysis using an unsupervised clustering method based on the expression of prognostic PRGs. Finally, all CRC patients were randomly categorized into training (n = 556) and testing (n = 553) sets at a ratio of 1:1, then used the former to construct the pyroptosis-related prognostic PRG_score. Briefly, based on pyroptosis-related prognostic genes, the Lasso Cox regression algorithm was used to minimize the risk of over-fitting using the “glmnet” R package. We analyzed the change trajectory of each independent variable and then used 10-fold cross-validation to establish a model. Candidate genes were selected using multivariate Cox analysis to establish a prognostic PRG_score in the training set.
The PRG_score was calculated as follows:
PRG_score = Σ(Expi * coefi)
where Coefi and Expi denote the risk coefficient and expression of each gene, respectively. Based on the median risk score, a total of 556 patients in the training set were divided into low-risk (PRG_score < median value) and high-risk (PRG_score > median value) groups and then subjected to Kaplan–Meier survival analysis. Then, principal component analysis (PCA) was performed using the “ggplot2” R package. Similarly, the testing and all sets were divided into low- and high-risk groups, each of which was subjected to Kaplan–Meier survival analysis and the generation of receiver operating characteristic (ROC) curves.
Tissue samples
Six pairs CRC and nearby non-tumor tissues were harvested from CRC patients at the Renmin Hospital of Wuhan University. The samples were preserved at −80°C till use. Written informed consents were offered by all individuals included in this study. The study was permitted by the Ethics Committee of the Renmin Hospital of Wuhan University.
RNA isolation and quantitative real-time polymerase chain reaction PCR (RT-qPCR)
Total RNA was extracted from CRC patient tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was synthesized using the total RNA and a PrimeScript RT reagent kit (Takara). SYBR-Green assays (Takara) were used to perform the RT-qPCR on a CFX-96 instrument (Bio-Rad Laboratories, Inc., USA). The data were compulated through the 2-ΔΔC t strategy, normalizing with GAPDH. The primer sequences used for qRT-PCR in this study are listed in Table S3.
Clinical correlation and stratification analyses of the prognostic PRG_score
Chi-square tests were used to explore the relationships between the PRG_score and the clinical characteristics (age, sex, tumor location, TNM stage, KRAS mutation, and BRAF mutation). To assess whether risk scores were independent of other available clinicopathological features, we subjected the training and testing sets to univariate and multivariate analyses. In addition, we performed a stratified analysis to determine whether the PRG_score retained its predictive ability in different subgroups according to age, sex, T stage, N stage, M stage, tumor stage, tumor location, KRAS mutation status, and BRAF mutation status.
Evaluation of immune status, microsatellite instability (MSI), and cancer stem cell (CSC) index between the high- and low-risk groups
To evaluate the proportions of TIICs in the TME, CIBERSORT was used to quantify the abundance of 22 infiltrating immune cells in heterogeneous samples in the low- and high-risk groups. We explored the associations between the fractions of 22 infiltrating immune cells and seven genes in the PRG_score. We also used boxplots to examine the differential expression levels of immune checkpoints between the low- and high-score groups. Furthermore, we analyzed the relationships between the two risk groups and MSI and CSC.
Mutation and drug susceptibility analysis
To determine the somatic mutations of CRC patients between high- and low-risk groups, the mutation annotation format (MAF) from the TCGA database was generated using the “maftools” R package. We also calculated the tumor mutation burden (TMB) score for each patient with CRC in the two groups. To explore differences in the therapeutic effects of chemotherapeutic drugs in patients in the two groups, we calculated the semi-inhibitory concentration (IC50) values of chemotherapeutic drugs commonly used to treat CRC using the “pRRophetic” package.
Establishment and validation of a nomogram scoring system
The clinical characteristics and risk score were used to develop a predictive nomogram using the “rms” package based on the outcome of the independent prognosis analysis. In the nomogram scoring system, each variable was matched with a score, and the total score was obtained by adding the scores across all variables of each sample.18 Time-dependent ROC curves for 3-, 5-, and 10-year survivals were used to assess the nomogram. Calibration plots of the nomogram were used to depict the predictive value between the predicted 3-, 5-, and 10-year survival events and the virtually observed outcomes.
Statistical analyses
All statistical analyses were performed using R version 4.1.0. Statistical significance was set at p < .05.
Results
Genetic and transcriptional alterations of PRGs in CRC
This study included a total of 48 PRGs. Summary analysis of the incidence of somatic mutations in these 48 PRGs showed a relatively high mutation frequency in the COAD cohort (Figure 1a). Of the 399 COAD samples, 297 (74.44%) had mutations in the PRGs (Figure 1a). Among them, TP53 had the highest mutation frequency (55%), followed by NLRP7, while five PRGs (CYCS, TNF, PYCARD, PRKACA, and CASP6) did not have any mutations. Compared to the COAD cohort, the READ cohort had a higher PRG mutation frequency (81.62%, 111/136 samples). Similarly, TP53 showed the highest mutation frequency, followed by NLRP7 (Figure 1b). As TP53 showed the highest mutation frequency, we evaluated the relationship between TP53 mutation and PRG expression. The results showed that the expression levels of 25 of the 48 PRGs were significantly associated with TP53 mutation status (Figure S2).
Figure 1.
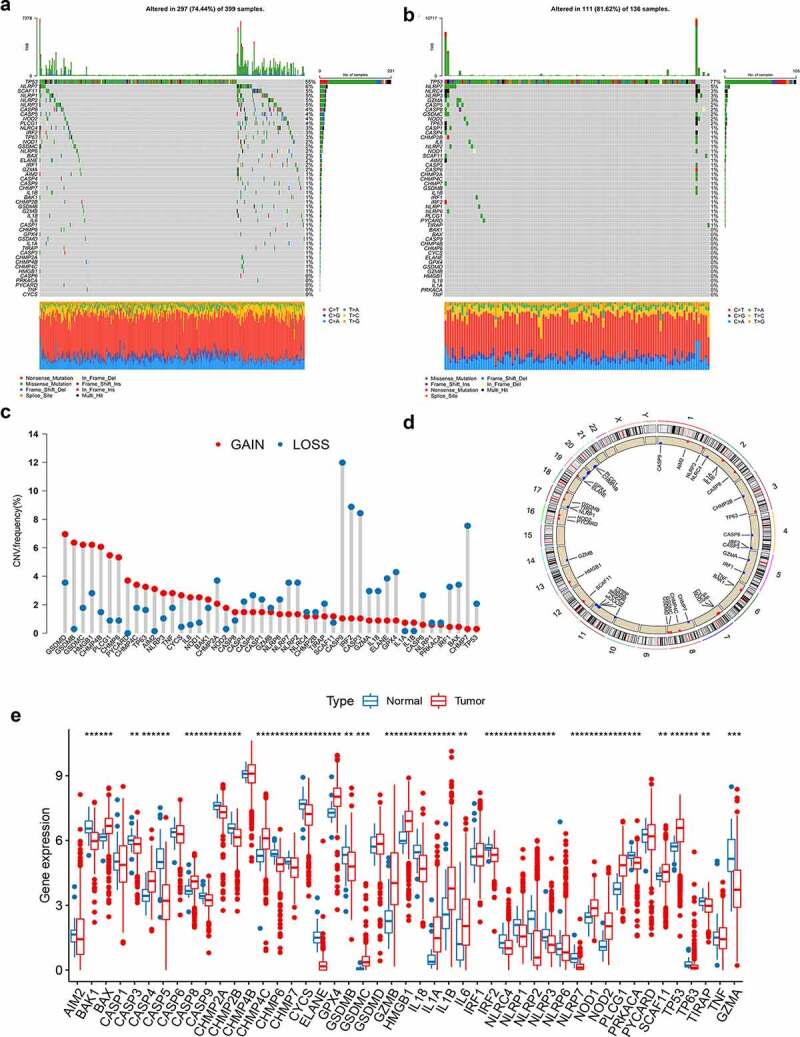
Genetic and transcriptional alterations of PRGs in CRC. (a-b) Mutation frequencies of 48 PRGs in 399 and 136 patients with COAD and READ, respectively, from the TCGA cohort. (c) Frequencies of CNV gain, loss, and non-CNV among PRGs. (d) Locations of CNV alterations in PRGs on 23 chromosomes. (e) Expression distributions of 48 PRGs between normal and CRC tissues. PRGs, pyroptosis-related genes; CRC, colorectal cancer; COAD, colon adenocarcinoma; READ, rectum adenocarcinoma; TCGA, The Cancer Genome Atlas; CNV, copy number variant
Next, we explored somatic copy number alterations in these PRGs and discovered prevalent copy number alterations in all 48 PRGs. Among them, GSDMD, GSDMB, GSDMC, HMGB1, CHMP4B, and PLCG1 had widespread copy number variation (CNV) increases, while CASP9, IRF2, CASP3, and CHMP7 showed CNV decreases (Figure 1c). Figure 1d shows the locations of the CNV alterations in the PRGs on their respective chromosomes. We further compared the mRNA expression levels between CRC and normal tissues and found that the expression levels of most PRGs were positively correlated with CNV alteration. PRGs with CNV loss, such as CASP9, IRF2, and CASP3, were expressed at lower levels in CRC samples compared to those in normal colorectal samples, while PRGs with CNV gain, such as GSDMC and HMGB1, were significantly elevated in CRC samples (Figure 1e), suggesting that CNV might regulate the mRNA expression of PRGs. However, some PRGs with CNV gain, such as GSDMB, showed downregulated mRNA expression, while other PRGs with high frequencies of CNV gain or loss showed no differences between tumor and normal samples. Thus, while CNV can explain many observed changes in PRG expression, CNV is not the only factor involved in the regulation of mRNA expression.19 Other factors, including DNA methylation and transcription factors, can also regulate gene expression.20,21 The results of our analysis showed a significant difference in both the genetic landscape and expression levels of PRGs between CRC and control samples, indicating the latent function of PRGs in CRC oncogenesis.
Identification of pyroptosis subtypes in CRC
The analytical process in this study is illustrated in Figure S1. To fully understand the expression pattern of PRG involved in tumorigenesis, 1109 patients from four eligible CRC cohorts (TCGA-COAD/READ, GSE39582, GSE17536, and GSE38832) were integrated in our study for further analysis. Detailed information on the 1109 CRC patients is presented in Table S1. The results of univariate Cox regression and Kaplan–Meier analysis revealed the prognostic values of 48 PRGs in patients with CRC (Table S4), and p < .05 was selected as the threshold for filtering. Next, we performed a multivariate Cox regression analysis on 12 prognostic PRGs, four of which (IRF1, CASP6, NLRP1, and NLRP6) were identified as independent predictive factors (Table 1). The comprehensive landscape of PRG interactions, regulator connections, and their prognostic value in patients with CRC patients was demonstrated in a pyroptosis network (Figure 2a and Table S5).
Table 1.
Multivariate Cox regression analysis of 4 PRGs associated with RFS in CRC patients
| id | HR | 95.0% CI | P value |
|---|---|---|---|
| IRF1 | 0.690585619 | 0.576–0.828 | P < .001 |
| CASP6 | 0.697606744 | 0.569–0.855 | P < .001 |
| NLRP1 | 1.315299061 | 1.059–1.634 | 0.013 |
| NLRP6 | 0.844306984 | 0.719–0.991 | 0.038 |
Figure 2.
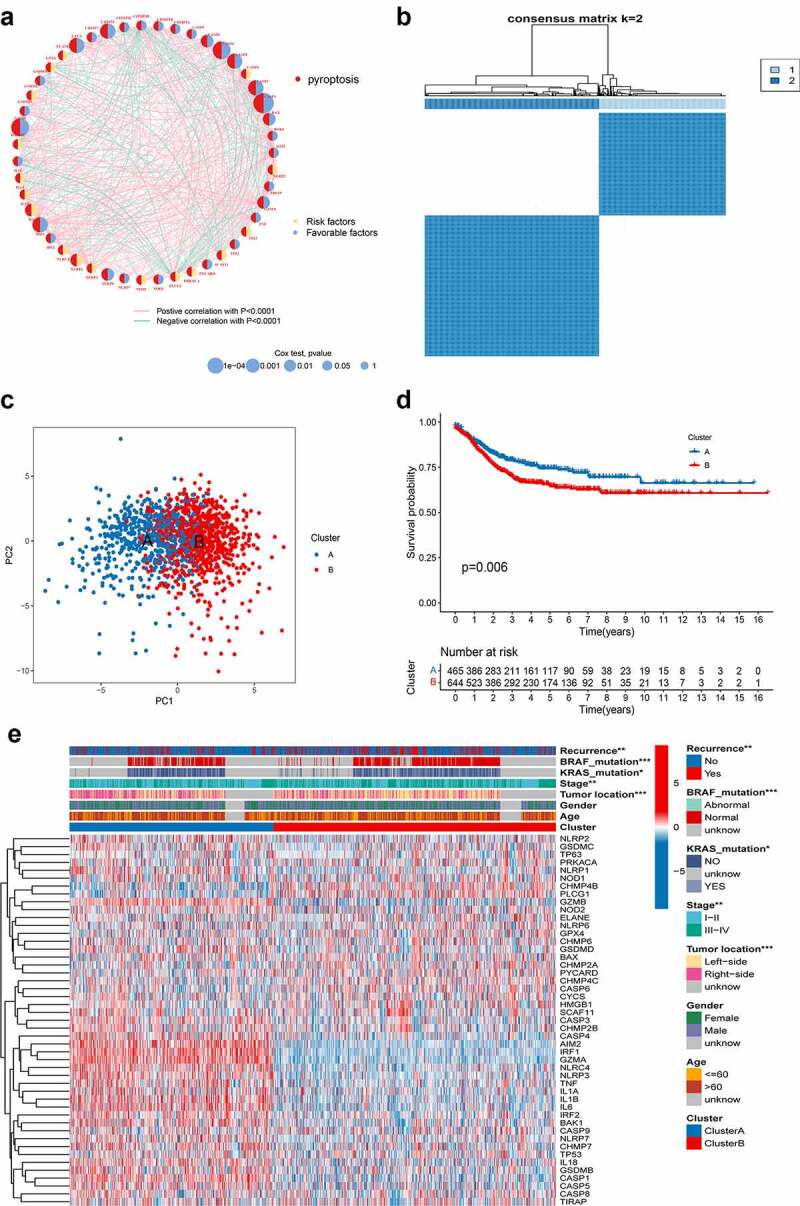
PRG subtypes and clinicopathological and biological characteristics of two distinct subtypes of samples divided by consistent clustering. (a) Interactions among PRGs in CRC. The line connecting the PRGs represents their interaction, with the line thickness indicating the strength of the association between PRGs. Green and pink represent negative and pink positive correlations, respectively. (b) Consensus matrix heatmap defining two clusters (k = 2) and their correlation area. (c) PCA analysis showing a remarkable difference in transcriptomes between the two subtypes. (d) Univariate analysis showing 48 PRGs related to the RFS time. (e) Differences in clinicopathologic features and expression levels of PRGs between the two distinct subtypes. PRG, pyroptosis-related gene; CRC, colorectal cancer; PCA, principal components analysis; RFS, recurrence-free-survival
To further explore the expression characteristics of PRGs in CRC, we used a consensus clustering algorithm to categorize the patients with CRC based on the expression profiles of the 48 PRGs (Figure S3). Our results showed that k = 2 appeared to be an optimal selection for sorting the entire cohort into subtypes A (n = 465) and B (n = 644) (Figure 2b). PCA analysis revealed significant differences in the pyroptosis transcription profiles between the two subtypes (Figure 2c). The Kaplan–Meier curves showed a longer RFS in patients with subtype A than that in patients with subtype B (log-rank test, p = .006; Figure 2d). Furthermore, comparisons of the clinicopathological features of the different subtypes of CRC revealed significant differences in PRG expression and clinicopathological characteristics (Figure 2e). As shown in Figure 2e, cluster A was preferentially related to left-sided CRC (p < .05), lower TNM stage (p < .05), without KRAS and BRAF mutations (p-value < 0.05), and lower recurrence risk (p < .05) compared to those in cluster B.
Characteristics of the TME in distinct subtypes
GSVA enrichment analysis showed that subtype A was significantly enriched in immune fully-activated pathways, including natural killer cell-mediated cytotoxicity, T and B cell receptor signaling pathway, antigen processing and presentation, cytokine receptor interaction, chemokine signaling pathway activation, RIG-I-like, NOD-like, and Toll-like receptor signaling pathways (Figure 3a; Table S6). To investigate the role of PRGs in the TME of CRC, we assessed the correlations between the two subtypes and 22 human immune cell subsets of every CRC sample using the CIBERSORT algorithm (Table S7). We observed significant differences in the infiltration of most immune cells between the two subtypes (Figure 3b). The infiltration levels of CD4 memory-activated T cells, CD8 + T cells, naive B cells, follicular helper T cells, activated NK cells, gamma delta T cells, M1 and M2 macrophages, resting mast cells, resting dendritic cells, eosinophils, and neutrophils were obviously higher in the subtype A than those in the subtype B, while resting CD4 memory T cells, memory B cells, plasma cells, regulatory T cells (Tregs), resting dendritic cells, resting NK cells, and activated mast cells had significantly lower infiltration in subtype A compared to those in subtype B. Similarly, analysis of two important immune checkpoints showed higher expression of PD1 and PD-L1 in subtype A (Figure 3c-d). We also evaluated the TME score (stromal score, immune score, and estimate score) of the two subtypes using the ESTIMATE package. For the TME score, higher stromal scores or immune scores represented higher relative contents of stromal cells or immunocytes in the TME, while estimate scores indicated the aggregation of stromal or immune scores in the TME. The results demonstrated higher TME scores in patients with subtype A (Figure 3e).
Figure 3.
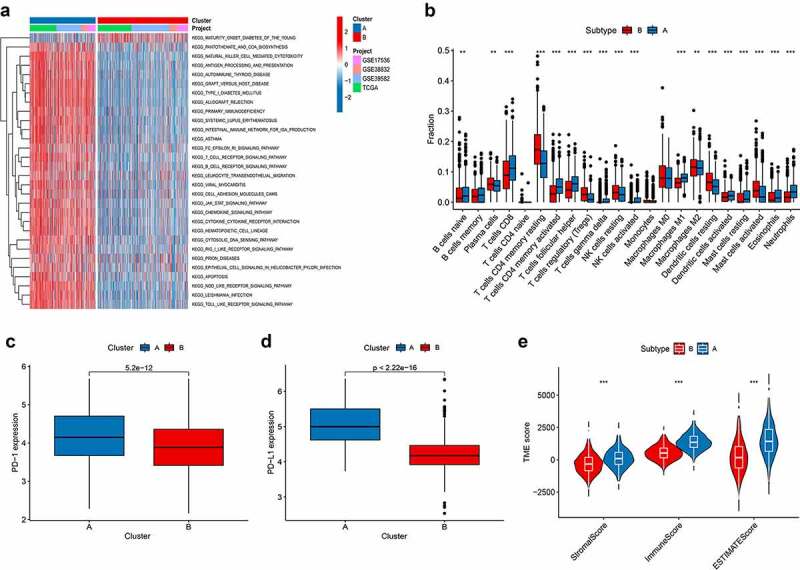
Correlations of tumor immune cell microenvironments and two CRC subtypes. (a) GSVA of biological pathways between two distinct subtypes, in which red and blue represent activated and blue inhibited pathways, respectively. (b) Abundance of 22 infiltrating immune cell types in the two CRC subtypes. (c-d) Expression levels of PD-1 and PD-L1 in the two CRC subtypes. (e-g) Correlations between the two CRC subtypes and TME score. CRC, colorectal cancer; GSVA, gene set variation analysis; TME, tumor microenvironment
Identification of gene subtypes based on DEGs
To explore the potential biological behavior of each pyroptosis pattern, we identified 409 pyroptosis subtype-related DEGs using the R package “limma,” and performed functional enrichment analysis (Figure 4a-B; Table S8). These pyroptosis subtype-related genes were significantly enriched in biological processes that were correlated with immunity (Figure 4a). KEGG analysis indicated enrichment of immune and cancer-related pathways (Figure 4b), suggesting that pyroptosis plays a vital role in the immune regulation of the TME. We then conducted univariate Cox regression analysis to identify the prognostic value of 409 subtype-related genes and screened out 129 genes related to RFS time (p < .05), which were used in the subsequent analysis (Table S9). To further validate this regulation mechanism, a consensus clustering algorithm was used to divide patients into three genomic subtypes based on prognostic genes; namely, gene subtypes A–C (Figure S4). Kaplan-Meier curves showed that patients with gene subtype B had the worst RFS, whereas patients in gene cluster C showed a favorable RFS (log-rank test, p < .001; Figure 4c). In addition, pyroptosis gene subtype B patterns were associated with advanced TNM stage, KRAS and BRAF mutations, and higher recurrence risk (Figure 4d). The three pyroptosis gene subtypes showed significant differences in PRG expression, consistent with the expected results of the pyroptosis patterns (Figure 4e).
Figure 4.
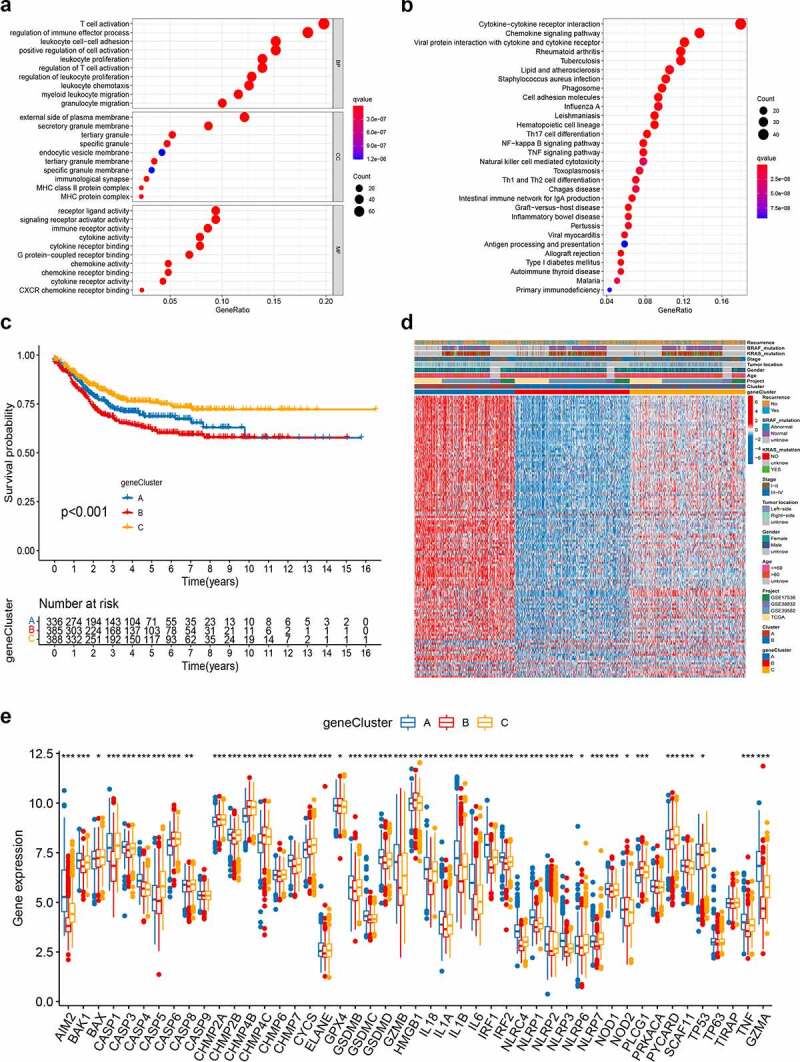
Identification of gene subtypes based on DEGs. (a-b) GO and KEGG enrichment analyses of DEGs among two pyroptosis subtypes. (c) Kaplan–Meier curves for RFS of the two gene subtypes (log-rank tests, p < .001). (d) Relationships between clinicopathologic features and the two gene subtypes. (e) Differences in the expression of 48 PRGs among the two gene subtypes. DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PRGs, PRGs, pyroptosis-related genes
Construction and validation of the prognostic PRG_score
The PRG_score was established based on the subtype-related DEGs. Figure 5a illustrates the distribution of patients in the two pyroptosis subtypes, three gene subtypes, and two PRG_score groups. First, we used the “caret package” in R to randomly classify the patients into training (n = 556) and testing (n = 553) groups at a ratio of 1:1. LASSO and multivariate Cox analyses for 129 pyroptosis subtype-related prognostic DEGs were performed to further select optimum prognostic signature. Followed by LASSO regression analysis, 20 RFS-associated genes remained according to the minimum partial likelihood deviance (Figure S5A-B). We then performed multivariate Cox regression analysis on 20 RFS-associated genes based on the Akaike information criterion (AIC) value to finally obtain seven (CXCL13, KLRD1, ICOS, MMP12, DPYD, ZBED2, and SLC2A3), including three high-risk genes (DPYD, ZBED2, and SLC2A3) and four low-risk genes (CXCL13, KLRD1, ICOS, and e) (Figure S5C). According to the results of the multivariate Cox regression analysis, the PRG_score was constructed as follows:
Figure 5.
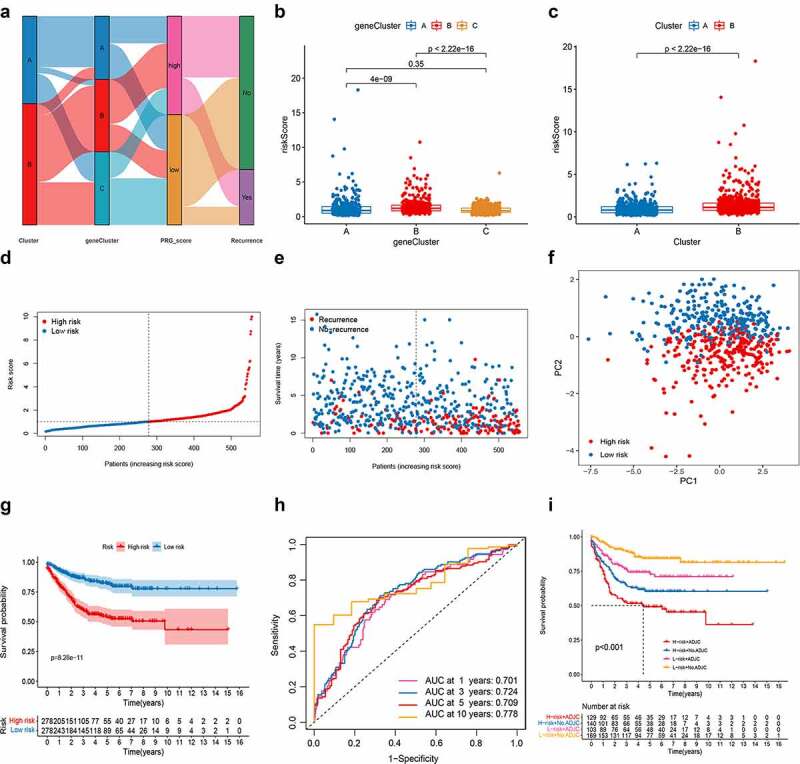
Construction of the PRG_score in the training set. (a) Alluvial diagram of subtype distributions in groups with different PRG_scores and survival outcomes. (b) Differences in PRG_score between gene subtypes. (c) Differences in PRG_score between pyroptosis subtypes. (d-e) Ranked dot and scatter plots showing the PRG_score distribution and patient survival status. (f) PCA analysis based on the prognostic signature. The high- and low-risk patients are represented by red and steel blue dots, respectively. (g) Kaplan–Meier analysis of the RFS between the two groups. (h) ROC curves to predict the sensitivity and specificity of 1-, 3-, 5-, and 10-year survival according to the PRG_score. (i) Survival analysis among four patient groups stratified by both PRG_score and treatment with adjuvant chemotherapy. ADJC, adjuvant chemotherapy. PCA, principal component analysis; RFS, recurrence-free survival; ROC, receiver operating characteristic
Risk score = (−0.1222* expression of CXCL13) + (−0.6623* expression of KLRD1) + (−0.3369* expression of ICOS) + (−0.1063* expression of MMP12) + (0.5528* expression of DPYD) + (0.2368* expression of ZBED2) + (0.3294* expression of SLC2A3).
We observed a significant difference in PRG_score between pyroptosis gene subtypes. The PRG_score of subtype C was the lowest, while that of subtype B was the highest, indicating that a low PRG_score may be closely related to immune activation-related features, while a high PRG_score may be related to stromal activation-related features (Figure 5b). More importantly, compared to subtype A, subtype B had a significantly higher PRG_score. The distributions of risk scores in the two subtypes are shown in Figure 5c. Patients with a PRG_score lower than the median risk score were categorized into the low-risk group (n = 278), whereas those with a PRG_score greater than the median risk score were placed in the high-risk group (n = 278). The distribution plot of the risk of PRG_score revealed that survival times decreased while recurrence rates increased with an increase in PRG_scores (Figure 5d-e). PCA analysis demonstrated discernible dimensions between the low- and high-PRG_score groups (figure 5f). The Kaplan–Meier survival curves revealed that patients with low scores had a significantly favorable overall survival compared to that in patients with high scores (log-rank test, p < .001; Figure 5g). In addition, the 1-, 3-, 5-, and 10-year survival rates of PRG_score were represented by AUC values of 0.701, 0.724, 0.709, and 0.778, respectively (Figure 5h). The PRG_score predicted 1- year survival with a 74% sensitivity and 54% specificity, 3- year survival with 75% sensitivity and 59% specificity, 5-year survival with a 72% sensitivity and 61% specificity, and 10- year survival with a 72% sensitivity and 65% specificity. We then specifically examined the ability of the PRG_score to predict the efficacy of adjuvant chemotherapy in patients with CRC. Patients with low scores showed significant therapeutic advantages among patients receiving adjuvant chemotherapy. Moreover, the predictive ability of the PRG_score was not affected by adjuvant chemotherapy. Regardless of whether chemotherapy was administered, the low-score group always showed a clear survival advantage (Figure 5i).
To validate the prognostic performance of the PRG_score, we calculated PRG_scores across internal (testing set) and three external validation groups (GSE39582, GSE17536, GSE38832) (Figures S6–9). The patients were also stratified into low- or high-risk groups according to the formula used for the training set. The PRG_scores, patient survival status, and PCA showing the variation tendencies of the low- and high-risk groups are, respectively, shown in Figures S6A-B, S7A-B, S8A-B, and S9A-B. Survival analysis revealed a significantly better prognosis in the low-risk group relative to that in the high-risk group (log-rank; p < .001; Figure S6-9 C). Analysis of the 1-, 3-, 5-, and 10-year prognostic prediction classification efficiencies showed that the PRG_score still had relatively high AUC values (Figure S6-9D), indicating that the PRG_score had excellent ability to predict the survival of CRC patients.
Validation of the expression levels of the seven PRGs use for the prognostic signature
The expression levels of seven prognostic genes were measured in six CRC tissues and six adjacent normal tissues by RT-qPCR. As shown in Figure S10, the expression levels of MMP12, ZBED2, and SLC2A3 were elevated while those of CXCL13, KLRD1, ICOS, and DPYD were downregulated in CRC tissues compared to the levels in the corresponding normal tissues.
Clinical correlation analysis and stratification analysis of the prognostic PRG_score
To investigate the impact of the PRG_score on clinical characteristics, we explored the correlation between PRG_score and different clinical features (age, sex, tumor location, TNM stage, KRAS mutation status, and BRAF mutation status). We observed significantly higher PRG_scores in patients in the stage III–IV subgroup relative to those in the stage I–II subgroup (p < .05; Figure S11A). To determine whether this prognostic PRG_score might independently predict RFS in patients with CRC, we combined the clinical features with the PRG_score of the prognostic PRG_score to perform univariate and multivariate analyses. As shown in Figure S11B-C, the TNM stage and PRG_score in the training set showed significant differences, with consistent results observed in the testing (Figure S11D-E), GSE39582 (Figure S11F-G), and GSE17536 (Figure S11H-I) groups. Moreover, a stratified analysis to evaluate whether the PRG_score retained its predictive ability in different subgroups, including age (≤60 and >60 years), sex (female and male), tumor location (left and right side), TNM stage (stage I–II and stage III–IV), and KRAS mutation (yes and no) showed significantly lower RFS in patients with high-risk scores compared to those in patients with low-risk scores for age (p < .001), sex (p = .014 in women and p < .001 in men), tumor location (p < .001 on the left side and p = .008 on the right side), TNM stage (p < .001), KRAS mutation (p = .002 for yes and p < .001 for no), and BRAF mutation (p = .043 for yes and p < .001 for no) (Figure S12).
Evaluation of TME and checkpoints between the high- and low-risk groups
We performed the CIBERSORT algorithm to assess the association between PRG_score and the abundance of immune cells. As shown in the scatter diagrams, the PRG_score was positively correlated with M0 macrophages, activated mast cells, M2 macrophages, neutrophils, and monocytes and negatively correlated with activated memory CD4 + T cells, CD8 + T cells, M1 macrophages, naive B cells, follicular helper T cells, plasma cells, activated dendritic cells, resting dendritic cells, gamma delta T cells, and activated NK cells (Figure 6a). A low PRG_score was also closely associated with a high immune score, whereas a high PRG_score was associated with a high stromal score (Figure 6b). We also assessed the relationship between the seven genes in the proposed model and the abundance of immune cells. We observed that most immune cells were significantly correlated with the seven genes (Figure 6c). In addition, we investigated the associations between immune checkpoints and our risk model. Figure 6d shows that 33 immune checkpoints were differentially expressed in the two groups, including PD-1, PD-L1, and CTLA-4.
Figure 6.
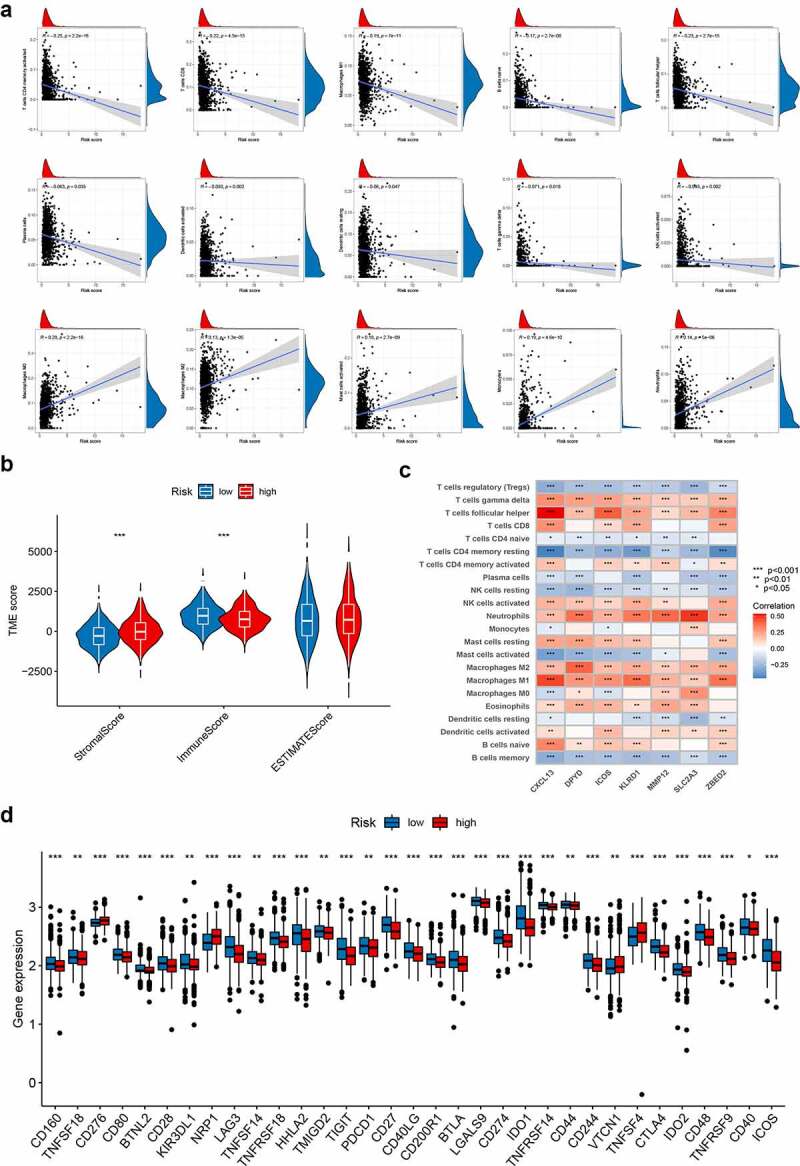
Evaluation of the TME and checkpoints between the two groups. (a) Correlations between PRG_score and immune cell types. (b) Correlations between PRG_score and both immune and stromal scores. (c) Correlations between the abundance of immune cells and seven genes in the proposed model. (d) Expression of immune checkpoints in the high and low-risk groups. TME, tumor microenvironment
Relationship of PRG_score with MSI and CSC index
Increasing evidence suggests that patients with high microsatellite instability (MSI-H) are more sensitive to immunotherapy and can benefit from immunotherapy drugs.22 Correlation analyses revealed that a low PRG_score was significantly correlated with MSI-H status, while a high PRG_score was associated with microsatellite stable (MSS) status (Figure 7a-b). To evaluate the influence of MSI status on RFS in patients with CRC, we performed survival analysis in the MSI and MSS groups. Although not significant, the MSI group showed a tendency for prolonged survival (p = .446; Figure S13A). However, the subsequent stratified survival analysis showed that the risk score could distinguish the survival of patients with CRC in both MSI and MSS subgroups and that the trend of survival advantage in the MSI group was reversed by the risk score (Figure S13B). In addition, we synthesized the PRG_score and CSC index values to assess the potential correlation between the PRG_score and CSC in CRC. Figure 7c shows the results of the linear correlation between PRG_score and CSC index. We concluded that PRG_score was negatively correlated with the CSC index (R = −0.31, p < .001), indicating that CRC cells with lower PRG_score had more distinct stem cell properties and a lower degree of cell differentiation (Figure 7c).
Figure 7.
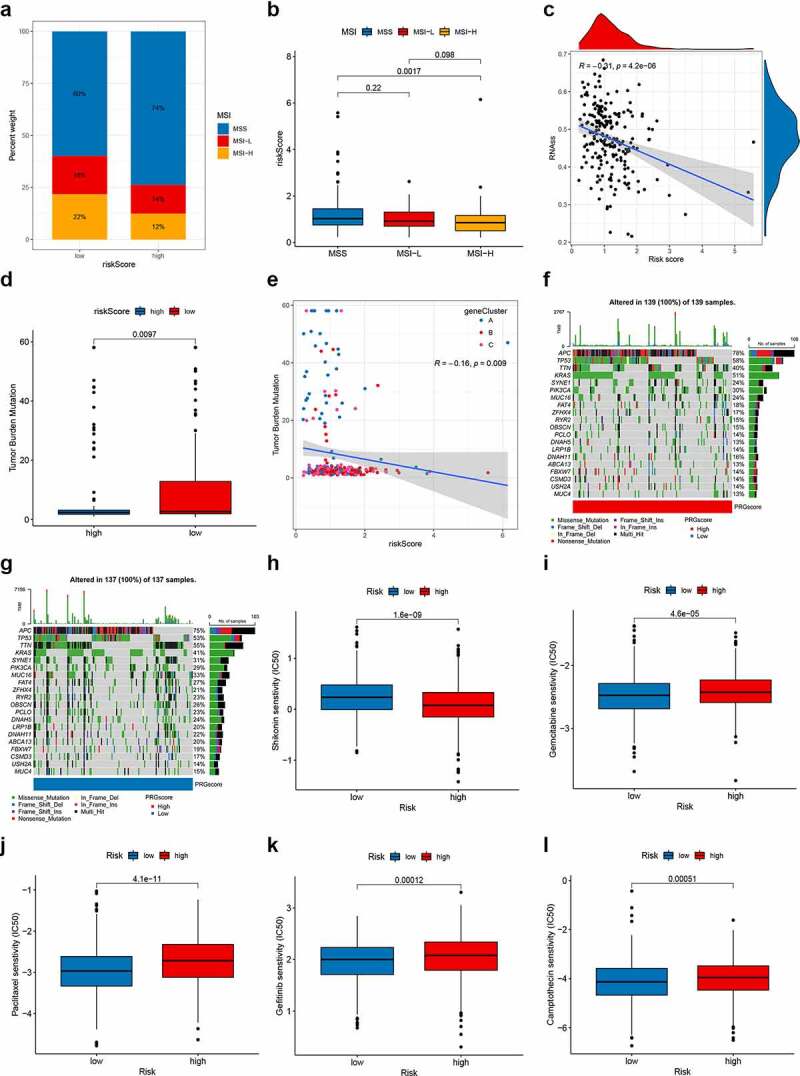
Comprehensive analysis of the PRG_score in CRC. (a-b) Relationships between PRG_score and MSI. (c) Relationships between PRG_score and CSC index. (d) TMB in different PRG_score groups. (e) Spearman correlation analysis of the PRG_score and TMB. (f-g) The waterfall plot of somatic mutation features established with high and low PRG_scores. Each column represented an individual patient. The upper barplot showed TMB, the number on the right indicated the mutation frequency in each gene. The right barplot showed the proportion of each variant type. (h-l) Relationships between PRG_score and chemotherapeutic sensitivity. CRC, colorectal cancer; MSI, microsatellite instability; CSC, cancer stem cell; TMB, tumor mutation burden
Mutation and drug susceptibility analysis
Accumulative evidence shows that patients with a high TMB may benefit from immunotherapy due to their higher numbers of neoantigens.23 Our analysis of the mutation data from the TCGA COAD/READ cohort showed a lower TMB in the high score group than that in the low score group (Figure 7d), implying that the low-risk group might benefit from immunotherapy. Spearman correlation analysis demonstrated that the PRG_score was negatively associated with the TMB (p = .009; Figure 7e). We then analyzed the distribution variations of the somatic mutations between two PRG_score groups in the TCGA-COAD cohort. The top ten mutated genes in the high- and low-risk groups were APC, TP53, TTN, KRAS, SYNE1, PIK3CA, MUC16, FAT4, ZFHX4, and RYR2 (figure 7f–G). Patients with a high PRG _score had markedly higher frequencies of APC, TP53, and KRAS mutations compared to those in patients with a low PRG _score. However, the exact opposite was observed regarding the mutation levels of TTN and MUC16 (figure 7f-g). We next selected chemotherapy drugs currently used for the treatment of CRC to evaluate the sensitivities of patients in the low- and high-risk groups to these drugs. Interestingly, we found that the patients in the high PRG_score group had lower IC50 value for shikonin, while IC50 values of chemotherapeutics such as gemcitabine, paclitaxel, gefitinib, and camptothecin were significantly lower in the patients with low PRG_score. Together, these results showed that PRGs were related to drug sensitivity (Figure 7h-l).
Development of a nomogram to predict survival
Considered the inconvenience clinical utility of PRG_score in predicting RFS in patients with CRC, a nomogram incorporating the PRG_score and clinicopathological parameters was established to predict the 1-, 3-, 5-, and 10-year RFS rates (Figure 8a). The predictors included PRG_score and patient stage. The results of our AUC experiments on the nomogram model showed higher accuracy for RFS at 1, 3, 5, and 10 years in the training set, testing set, and two external validation sets (Figure 8b-e). Our comparison of the predictive accuracy of the nomogram with that of the TNM stage in the five sets (Figure S14) showed 1-, 3-, 5-, and 10-year AUC values of the nomogram in the training set of 0.801, 0.763, 0.760, and 0.818, respectively, whereas those of the TNM stage were 0.740, 0.706, 0.703, and 0.683, respectively (Figure S14A–D). In the testing set, the 1-, 3-, 5-, and 10-year AUC values of the nomogram were 0.788, 0.758, 0.739, and 0.801, respectively, while those of the TNM were 0.734, 0.710, 0.681, and 0.693, respectively (Figure S14E–H). Furthermore, the AUC values of the nomogram in two external validation sets (GSE29582 and GSE17536) were also higher than that of the TNM stage (Figure S14 I–O), suggesting that the nomogram exhibited superior survival predictive ability compared to the TNM stage. The subsequent calibration plots suggested that the proposed nomogram had a similar performance in both the training and testing sets compared to an ideal model (figure 8f–I).
Figure 8.
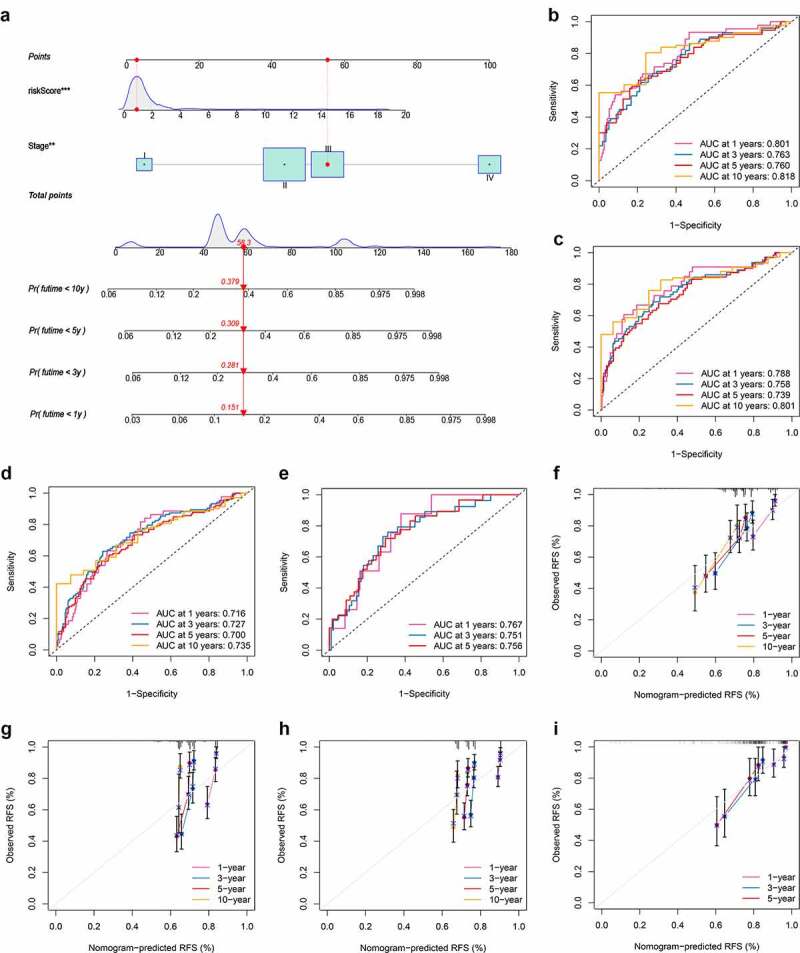
Construction and validation of a nomogram. (a) Nomogram for predicting the 1-, 3-, 5-, and 10-year RFS of CRC patients in the training set. (b-e) ROC curves for predicting the 1-, 3-, 5-, and 10-year ROC curves in the training, testing, GSE29582, and GSE17536 sets. (f-i) Calibration curves of the nomogram for predicting of 1-, 3-, 5-, and 10-year RFS in the training, testing, GSE29582, and GSE17536 sets. RFS, recurrence-free survival; CRC, colorectal cancer; ROC, receiver operating characteristic
Discussion
Numerous studies have revealed the indispensable role of pyroptosis in innate immunity and antitumor effects.24,25 However, most studies have focused on a single PRG or a single type of TME cell; thus, the overall effect and TME infiltration characteristics mediated by the combined effects of multiple PRGs have not yet been fully elucidated. The results of the present study revealed global alterations in PRGs at the transcriptional and genetic levels in CRC. We identified two distinct molecular subtypes based on 48 PRGs. Compared to patients with subtype A, patients with subtype B had more advanced clinicopathological features and worse RFS. The characteristics of the TME also differed significantly between the two subtypes. The CRC subtypes were also characterized by a significant immune activation, including antigen processing and presentation, T and B cell receptor signaling pathway, natural killer cell-mediated cytotoxicity, RIG-I-like, NOD-like, and the Toll-like receptor signaling pathways. Furthermore, differences in mRNA transcriptomes between different pyroptosis subtypes were significantly related to PRG and immune-related biological pathways. We identified three gene subtypes d based on the DEGs between the two pyroptosis subtypes. Thus, our findings indicate that PRGs might serve as a predictor for evaluating the clinical outcome and immunotherapy response of CRC. Therefore, we constructed the robust and effective prognostic PRG_score and demonstrated its predictive ability. The expression levels of seven genes included the PRG_score in CRC tissues were also explored. The pyroptosis patterns characterized by immune activation and inhibition showed lower and higher PRG_scores, respectively. Patients with low- and high-risk PRG_scores showed significantly different clinicopathological characteristics, prognosis, mutation, TME, immune checkpoints, MSI, CSC index, and drug susceptibility. Finally, by integrating the PRG_score and tumor stage, we established a quantitative nomogram, which further improved the performance and facilitated the use of the PRG_score. The prognostic model can be used for prognosis stratification of patients with CRC, will assist in better understanding the molecular mechanism of CRC, and will provide new ideas for targeted therapies.
The prognosis of CRC after conventional chemotherapy is poor, with higher levels of tumor-infiltrating lymphocytes, tumor neoantigens, and checkpoints. Despite recent advances in immunotherapy, patients with CRC still show heterogeneity in their outcomes, highlighting the crucial role of TME in CRC tumorigenesis and progression. Immune cells, such as granulocytes, lymphocytes, and macrophages are major cellular components of TME. These cells participate in various immune responses and activities, such as the inflammatory response coordinated by tumors to promote survival.26 The TME that surrounds tumor cells comprises TIICs, blood vessels, extracellular matrix (ECM), fibroblasts, lymphocytes, bone marrow-derived inflammatory cells.27 Evidence has also shown the significant effects of the TME on tumor development, progression, and therapeutic resistance.28 In the present study, the pyroptosis pattern characterized by immune inhibition (subtype B) was associated with a higher PRG_score, while the pattern characterized by immune activation (subtype A) was associated with a lower PRG_score. We discovered that the characteristics of the TME and the relative abundance of 22 TIICs differed significantly between the two molecular subtypes and different PRG_scores. This finding suggests the critical role of PRGs in CRC progression. Increasing evidence has shown that effector T cells, memory T cells, and T cell differentiation play a vital role in the immune defense of CRC.29 Gamma delta T cells can effectively recognize and kill CRC cells, thereby suppressing tumor progression via multiple mechanisms.30 The densities of tumor-infiltrating T cells in CRC tissues were higher than those in normal tissues, with higher densities indicating a good prognosis.30–32 Subtype A and low PRG_score, with a better prognosis, showed higher infiltration of activated memory CD4+ and CD8 + T cells and gamma delta T cells, suggesting that they play a positive role in CRC development. The infiltration of Tregs, which suppress the anti-cancer immune response, was associated with poor prognosis.33 This corresponds to our finding of more Tregs in the TME for patients with subtype B and high PRG_scores compared to those in the low-risk group. Recent studies revealed that B cells also participate in the immune response.34,35 Petitprez et al.36 suggested that B cell enrichment was the strongest prognostic factor for prolonged survival and was positively correlated with the response to PD-1 blockade in soft-tissue sarcomas. Meanwhile, Helmink et al. reported that the levels of the B cell-related genes MZB1, JCHAIN, and IGLL5 were markedly higher in patients who responded to immune checkpoint blockade compared to those in non-responders.35 Moreover, tumor-infiltrating B cells were associated with a favorable prognosis in CRC.37,38 In metastatic CRC, patients with high B cell infiltration have a significantly lower risk of disease recurrence and prolonged overall survival.38 The results of these studies demonstrated that B cells are not just bystanders in anti-tumor immunotherapy; rather, B cells provide a new target for immunotherapy and could be a strong weapon against tumors. In our study, we observed no significant difference in memory B cell infiltration between the two subtypes and PRG_score groups, while the abundance of naive B cells in subtype B and high PRG_score with worse overall survival were significantly lower than those in subtype A. Therefore, infiltration of B cell inhibited tumor progression in CRC, consistent with the findings of previous studies.37,38 Tumor-associated macrophages are categorized into two main phenotypes: M1 macrophages (which inhibit cancer progression) and M2 macrophages (which promote cancer progression). M1 macrophages produce type I pro-inflammatory cytokines and have anti-tumor function.39 In this study, macrophages M1 were higher in the low PRG_score group, suggesting that patients with low PRG_ scores might benefit from immunotherapy. M2 macrophages are immunosuppressive, contribute to the matrix-remodeling, and hence favor tumor growth.39,40 CRC has a high level of MMP-9, which can degrade collagen in the type IV basement membrane, thereby promoting metastasis.41 Previous researches have demonstrated that M2 macrophages are related to EMT and the infiltration of M2 macrophages in TME enhances the metastasis of CRC.42,43 Furthermore, high tumor stromal density of M2 macrophages have been associated with worse cancer-specific survival in patients with CRC, and patients with high M1:M2 density ratio in tumor stroma have a higher survival rate.44 Consistent with previous publications, we noticed an increased infiltration of M1 macrophages in subtype A and low PRG_score groups with a favorable prognosis, while increased infiltration of M2 macrophages and high PRG_score group in subtype B with a worse prognosis.
With in-depth research on tumor immunology and molecular biology, immunotherapy has provided a new direction for tumor treatment. This immunotherapy includes immune checkpoint inhibitors (ICIs), therapeutic antibodies, and cell therapy. The research of ICIs targeting CTLA-4, PD-1, and PD-L1 is blooming and clinical studies have demonstrated their safety and efficacy.45,46 ICIs have recently been used to treat CRC.47 In the present study, we observed higher expression levels of PD-1 and PD-L1 in the subtype A and low PRG_score groups. Patients with MSI showed a higher response to PD-1 treatment. Mismatch repair defective (dMMR) CRC accounts for 14% of all CRCs.48 The presence of dMMR-MSI-H disease is prognostic, as the recurrence risk of dMMR-MSI-H tumors is lower than that of dMMR-MSI-L tumors, with a hazard ratio for overall survival associated with MSI of <1.49 In the present study, the proportion of patients with MSI-H was higher in the subtype A and low PRG_score groups with a poor prognosis. The expression levels of PD-1, PDL-1, and CTLA4 were significantly upregulated in patients with dMMR-MSI-H.50 Thus, dMMR-MSI-H CRC may respond well to immune checkpoint blockade. Immune checkpoint therapy was approved by regulatory agencies in 2017 for the treatment of severely mutated CRC tumors (dMMR or MSI-H).22 Nivolumab (anti-PD-1) provides clinical benefits for previously treated dMMR/MSI-H metastatic CRC, including improved objective response rates, disease control rates, and 12-month overall survival. Furthermore, nivolumab plus ipilimumab can improve these outcomes (43). Tremelimumab (anti-CTLA-4) was well tolerated by patients with CRC who do not respond well to other immunotherapies.51 We concluded that patients with low PRG_score; higher expression of PD-1, PD-L1, and CTLA-4 expression; and MSI-H might be inclined to respond to immune checkpoint blockade.
This study had several limitations. First, all analyses were conducted solely on data from public databases, and all samples used in our study were obtained retrospectively. Therefore, an inherent case selection bias may have influenced the results. Large-scale prospective studies and additional in vivo and in vitro experimental studies are needed to confirm our findings. Furthermore, data on some important clinical variables such as surgery, neoadjuvant chemotherapy, and chemoradiotherapy were unavailable for analysis in most datasets, which may have affected the prognosis of the immune response and pyroptosis state.
Conclusions
Our comprehensive analysis of PRGs revealed an extensive regulatory mechanism by which they affect the tumor-immune-stromal microenvironment, clinicopathological features, and prognosis. We also determined the therapeutic liability of PRGs in targeted therapy and immunotherapy. These findings highlight the crucial clinical implications of PRGs and provide new ideas for guiding personalized immunotherapy strategies for patients with CRC.
Supplementary Material
Acknowledgments
None.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Abbreviations
GSDMD: gasdermin D; GSDMC: gasdermin C; GSDME: gasdermin E; PRGs: pyroptosis-related genes; TME: tumor microenvironment; CRC: colorectal cancer; MSI: microsatellite instability; CSC: cancer stem cell; RFS: recurrence-free survival; MSI-H: microsatellite instability-high; TIICs: tumor-infiltrating immune cells; TCGA: The Cancer Genome Atlas; GEO: gene expression omnibus; CDF: cumulative distribution function; DEGs: Differentially Expressed Genes; ROC: receiver operating characteristic; FPKM: Fragments Per Kilobase Million; GSVA: Gene set variation analysis; TMB: tumor mutation burden; CNV: copy number variation; ICIs: immune checkpoint inhibitors; ssGSEA: single-sample gene set enrichment analysis; PCA: principal component analysis; MAF: mutation annotation format; AIC: Akaike information criterion; MSS: microsatellite stable; TLS: tertiary lymphoid structures; dMMR: Mismatch repair-defective.
Availability of data and material
The datasets analyzed for this study can be found in the TCGA-COAD/READ project (http://www.cancer.gov/tcga) and GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE39582/GSE17536/GSE38832).
Disclosure statement
The authors report no conflict of interest.
Author contributions
WS, JR, and TF conceived the project. WS, JR, RSX, CK, and TF contributed to data acquisition, analysis and interpretation, and manuscript writing. WS and RSX conducted the experiments and revised the manuscript. All authors read and approved the submitted manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Fang Y, Tian S, Pan Y, Li W, Wang Q, Tang Y, Yu T, Wu X, Shi Y, Ma P, et al. Pyroptosis: a new frontier in cancer. Biomed Pharmacother. 2020;121:109595. Epub 2019/ 11/12. doi: 10.1016/j.biopha.2019.109595. [DOI] [PubMed] [Google Scholar]
- 2.Kovacs SB, Miao EA.. Gasdermins: effectors of pyroptosis. Trends Cell Biol. 2017;27(9):673–17. doi: 10.1016/j.tcb.2017.05.005. Epub 2017/ 06/18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruan J, Wang S, Wang J. Mechanism and regulation of pyroptosis-mediated in cancer cell death. Chem Biol Interact. 2020;323:109052. Epub 2020/ 03/15. doi: 10.1016/j.cbi.2020.109052. [DOI] [PubMed] [Google Scholar]
- 4.Wang YY, Liu XL, Zhao R. Induction of pyroptosis and its implications in cancer management. Front Oncol. 2019;9:971. Epub 2019/ 10/17. doi: 10.3389/fonc.2019.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu LS, Liu Y, Wang XW, Xu B, Lin YL, Song Y, Dong Y, Liu JL, Wang XJ, Liu S, et al. LPS enhances the chemosensitivity of oxaliplatin in HT29 cells via GSDMD-mediated pyroptosis. Cancer Manag Res. 2020;12:10397–10409. Epub 2020/ 10/30. doi: 10.2147/cmar.S244374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miguchi M, Hinoi T, Shimomura M, Adachi T, Saito Y, Niitsu H, Kochi M, Sada H, Sotomaru Y, Ikenoue T, et al. Gasdermin C is upregulated by inactivation of transforming growth factor β receptor type II in the presence of mutated apc, promoting colorectal cancer proliferation. PloS One. 2016;11(11):e0166422. Epub 2016/ 11/12. doi: 10.1371/journal.pone.0166422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Li S, Qi J, Chen Z, Wu Y, Guo J, Wang K, Sun X, Zheng J. Cleavage of GSDME by caspase-3 determines lobaplatin-induced pyroptosis in colon cancer cells. Cell Death Dis. 2019;10(3):193. doi: 10.1038/s41419-019-1441-4. Epub 2019/ 02/26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orning P, Lien E, Fitzgerald KA. Gasdermins and their role in immunity and inflammation. J Exp Med. 2019;216(11):2453–2465. doi: 10.1084/jem.20190545. Epub 2019/ 09/25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erkes DA, Cai W, Sanchez IM, Purwin TJ, Rogers C, Field CO, Berger AC, Hartsough EJ, Rodeck U, Alnemri ES, et al. Mutant BRAF and MEK inhibitors regulate the tumor immune microenvironment via pyroptosis. Cancer Discov. 2020;10(2):254–269. Epub 2019/ 12/05. doi: 10.1158/2159-8290.Cd-19-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Runa F, Hamalian S, Meade K, Shisgal P, Gray PC, Kelber JA. Tumor microenvironment heterogeneity: challenges and opportunities. Current Molecular Biology Reports. 2017;3(4):218–229. doi: 10.1007/s40610-017-0073-7. Epub 2018/ 02/13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pottier C, Wheatherspoon A, Roncarati P, Longuespée R, Herfs M, Duray A, Delvenne P, Quatresooz P. The importance of the tumor microenvironment in the therapeutic management of cancer. Expert Rev Anticancer Ther. 2015;15(8):943–954. doi: 10.1586/14737140.2015.1059279. Epub 2015/ 06/23 [DOI] [PubMed] [Google Scholar]
- 12.Lee SS, Cheah YK. The interplay between micrornas and cellular components of tumour microenvironment (TME) on Non-Small-Cell Lung Cancer (NSCLC) Progression. Journal of Immunology Research. [2019:3046379. Epub 2019/ 04/05]. 2019:. doi: 10.1155/2019/3046379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pachter L Models for transcript quantification from RNA-seq. arXiv.org., 2011. http://arxiv.org/abs/1104.3889. Accessed 12 February 2021. [Google Scholar]
- 14.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17(1):13. Epub 2016/ 01/28. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Y, Dai Q, Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discovery. 2021;7(1):71. doi: 10.1038/s41420-021-00451-x. Epub 2021/ 04/09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. Epub 2015/ 03/31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2008;26(8):1364–1370. doi: 10.1200/jco.2007.12.9791. Epub 2008/ 03/08 [DOI] [PubMed] [Google Scholar]
- 19.Sebestyén E, Singh B, Miñana B, Pagès A, Mateo F, Pujana MA, Valcárcel J, Eyras E. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016;26(6):732–744. doi: 10.1101/gr.199935.115. Epub 2016/ 05/20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch A, Joosten SC, Feng Z, de Ruijter TC, Draht MX, Melotte V, Smits KM, Veeck J, Herman JG, Van Neste L, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15(7):459–466. Epub 2018/ 04/19. doi: 10.1038/s41571-018-0004-4. [DOI] [PubMed] [Google Scholar]
- 21.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. the human transcription factors. Cell. 2018;172(4):650–665. doi: 10.1016/j.cell.2018.01.029. Epub 2018/ 02/10 [DOI] [PubMed] [Google Scholar]
- 22.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–375. doi: 10.1038/s41575-019-0126-x. Epub 2019/ 03/20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. Epub 2014/ 11/20. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Wang Y, Ding J, Wang C, Zhou X, Gao W, Huang H, Shao F, Liu Z. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature. 2020;579(7799):421–426. doi: 10.1038/s41586-020-2079-1. Epub 2020/ 03/20 [DOI] [PubMed] [Google Scholar]
- 25.Tsuchiya K. Switching from apoptosis to pyroptosis: gasdermin-elicited inflammation and antitumor immunity. Int J Mol Sci. 2021;22(1):426. doi: 10.3390/ijms22010426. Epub 2021/ 01/08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seager RJ, Hajal C, Spill F, Kamm RD, Zaman MH. Dynamic interplay between tumour, stroma and immune system can drive or prevent tumour progression. Converg Sci Phys Oncol. 2017;3(3):034002. doi: 10.1088/2057-1739/aa7e86. Epub 2017/ 01/01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15(11):669–682. doi: 10.1038/nri3902. Epub 2015/ 10/17 [DOI] [PubMed] [Google Scholar]
- 28.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–4566. doi: 10.1158/0008-5472.Can-18-3962. Epub 2019/ 07/28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564(7735):268–272. Epub 2018/ 11/28. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 30.Ma R, Yuan D, Guo Y, Yan R, Li K. Immune effects of γδ T cells in colorectal cancer: a review. Front Immunol. 2020;11:1600. Epub 2020/ 10/06. doi: 10.3389/fimmu.2020.01600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuwahara T, Hazama S, Suzuki N, Yoshida S, Tomochika S, Nakagami Y, Matsui H, Shindo Y, Kanekiyo S, Tokumitsu Y, et al. Intratumoural-infiltrating CD4 + and FOXP3 + T cells as strong positive predictive markers for the prognosis of resectable colorectal cancer. Br J Cancer. 2019;121(8):659–665. Epub 2019/ 09/07. doi: 10.1038/s41416-019-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Governa V, Trella E, Mele V, Tornillo L, Amicarella F, Cremonesi E, Muraro MG, Xu H, Droeser R, Däster SR, et al. The interplay between neutrophils and cd8+ t cells improves survival in human colorectal cancer. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2017;23(14):3847–3858. Epub 2017/ 01/22. doi: 10.1158/1078-0432.Ccr-16-2047. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27(1):109–118. doi: 10.1038/cr.2016.151. Epub 2016/ 12/21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, Johansson I, Phung B, Harbst K, Vallon-Christersson J, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. Epub 2020/ 01/17. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 35.Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, Yizhak K, Sade-Feldman M, Blando J, Han G, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. Epub 2020/ 01/17. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petitprez F, De Reyniès A, Keung EZ, Chen TW, Sun CM, Calderaro J, Jeng YM, Hsiao LP, Lacroix L, Bougoüin A, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. Epub 2020/ 01/17. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 37.Berntsson J, Nodin B, Eberhard J, Micke P, Jirström K. Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer. Int J Cancer. 2016;139(5):1129–1139. doi: 10.1002/ijc.30138. Epub 2016/ 04/14 [DOI] [PubMed] [Google Scholar]
- 38.Meshcheryakova A, Tamandl D, Bajna E, Stift J, Mittlboeck M, Svoboda M, Heiden D, Stremitzer S, Jensen-Jarolim E, Grünberger T, et al. B cells and ectopic follicular structures: novel players in anti-tumor programming with prognostic power for patients with metastatic colorectal cancer. PloS One. 2014;9(6):e99008. Epub 2014/ 06/07. doi: 10.1371/journal.pone.0099008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y, Yu Y, Wang X, Zhang T. Tumor-associated macrophages in tumor immunity. Front Immunol. 2020;11:583084. Epub 2020/ 12/29. doi: 10.3389/fimmu.2020.583084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sousa S, Brion R, Lintunen M, Kronqvist P, Sandholm J, Mönkkönen J, Kellokumpu-Lehtinen PL, Lauttia S, Tynninen O, Joensuu H, et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res. 2015;17(1):101. Epub 2015/ 08/06. doi: 10.1186/s13058-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng ZS, Huang Y, Cohen AM, Guillem JG. Prediction of colorectal cancer relapse and survival via tissue RNA levels of matrix metalloproteinase-9. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 1996;14(12):3133–3140. doi: 10.1200/jco.1996.14.12.3133. Epub 1996/ 12/01 [DOI] [PubMed] [Google Scholar]
- 42.Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, He XW, Wu XJ, Xie D, Wu XR, et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10(11):829. Epub 2019/ 11/07. doi: 10.1038/s41419-019-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin X, Wang S, Sun M, Zhang C, Wei C, Yang C, Dou R, Liu Q, Xiong B. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J Hematol Oncol. 2019;12(1):20. doi: 10.1186/s13045-019-0708-7. Epub 2019/ 02/28 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Väyrynen JP, Haruki K, Lau MC, Väyrynen SA, Zhong R, Dias Costa A, Borowsky J, Zhao M, Fujiyoshi K, Arima K, et al. The prognostic role of macrophage polarization in the colorectal cancer microenvironment. Cancer Immunol Res. 2021;9(1):8–19. Epub 2020/ 10/08. doi: 10.1158/2326-6066.Cir-20-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yaghoubi N, Soltani A, Ghazvini K, Hassanian SM, Hashemy SI. Hashemy SI. PD-1/ PD-L1 blockade as a novel treatment for colorectal cancer. Biomed Pharmacother. 2019;110:312–318. Epub 2018/ 12/07. doi: 10.1016/j.biopha.2018.11.105. [DOI] [PubMed] [Google Scholar]
- 46.Rotte A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J Exp Clin Cancer Res. 2019;38(1):255. doi: 10.1186/s13046-019-1259-z. Epub 2019/ 06/15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marin-Acevedo JA, Dholaria B, Soyano AE, Knutson KL, Chumsri S, Lou Y. Next generation of immune checkpoint therapy in cancer: new developments and challenges. J Hematol Oncol. 2018;11(1):39. doi: 10.1186/s13045-018-0582-8. Epub 2018/ 03/17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward R, Meagher A, Tomlinson I, O’Connor T, Norrie M, Wu R, Hawkins N. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48(6):821–829. doi: 10.1136/gut.48.6.821. Epub 2001/ 05/19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2005;23(3):609–618. doi: 10.1200/jco.2005.01.086. Epub 2005/ 01/22 [DOI] [PubMed] [Google Scholar]
- 50.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. Epub 2014/ 11/02. doi: 10.1158/2159-8290.Cd-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung KY, Gore I, Fong L, Venook A, Beck SB, Dorazio P, Criscitiello PJ, Healey DI, Huang B, Gomez-Navarro J, et al. Phase II study of the anti-cytotoxic T-lymphocyte-associated antigen 4 monoclonal antibody, tremelimumab, in patients with refractory metastatic colorectal cancer. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2010;28(21):3485–3490. Epub 2010/ 05/26. doi: 10.1200/jco.2010.28.3994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed for this study can be found in the TCGA-COAD/READ project (http://www.cancer.gov/tcga) and GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE39582/GSE17536/GSE38832).


