ABSTRACT
Plant response to light is a complex and diverse phenomenon. Several studies have elucidated the mechanisms via which light and hormones regulate hypocotyl growth. However, the hormone-dependent ultraviolet-B (UV-B) response in plants remains obscure. Involvement of gibberellins (GAs) in UV-B-induced hypocotyl inhibition and its mechanisms in Arabidopsis thaliana were investigated in the present research. UV-B exposure remarkably decreased the endogenous GA3 content through the UV RESISTANCE LOCUS 8 (UVR8) receptor pathway, and exogenous GA3 partially restored the hypocotyl growth. UV-B irradiation affected the expression levels of GA metabolism-related genes (GA20ox1, GA2ox1 and GA3ox1) in the hy5-215 mutant, resulting in increased GA content.ELONGATED HYPOCOTYL 5 (HY5) promoted the accumulation of DELLA proteins under UV-B radiation; HY5 appeared to regulate the abundance of DELLAs at the transcriptional level under UV-B. As a result, the GA3 content decreased, which eventually led to the shortening of the hypocotyl. To conclude, the present study provides new insight into the regulation of plant photomorphogenesis under UV-B.
KEYWORDS: GA, UV-B, hypocotyl elongation, DELLA, HY5, UVR8
1. Introduction
Sunlight is one of the necessary environmental cues that regulate plant growth and development. It provides energy during photosynthesis and plays a vital role in plant growth and development. However, plants are inevitably exposed to ultraviolet (UV) radiation during photosynthesis.1 Ultraviolet is divided by wavelength into three groups: UV-A (315–400 nm), UV-B (280–315 nm), and UV-C (200–280 nm). UV-B is an vital environmental cue for plants.1,2 High intensity and relatively short wavelength of UV-B light induce stress response in plants, such as DNAdamage, ROS (reactive oxygen species) accumulation and senescence.3–6 Nevertheless, low intensity and long wavelength UV-B light positively regulate various developmental processes in plants, such as photomorphogenesis, shade avoidance response, phototropism, leaf growth, secondary metabolites such as anthocyanin and flavonoid, etc.4–7
UV RESISTANCE LOCUS 8 (UVR8) is the photoreceptor, which senses UV-B triggering specific responses and regulating plant resistant to adversity.8–12 UVR8 initiates UV-B signaling pathway in Arabidopsis by dissociating the UVR8 dimer to monomers, which get transported from the cytoplasm to the nucleus.8,9,13 In the nucleus, the UVR8 monomer interacts with CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) through the C-terminal C27 domain, stabilizing the transcription factor ELONGATED HYPOCOTYL 5 (HY5) and initiating the expression of downstream UV-B responsive genes. At the same time, HY5 transcription factor binds to the promoter of COP1 to regulate the expression of COP1 gene. UV-B induces the expression of the WD40 repeat sequence/repeat proteins RUP1 (REPRESSOR OF UV-B PHOTOGENESIS 1) and RUP2 (REPRESSOR OF UV-B PHOTOGENESIS 2) in a HY5-dependent manner. RUP1 and RUP2 directly interact with UVR8, promote the conversion of UVR8 monomer to dimer, and balance the UV-B signaling pathway.13–23 It has been reported that B-Box ZINC FINGER PROTEIN24/SALT TOLERANCE (BBX24/STO), a negative regulator of UV-B signaling pathway, does not directly interact with UVR8, but interacts with HY5 to reduce its accumulation and inhibit its transcriptional activity.24,25
A highly intricate regulatory system of plant requires the intersection of the UV-B signaling pathway with a multiplicity of other signaling pathways. Phytohormones are signaling molecules that play a indispensable role in intrinsic plant developmental network, as well as in the modulation of such programs in response to biotic and abiotic stresses.26–29 Meanwhile, specific hormones have been shown to affect plant growth. A key hormone for plant growth and development is gibberellic acid (GA).30–32 GA induces seed germination, cell growth and elongation, and pollen maturity, enabling the plants to switch from the vegetative to reproductive phase.26,33 Until now, scientists have isolated and identified 130 gibberellins from vascular plants, fungi, and bacteria. Exogenous gibberellins (GAs) could accelerate dormancy release to some extent, and different kinds of GAs showed inconsistent effects in various plants. GA3 could robustly accelerate bud dormancy release, induced faster bud burst, higher shoot and more flowers in per plant among them.26,34,35 SD1 (Semi-dwarf 1) was first identified as the key gene of gibberellin 20-oxidase (GA20ox) controlling gibberellin biosynthesis in rice,36 which is related to plant phenotype and yield. The loss-of-function mutant allele sd1 resulted in a semi-dwarf phenotype and significantly increased yield in rice plants.37 Besides, GA20ox and gibberellin 3-oxidase (GA3ox) catalyze the inactive GA into bioactive GA, and the gibberellin 20-oxidase 1 (GA20ox1) involved in the last steps of gibberellin biosynthesis inactivating GA signal.33,38
DELLA proteins act as central negative regulators in the GA signaling pathway.38–42 In Arabidopsis, the GA receptor GA INSENSITIVE DWARF1 (GID1) perceives the GA cues and present a hydrophobic surface for DELLA binding, subsequently promotes the recognition of DELLAs by the SCFSLY1 complex, This complex further recruits the SCFSLY1 complex.43 DELLA proteins are then poly-ubiquitinated and finally degraded via the 26S-proteasome pathway. Some studies revealed that UV-B inhibits growth-promoting hormones and promotes abiotic stress-induced defensive hormones.44 Low dose UV-B-induced morphological changes are related to the dynamic balance in phytohormones (auxins, brassinosteroids, and gibberellins).45 Meanwhile, transcriptome studies showed that UV-B induced GA2ox2 and GA2ox8 in Arabidopsis, which inactivate GAs.46 The UV-B radiation also upregulated GA2ox1 expression in a UVR8 and HY5/HYH(HY5 HOMOLOG)-dependent manner, stabilizing DELLA proteins and destabilizing PHYTOCHROME INTERACTING FACTORS (PIFs), thereby influencing all aspects of plant physiology, including hypocotyl inhibition.47 Meanwhile, the GA metabolism and signal transduction in the shoot tips and young leaves of peas regulated the UV-B-induced inhibition of shoot elongation and leaf expansion.48 These earlier findings indicated the role of UV-B in regulating plant GA. Although several interactions between light and GA signals have been reported, the role of GA in UV-B response and the effect of transcription factors in UV-B signals on GA are not fully understood. Here, we use hy5, uvr8, rga-24 and sto mutants to elucidate the mechanism of GA regulating UV-B-induced hypocotyl growth inhibition in Arabidopsis thaliana from phenotypic, hormone, gene and protein levels. This study confirmed that DELLA is involved in UV-B photomorphogenesis, while HY5 regulates GA signaling pathways to regulate hypocotyl growth and promote RGA protein accumulation.
2. Materials and methods
2.1. Plant materials and growth conditions
Arabidopsis thaliana wild-type Columbia (Col-0) and its mutants bbx24/sto, cop1-435, hy5-21536, Landsberg erecta (Ler) and its mutant uvr8-2, rga-2434 seeds were vernalized at 4°C in dark for 4 d. These seeds were surface-sterilized with 30% (v/v) commercial bleach for 10 min, washed with sterile water, then sown on 1/2 Murashige and Skoog (MS) medium containing 0.8% agar and 2% sucrose. Seedings were grown under white light (Philips TLD30W/865 tubes; Quantitherm Light Meter, Hansatech, UK) for several days, the UV-B treatment was performed by narrowband UV-B (Philips TL20W/01RS; iHR550 spectroradiometer, Horiba Jobin Yvon, Japan). Overall intensity measured using a LI-COR 250A light meter (International Light CALIBRATION UVB; IL-1700; Tektronix). The growth chamber was maintained at a 16 h (long day) photoperiod at 22°C. The lamps were filtered through cellulose acetate film to block potential UV-C radiations for the experimental group (+UV-B) and through Mylar film to remove both UV-B and UV-C radiations for the control group (-UV-B). UV-B spectral irradiance was calibrated to 300 nm by weighted calculation of the plant response spectrum to obtain biological effective irradiance (UV-BBE).49 The UV-B irradiance used in the experiment was 0.6 W•m2 .
2.2. GA treatments
Arabidopsis seeds Ler were sown in 1/2 MS medium containing different concentrations of GA3 (0, 1, 5, 10, and 20 µmol/L; Sigma-Aldrich),which were maintained under white light for 4 d to germinate before allowing to grow in the absence or presence of UV-B for 3 d. ImageJ software (https://imagej.nih.gov/ij/) was used to measure the hypocotyl length and calculate the hypocotyl length ratio (+UV-B/-UV-B).
2.3. RNA extraction and real-time qPCR
Arabidopsis seedlings (Col/WT, hy5, bbx24, Ler, and uvr8) were initially grown for 7 d in white light before exposure to ±UV-B for 4 h. Seedlings were harvested and ground immediately in liquid nitrogen. Total RNA was extracted using RNAiso Plus Kit (Takara, Japan), following the manufacturer’s instructions. The complementary DNA (cDNA) was synthesized using PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time; Takara Biotechnology, China). RT-qPCR was performed in optical 96-well plates using Platinum® SYBR® Green qPCR SuperMix-UDG Kit (Invitrogen, USA) on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, USA) and RT-PCR was performed with three technical replicates for each sample. Primers used for qRT-PCR are listed in Table 1. Data were analyzed using the SDS 2.2.1 software (Applied Biosystems, USA). Three technical replicates and three biological replicates were analyzed.
Table 1.
Primer sequence for the real-time PCR
| AGI No. | Gene | Primers (5ʹ→3ʹ) | |
|---|---|---|---|
| At3G18780 | Actin 2 | GCTCTTCAGGAGCAATACGAAG GTTGGGATGAACCAGAAGGA |
|
| At5G51810 | GA20ox2 | TCCAACGATAATAGTGGCT TTGGCATGGAGGATAATGA |
|
| At4G25420 | GA20ox1 | TATCGAACGATAGATACAAGAGC TATCTTCTTGATGTGATGCTGTC |
|
| At1G15550 | GA3ox1 | CCATTCACCTCCCACACTCT GCCAGTGATGGTGAAACCTT |
|
| At1G78440 | GA2ox1 | CGGTTCGGGTCCACTATTTC ACCTCCCATTTGTCATCACCTG |
|
| At5G50915 | bHLH-137 | ACATTCACGTTCGAGC CGGAGCCAAAGTCATAG |
|
| At5G41315 | GL3 | CAATCTCAACGACTCCTCCAAGG AATGCCTGGACGGACGTTTG |
|
| At5g18800 | RGA | CAAGGTTATCGTGTGGAGGAGAG CAAGCGGAGGTGGTAATGAGTG |
|
| At5g12250 | GAI | CTATGCTCACCGACCTTAATCCT TGGTTAGACGAAGAAGCCGAAT |
|
| At4g20890 | RGL1 | ATCGGTTCACGGAATCGCTACAT ACATCACTCGGTCTTGGCTTGG |
|
| At3g62250 | RGL2 | CCGTGGTGCTCGTTGACTCT TTCTCCTGGTGAATCGCCTCAG |
|
| At5g11260 | RGL3 | AGCAATAGTAACAGCAACAAGAGGA GATAAGCACCACGGAACGAGT |
|
2.4. Extraction and quantification of plant hormones
The wild-type Ler and uvr8-2 mutant plants were grown for 7 d in white light and exposed to UV-B radiation for 4 h. Approximately 0.5 g of the seedlings were ground in liquid nitrogen, mixed with 5 mL extraction buffer composed of isopropanol/hydrochloric acid, and shaken at 4°C for 30 min. Then, 10 mL of dichloromethane was added to this mixture and kept shaking for 30 min at 4°C. The lower organic phase, obtained after centrifugation at 13,000 rpm for 5 min at 4°C was dried under nitrogen and dissolved in 400 mL of methanol (0.1% formic acid). The solution was passed through a 0.22-μm filter membrane and analyzed by high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS).
HPLC analysis of the extracted hormones was performed using a ZORBAXSB-C18 System (Agilent Technologies, USA). Different GA3 standard solutions (0.1, 0.2, 0.5, 2, 5, 20, 50, and 200 ng/mL) were prepared using methanol (0.1% formic acid) as the solvent; two replicates were maintained for each standard concentration. The chromatographic separation was carried out under the following conditions: chromatographic column, Poroshell 120 SB-C18 reversed-phase chromatographic column (2.1 mm × 150 mm, 2.7 µm); column temperature, 30°C; Mobile phase, (A:B) methanol/0.1% formic acid:water/0.1% formic acid; and injection volume, 2 μL. The following gradient was used to elute: 0–1 min, 20% A; 1–9 min, A increased to 80%; 9–10 min, 80% A; 10–10.1 min, A decreased to 20%; 10.1–15 min, 20% A. Mass spectrometry conditions were as follows: air curtain gas, 15 psi; spray voltage, 4500 V; atomization pressure, 65 psi; auxiliary pressure, 70 psi; and atomization temperature, 400°C.
2.5. Western blotting
Arabidopsis seedlings (Col, hy5-215, bbx24/sto, and rga-24) were grown under white light for 7 d and exposed to UV-B radiation for 4 h. Total proteins were extracted from Arabidopsis seedlings by extraction buffer (100 mM Tris-HCl pH 8, 4 M urea, 5% w/v SDS, 15% v/v glycerol, 10 mM β-ME, and 30 μL/mL protease inhibitor cocktail; P95599, Sigma-Aldrich) and protein lysate were centrifuged at 12000 g in 4°C for 15 min after being put on ice for 10 min. Then, taking supernatant and added with SDS/PAGE sample buffer [4×; 8% w/v SDS, 0.4% w/v bromophenol blue, 40% v/v glycerol, 200 mM Tris·HCl pH 6.8, and 400 mM β-ME] in a ratio of 4:1. The protein samples were heated for 10 min at 95°C. Aliquots containing 30 μg of total protein were loaded onto 10% SDS/PAGE gels and blotted onto nitrocellulose membrane (Bio-Rad, America). The membranes were blocked with TBST (137 mM NaCl, 20 mM Tris, 1% Tween-20, pH 7.6) containing 5% skim fat milk at room temperature (RT) for 2 h. Later, the membranes were incubated at 4°C overnight with the anti-RGA (PHYTO AB, USA) and anti-actin (Cell Signaling Technology, USA) primary antibody. Subsequently, the PVDF films were washed three times for 10 min with TBST and incubated with HRP-conjugated secondary anti-rabbit IgG antibody (1/10,000 dilution, Bioss, Beijing) for 1 h and washed three times again. Chemiluminescent signals were generated by using Pierce ECL Western blotting substrate (Thermo Scientific) and ChemiDocTM Touch Imaging System (Bio-Rad).
2.6. Statistical analysis
The Origin 2018 was used for statistical analysis and graphing. Statistical analyses were carried out using SPSS version 23.0 (IBM) by one-way analysis of variance (ANOVA). Tukey least significant difference test was used to deduce statistically significant differences between mean values (P < 0.05).
3. Results
3.1. UV-B radiation affects hypocotyl growth in Arabidopsis
The hypocotyls of Arabidopsis seedlings were significantly shortened after exposure to UV-B radiation; Noteworthy, the mutant hy5, uvr8, rga-24, showed longer hypocotyl length than the wild type, while bbx24/sto showed a shorter hypocotyl (Figure 1a). After UV-B treatment, the hypocotyles of hy5 mutant were 109% longer than the wild-type Col and the hypocotyls of uvr8, rga-24 mutants were 55% and 29% longer than wild-type Ler, respectively; while the hypocotyles of sto mutant were 30% shorter than the wild-type Col (Figure 1(b,c)). These results indicate that RGA may positively participate in the UV-B induced photomorphogenesis, like HY5 and UVR8, while BBX24 is a negative regulator in UV-B photomorphogenesis, which is consistent with our previous studies.24,25
Figure 1.
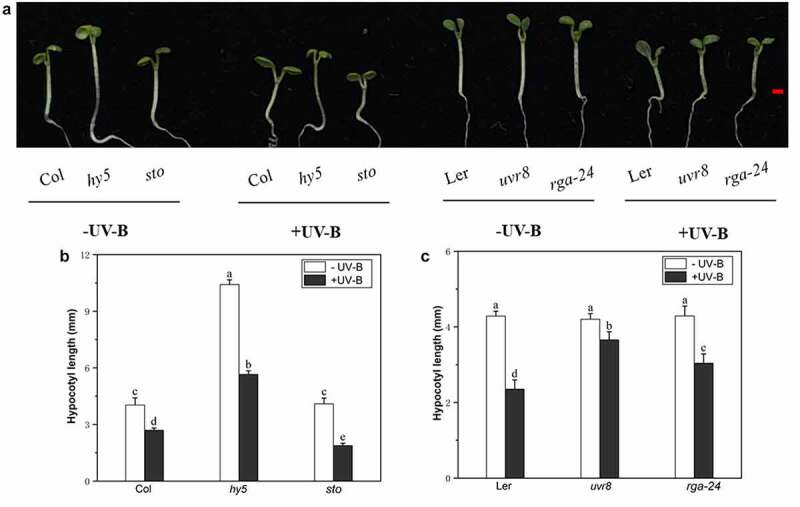
UV-B exposure inhibits Arabidopsis hypocotyl elongation. The uvr8, rga-24, hy5, bbx24, wild type (Col), and wild type (Ler) seedlings were grown under white light for 4 d and subsequently exposed to UV-B radiation for 3 d
(a) Phenotype of hypocotyl elongation of wild type (Col-0), stoand hy5 mutant. (b–c). Hypocotyl length of uvr8, rga-24, hy5, sto, wild type (Col), and wild-type (Ler) seedlings grown in the presence or absence of UV-B radiation. The hypocotyl length was measured using ImageJ software. Different lowercase letters indicate statistically significant differences between seedlings grown in the presence or absence of UV-B radiation (Tukey’s test; P < 0.05).
3.2. GA3 partially restores UV-B induced hypocotyl growth inhibition
GA has been linked to hypocotyl elongation in Arabidopsis under white light.33 In this study, the role of GA in UV-B-mediated hypocotyl growth inhibition was investigated in wild type (Ler). Under UV-B radiation, the hypocotyl growth promotion in the experimental group treated with GA3 was significantly higher than in the Mock group (Figure 2(a,b)). The relative hypocotyl growth rate was 66.4% (compared with -UV-B) without GA3 treatment. After GA3 application, under UV-B radiation, the relative hypocotyl growth rate gradually increased. At 5 μM, the GA3 effect was the most significant, and the relative hypocotyl growth-promoting rate reached 80.93%. At concentrations higher than 5 μM, the relative hypocotyl growth rate decreased gradually. At 20 μM, the relative hypocotyl growth rate was 66.3%, close to that without GA3 (Figure 2(c)). These results suggest the role of GA in UV-B-induced photomorphogenesis.
Figure 2.
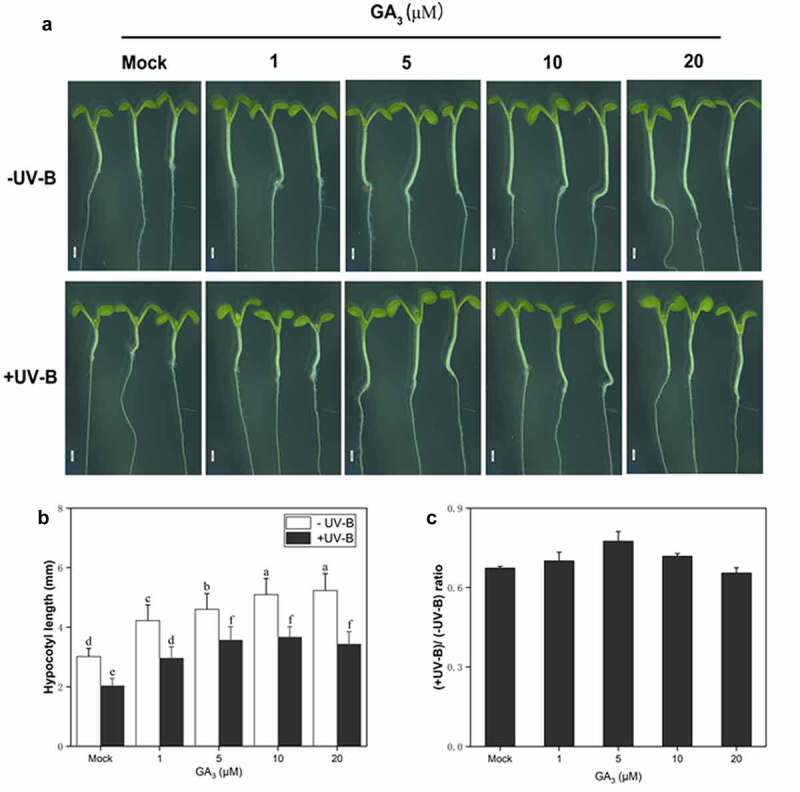
GA3 partly restores the hypocotyl growth of Col seedlings under UV-B light
(a). Wild-type (Col) seedlings grown under white light for 4 d and exposed to UV-B radiation for 3 d (scale bar = 1 mm); (b). The bar graph shows the hypocotyl length measured by ImageJ software. (c). The ratio (+UV-B)/(-UV-B) of hypocotyl length. Data are presented as mean ± standard error (n = 3; each replicate had at least 12 seedlings). Different lowercase letters indicate statistically significant differences between seedlings grown in the presence or absence of UV-B radiation (Tukey’s test; P < 0.05).
3.3. UV-B influences the expression of GA metabolic genes require UVR8
Phytohormones play a key role in the morphology and metabolism of plants. In this study, GA3 content in wild-type (Ler) and uvr8 seedlings under ±UV-B was analyzed to explore the effect of UV-B on GA metabolism and the role of UV-B receptor UVR8 in UV-B-induced phenotype in Arabidopsis. Under UV-B radiation, the GA3 content in Ler was reduced by 74.19% compared with the -UV-B, while no significant change was observed in the uvr8-2 (Figure 3(a)). After UV-B treatment, three critical genes involved in GA metabolism were analyzed; GA3ox1 and GA2ox2 genes, regulating GA synthesis, were downregulated by 57% and 84%, respectively, (Figure 3(b,c)), while GA2ox1 gene, regulating GA inactivation, was upregulated 40 folds in the wild type (Figure 3(d)). However, no significant difference was observed in the uvr8 mutant. These results prove that UV-B significantly reduced the endogenous GA levels in plants, and the decrease depends on the receptor UVR8.
Figure 3.
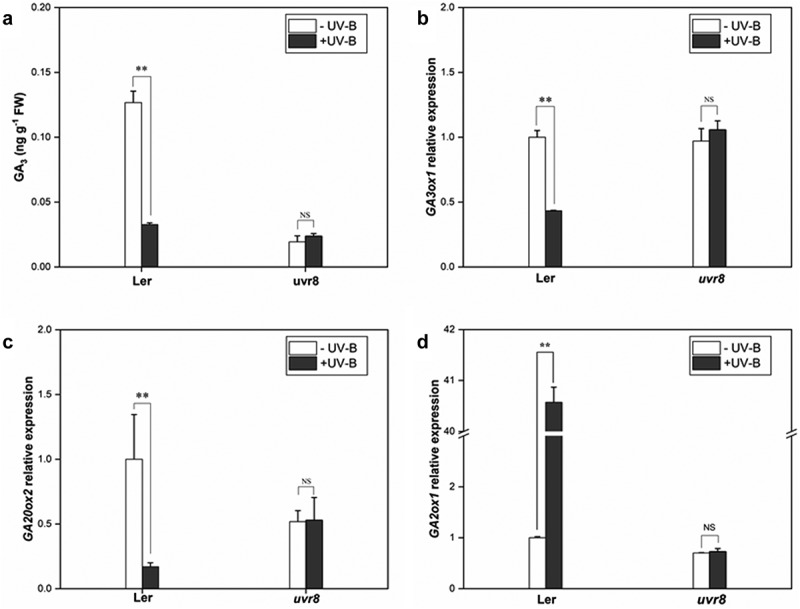
UV-B influences the expression of GA metabolic genes via UVR8. Wild type (Col) and uvr8-2 seedlings were grown in white light for 7 days and exposed to ±UV-B for 4 h
(a). GA3 content in Arabidopsis seedlings; (b–d). mRNA levels of GA metabolism-related genes (GA20ox2, GA3ox1, and GA2ox1) quantified by real-time PCR. Error bars represent the standard error (SE) of the mean of three biological replicates. ‘**’ indicates statistically significant differences at P < 0.01; ‘NS’ indicates no significant difference.
3.4. UV-B affects the expression of DELLA-responsive genes in hy5 mutant
DELLA is a negative regulator of GA signaling. UV-B radiation significantly reduces endogenous GA levels in plants and results in DELLA protein accumulation. In order to prove that DELLA is involved in the photomorphogenesis of UV-B, and to explore which key gene in the UV-B signaling pathway affects the expression of DELLA protein, the expression levels of bHLH-137 (Basic helix-loop-helix-137) and GL3(GLABRA3) genes in wild type, bbx24 mutant, and hy5-215 mutant were analyzed to determine whether HY5 or BBX24 affects DELLA accumulation under UV-B radiation. In this study, no significant differences in the expression levels of bHLH-137 and GL3 genes were observed between wild-type and hy5-215 mutants without UV-B treatment (Figure 4). With UV-B treatment, the two genes’ expression levels were significantly downregulated; GL3 was downregulated by 55.4% and 67%, while bHLH-137 was downregulated by 18.1% and 62.6% in the wild type and the hy5 mutant, respectively. However, no significant difference was observed in bbx24. The changes in the expression levels of these two genes indicate that HY5 may affect the expression of DELLA-responsive genes under UV-B treatment.
Figure 4.
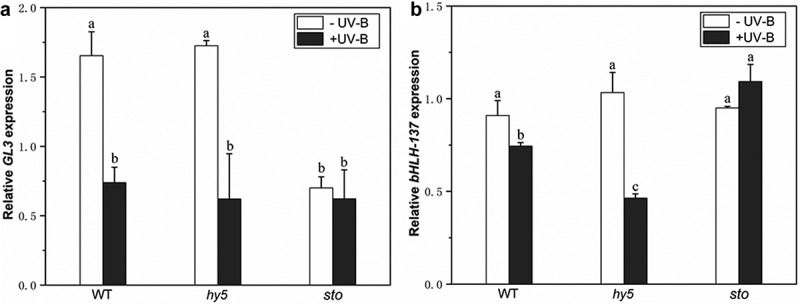
UV-B affects the expression of DELLA-responsive genes via HY5. Arabidopsis seedlings were grown in white light for 7 d and exposed to ±UV-B for 4 h
(a–b). Relative expression levels of GL3 and bHLH-137 genes in wild type, hy5, and bbx24 mutants detected by real-time PCR. Error bars represent the standard error (SE) of the mean of three biological replicates. Different lowercase letters indicate statistically significant differences in gene expression levels (Tukey’s test; P < 0.05).
3.5. HY5 promotes the accumulation of DELLA protein under UV-B radiation
The expression levels of three key genes, GA20ox1, GA2ox1, and GA3ox1, in response to GA signal in wild type and hy5 mutant with UV-B exposure were analyzed to explore the relationship between HY5 and DELLA protein. With UV-B exposure, GA2ox1 gene expression level in the wild type was significantly upregulated 2.2-fold compared with the hy5 mutant (Figure 5(a)), while GA20ox1 and GA3ox1 gene expression levels were significantly reduced by 23% and 12%, respectively, compared with the hy5 mutant (Figure 5(b,c)).
Figure 5.
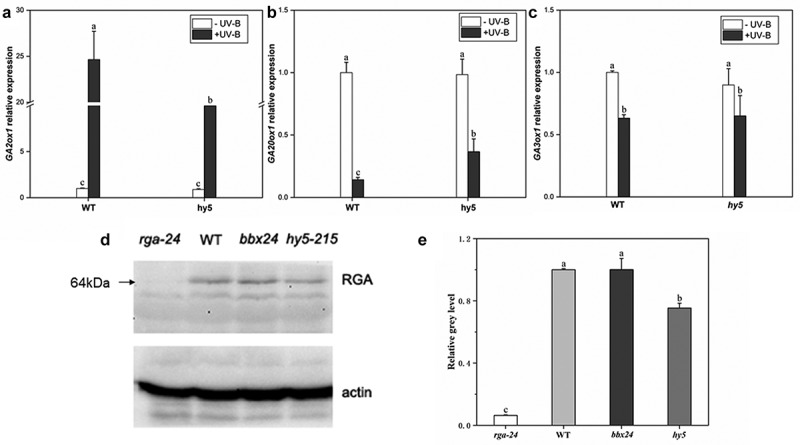
HY5 promotes RGA accumulation under UV-B irradiation. Seedlings were grown in white light for 7 d and exposed to ±UV-B for 4 h
(a–c). mRNA levels of GA metabolism-related genes (GA20ox1, GA3ox1, and GA2ox1) in wild type (WT) and hy5 mutant were quantified by real-time PCR; (d). RGA protein levels in WT, bbx24, rga-24, and hy5-215 after UV-B treatment; and (e). Gray value of RGA protein band (a). Error bars represent the standard error (SE) of the mean of three biological replicates. Different lowercase letters indicate statistically significant differences between the gene expression levels (Tukey’s test; P < 0.05).
RGA(REPRESSOR OF GA) protein is one of the most important members of the DELLA protein family. The levels of RGA protein in rag-24, WT, and hy5-215 under UV-B were analyzed by Western blotting (Figure 5(d)) and using ImageJ software (Figure 5(e)) to investigate the correlation between HY5 and DELLA. After UV-B exposure, RGA protein content was significantly lower in the hy5-215 than the WT; however, the difference in RGA protein content between WT and bbx24 was insignificant. These results indicate that HY5 but not BBX24 is required for the accumulation of the DELLA protein, RGA after exposure to UV-B radiation.
3.6. HY5 regulates RGA protein abundance at the transcriptional level under UV-B radiation
The expression levels of five members of the DELLA protein family (GAI (REPRESSOR OF gai-3), RGA, RGL1 (RGA-LIKE1), RGL2 (RGA-LIKE2), and RGL3 (RGA-LIKE1)) in Arabidopsis were analyzed to investigate whether HY5 promotes DELLA protein accumulation at the transcriptional level. Without UV-B treatment, the expression levels of five genes in the hy5 mutant were the same as those in the wild type (Figure 6). However, after UV-B treatment, the expression levels of the DELLA genes in the hy5 mutant were significantly downregulated compared with the wild type (Figure 6(a,d)). Among them, the expression level of GAI decreased by 31% compared with the wild type (Figure 6(a)), the RGL3 gene decreased by 32% (Figure 6(b)), the RGA gene by 16% (Figure 6(c)), and the RGL1 gene by 37% (Figure 6(d)), while no significant difference (26% decrease) was observed in the RGL2 gene (Figure 6(e)). These results suggest that HY5 may regulate DELLA protein abundance under UV-B at the transcriptional level.
Figure 6.
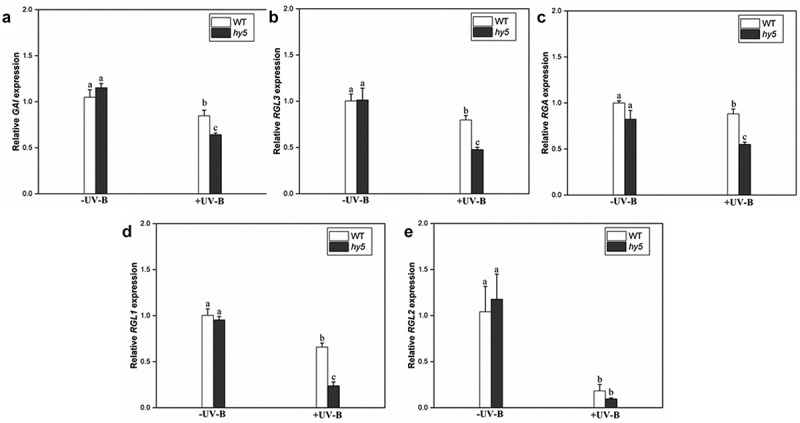
Transcript levels of DELLA genes in wild-type WT and hy5-215 mutant quantified by qRT-PCR
Seedlings were grown in white light for 7 d and exposed to ±UV-B for 4 h. Relative expression levels of (a) GAI gene; (B) RGL3 gene; (c) RGA gene; (d) RGL1 gene; and (e) RGL2 gene. Error bars represent the standard error (SE) of the mean of three biological replicates. Different lowercase letters indicate statistically significant differences between the gene transcript levels (Tukey’s test; P < 0.05).
4. Discussion
4.1. UV-B induces photomorphogenesis in Arabidopsis thaliana
Multiple endogenous hormone pathways and external environmental signals jointly regulate the growth of hypocotyls. UV-B, an inevitable environmental cue, induces various morphological changes such as inhibition of hypocotyl, stem and leaf growth, branch formation, and variation in root–shoot ratio. In this study, UV-B, at the dose used in the phenotypic experiments (0.6 W•m2), did not cause any visible damage (Figure 1(a)), consistent with the role of UV-B in signaling photomorphogenesis.50
In recent years, the interaction between UV-B and plant hormones has attracted the attention of researchers. In this study, exogenous GA3 significantly promoted the hypocotyl growth of Arabidopsis seedlings under UV-B, and GA3 partly restored the hypocotyl growth of seedlings under UV-B light (Figure 2). Roro found that UV-B treatment reduced the active GA1 content of the root tip and young leaves of wild-type pea plants.48 Therefore, the plant endogenous GA3 content in Ler and uvr8 exposed to UV-B were analyzed, GA3 content significantly decreased in Ler but not in the uvr8 mutant. This observation indicated the role of UV-B signal receptor UVR8 in reducing the endogenous GA3 content of wild-type Arabidopsis after UV-B treatment (Figure 3(a)).
To further investigate the role of GA in UV-B-mediated photomorphogenesis, the changes in the expression levels of GA metabolism-related genes in Ler and uvr8-2 mutant after UV-B treatment were analyzed. In wild-type Ler, the expression levels of genes related to GA degradation (GA2ox1) were significantly upregulated, and that of genes related to GA synthesis (GA20ox2 and GA3ox1) were downregulated; however, no significant difference was observed in the uvr8 mutant (Figure 3(b-d)). The results indicated that UV-B degrades endogenous GA3 by increasing the transcription level of GA2ox1 and inhibiting the transcription level of GA20ox2 and GA3ox1 at the same time. The degradation of endogenous GA3 depends on the UVR8 receptor. Hectors found that GA3ox1 expression level was reduced after exposure to low UV-B dose,45 Hayes detected GA2ox1 gene transcript levels in wild-type Ler and uvr8 mutants47; however, only a minor change was detected in the uvr8 mutant, while the gene upregulated in the wild-type Ler after UV-B exposure. But both of them only verified the relationship between GA and UVR8 under UV-B radiation at the genetic level. The present study demonstrated that UVR8 photoreceptor is involved in reducing endogenous GA3 content after UV-B treatment from hormone level and transcription level. These results suggest that UV-B-induced photomorphogenesis is closely related to GA at the metabolic level, and GA plays a role in UV-B-induced morphogenesis.
4.2. Potential correlation between HY5 and DELLA proteins under UV-B radiation
Phytohormones, especially GA, are known to influence the plant growth cycle. HY5 and DELLA are transcription factors regulating UV-B and GA signaling pathways.41,51 HY5 transmits the signals and regulates the expression of downstream genes, promoting UV-B photomorphogenesis. Meanwhile, BBX24 antagonizes HY5 and regulates UV-B photomorphogenesis,24 and GA triggers DELLA degradation to promote plant growth.47 However, the role of DELLAs in UV-B signaling remains unknown. In the present study, rga-24, uvr8, and hy5 mutants all exhibited long hypocotyl phenotypes under UV-B treatment, while sto exhibited short hypocotyl phenotypes. Suggesting the role of RGA (one of the DELLAs) in positively regulating the UV-B pathway, DELLA may be a positive regulator in UV-B light morphology (Figure 1).
Under UV-B treatment, the decrease in GA content appeared to have disturbed the dynamic balance between GA and DELLA, resulting in the accumulation of DELLA protein.39,41,47 After exposure to UV-B radiation, the expression levels of DELLA response genes bHLH-137 and GL3, were significantly downregulated in wild type and hy5 mutant, but no significant change in bbx24 (Figure 4). These observations suggest that the transcription level of DELLA in the hy5 mutant is decreased, indicating that UV-B affects the expression level of the DELLA gene which may be associated with HY5 rather than BBX24. Further, it was found that the transcript levels of GA20ox1 and GA3ox1 genes in hy5 mutant were higher than those in the wild type after UV-B exposure, while that of GA2ox1 was lower (Figure 5(a,c)). It results that GA content in hy5 mutant increases under UV-B radiation, while the expression of DELLA is inhibited (Figure 5(d)). Western blotting showed that RGA protein expression level was significantly lower in the hy5 mutant than in the wild type, while not much difference was evident in bbx24, suggesting that HY5 promoted the accumulation of DELLA protein under UV-B radiation (Figure 5(e)). The transcript levels of five DELLA genes (GAI, RGA, RGL1, RGL2, and RGL3) in the wild type and hy5 mutant were analyzed to determine whether HY5 regulates DELLA abundance. The transcript levels of these five DELLA genes in the hy5 mutant were lower than those in the wild type (Figure 6), indicating the role of HY5 in regulating DELLA protein abundance may at the transcriptional level. These results are consistent with the elongated hypocotyl phenotype of hy5 mutant seedlings under UV-B radiation. Thus, UV-B exposure inhibited hypocotyl growth due to DELLA protein accumulation through HY5 and decreased endogenous GA3 content in Arabidopsis.
5. Conclusion
In Arabidopsis, UV-B inhibited hypocotyl growth, which was partly restored by exdogenous GA3. Exposure to UV-B radiation reduced endogenous GA3 content via UVR8 receptor and triggered DELLA protein accumulation, disrupting the GA and DELLA balance. Specifically, HY5 promoted RGA protein accumulation under UV-B irradiation, which ultimately led to hypocotyl inhibition (see the proposed working model in Figure 7).
Figure 7.
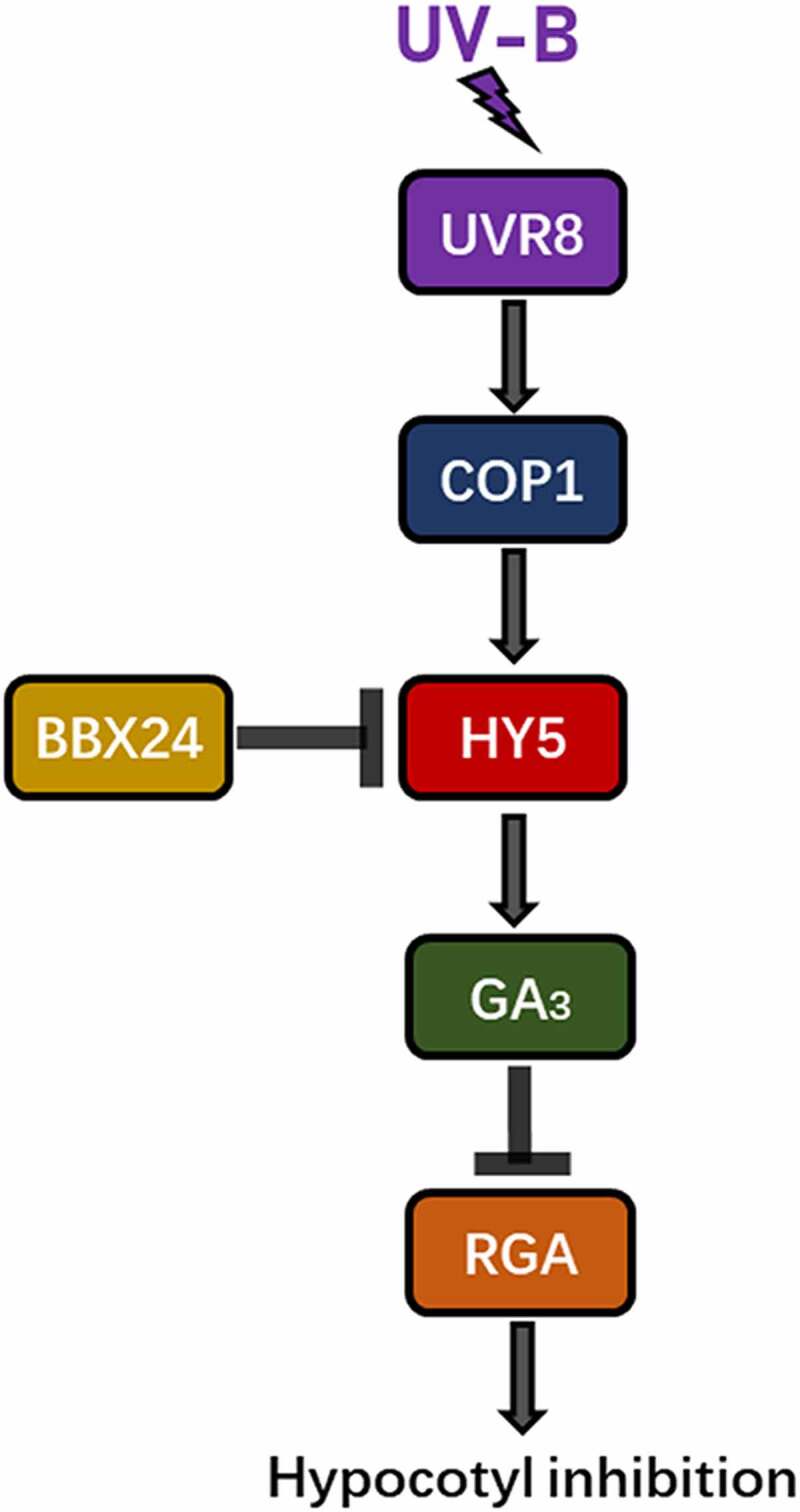
Model for GA involvement in UV-B-induced inhibition of hypocotyl elongation in Arabidopsis.
UV-B exposure causes UVR8 to bind with COP1 and regulates the transcription of the downstream gene HY5, promoting the accumulation of RGA protein (one of the most important member of DELLA family), reducing GA3 content, and ultimately inhibiting hypocotyl elongation. BBX24 negatively regulates the expression of HY5. Modified from Hayes.47
Acknowledgments
We would like to thank Professor Lars Olof Björn from Lund University, Sweden, for his guidance and advice. Thanks to Professor M.A.K. Jansen form University College Cork, Ireland, for providing seeds of Arabidopsis thalianamutant uvr8-2 and Professor Xiaojing Wang from South China Normal University for providingrga-24mutant. We would like to thank Professor Paul Giller from University College Cork, Ireland, for the English editing of the manuscript.
Appendix A.
Abbreviations
| GA | gibberellin |
| GA3 | Gibberelic acid |
| UVR8 | UV RESISTANCE LOCUS 8 |
| GAs | Exogenous gibberellins |
| COP1 | CONSTITUTIVELY PHOTOMORPHOGENIC 1 |
| HY5 | ELONGATED HYPOCOTYL 5 |
| RUP1/2 | REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1/2 |
| GA20ox1 | gibberellin 20-oxidase 1 |
| bHLH-137 | Basic helix-loop-helix-137 |
| GL3 | GLABRA3 |
| GA20ox2 | Gibberellin 20-oxidase 2 |
| GA2ox1 | Gibberellin 2-oxidase 1 |
| GA3ox1 | Gibberellin 3-oxidase 1 |
| PIFs | PHYTOCHROME INTERACTING FACTORS |
| BBX24/STO | B-Box ZINC FINGER PROTEIN24/SALT TOLERANCE |
| GID1 | GA-INSENSIVE DWARF1 |
| LIP1–LONG1 | orthologs of COP1–HY5 in Arabidopsis |
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was financially supported by the National Natural Science Foundation of China [grant no. 31670266 and 41976082]; the Natural Science Foundation of Guangdong Province [grant no. 2017A030313115 and 2021A1515010426]; the Undergraduate Students Scientific Research Foundation of South China Normal University. The research was also supported by Guangzhou Science and Technology Planning Project (No.2022010630080037).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contribution statement
T.M., D.L. and S.L. designed the experiments; T.M., D.L., Z.H. and Y.H. performed the experiments; T.M. and D.L. wrote the draft of the manuscript. S.L. and Y.W. corrected and revised the draft. Funding acquisition and project administration were done by S.L. and Y.W. All authors have read and agreed to the submitted version of the manuscript.
References
- 1.Yin R, Ulm R.. How plants cope with UV-B: from perception to response. CurrOpin Plant Biol. 2017;37:1–11. doi: 10.1016/j.pbi.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Heijde M, Ulm R.. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012;17(4):230–237. doi: 10.1016/j.tplants.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Casati P, Walbot V. Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant Physiol. 2004;136(2):3319–3332. doi: 10.1104/pp.104.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aphalo PJ, Jansen MA, McLeod AR, Urban O. Ultraviolet radiation research: from the field to the laboratory and back. Plant Cell Environ. 2015;38(5):853–855. doi: 10.1111/pce.12537. [DOI] [PubMed] [Google Scholar]
- 5.Kim BM, Rhee JS, Lee KW, et al. UV-B radiation-induced oxidative stress and p38 signaling pathway involvement in the benthic copepod Tigriopus japonicus. Comp BiochemPhysiol C ToxicolPharmacol. 2015;167:15–23. doi: 10.1016/j.cbpc.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 6.González Besteiro MA, Bartels S, Albert A, Ulm R. Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 2011;68(4):727–737. doi: 10.1111/j.1365-313X.2011.04725.x. [DOI] [PubMed] [Google Scholar]
- 7.Ulm R, Nagy F. Signalling and gene regulation in response to ultraviolet light. CurrOpin Plant Biol. 2005;8(5):477–482. doi: 10.1016/j.pbi.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Kaiserli E, Jenkins GI. UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell. 2007;19(8):2662–2673. doi: 10.1105/tpc.107.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. Perception of UV-B by the Arabidopsis UVR8 Protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 10.Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335(6075):1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Zhang J, Yang P, Deng H, Wang J, et al.Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484(7393):214–219. Published. 2012 Feb 29. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 12.O’Hara A, Jenkins GI. In vivo function of tryptophans in the Arabidopsis UV-B photoreceptor UVR8. Plant Cell. 2012;24(9):3755–3766. doi: 10.1105/tpc.112.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin L, Dong H, Yang G, Yin R. The C-terminal 17 amino acids of the photoreceptor UVR8 is involved in the fine-tuning of UV-B signaling. J Integr Plant Biol. 2020;62(9):1327–1340. doi: 10.1111/jipb.12977. [DOI] [PubMed] [Google Scholar]
- 14.Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28(5):591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, O’Hara A, Smith BO, Christie JM, Jenkins GI. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci U S A. 2012;109(40):16366–16370. doi: 10.1073/pnas.1210898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Ouyang X, Yang P, Lau OS, Chen L, Wei N, Deng XW. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci U S A. 2013;110(41):16669–16674. doi: 10.1073/pnas.1316622110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Yang P, Ouyang X, Chen L, Deng XW, Copenhaver GP. Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet. 2014;10(3):e1004218. doi: 10.1371/journal.pgen.1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin R, Arongaus AB, Binkert M, Ulm R. Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell. 2015;27(1):202–213. doi: 10.1105/tpc.114.133868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau K, Podolec R, Chappuis R, Ulm R, Hothorn M. Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 2019;38(18):e102140. doi: 10.15252/embj.2019102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin R, Skvortsova MY, Loubéry S, Ulm R. COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci U S A. 2016;113(30):E4415–E4422. doi: 10.1073/pnas.1607074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci U S A. 2013;110(3):1113–1118. doi: 10.1073/pnas.1214237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren H, Han J, Yang P, Mao W, Liu X, Qiu L, Qian C, Liu Y, Chen Z, Ouyang X, et al. Two E3 ligases antagonistically regulate the UV-B response in Arabidopsis. Proc Natl Acad Sci U S A. 2019;116(10):4722–4731. doi: 10.1073/pnas.1816268116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heilmann M, Jenkins GI. Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol. 2013;161(1):547–555. doi: 10.1104/pp.112.206805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L, Wang Y, Li QF, Björn LO, He JX, Li SS. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 2012;22(6):1046–1057. doi: 10.1038/cr.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyu G, Li D, Li S, Hu H. STO and GA negatively regulate UV-B-induced Arabidopsis root growth inhibition. Plant Signal Behav. 2019;14(12):1675471. doi: 10.1080/15592324.2019.1675471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olszewski N, Sun TP, Gubler F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14(Suppl):S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleet CM, Sun TP. A DELLAcate balance: the role of gibberellin in plant morphogenesis. CurrOpin Plant Biol. 2005;8(1):77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Yu Z, Duan X, Luo L, Dai S, Ding Z, Xia G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020;25(11):1117–1130. doi: 10.1016/j.tplants.2020.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Ku YS, Sintaha M, Cheung MY, Lam HM. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci. 2018;19(10):3206. doi: 10.3390/ijms19103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao QP, Zhu JD, Li NN, Wang XN, Zhao X, Zhang X. Cryptochrome-mediated hypocotyl phototropism was regulated antagonistically by gibberellic acid and sucrose in Arabidopsis. J Integr Plant Biol. 2020;62(5):614–630. doi: 10.1111/jipb.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Terzaghi W, Gong Y, Li C, Ling -J-J, Fan Y, Qin N, Gong X, Zhu D, Deng XW, et al.Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat Commun. 2020;11(1):1592. Published. 2020 Mar 27. doi: 10.1038/s41467-020-15394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao Z, He S, Xu F, Wei X, Jiang L, Liu Y, Wang W, Li T, Xu P, Du S, et al. Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol. 2020;225(2):848–865. doi: 10.1111/nph.16194. [DOI] [PubMed] [Google Scholar]
- 33.Claeys H, De Bodt S, Inzé D. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 2014;19(4):231–239. doi: 10.1016/j.tplants.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Yuxi Z, Yanchao Y, Zejun L, Tao Z, Feng L, Chunying L, Shupeng G. GA3 is superior to GA4 in promoting bud endodormancy release in tree peony (Paeonia suffruticosa) and their potential working mechanism. BMC Plant Biol. Published. 2021;21(1):323. 2021 Jul 5 doi: 10.1186/s12870-021-03106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P, Zheng T, Zhang Z, Liu W, Qiu L, Wang J, Cheng T, Zhang Q. Integrative Identification of Crucial Genes Associated With Plant Hormone-Mediated Bud Dormancy in Prunus mume. Front Genet. 2021; 12:698598. 2021 Jul 6. Published: doi: 10.3389/fgene.2021.698598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Zheng L, Wang X, Hu Z, Zheng Y, Chen Q, Hao X, Xiao X, Wang X, Wang G, et al. Comprehensive expression analysis of Arabidopsis GA2-oxidase genes and their functional insights. Plant Sci. 2019;285:1–13. doi: 10.1016/j.plantsci.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12(6):797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- 38.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21(9):R338–R345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 39.Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development. 2004;131(5):1055–1064. doi: 10.1242/dev.00992. [DOI] [PubMed] [Google Scholar]
- 40.Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19(10):3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davière JM, Achard P, Pivotal A. Role of DELLAs in regulating multiple hormone signals. Mol Plant. 2016;9(1):10–20. doi: 10.1016/j.molp.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Li K, Yu R, Fan LM, Wei N, Chen H, Deng XW. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat Commun. 2016;7(1):11868. doi: 10.1038/ncomms11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dill A, Thomas SG, Hu J, Steber CM, Sun TP. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 2004;16(6):1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanhaelewyn L, Prinsen E, Van Der Straeten D, Vandenbussche F. Hormone-controlled UV-B responses in plants. J Exp Bot. 2016;67(15):4469–4482. doi: 10.1093/jxb/erw261. [DOI] [PubMed] [Google Scholar]
- 45.Hectors K, Prinsen E, De Coen W, Jansen MAK, Guisez Y. Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation show specific changes in morphology and gene expression in the absence of stress symptoms. New Phytol. 2007;175(2):255–270. doi: 10.1111/j.1469-8137.2007.02092.x. [DOI] [PubMed] [Google Scholar]
- 46.Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci U S A. 2004;101(5):1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayes S, Velanis CN, Jenkins GI, Franklin KA. UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci U S A. 2014;111(32):11894–11899. doi: 10.1073/pnas.1403052111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roro AG, Dukker S, Melby TI, Solhaug KA, Torre S, Olsen JE. UV-B-induced inhibition of stem elongation and leaf expansion in Pea depends on modulation of Gibberellin metabolism and intact Gibberellin signalling. J Plant Growth Regul. 2017;36(3):680–690. doi: 10.1007/s00344-017-9671-0. [DOI] [Google Scholar]
- 49.Caldwell MM, Ballaré CL, Bornman JF, Flint SD, Björn LO, Teramura AH, Kulandaivelu G, Tevini M. Terrestrial ecosystems, increased solar ultraviolet radiation and interactions with other climatic change factors. Photochem. Photobiol. Sci 2003;2(1):29–38. doi: 10.1039/b211159b. [DOI] [PubMed] [Google Scholar]
- 50.Jenkins GI. Photomorphogenic responses to ultraviolet-B light. Plant Cell Environ. 2017;40(11):2544–2557. doi: 10.1111/pce.12934. [DOI] [PubMed] [Google Scholar]
- 51.Ang LH, Deng XW. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6(5):613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]


