Abstract
This study was the first report complete chloroplast genome of Aster batangensis (Astereae: Asteraceae), the perennial herb endemic to China. The plastid genome of Aster batangensis include 132 unique genes, with 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among these genes, 21 duplicate genes, including10 protein-coding genes, 7 tRNA genes, and 4 rRNA genes were detected. The complete genome size of Aster batangensis has a typical quadripartite circular structure with 152,605 bp in total length, consisting a large single copy (LSC) of 84,351 bp and a small single copy (SSC) of 18,212 bp, separated by a pair of invested repeats (IR) of 25,021 bp. The average GC content of whole plastome sequence is 37.3%, and the LSC, SSC and IR regions is 35.3%, 31.3%, and 43.0%, respectively. The phylogenetic analysis by the maximum likelihood method showed that A. batangensis was closely related to the other members of Astereae (e.g. Aztecaster matudae, Conyza bonariensis, Lagenophora cuchumatanica, Baccharis tricuneata, Baccharis genistelloides)
Keywords: Aster batangensis, complete chloroplast genome, phylogenomics analysis
The genus Aster is one of the most diverse genera in the tribe Astereae, family Asteraceae, including about 152 species (Nesom 1994; Chen et al. 2011). The species of Aster batangensis Bureau and Franchet 1891 (Asteraceae, Astereae) is a perennial herb and it is endemic to southwestern China (Sichuan, Xizang, Yunnan) (Chen et al. 2011). It has ecological and medicinal value in western China. So far, the complete chloroplast genome of A. batangensis has not yet been published. Genetic knowledge of A. batangensis would provide information for protection of this wild germplasm resource. Here, we obtained the complete plastome of A. batangensis by Illumina sequencing technology. The complete plastome reported here will contribute to the further studies on the phylogenetic analysis of genus Aster and its related genera.
Fresh leaves of A. batangensis were collected from Bowo village (101°07′11ʺE, 28°45′′30ʺN), Muli county, Sichuan Province, China. A specimen was deposited at the botany herbarium of Sichuan Normal University, SCNU (contact person: Associate Professor, Dr. Zhixi Fu and Email: fuzx2017@sicnu.edu.cn) under the voucher number Z.X. Fu 4061. High-quality total genomic DNA was extracted from ca. 6 cm2 sections of the silica-dried leaf using improved Tiangen Plant Genomic DNA Kits, add the 4 μl RNAseA and 20 μl Proteinase K after incubated (65 °C). Total DNA was directly constructed short-insert of 150 bp in length libraries and sequenced on the Illumina Genome Analyzer (Hiseq 2000) based the manufacturer’s protocol (Illumina, San Diego, CA, USA) by ORI-GENE, Beijing. Generally, more than 3.8 Gb of data was obtained for complete chloroplast genome of A. batangensis. De novo assembled in CLC Genomic Workbench v11 (CLC Bio, Aarhus, Denmark) and consensus sequence in Geneious R11.1.5 (Biomatters Ltd., Auckland, New Zealand) with referenced chloroplast genome sequence of Conyza bonariensis (Accession: KX792499). The chloroplast genome were annotated using a web-based annotation program GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) and editing by manual and imagining with OGDraw v1.2 (Lohse et al. 2013). We also developed the HMMER (Wheeler and Eddy 2013), tRNAscan-SE version 2.0.6 (Lowe and Eddy 1997) program which as the part of CHLOROBOX web toolbox (https://chlorobox.mpimp-golm.mpg.de/geseq.html) for checking the annotation via same reference genome. Final, the raw sequence data (SRA) and complete chloroplast genome (GenBank) of A. batangensis was submitted to NCBI.
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MZ292735. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA729213, SRP319392 and SAMN19114282 (SRS8948579) respectively. The complete genome size of Aster batangensis has a typical quadripartite circular structure with 152,605 bp in total length, consisting a large single-copy (LSC) of 84,351 bp and a small single-copy (SSC) of 18,212 bp, separated by a pair of inverted repeats (IR) of 25,021 bp. The average GC content of whole plastome sequence is 37.3%, and the LSC, SSC and IR regions is 35.3%, 31.3%, and 43.0%, respectively. The plastid genome of Aster batangensis include 132 unique genes, with 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among these genes, 21 duplicate genes, including10 protein-coding genes, 7 tRNA genes, and 4 rRNA genes were detected.
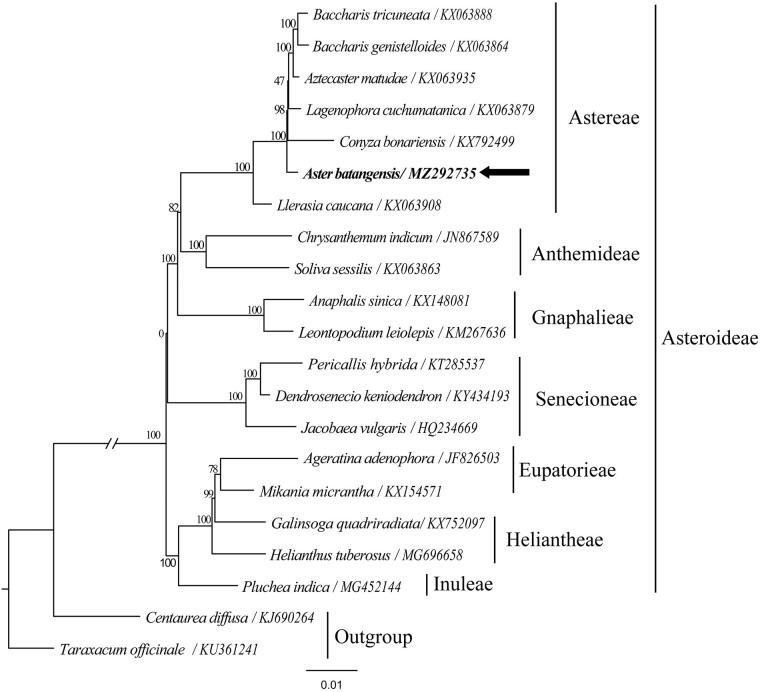
To identify the phylogenetic position of A. batangensis, we used a total of 20 additional complete cp genomes of the family Asteraceae and two outgroup taxa to clarify the phylogenetic position of A. batangensis (Figure 1). All of the cp genome sequences were aligned in MAFFT (Katoh and Standley 2013). A maximum likelihood analysis based on the GTRGAMMA model was performed with RAxML method on the CIPRES (Stamatakis et al. 2008; Miller et al. 2010) using 1000 bootstrap replicates. The maximum likelihood method (ML) result showed that A. batangensis was closely related to the other members of Astereae (e.g. Aztecaster matudae, Conyza bonariensis, Lagenophora cuchumatanica, Baccharis tricuneata, Baccharis genistelloides) (Bootstrap support = 100, Figure 1). The complete cp genome sequence of A. batangensis will be the valuable resource for future studies on taxonomy and phylogeny of family Asteraceae and provides useful molecular data for further phylogenetic and evolutionary analysis.
Figure 1.
A maximum-likelihood (ML) tree inferred from 31 chloroplast genomes in Asteraceae (the support value are indicated on the branches). The position of Aster batangensis is in bold.
Funding Statement
This work was supported by the National Natural Science Foundation of China under Grant [32000158], the Talent Engineering Research Initiation Project of Chengdu University under Grant [2081920047], the Southwest Minzu University Talent Supporting Funds and the Fundamental Research funds for the Central University, Southwest Minzu University under Grant [2021PTJS36], The Special Research Project of National Tradition Chinese Medicine Industy, the Fourth National Survey on Chinese Material Medica Resources under Grant [GZY-KJS-2018-004], The Sichuan Science and Technology Research Projects of Traditional Chinese Medicine under Grant [2018PC005]; the project of Sustainable Development Research Center of Resources and Environment of Western Sichuan under Grant [2020CXZYHJZX03]; the Philosophy and Social Science Key Research Base Project of Sichuan Province, Sichuan Nationalities and Mountain Economy Development Research Center under Grant [SDJJ1907].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ292735. The associated BioProject, SRA, and Bio-Sample numbers are RJNA729213, SRP319392 and SAMN19114282 (SRS8948579) respectively.
References
- Chen YL, Brouillet L, Semple JC.. 2011. Aster L. In: Wu ZY, Raven PH, editors. Flora of China vols. 20–21. St. Louis: Science Press; Beijing & Missouri Botanical Garden Press; p. 574–632. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R.. 2013. Organellar Genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and vsualizing expression data sets. Nucl Acids Res. 41:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the gateway computing environments workshop (GCE); New Orleans (LA): IEEE; p. 1–8. [Google Scholar]
- Nesom GL. 1994. Review of the taxonomy of Aster sensu lato (Asteraceae: Astereae), emphasizing the New World species. Phytologia. 77:14–297. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57(5):758–771. [DOI] [PubMed] [Google Scholar]
- Wheeler TJ, Eddy SR.. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 29(19):2487–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ292735. The associated BioProject, SRA, and Bio-Sample numbers are RJNA729213, SRP319392 and SAMN19114282 (SRS8948579) respectively.