Abstract
Critically ill patients often develop symptoms of sepsis and therefore require microbiological tests for bacteremia that use conventional blood culture (BC) techniques. However, since these patients frequently receive early empirical antibiotic therapy before diagnostic procedures are completed, examination by BC can return false-negative results. We therefore hypothesized that PCR could improve the rate of detection of microbial pathogens over that of BC. To test this hypothesis, male Wistar rats were challenged intravenously with 106 CFU of Escherichia coli. Blood was then taken at several time points for detection of E. coli by BC and by PCR with E. coli-specific primers derived from the uidA gene, encoding β-glucuronidase. In further experiments, cefotaxime (100 or 50 mg/kg of body weight) was administered intravenously to rats 10 min after E. coli challenge. Without this chemotherapy, the E. coli detection rate decreased at 15 min and at 210 min after challenge from 100% to 62% of the animals with PCR and from 100% to 54% of the animals with BC (P, >0.05). Chemotherapy decreased the E. coli detection rate at 25 min and at 55 min after challenge from 100% to 50% with PCR and from 100% to 0% with BC (P, <0.05). Thus, at clinically relevant serum antibiotic levels, PCR affords a significantly higher detection rate than BC in this rat model. The results suggest that PCR could be a useful adjunct tool supplementing conventional BC techniques in diagnosing bacteremia.
Episodes of sepsis are common and frequently life-threatening (12) complications, occurring in 44.8% of all critically ill patients (38). By definition (4), sepsis is caused by microbial infection. However, bacteremia, i.e., the presence of microorganisms in the bloodstream, is diagnosed in only approximately 4 to 12% of all blood cultures (BC) (26, 37). Escherichia coli is one of the most common microorganisms isolated during hospital-acquired bacteremia in intensive care patients (2, 21, 38). Several factors may contribute to the low bacteremia detection rate. First, some species of bacteria are difficult to culture (9), and some, such as Mycobacterium spp. (5, 11, 15) or Bartonella spp. (6), grow very slowly. Second, bacteremia is often transient (29), meaning that the number of viable microorganisms circulating in the blood decreases rapidly after the onset of bacteremia due to phagocytic and other host defense mechanisms. Finally, up to 65% of intensive care patients showing symptoms of sepsis are already under antimicrobial treatment, thus compromising the results of microbiological culture techniques (8, 25, 31, 37).
Since the emergence of bacteremia is associated with high mortality rates and financial costs as well as a longer hospital stay (7, 27, 30), independent of the severity of the underlying disease (3, 33), improved techniques for diagnosing bacteremia are needed. We hypothesized that amplification of bacterial DNA by PCR could overcome some of the problems associated with conventional BC techniques. The potential of PCR technology to recover intracellular or nongrowing microorganisms may be useful for diagnosing bacteremia. Although PCR has already been successfully applied in the detection of fastidious microorganisms (22, 43) and, in particular, for detecting bacteremia (23, 28, 44), a direct comparison of BC techniques with PCR with respect to detection levels and time course during bacteremia has yet to be carried out. Therefore, we assessed these parameters for experimental bacteremia induced by E. coli in rats.
MATERIALS AND METHODS
Rat model of bacteremia.
Male Wistar rats (350 to 500 g) were anesthesized by an initial intramuscular injection of ketamine at 80 mg/kg of body weight (bw) and xylazine at 4 mg/kg of bw, separately. Following this injection, both jugular veins were cannulated with a 26-gauge cannula (Abbott, Wiesbaden, Germany) under aseptic conditions. One cannula was used exclusively for the collection of blood samples. Bacteremia was induced via the other cannula by injection of 100 μl of a suspension of E. coli (ATCC 11229; 107 CFU/ml in 0.9% saline). E. coli was chosen to represent nonfastidious pathogens for comparing the sensitivities of BC and PCR because it is the most frequent gram-negative pathogen causing bacteremia. We collected 2.2-ml blood samples from 19 animals (group I) 5 min before bacterial challenge and at 5, 15, 150, 180, and 210 min afterward. One blood sample from each of 16 animals surviving 210 min was analyzed at each time point, with the exception of time points 15, 150, and 210 min after bacterial challenge, when analysis for E. coli was limited to 10, 15, and 13 rats, respectively, due to insufficient blood sample volumes. One milliliter of whole blood was immediately inoculated into the BC medium, and 1 ml was stored in EDTA tubes (Sarstedt, Nürmbrecht, Germany) for PCR analysis; 100 μl of each sample was directly plated in duplicate on sheep blood agar plates for the quantitation of bacteria.
From an additional 20 animals, 2.0-ml blood samples were collected before bacterial challenge and at 5, 25, 40, and 55 min afterward for detecting E. coli by BC and PCR. Ten animals (group II) received 100 mg of cefotaxime/kg of bw 10 min after bacterial challenge via the cannula used to inject E. coli. The other 10 animals (group III) received 50 mg of cefotaxime/kg of bw. At each time point, one blood sample was analyzed from each of 10 animals. From three animals in each group, an additional 0.5 ml of blood was taken and stored in an EDTA tube (Sarstedt) to check serum cefotaxime concentrations. The experiments were approved by the Animal Ethics Committee of the University of Tübingen, Tübingen, Germany.
In order to detect E. coli by BC, 1-ml blood samples were inoculated into BACTEC Peds Plus F medium (Becton Dickinson, Sparks, Md.), which contains resin for neutralizing antimicrobial agents (13, 31, 45) and is designed for an inoculum of 1 to 3 ml. The vials were incubated for 7 days in a shaker incubator at 37°C. After 48 h, the vials were punctured under sterile conditions, and 100 μl was subcultured on sheep blood (5%) agar in duplicate for 24 h (5% CO2). If bacterial growth was negative, the vials were incubated for another 5 days, followed by subculturing as described above.
PCR.
A 1-ml blood sample was divided into three aliquots of 300 μl each, which were transferred into sterile cups. Erythrocytes were lysed, and DNA was extracted according to the manufacturer’s protocol with a Puregene DNA isolation kit (Biozym Diagnostik GmbH, Hessisch Oldendorf, Germany) for whole blood. Two pairs of oligonucleotide primers (one nested in the other) were derived from the uidA gene of E. coli, encoding β-glucuronidase specific for E. coli and Shigella spp. Oligonucleotides P1 (5′-ATC ACC GTG GTG ACG CAT GTC GC-3′) and P2 (5′-CAC CAC GAT GCC ATG TTC ATC TGC-3′) were used in the first round of PCR to amplify a 486-bp fragment (19). Oligonucleotides P3 (5′-TAT GAA CTG TGC GTC ACA GCC-3′) and P4 (5′-CAT CAG CAC GTT ATC GAA TCC-3′) were used in the nested PCR for the amplified products from the primary PCR to amplify a 186-bp fragment. For DNA amplification, 20.4 μl of reaction mixture was added to 29.6 μl of purified DNA. The master mixture for the first round of PCR included the following: 10 mM Tris-HCl, 50 mM KCl, 4.5 mM MgCl2, 600 μM each deoxynucleoside triphosphate (dNTP) (Amersham Pharmacia Biotech, Freiburg, Germany), 0.6 μM each primer, and 1 U of Taq DNA polymerase (Amersham Pharmacia Biotech). Each reaction mixture was overlaid with 1 or 2 drops of mineral oil. PCR amplifications were performed with a Hybaid Omni Gene Temperature Cycler (MWG Biotech, Ebersberg, Germany).
The primary PCR amplification consisted of an initial denaturation step at 95°C for 5 min; 35 cycles at 95°C for 30 s, 50°C for 1 min, and 72°C for 1 min; and a final elongation phase at 72°C for 5 min. After the first reaction, 1 μl of the amplified product was added to 49 μl of the second master mixture. The master mixture for the nested PCR contained the following: 10 mM Tris-HCl, 50 mM KCl, 4 mM MgCl2, 400 μM each dNTP (Amersham Pharmacia Biotech), 0.4 μM each primer, 10 μl of Q-solution (Qiagen, Hilden, Germany), and 1 U of Taq DNA polymerase (Amersham Pharmacia Biotech). The nested PCR amplification was performed with an initial denaturation step at 95°C for 5 min; 35 cycles at 95°C for 30 s, 50°C for 1 min, and 72°C for 1 min; and a final elongation phase at 72°C for 5 min.
Each time a PCR was performed, we also ran a positive control (bacterial DNA), a negative control (rat DNA), and a reagent control (all PCR reagents without DNA) to evaluate the success of amplification, the specificity of the reaction, and the purity of the reagents, respectively. The product of the nested PCR was electrophoresed with 2 μl of gel loading buffer (0.25 g of bromphenol blue and 40 g of saccharose dissolved in 100 ml of distilled water) through a 2% agarose gel at 110 V for 40 min in Tris-acetate–EDTA buffer). Molecular size markers (Promega, Madison, Wis.) were run concurrently. The gel, stained with ethidium bromide (0.5 μg/ml), was examined under UV light for the presence of a 186-bp band and photographed for documentation.
In order to assess the sensitivity of a specific PCR, 1 ml of rat blood was spiked with E. coli (106 to 100 CFU) and immediately processed as described above.
Verification of correct fragment production by the PCR was performed by Southern hybridization. PCR fragments were Southern blotted onto positively charged nylon membranes (Boehringer GmbH, Mannheim, Germany) by use of a Turbo Blotter (Schleicher & Schuell, Inc., Keene, N.H.) as prescribed by the manufacturer for alkaline transfer. The DNA was cross-linked to the damp membranes by use of a GS Gene Linker UV chamber (BioRad Laboratories, Hercules, Calif.) according to the instructions of the provider. The gene probe used for hybridization was a digoxigenin-UTP-labeled (Boehringer) 186-bp fragment produced by PCR, resolved on a 2% agarose gel, and purified with a PCR product purification kit (Qiagen). The standard nylon membrane prehybridization and hybridization protocol was performed under stringent conditions at 65°C. Probed membranes were exposed to Hyperfilm ECL (Amersham Life Science, Little Chalfont, England).
Analysis of cefotaxime levels in serum.
Cefotaxime concentrations were measured by high-performance liquid chromatography with a UV detection system by the method reported by Dell et al. (10).
Statistical analysis.
In order to detect any relevant difference between the PCR and the BC methods with respect to bacteremia detection as a function of time, we chose confidence intervals of 95% (P, <0.05). The minimum number of experiments needed to establish a difference of at least 30% between the two techniques for group I was calculated to be 20. A preliminary data analysis was scheduled after the completion of 12 experiments. For groups II and III, the confidence intervals were also 95%. The minimum number of experiments needed to reveal a difference of at least 80% between the two methods for groups II and III was 10 (1).
RESULTS
For the detection of E. coli DNA in rat blood samples, PCR was performed with primers for the uidA gene of E. coli (19). PCR products from rat blood samples were verified to be specific for the target gene by Southern hybridization with a dUTP-labeled probe for uidA. No differences in the sizes of the PCR fragments from rat blood samples on agarose gels and the hybridization product on the Southern blot were observed (data not shown).
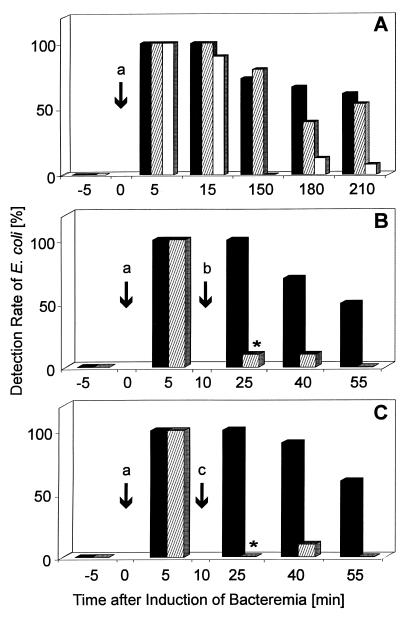
The sensitivity of PCR and BC for detecting E. coli in whole rat blood was 1 CFU/ml (data not shown). Before bacterial challenge, BC, the direct plating method, and PCR all returned negative results, indicating the absence of bacterial contamination during the animal experiment and during the PCR procedure (Fig. 1A). After bacterial challenge, the rates of detection of E. coli by the three methods decreased in a time-dependent manner (Fig. 1A). Even after 210 min, the E. coli PCR was positive for 61% of the 13 blood samples, whereas BC with the BACTEC system and the direct plating method were positive for 53 and 8% of the samples, respectively. No significant difference was observed between PCR and BC for the detection of E. coli at any of the examination times, but the rate of detection by the direct plating method was noticeably lower than that by either PCR or BC. Quantitatively, the direct plating method showed mean numbers of bacteria per milliliter of 27, 15, 0.0, 0.6, and 0.4 at 5, 15, 150, 180, and 210 min after E. coli challenge.
FIG. 1.
Time course of E. coli detection in experimental bacteremia in rats by PCR, BC, and the direct plating method. (A) Sixteen rats were challenged intravenously with 106 CFU of E. coli at time zero (arrow a) without further intervention. (B and C) Another 10 rats each received 100 mg (B) (arrow b) or 50 mg (C) (arrow c) of cefotaxime/kg of bw 10 min after bacterial challenge. For E. coli detection, blood was collected 5 min before challenge and at the indicated times after challenge. E. coli was detected by PCR (A, B, and C) (filled bars), BC (A, B, and C) (hatched bars), and the direct plating method (A) (open bars). Bars represent the percentages of E. coli-positive blood samples. ∗, P < 0.05.
As expected, the sensitivity of BC decreased significantly during experimental E. coli bacteremia in rats receiving clinically relevant doses of cefotaxime (Fig. 1B and C). In antibiotic treatment groups II and III, peak serum cefotaxime levels ranged between 121 and 137 mg/liter and between 94 and 86 mg/liter 15 min after injection, respectively (data not shown). When blood was taken 15 min after cefotaxime was administered intravenously, i.e., 25 min after bacterial challenge, only 10% of the samples were found positive by BC, whereas 100% of the samples were found positive by PCR (Fig. 1B and C). Data analysis established an 80% higher detection rate for the PCR method 25 min after bacterial challenge with both cefotaxime doses. At subsequent sampling times, the advantage of the PCR technique was also evident but did not reach the 80% range.
DISCUSSION
BC is considered the “gold standard” for detecting microorganisms in the blood (29, 37). Nevertheless, many clinicians and microbiologists are concerned that BC for critically ill patients receiving antimicrobial chemotherapy could indicate lower rates of bacteremia than are actually present (8, 34). Antibiotic-binding BC devices, such as the BACTEC Plus F system, were developed in response to these concerns (13, 31, 41). Still, the rate of detection of bacteria in blood specimens has not changed substantially. The results of the present study strongly suggest that the clinicians’ suspicions have a scientific basis. In an experimental rat model for E. coli bacteremia, a significantly higher rate of detection of E. coli was achieved with PCR technology than with BC when the animals were treated with clinically relevant levels of cefotaxime.
Interestingly, this difference in the detection rate between PCR and BC was not due to a higher sensitivity of our PCR than of BC. Regardless of whether PCR or BC is used, the ability to detect bacteremia depends on the presence of at least one microorganism in the blood being sampled. When tested in vitro, both methods detected as little as 1 CFU of E. coli per ml of blood, a volume which is normally used for diagnosing bacteremia, particularly in neonatal intensive care units (26). Furthermore, no significant difference in E. coli detection rates was observed between PCR and BC at any time point during experimental bacteremia in animals without antibiotic treatment. The sensitivity of our PCR was comparable to that previously reported for PCR used to detect bacteremia (17, 23, 28, 32, 35, 44). To our knowledge, however, minimal detection levels of BC systems in vitro have yet to be reported. Since the number of pathogens is less than 1 microorganism per ml of blood in 62% of all adult patients with E. coli bacteremia (37) and can be less than 0.04 organism per ml of blood (18), large blood volumes (20 to 30 ml) have to be used in order to avoid false-negative results. Thus, even though the PCR method was designed for smaller volumes from the very outset, it has to be adapted accordingly. This problem might be solved in the near future, because the rapidly increasing clinical use of PCR technology to detect pathogens which could not be isolated by conventional methods (5, 15, 24) has already prompted the development of a variety of different DNA isolation kits.
Our finding that BC is much less efficient than PCR in detecting bacteremia during antimicrobial treatment is most probably due to the killing of E. coli by cefotaxime; killed bacteria are not detectable by BC, while PCR detects bacterial DNA independently of viability.
Similarly, in a rabbit model of endocarditis, recovery rates for E. coli and other bacteria were reduced during antimicrobial treatment (25). In addition, the higher sensitivity of PCR technology than of conventional BC techniques is due to the fact that intracellular or phagocytosed microorganisms are also detectable by PCR.
In order to quantify bacterial numbers in E. coli-challenged rat blood, the direct plating method was used. Direct plating corroborated the results obtained by PCR and BC with respect to the rapid clearance of E. coli from the bloodstream. The lower sensitivity of the direct plating method compared with PCR or BC may be due to its smaller sample volume and a lack of dilution of bacterial growth-inhibiting factors in the blood, which are known to influence bacterial recovery rates in BC (29, 37). Thus, since direct plating is clearly less sensitive than BC in detecting E. coli in blood samples, BC is a better standard than direct plating for the evaluation of PCR technology (20).
From the clinical point of view, it is vital to develop improved techniques for diagnosing bacteremia, because the emergence of bacteremia is of substantial prognostic and therapeutic importance (2, 16, 39). The occurrence of secondary bacteremia (14) is a signal that the host’s defenses have failed to contain an infection at its primary site or that the physician has failed to control the infectious process (40, 42). Thus, the detection of bacteria in the patient’s blood, regardless of whether they are still viable or have been killed by antibiotics, implies that the treatment regimen might be insufficient and has to be augmented.
Extracellular bacterial DNA has been considered to be immunologically inert in mammals. However, it was shown recently that bacterial DNA has substantial immunostimulatory properties comparable to those of endotoxin (36) and that its presence can cause sepsis-like symptoms in mice (36). Thus, methods for detecting DNA might improve our understanding of the frequently life-threatening systemic inflammatory response syndrome (4) in intensive care patients. In summary, our results suggest that PCR could prove to be a useful adjunct tool supplementing conventional BC techniques in diagnosing bacteremia.
ACKNOWLEDGMENTS
We are indebted to W. Hoffmann and T. Gottwald of the University of Tübingen for technical assistance concerning the rat model; K.-J. Plaueln of Hoechst-Marrion-Roussel, Frankfurt, Germany, for the kind gift of cefotaxime; M. Ulrich, C. Goerke, P. Krüger, and C. Wolz for discussions concerning the establishment of PCR technology; C. Meisner for assistance in the statistical evaluation of our data; D. Isaacman for reading the manuscript; and D. Blaurock for language corrections in the manuscript.
REFERENCES
- 1.Altman D G. Comparing groups—categorical data. In: Altmann D G, editor. Practical statistics for medical research. London, England: Chapman & Hall, Ltd.; 1991. pp. 229–276. [Google Scholar]
- 2.Bachur R, Caputo G L. Bacteremia and meningitis among infants with urinary tract infections. Pediatr Emerg Care. 1995;11:280–284. doi: 10.1097/00006565-199510000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Bates D W, Pruess K E, Lee T H. How bad are bacteremia and sepsis? Arch Intern Med. 1995;155:593–598. [PubMed] [Google Scholar]
- 4.Bone R C, Balk R A, Cerra F B, Dellinger R P, Fein A M, Knaus W A, Schein R M H, Sibbald W J. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 5.Bottger E C, Teske A, Kirschner P, Bost S, Chang H R, Beer V, Hirschel B. Disseminated Mycobacterium genevase infection in patients with AIDS. Lancet. 1992;340:76–80. doi: 10.1016/0140-6736(92)90397-l. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S A, Rooney J A, Manzewitsch P, Regnery R L. Isolation of Bartonella (Rochalimaea) henselae: effects of methods of blood collection and handling. J Clin Microbiol. 1997;35:544–547. doi: 10.1128/jcm.35.3.544-547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowe M, Ispahani P, Humphreys H, Kelley T, Winter R. Bacteraemia in the adult intensive care unit of a teaching hospital in Nottingham, UK, 1985–1996. Eur J Clin Microbiol Infect Dis. 1998;17:377–384. doi: 10.1007/BF01691564. [DOI] [PubMed] [Google Scholar]
- 8.Darby J M, Linden P, Pasculle W, Saul M. Utilization and diagnostic yield of blood cultures in a surgical intensive care unit. Crit Care Med. 1997;25:989–994. doi: 10.1097/00003246-199706000-00016. [DOI] [PubMed] [Google Scholar]
- 9.Davies S, Eggington R. Recovery of Mycoplasma hominis from blood culture media. Med Lab Sci. 1991;48:110–113. [PubMed] [Google Scholar]
- 10.Dell D, Chamberlain J, Coppin F. Determination of cefotaxime and desacetylcefotaxime in plasma and urine by high-performance liquid chromatography. J Chromatogr. 1981;226:431–440. doi: 10.1016/s0378-4347(00)86077-6. [DOI] [PubMed] [Google Scholar]
- 11.Esteban J, Molleja A, Fernandez-Roblas R, Soriano F. Number of days required for recovery of mycobacteria from blood and other samples. J Clin Microbiol. 1998;36:1456–1457. doi: 10.1128/jcm.36.5.1456-1457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagon J Y, Novara A, Stephan F, et al. Mortality attributable to nosocomial infections in the ICU. Infect Control Hosp Epidemiol. 1994;15:428–434. doi: 10.1086/646946. [DOI] [PubMed] [Google Scholar]
- 13.Fuller D D, Davis T E. Comparison of Bactec plus Aerobic/F, Anaerobic/F, Peds Plus/F and Lytic/F media with and without fastidious organism supplement to conventional methods for culture of sterile body fluids. Diagn Microbiol Infect Dis. 1997;29:219–225. doi: 10.1016/s0732-8893(97)00164-8. [DOI] [PubMed] [Google Scholar]
- 14.Garner J S, Jarvis W R, Emori T G, Horan T C, Hughes J M. CDC definitions for nosocomial infections. Am J Infect Control. 1988;16:128–140. doi: 10.1016/0196-6553(88)90053-3. [DOI] [PubMed] [Google Scholar]
- 15.Haas W H, Kirschner P, Ziesing S, Bremer H J, Bottger E C. Cervical lymphadenitis in a child caused by a previously unknown mycobacterium. J Infect Dis. 1993;167:237–240. doi: 10.1093/infdis/167.1.237. [DOI] [PubMed] [Google Scholar]
- 16.Isaacman D J. Strategies for improving the detection of bacteremia in children. Infect Dis Clin Pract. 1998;7:28–31. [Google Scholar]
- 17.Isaacman D J, Zhang Y, Rydquist-White J, Wadowsky R M, Post J C, Ehrlich G D. Identification of a patient with Streptococcus pneumoniae bacteremia and meningitis by the polymerase chain reaction (PCR) Mol Cell Probes. 1995;9:157–160. doi: 10.1006/mcpr.1995.0026. [DOI] [PubMed] [Google Scholar]
- 18.Jonsson B, Nyberg A, Henning C. Theoretical aspects of detection of bacteraemia as a function of the volume of blood cultured. APMIS. 1993;101:595–601. doi: 10.1111/j.1699-0463.1993.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 19.Juck D, Ingram J, Prevost M, Coallier J, Greer C. Nested PCR protocol for the rapid detection of Escherichia coli in potable water. Can J Microbiol. 1996;42:862–866. doi: 10.1139/m96-110. [DOI] [PubMed] [Google Scholar]
- 20.Kane T D, Johnson S R, Alexander J W, Babcock G F, Ogle C K. Detection of intestinal bacterial translocation using PCR. J Surg Res. 1996;63:59–63. doi: 10.1006/jsre.1996.0223. [DOI] [PubMed] [Google Scholar]
- 21.Kiehn T E, Wong B, Edwards F F, Armstrong D. Comparative recovery of bacteria and yeasts from lysis-centrifugation and a conventional blood culture system. J Clin Microbiol. 1983;18:300–304. doi: 10.1128/jcm.18.2.300-304.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kox L F F, Rhienthong D, Miranda A M, Udomsantisuk N, Ellis K, van Leeuwen J, van Heusden S, Kuijper S, Kolk A H J. A more reliable PCR for detection of Mycobacterium tuberculosis in clinical samples. J Clin Microbiol. 1994;32:672–678. doi: 10.1128/jcm.32.3.672-678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ley B E, Linton C J, Bennett D M C, Jalal H, Foot A B M, Millar M R. Detection of bacteraemia in patients with fever and neutropenia using 16S RNA gene amplification by polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1998;17:247–253. doi: 10.1007/BF01699981. [DOI] [PubMed] [Google Scholar]
- 24.Martin A B, Webber S, Fricker J, Jaffe R, Demmler G, Kearney D, Zhang Y-H, Bodurtha J, Gelb B, Ni J, Bricker T, Towbin J A. Acute myocarditis: rapid diagnosis by PCR in children. Circulation. 1994;90:330–339. doi: 10.1161/01.cir.90.1.330. [DOI] [PubMed] [Google Scholar]
- 25.McKenzie R, Reimer L G. Effect of antimicrobials on blood cultures in endocarditis. Diagn Microbiol Infect Dis. 1987;8:165–172. doi: 10.1016/0732-8893(87)90167-2. [DOI] [PubMed] [Google Scholar]
- 26.Paisley J W, Lauer B A. Pediatric blood cultures. Clin Lab Med. 1994;14:17–30. [PubMed] [Google Scholar]
- 27.Pittet D, Tarara D, Wenzel R P. Nosocomial bloodstream infection in critically ill patients. Extra length of stay, extra costs, and attributable mortality. JAMA. 1994;271:1598–1601. doi: 10.1001/jama.271.20.1598. [DOI] [PubMed] [Google Scholar]
- 28.Rattanathongkom A, Sermswan R W, Wongratanacheewin S. Detection of Burkholderia pseudomallei in blood samples using polymerase chain reaction. Mol Cell Probes. 1997;11:25–31. doi: 10.1006/mcpr.1996.0072. [DOI] [PubMed] [Google Scholar]
- 29.Reimer L G, Wilson M L, Weinstein M P. Update on detection of bacteremia and fungemia. Clin Microbiol Rev. 1997;10:444–465. doi: 10.1128/cmr.10.3.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rello J, Richart M, Mirelis B, et al. Nosocomial bacteremia in a medical-surgical intensive care unit: epidemiologic characteristics and factors influencing mortality in 111 episodes. Intensive Care Med. 1994;20:94–98. doi: 10.1007/BF01707661. [DOI] [PubMed] [Google Scholar]
- 31.Rohner P, Pepey B, Auckenthaler R. Advantage of combining resin with lytic Bactec blood culture media. J Clin Microbiol. 1997;35:2634–2638. doi: 10.1128/jcm.35.10.2634-2638.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudolph K M, Parkinson A J, Black C M, Mayer L W. Evaluation of polymerase chain reaction for diagnosis of pneumococcal pneumonia. J Clin Microbiol. 1993;31:2661–2666. doi: 10.1128/jcm.31.10.2661-2666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith R L, Meixler S M, Simberkoff M S. Excess mortality in critically ill patients with nosocomial bloodstream infections. Chest. 1991;100:164–167. doi: 10.1378/chest.100.1.164. [DOI] [PubMed] [Google Scholar]
- 34.Smith S M, Eng R H K. In vitro evaluation of the Bactec resin-containing blood culture bottle. J Clin Microbiol. 1983;17:1120–1126. doi: 10.1128/jcm.17.6.1120-1126.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song J-H, Cho H, Park M Y, Na D S, Moon H B, Pai C H. Detection of Salmonella typhi in the blood of patients with typhoid fever by polymerase chain reaction. J Clin Microbiol. 1993;31:1439–1443. doi: 10.1128/jcm.31.6.1439-1443.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sparwasser T, Miethke T, Lipford G, Borschert K, Häcker H, Heeg K, Wagner H. Bacterial DNA causes septic shock. Nature. 1997;386:336–337. doi: 10.1038/386336a0. [DOI] [PubMed] [Google Scholar]
- 37.Spencer R S. Blood cultures: where do we stand? J Clin Pathol. 1988;41:668–670. doi: 10.1136/jcp.41.6.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent J-L, Bihari D J, Suter P M, Bruining H A, White J, Nicolas-Chanoin M-H, Wolff M, Spencer R C, Hemmer M. The prevalence of nosocomial infections in intensive care units in Europe. JAMA. 1995;274:639–644. [PubMed] [Google Scholar]
- 39.Washington J A. Blood cultures: an overview. Eur J Clin Microbiol Infect Dis. 1989;8:803–806. doi: 10.1007/BF02185852. [DOI] [PubMed] [Google Scholar]
- 40.Weinstein M P. Current blood culture methods and systems: clinical concepts and interpretation of results. Clin Infect Dis. 1996;23:40–46. doi: 10.1093/clinids/23.1.40. [DOI] [PubMed] [Google Scholar]
- 41.Wilson M L, Weinstein M P. General principles in the laboratory detection of bacteremia and fungemia. Clin Lab Med. 1994;14:69–82. [PubMed] [Google Scholar]
- 42.Yagupsky P, Nolte F S. Quantitative aspects of septicemia. Clin Microbiol Rev. 1990;3:269–279. doi: 10.1128/cmr.3.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita Y, Kohno S, Koga H, Tomono K, Kaku M. Detection of Bacteroides fragilis in clinical specimens by PCR. J Clin Microbiol. 1994;32:679–683. doi: 10.1128/jcm.32.3.679-683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Isaacman D H, Wadowsky R M, Rydquist-White J, Post J C, Ehrlich G D. Detection of Streptococcus pneumoniae in whole blood by PCR. J Clin Microbiol. 1995;33:596–601. doi: 10.1128/jcm.33.3.596-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegler R, Johnscher I, Martus P, Lenhardt D, Just H-M. Controlled clinical laboratory comparison of two supplemented aerobic and anaerobic media used in automated blood culture systems to detect bloodstream infections. J Clin Microbiol. 1998;36:657–661. doi: 10.1128/jcm.36.3.657-661.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]