ABSTRACT
Indole-3-acetaldoxime (IAOx) and phenylacetaldoxime (PAOx) are precursors for the growth hormones indole-3-acetic acid (IAA) and phenylacetic acid (PAA) and the defense compounds glucosinolates in Brassicales. Our recent work has shown that Arabidopsis transgenic lines overexpressing AtCYP79A2, a PAOx-production enzyme, accumulate the PAOx-derived compounds benzyl glucosinolate and PAA. Here we report that they also accumulate the benzyl glucosinolate hydrolysis products benzyl isothiocyanate and benzyl cyanide, which indicates that the turnover of benzyl glucosinolate can occur in intact tissues. Myrosinases or β-glucosidases are known to catalyze glucosinolate breakdown. However, transcriptomics analysis detected no substantial increase in expression of known myrosinases or putative β-glucosidases in AtCYP79A2 overexpressing lines. It was previously shown that accumulation of aldoximes or their derivatives represses the phenylpropanoid pathway. For instance, ref2 mutant having a defect in one of the aldoxime catabolic enzymes decreases phenylpropanoid production. Considering that AtCYP79A2 is not expressed in most organs under optimal growth condition, ref2 accumulates aliphatic aldoximes but not PAOx. Interestingly, overexpression of AtCYP79A2 in ref2 resulted in a further decrease in sinapoylmalate content compared to ref2. This indicates that accumulation of PAOx has an additive effect on phenylpropanoid pathway suppression mediated by other aldoximes.
KEYWORDS: Aldoximes, glucosinolates, phenylpropanoids, benzyl isothiocyanate
The amino acid derived aldoximes are hub metabolites in plants that branch into several pathways important for growth and defense. Tryptophan-derived indole-3-acetaldoxime (IAOx) and phenylalanine-derived phenylacetaldoxime (PAOx), are precursors of the growth regulating auxins indole-3-acetic acid (IAA) and phenylacetic acid (PAA) respectively.1–5 Besides serving as auxin precursors, IAOx and PAOx are also intermediates of various defense-related compounds6–8 including Brassicales-specific glucosinolates in Arabidopsis. In addition to their role as metabolic intermediates, the accumulation of aldoximes also represses phenylpropanoid biosynthesis via accelerated degradation of phenylalanine ammonia-lyase (PAL), the first step of the phenylpropanoid biosynthesis pathway.5,9
Aldoximes are synthesized by cytochrome P450 monooxygenases belonging to the 79 family in most plants. In Arabidopsis, PAOx is made from phenylalanine by the enzyme CYP79A2 (At5g05260).8 PAOx can then be catalyzed by two enzymes, REF2 (CYP83A1) and REF5 (CYP83B1) to produce its aci-nitro intermediate which is further converted to produce benzyl glucosinonate (Figure 1a).5,10 Although both REF2 and REF5 function redundantly to catalyze various aldoximes, REF5 has higher activity toward IAOx whereas REF2 is a major enzyme for aliphatic aldoximes. Previously, we have shown that Arabidopsis plants that overexpress CYP79A2 in the wild-type and ref2 genetic backgrounds accumulate high levels of the PAOx-derived benzyl glucosinolate although as expected the accumulation is higher in the wild-type background (ox-1/WT, ox-2/WT) than in the ref2 background (ox-21/ref2, ox-22/ref2).5 Interestingly, we found that CYP79A2 overexpression lines accumulate benzyl glucosinolate hydrolysis products such as benzyl isothiocyanate and benzyl cyanide in intact seedlings. We analyzed metabolites in undamaged 2-week old whole seedlings following published protocol.5,11 Sterilized seeds were vertically grown on agar plates containing Murashige and Skoog (MS) media12 under long day condition (16 hour light/ 8 hour dark) for two weeks and then sampled in liquid nitrogen immediately. To quantify PAA, benzyl isothiocyanate and benzyl cyanide, we used a method adapted from Schmelz et.al.,11 Briefly, tissues were flash frozen in liquid nitrogen and then 100 mg were extracted in 300 µl of H2O:1-propanol:HCL(1:2:0.005) spiked with 100ng of d7-PAA as an internal standard. Then, samples were homogenized and further extracted and analyzed as stated in a published paper.11 Benzyl glucosinolate was quantified as following published protocol.5 The ions monitored and retention times for the compounds were as follows: PAA (151 m/z, RT 6.76 min), d7-PAA (158 m/z, RT 6.71 min), BC (118 m/z, 6.82 min), BTIC (150 m/z, RT 8.92 min). Compounds are confirmed and quantified using authentic standards. As shown in Figure 1b–e, whole seedlings overexpressing CYP79A2 increase PAA and benzyl glucosinolate compared to wild type similarly to soil-grown plants and were also found to produce benzyl glucosinolate hydrolysis products, benzyl isothiocyanate and benzyl cyanide. The activation of glucosinolates into cytotoxic products such as benzyl isothiocyanate is initiated by β-glucosidases known as myrosinases when pathogen/insect attack disrupts specialized cells or subcellular compartments separating glucosinolates from these enzymes.13–15 However, several studies indicate that aliphatic or indole glucosinolates may also undergo this turnover even in the absence of tissue disruption.13,15–19 The accumulation of benzyl isothiocyanate and benzyl cyanide, two products of benzyl glucosinolate hydrolysis, in intact Arabidopsis seedlings grown in aseptic condition supports in vivo turnover of benzyl glucosinolate (Figure 1d,e). Current dogma indicates that while a group of myrosinases known as ‘classical myrosinases’ (TGG1-6) are mainly involved in tissue disruption-mediated glucosinolate degradation, other β-glucosidases categorized as atypical myrosinases likely function in glucosinolate turnover not resulting from tissue disruption.13,15–20 To identify β-glucosidases that might function in benzyl glucosinolate turnover, the transcriptomes5 of wild-type and ox-2/WT were analyzed to determine if any known or candidate β-glucosidases were upregulated in the benzyl isothiocyanate-accumulating lines. However, TGG 3 ~ 6 did not express in our sample whereas TGG1 and TGG2 were expressed but not transcriptionally induced in ox-2/WT (Figure 1f). Nine putative β-glucosidases were expressed in our sample and PYK10, BGU19, and BGLU33 were slightly upregulated in ox-2/WT. However, the remaining β-glucosidases were down-regulated (Figure 1f). A recent study has shown that BGLU28 and BGLU30 are responsible for glucosinolate breakdown under sulfur deficiency.19 In our samples, BGLU30 was not expressed and BGLU28 was down-regulated (Figure 1f), which further supports their roles in sulfur deficient condition as our samples were grown in sulfur sufficient media.12,19 More work is needed to identify which enzymes are participating in benzyl glucosinolate turnover in Arabidopsis.
Figure 1.
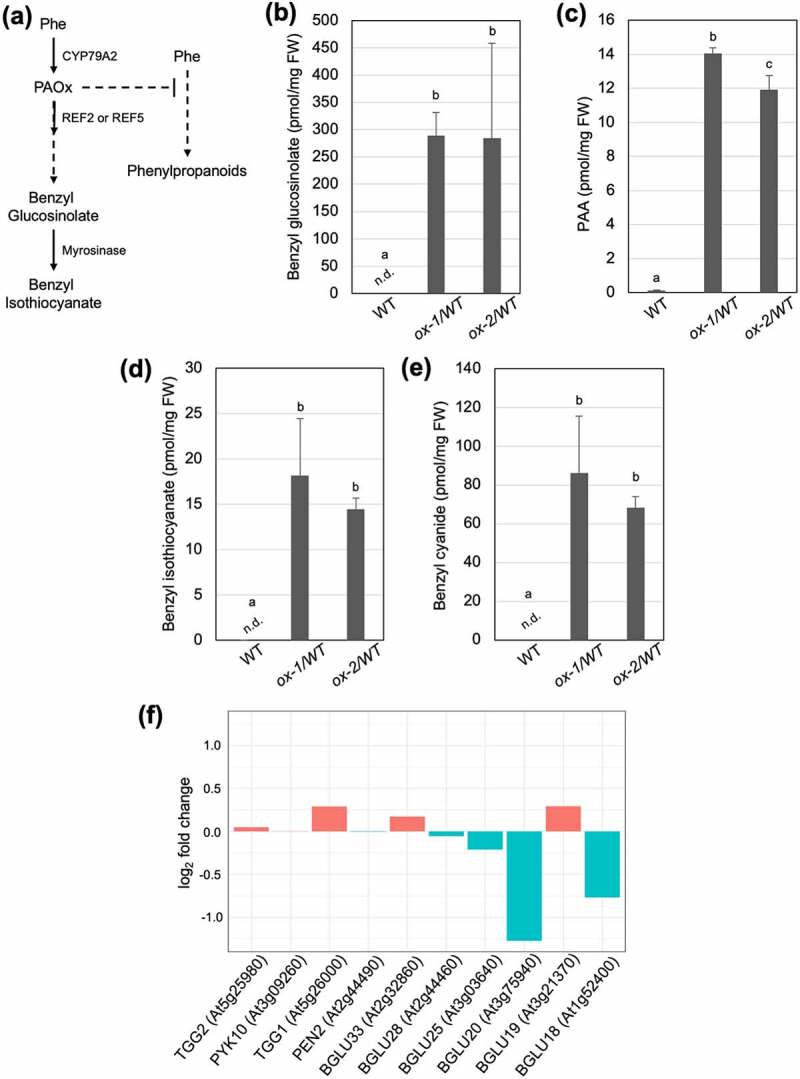
Glucosinolate turnover occurs in vivo without tissue disruption. (a) A schematic diagram of PAOx metabolism and benzyl glucosinolate degradation into benzyl isothiocyanate in Arabidopsis thaliana. In addition to the shown pathways, PAOx can also act as a precursor for PAA. (b) Benzyl glucosinolate, (c) PAA, (d) benzyl isothiocyanate, and (e) benzyl cyanide content of WT, ref2, and CYP79A2 overexpression lines. All metabolite data was collected from 2-week-old whole seedlings grown on MS plates. Benzyl glucosinolate content was determined via HPLC, while all other metabolites were analyzed via GC-MS. Data represents mean ± SD (n = 3). The means were compared by one-way ANOVA and statistically significant differences (P < .05) were identified by Tukey’s test and are indicated by letters to represent differences among groups. (f) Expression levels (log2 fold change) of myrosinase and β-glucosidase genes in CYP79A2 overexpression line ox-2 compared with wild-type. TGG3, 4, 5, and 6 were not expressed in our samples. Expression levels were calculated using published RNAseq data (PRJNA682862).5
Despite differences in benzyl glucosinolate content dependent on the genetic background, the CYP79A2 overexpression lines accumulated PAA to a similar degree.5 It was therefore unexpected that CYP79A2 overexpression in the ref2 background resulted in more severe developmental changes than its overexpression in the wild-type background.5 The ox-21/ref2 and ox-22/ref2 lines showed small dark green leaves compared to ox-1/WT and ox-2/WT lines (Figure 2a). Given their similarity in PAA content, this difference in growth may be due to an alteration in phenylpropanoid content rather than auxin accumulation (Figure 1a). The ref2 mutant accumulates less phenylpropanoids and has an altered lignin profile, possibly due to the accumulation of aldoximes derived from mostly aliphatic amino acids.10 Since CYP79A2 is not expressed in most organs, it is unlikely that this alteration in phenylpropanoid biosynthesis and profile is due to PAOx in the ref2 background. This circumstance gave us the opportunity to determine how the accumulation of various aldoximes (PAOx and aliphatic aldoximes) impacts phenylpropanoid metabolism. To determine whether different types of aldoximes have an additive effect on phenylpropanoid repression, the level of sinapoylmalate (a major phenylpropanoid of Arabidopsis leaves) was determined in ox-21/ref2 and ox-22/ref2. A statistically significant decrease in sinapoylmalate content was observed in ox-21/ref2 and ox-22/ref2 compared to ref2 (Figure 2b), which suggests that the accumulation of multiple aldoximes has an additive effect on repression of phenylpropanoid biosynthesis. Given that aldoxime-mediated phenylpropanoid repression includes accelerated degradation of PAL9 which can affect the production of all phenylpropanoids including lignin, a pivotal structure compound, this combined repression in phenylpropanoid biosynthesis by both PAOx and aliphatic aldoximes may contribute to the altered growth phenotype of ox-21/ref2 and ox-22/ref compared to ox-1/WT and ox-2/WT. Further study awaits to determine if the repression caused by PAOx and aliphatic aldoximes acts synergistically with IAOx-mediated phenylpropanoid repression and to identify aldoxime-mediated repression mechanisms besides PAL degradation.
Figure 2.
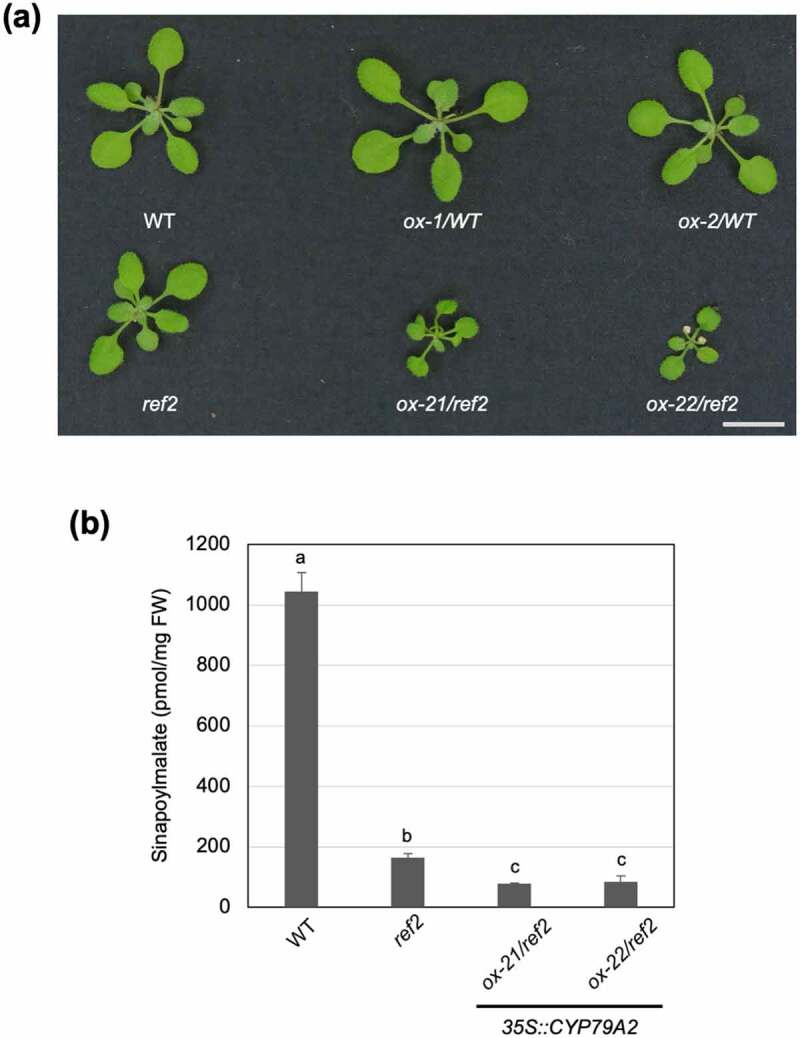
Aldoxime-derived repression of phenylpropanoid biosynthesis is additive. (a) Representative 2-week-old CYP79A2 overexpression lines in the WT and ref2 genetic backgrounds compared with their controls. Bar, 1 cm. (b) Sinapoylmalate content of WT, ref2, and CYP79A/ref2 overexpression lines (ox-21, ox-22). Sinapoylmalate in 2-week-old whole aerial parts was analyzed via HPLC. Data represents mean ± SD (n = 4). The means were compared by one-way ANOVA and statistically significant differences (P < .05) were identified by Tukey’s test and indicated by letters to represent differences among groups
Acknowledgments
We thank ICBR Proteomics & Mass Spectrometry Facility at UF with LTS operation.
Funding Statement
This work was supported by the United States Department of Agriculture (USDA)-National Institute of Food and Agriculture Hatch (005681) and a startup fund from the Horticultural Sciences Department and Institute of Food and Agricultural Sciences at the University of Florida to JK. The United States Department of Agriculture (USDA)-Agricultural Research Service Project number 6036-11210-001-00D and the USDA- National Institute of Food and Agriculture-Specialty Crop Research Initiative grant 2018-51181-28419 to AB
Disclosure statement
No potential conflicts of interest were disclosed.
References
- 1.Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:1–4. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Y. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002;16:3100–3112. doi: 10.1101/gad.1035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugawara S, Hishiyama S., Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H. Biochemical analyses of índole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. PNAS. 2009;106:5430–5435. doi: 10.1073/pnas.0811226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoi Y, Tanaka K, Cook SD, Hayashi KI, Kasahara H. GH3 auxin-amido synthetases alter the ratio of indole-3-acetic acid and phenylacetic acid in arabidopsis. Plant Cell Physiol. 2020;61:596–605. doi: 10.1093/pcp/pcz223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez VC, Dai R, Bai B, Tomiczek B, Askey BC, Zhang Y, Rubin GM, Ding Y, Grenning A, Block AK, Kim J. Aldoximes are precursors of auxins in Arabidopsis and maize. New Phytologist 231, 1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- 7.Glawischnig E, Hansen BG, Olsen CE, Halkier BA. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. PNAS. 2004;101:8245–8250. doi: 10.1073/pnas.0305876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittstock U, Halkier BA. Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. Catalyzes the conversion of L-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J Biol Chem. 2000;275:14659–14666. doi: 10.1074/jbc.275.19.14659. [DOI] [PubMed] [Google Scholar]
- 9.Kim JI, Zhang X, Pascuzzi PE, Liu C, Chapple C. Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytol. 2020;225:154–168. doi: 10.1111/nph.16108. [DOI] [PubMed] [Google Scholar]
- 10.Hemm MR, Ruegger MO, Chapple C. The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell. 2003;15:179–194. doi: 10.1105/tpc.006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J L. 2004;39(5):790–808. doi: 10.1111/j.1365-313X.2004.02168.x. [DOI] [PubMed] [Google Scholar]
- 12.Murashige T, Skoog F, Revised A. Medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 13.Wittstock U, Kurbach E, Herfurth AM, Stauber EJ. Glucosinolate breakdown. Advances in Botanical Research. 80:125–169.
- 14.Bones AM, Rossiter JT. The myrosinase-glucosinolate system, its organisation and biochemistry. Physiol Plant. 1996;97:194–208. doi: 10.1111/j.1399-3054.1996.tb00497.x. [DOI] [Google Scholar]
- 15.Sugiyama R, Hirai MY. Atypical myrosinase as a mediator of glucosinolate functions in plants. Front Plant Sci. 2019;10:1008. doi: 10.3389/fpls.2019.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen B, Chen S, Hansen C, Olsen C, Halkier B. Composition and content of glucosinolates in developing. Arabidopsis thaliana. Planta. 2002;214:562–571. [DOI] [PubMed] [Google Scholar]
- 17.Jeschke V, Weber K, Moore SS, Burow M. Coordination of glucosinolate biosynthesis and turnover under different nutrient conditions. Front Plant Sci. 2019;10. doi: 10.3389/fpls.2019.01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittstock U, Burow M. Glucosinolate breakdown in arabidopsis: mechanism, regulation and biological significance. Arabidopsis Book. 2010;8. doi: 10.1199/tab.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama R, et al. Retrograde sulfur flow from glucosinolates to cysteine in Arabidopsis thaliana. PNAS. 2021;118:22. doi: 10.1073/pnas.2017890118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harun S, Abdullah-Zawawi M-R, Goh -H-H, Mohamed-Hussein Z-A. A comprehensive gene inventory for glucosinolate biosynthetic pathway in arabidopsis thaliana. J Agric Food Chem. 2020;68(28):7281–7297. doi: 10.1021/acs.jafc.0c01916. [DOI] [PubMed] [Google Scholar]


