Abstract
We herein describe rapid and accurate clinical testing for COVID-19 by nicking and extension chain reaction system-based amplification (NESBA), an ultrasensitive version of NASBA. The primers to identify SARS-CoV-2 viral RNA were designed to additionally contain the nicking recognition sequence at the 5′-end of conventional NASBA primers, which would enable nicking enzyme-aided exponential amplification of T7 RNA promoter-containing double-stranded DNA (T7DNA). As a consequence of this substantially enhanced amplification power, the NESBA technique was able to ultrasensitively detect SARS-CoV-2 genomic RNA (gRNA) down to 0.5 copies/μL (= 10 copies/reaction) for both envelope (E) and nucleocapsid (N) genes within 30 min under isothermal temperature (41 °C). When the NESBA was applied to test a large cohort of clinical samples (n = 98), the results fully agreed with those from qRT-PCR and showed the excellent accuracy by yielding 100% clinical sensitivity and specificity. By employing multiple molecular beacons with different fluorophore labels, the NESBA was further modulated to achieve multiplex molecular diagnostics, so that the E and N genes of SARS-CoV-2 gRNA were simultaneously assayed in one-pot. By offering the superior analytical performances over the current qRT-PCR, the isothermal NESBA technique could serve as very powerful platform technology to realize the point-of-care (POC) diagnosis for COVID-19.
Keywords: COVID-19, SARS-CoV-2, qRT-PCR, Isothermal amplification, NESBA, NASBA
1. Introduction
Since the outbreak of novel coronavirus disease 2019 (COVID-19) in December 2019, it has rapidly spread across the globe and was declared pandemic by the World Health Organization (WHO) on March 11th, 2020 (Wang et al., 2020; Yan et al., 2020; Zhu et al., 2020). As of 21 September 2021, the pandemic of COVID-19 has infected over 228 millions and killed over four millions, unprecedently threatening human health and causing tremendous economic loss on the global community (World Health Organization, 2021).
Without question, timely and aggressive testing is essential in order to effectively contain the spread of COVID-19. The current gold standard for COVID-19 testing is quantitative reverse transcription-polymerase chain reaction (qRT-PCR) in which the viral RNA of SARS-CoV-2 is first reverse transcribed into complementary DNA (cDNA) followed by the qPCR reaction using the cDNA template (Guo et al., 2020; Lu et al., 2020; Tahamtan and Ardebili, 2020). This technique is highly sensitive and could identify the viral RNA even from the patients at the early stage before the onset of symptoms. However, it is relatively slow and requires costly and bulky instrument for precise temperature control and skilled personnel, which significantly restrict its use only to centralized clinics or laboratories and preclude prompt patient triage elsewhere (Cui and Zhou, 2020; Park et al., 2020; Zhu et al., 2020).
In resource-limited settings where challenges with molecular diagnostics exist, direct antigen testing based on lateral flow immunoassay offers an alternative option for the diagnosis of COVID-19 (Sheridan, 2020). This technique is very rapid, simple, and cost-effective, but can not very effectively diagnose the patients with low viral titers and may lead to false-negative results, due to low sensitivity (Liu and Rusling, 2021; Mak et al., 2020). Therefore, the direct antigen testing is used only for first screening where the qRT-PCR is not feasible and the qRT-PCR is normally recommended for the confirmation of COVID-19 cases (Habibzadeh et al., 2021; Lee et al., 2020; Peeling et al., 2021).
To advance the molecular diagnostics and facilitate point-of-care (POC) molecular diagnostics in resource-limited settings, isothermal nucleic acid amplification methods such as nucleic acid sequence-based amplification (NASBA) (Compton, 1991), rolling circle amplification (RCA) (Lizardi et al., 1998), loop-mediated isothermal amplification (LAMP) (Notomi et al., 2000), nicking enzyme amplification reaction (NEAR) (Van Ness et al., 2003), and recombinase polymerase amplification (RPA) (Piepenburg et al., 2006) and so on (Jia et al., 2010; Jung et al., 2010; Lee et al., 2020b; Song et al., 2021; Vincent et al., 2004) have emerged as a compelling alternative to the conventional thermocycling-based amplification method. Some of them have been successfully integrated with CRISPR/Cas system to develop ultrasensitive strategies to identify SARS-CoV-2 viral RNA such as specific high sensitivity enzymatic reporter unlocking (SHERLOCK) and DNA endonuclease targeted CRISPR trans reporter (DETECTR) (Broughton et al., 2020; Chen et al., 2018; Kellner et al., 2019; Patchsung et al., 2020).
Of these, NASBA is one of the most representative isothermal amplification strategies, which was designed to detect genomic RNA (gRNA) targets. In the NASBA reaction, target RNA is first reverse transcribed and its opposite strand is synthesized to produce T7 promotor-containing double stranded DNA (T7DNA). T7 RNA polymerase (T7RP) then promotes the transcription from the T7 RNA promoter region within the T7DNA, consequently producing numerous antisense RNA amplicons, which could produce fluorescent signal upon binding to a specific molecular beacon (MB) probe. The NASBA has been very intensively utilized to detect target RNA molecules under an isothermal condition but the key intermediate component of NASBA, T7DNA, is just produced linearly and not in an exponential manner and thus the final amplification efficiency is not sufficiently high enough to enable highly sensitive detection of target RNA.
To overcome this limitation and advance the NASBA technique, we recently developed a novel isothermal nucleic acid amplification method, termed nicking and extension chain reaction system-based amplification (NESBA) (Ju et al., 2021). By incorporating the nicking recognition sequence to the primer set to enable the nicking enzyme (NE)-aided exponential amplification, the NESBA achieved remarkably high sensitivity under an isothermal condition, which significantly outperforms NASBA. In this work, we applied the NESBA technique to test a large cohort of clinical samples for target envelope (E) and nucleocapsid (N) genes of SARS-CoV-2 and successfully confirmed all the COVID-19 cases.
2. Materials and methods
2.1. Materials
All DNA oligonucleotides used in this study were synthesized and purified with high performance liquid chromatography (HPLC) by Bioneer® (Daejeon, Korea). The sequences of the oligonucleotides are listed in Table S1. Nt.AlwI, 10 × NEBuffer™ 2.1 (100 mM Tris-HCl, pH 7.9, 500 mM NaCl, 100 mM MgCl2, and 1 mg/ml BSA), ribonucleotide solution mixture (rNTPs), deoxynucleotide solution mixture (dNTPs), DNase I (RNase-free) were purchased from New England Biolabs Inc. (Beverly, MA, USA). NASBA enzyme cocktail (Avian myeloblastosis virus reverse transcriptase (AMV RT), RNase H, and T7 RNA polymerase in high molecular weight sugar matrix), and 3 × NASBA reaction buffer (120 mM Tris-HCl, pH 8.5, 210 mM KCl, 36 mM MgCl2, 30 mM DTT, and 45% DMSO) were purchased from Life Science Advanced Technologies Inc. (St Petersburg, FL, USA). The gRNAs of SARS-CoV-2 (BetaCoV/Korea/KCDC03/2020), MERS-CoV, and HCoV-NL63 were provided by the National Culture Collection for Pathogens (NCCP, Cheongju, Korea), and those of HCoV-229E and HCoV-OC43 were purchased from the Korea Bank for Pathogenic Viruses (KBPV, Seoul, Korea). Plasmid controls and gBlock® gene fragments of E and N genes of SARS-CoV and HCoV-HKU-1 were purchased from Intergrated DNA Technologies Inc. (Coralville, IA, USA). Ultrapure DNase/RNase-free distilled water (DW) purchased from Bioneer® was used in all experiments. All other chemicals were of analytical grade and used without further purification.
2.2. In vitro transcription (IVT) from the DNA templates
For the production of RNA for E and N genes of SARS-CoV and HCoV-HKU-1 from DNA templates, the PCR mixture (20 μL) was prepared to contain 2 μL IVT primer set (10 μM each), 10 μL KAPA HiFi HotStart Ready Mix (2 ×) (KAPA Biosystems, MA, USA), and 104 copies/μL of plasmid controls or gBlock® gene fragments. The PCR was then performed for 3 min at 95 °C, followed by 35 cycles of 20 s at 98 °C, 15 s at 60 °C, and 30 s at 72 °C, and further incubation for 2 min at 72 °C. After the completion of the reaction, the PCR product was analyzed by 1% agarose gel electrophoresis and purified by using Wizard® SV Gel and PCR Clean-Up System (Promega, WI, USA), and applied to HiScribe™ T7 High Yield T7 RNA synthesis mix (New England Biolabs Inc., MA, USA) with the recommended composition of T7 RNA polymerase mix, rNTPs, and reaction buffer. The IVT reaction mixture was incubated for 3 h at 37 °C, followed by DNase I treatment for 30 min to prevent DNA contamination. Finally, the RNA product was purified by using Monarch® RNA cleanup kit (New England Biolabs Inc., MA, USA) according to the manufacturer's protocol. Its concentration was determined by using NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific, MA, USA).
2.3. The NESBA reaction procedure for SARS-CoV-2 detection
Prior to the assay, 30 μM of gene-specific MB in 1 × NESBA reaction buffer (45 mM Tris-HCl (pH 8.5), 70 mM KCl, 25 mM NaCl, 17 mM MgCl2, 10 mM DTT, 15% DMSO, and 50 μg/mL BSA) was heated up to 95 °C for 5 min followed by slow cooling down to 25 °C, and further incubated at 25 °C for 30 min.
The solution for NESBA (20 μL) was prepared by mixing 2.8 μL rNTPs (25 mM each), 1.4 μL dNTPs (10 mM each), 1.4 μL NESBA primer set (20 μM each), 0.2 μL MB (30 μM), 7.7 μL NESBA reaction buffer (2.6 ×), 5 μL NASBA enzyme cocktail (4 ×), 0.5 μL Nt.AlwI (10 U/μL), and 1 μL of viral RNA. The prepared solution was then incubated at 41 °C for 1 h and fluorescent signals from each MB were measured every 1 min by using CFX Connect™ Real-Time System (Bio-Rad, CA, USA).
2.4. RT-PCR
The reverse transcription (RT)-PCR was conducted on a C1000™ thermal cycler (Bio-rad, CA, USA) in a 20 μL solution containing 4 μL reaction buffer for RT (5 ×), 0.6 μL RevertAid RT (200 U/μL), 2 μL PCR reaction buffer (10 ×), 1.4 μL dNTPs (2.5 mM each), 1.4 μL primer set (20 μM each), 0.5 μL i-StarmaxⅡ™ DNA polymerase (5 U/μL), and 1 μL gRNA solution. T7DNA (NESBA), T7DNA-1 (NESBA), T7DNA-2 (NESBA), and T7DNA (NASBA) were produced by employing NESBA and NASBA primers (T7DNA (NESBA): NESBA primer set, T7DNA-1 (NESBA): NESBA forward primer and NASBA reverse primer, T7DNA-2 (NESBA): NASBA forward primer and NEBSA reverse primer, T7DNA (NASBA): NASBA primer set). RT was first carried out for 30 min at 50 °C and the PCR was performed for 10 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 45 s at 50 °C, and 60 s at 72 °C, and further incubation for 5 min at 72 °C. After the completion of the reaction, the RT-PCR products were analyzed by agarose gel electrophoresis. The RT-PCR products were also purified from the product solution by using a Wizard® SV Gel and PCR Clean-Up System (Promega, WI, USA), and their concentrations were determined by using NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific, MA, USA). The purified RT-PCR products were used as the markers on lane M1∼M4.
2.5. Gel electrophoresis analysis
For agarose gel electrophoresis, a 5 μL aliquot of the reaction solution was resolved on 2% agarose gel containing EtBr at a constant voltage of 135 V for 45 min using 1 × TBE as the running buffer. Gels were scanned using UV transilluminator (Bio-rad, CA, USA).
2.6. Clinical sample testing with NESBA
A total of 98 clinical samples including nasopharyngeal swab and sputum were collected from individuals with suspected SARS-CoV-2 infection and placed into the universal transport medium (UTM) (Asanpharm, Seoul, Korea) by Severance hospital (IRB approval number: 4-2020-0465; Seoul, Korea). Severance hospital was approved by Korea Centers for Disease Control and Prevention (KCDC) as a biosafety level-3 (BL-3) facility in accordance with institutional biosafety requirements. The gRNAs of the clinical samples were extracted by using AdvanSure™ Nucleic Acid R kit (LG chem, Seoul, Korea) according to the manufacturer's protocol. The gRNAs were then assayed with the NESBA and the results were compared with those from qRT-PCR to verify its clinical applicability.
2.7. The qRT-PCR procedure for clinical sample analysis
The qRT-PCR was set up and carried out according to the manufacturer's protocol of Allplex™ 2019-nCoV assay kit (Seegene, Seoul, Korea). RT was first carried out for 20 min at 50 °C and the PCR was performed for 15 min at 95 °C, followed by 45 cycles of 15 s at 94 °C and 30 s at 58 °C. The fluorescent signal was measured every cycle with CFX96™ Real-Time System (Bio-Rad, CA, USA). Based on Ct values (i.e. threshold cycle) determined by built-in system software, the diagnostic decision was made on each clinical sample.
3. Results and discussion
3.1. Overall procedure of the NESBA for SARS-CoV-2 detection
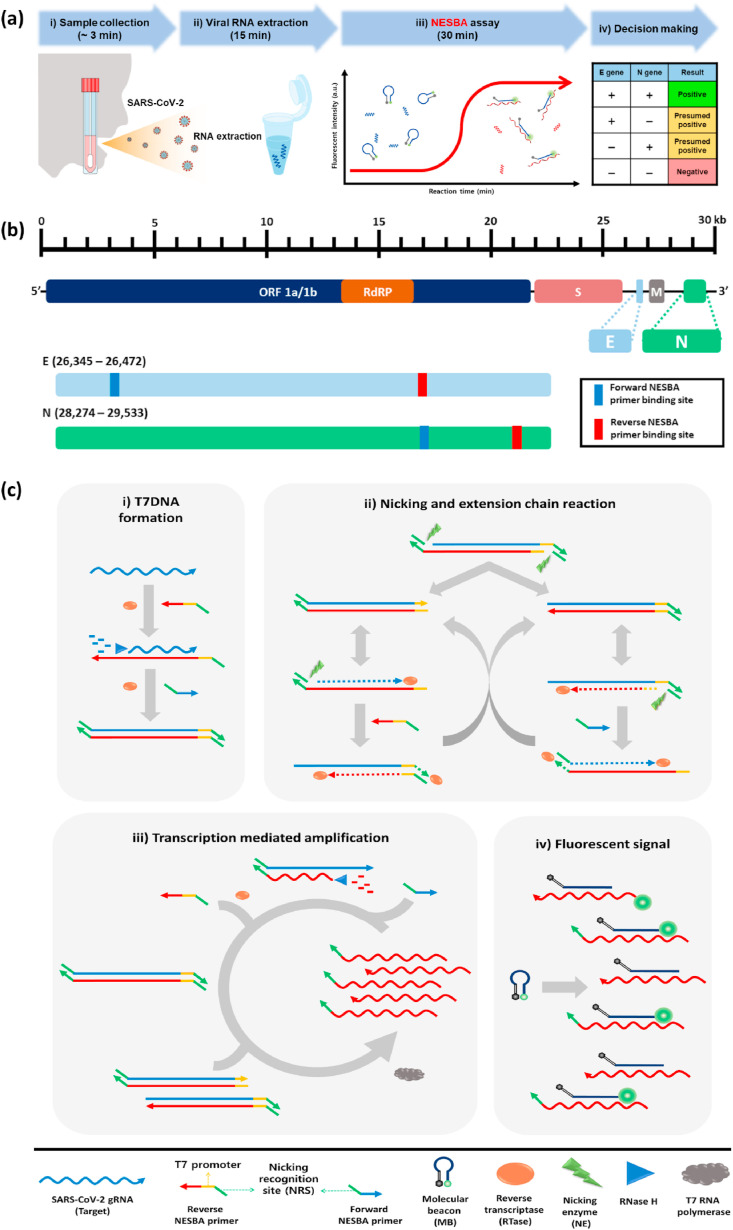
The overall procedure of the COVID-19 diagnosis based on the NESBA is illustrated in Fig. 1 . The workflow consists of four sequential steps as follows: i) collection of clinical specimens (∼3 min), ii) extraction of viral RNA (15 min), iii) NESBA assay for E and N genes of SARS-CoV-2 gRNA (30 min), and iv) decision making based on assay results (Fig. 1(a)). The target regions of E and N genes were set based on SARS-CoV-2 genome map (GenBank accession number: MN908947) (Fig. 1(b)) and the NESBA reaction mainly consists of four interconnected reactions: i) formation of T7 promotor-containing double-stranded DNA, ii) nicking and extension chain reaction-based exponential amplification of T7DNAs, iii) transcription-mediated RNA amplification, and iv) molecular beacon probe-based fluorescent enhancement (Fig. 1(c)). Particularly, to enable the exponential amplification of T7DNAs, the NESBA primer sets for E and N genes were designed to contain nicking recognition sequence (NRS) at the 5’ end of the primer sets and T7 promoter sequence for reverse NESBA primers.
Fig. 1.
The overall procedure of the COVID-19 diagnosis by NESBA. (a) The workflow of the COVID-19 diagnosis. (b) The SARS-CoV-2 genome map (GenBank accession number: MN908947) and target regions of the NEBSA. (c) Schematic illustration of the NESBA for target RNA detection. The arrow indicates the 3′ end of the nucleic acid strand.
In the presence of SARS-CoV-2 gRNA, the reverse NESBA primer binds to the target RNA and is reverse-transcribed to produce cDNA/RNA hybrid by reverse transcriptase (RTase). After RNA degradation by RNase H, forward NESBA primer binds to the cDNA and is extended to form T7DNA by the intrinsic DNA polymerase activity of the RTase. The nicking enzyme then recognizes the double-stranded NRSs (dsNRSs) of the T7DNA and cleaves the site four bases downstream to the dsNRS, where the nicking and extension chain reaction with displacement is repeated to generate a large amount of templates for the counterpart primers. After extension of the counterpart primer, the dsNRS is also formed to initiate another cycle, which provides templates for the opposite primers. In this way, the nicking and extension chain reaction repeatedly proceeds to exponentially amplify the T7DNAs.
The amplified T7DNAs produce a large amount of RNA amplicons utilizing T7 RNA polymerase-catalyzed transcription, whose sequence is complementary to MB. This leads to the opening of MBs and concomitant fluorescent signal enhancement from FAM. Importantly, the RNA amplicons also serve as a template for the production of T7DNAs, accelerating the amplification.
3.2. Feasibility of the NESBA for SARS-CoV-2 detection
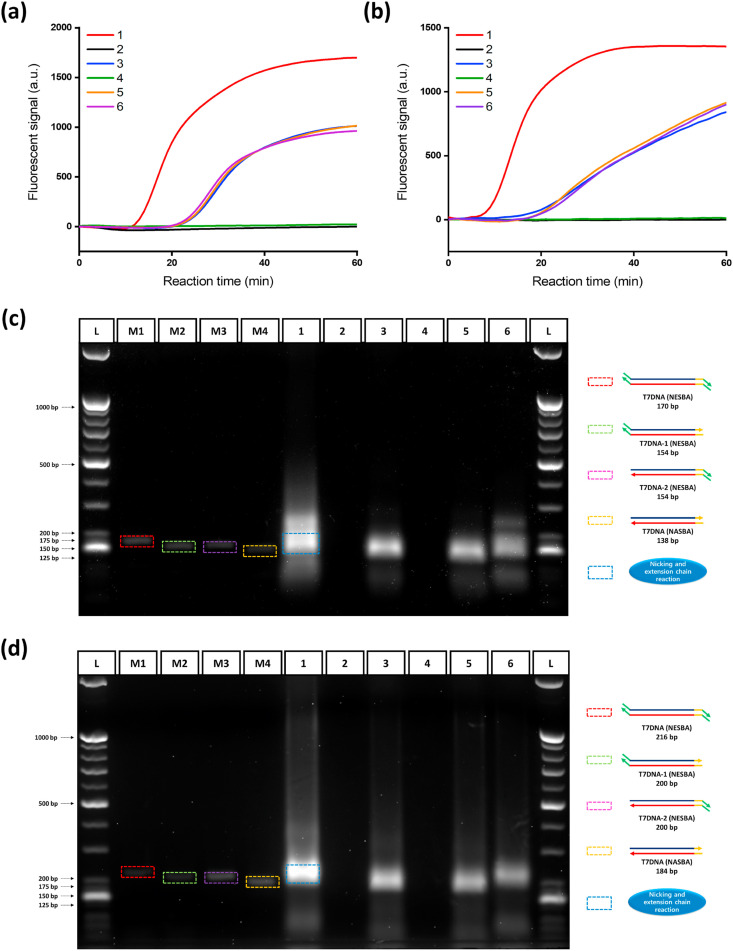
We first validated the feasibility of the NESBA by conducting the reactions for E and N genes of SARS-CoV-2 gRNA and monitoring the real-time fluorescent signals produced from gene-specific MBs binding to the amplified RNA amplicon under various combinations of key reaction components (Fig. 2 (a) and (b)). The highly enhanced fluorescent signal was observed when all key components including the NESBA primer set and NE together with target gRNA were applied (curve 1), whereas no fluorescent signal was observed without target gRNA (curve 2). In the absence of one or more key components, the reactions also produced fluorescent signal to some extent from the target, but the signal intensity was quite lower than that from the complete NESBA reaction because the reactions are not able to bring about the nicking and extension chain reaction (curves 3, 5, and 6). We additionally conducted agarose gel electrophoresis analysis for the products obtained from the NESBA reactions to further support the fluorescent results (Fig. 2(c) and (d)). For this analysis, we priorly conducted the RT-PCR reactions by employing a NESBA, NESBA/NASBA combined, or NASBA primer set to obtain the possible T7DNA intermediates products, which include T7DNA (NESBA) having the nicking site at both ends, T7DNA-1 (NESBA) and T7DNA-2 (NESBA) having the nicking site at only one end, and T7DNA (NASBA) without any nicking site. The produced four T7DNA intermediate products were then used as markers (M1-M4) for the analysis of the NESBA reaction products. As a result presented in Fig. 2(c) and (d), only lane 1 containing all NESBA components (target gRNA, a NESBA primer set, NASBA enzyme cocktail, and NE) showed the bands corresponding to the three key intermediate products of the NESBA reaction including T7DNA (NESBA) (M1), T7DNA-1 (NESBA) (M2), and T7DNA-2 (NESBA) (M3) while none of them was observed without target gRNA (lane 2). On the other hand, the conventional NASBA reaction using a NASBA primer set correctly produced the key intermediate T7DNA product without any nicking site at a slightly lower position only in the presence of target gRNA (lane 3). When NE was additionally added to the NASBA reaction, there was no change for the T7DNA product band (lane 5), indicating that a NESBA primer set is essential to produce the intermediate T7DNA products containing the nicking site. Very importantly, the T7DNA product bands obtained from the complete NESBA reaction (lane 1) were much more intense than those from the NASBA reaction (lane 3) and the incomplete NESBA reactions omitting either a NESBA primer set (lane 5) or NE (lane 6). All these results clearly confirm that the NESBA reaction is properly initiated by target gRNA and both a NESBA primer set containing nicking site at 5’ end and NE are quite essential to achieve the highly enhanced amplifying capability of the NESBA reaction, as envisioned in Fig. 1(c).
Fig. 2.
Feasibility test of the NESBA for E and N genes of SARS-CoV-2 gRNA. Time-dependent fluorescent signals produced from gene-specific MBs during the NESBA reaction for (a) E and (b) N genes. Agarose gel electrophoresis image of the products obtained after 45 min NESBA reaction for (c) E and (d) N genes (1: Target gRNA + E(I) or N(I)-NESBA primer set + NASBA enzyme cocktail + NE, 2: E(I) or N(I)-NESBA primer set + NASBA enzyme cocktail + NE, 3: Target gRNA + E or N-NASBA primer set + NASBA enzyme cocktail, 4: E or N-NASBA primer set + NASBA enzyme cocktail, 5: Target gRNA + E or N-NASBA primer set + NASBA enzyme cocktail + NE, and 6: Target gRNA + E(I) or N(I)-NESBA primer set + NASBA enzyme cocktail). The final concentration of SARS-CoV-2 gRNA is 5 × 105 copies/μL. M1-M4 are markers for band analysis (L: 25/100 bp mixed DNA ladder, M1: T7DNA (NESBA), M2: T7DNA-1 (NESBA), M3: T7DNA-2 (NESBA), M4: T7DNA (NASBA). We obtained the representative lines in (a) and (b) from the triplicate experiments and the reproducibility of the experiments were confirmed by the CV values for Tt in Table S5, which were all smaller than 5%. Tt is the time when the fluorescent signal reaches the threshold (100 a.u.) and CV is calculated as (Standard deviation of Tt)/(Mean of Tt) × 100. The final concentrations of markers used for M1-M4 are 100 nM.
3.3. Sensitivity of the NESBA for SARS-CoV-2 detection
To maximize the performance of the NESBA reaction, the NESBA primer set was optimized by monitoring real-time fluorescent signals produced from the assay reactions using differently designed NESBA primer sets. As presented in Figs. S2(a) and (b), E(I)- and N(I)-NESBA primer sets resulted in the most efficient amplification for E and N genes of SARS-CoV-2 gRNA and were employed for further experiments.
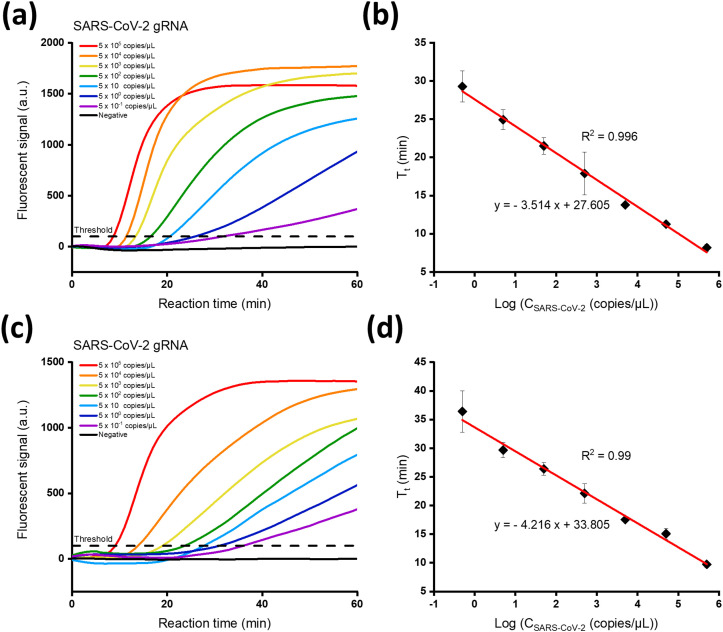
To determine the sensitivity of the proposed strategy, SARS-CoV-2 gRNA at varying concentrations in the range from 5 × 10−1 to 5 × 105 copies/μL was subjected to the reaction and the produced fluorescent signals were measured. As shown in Fig. 3 (a) and (c), the threshold time (Tt), defined as the time when the fluorescent signal reaches the threshold (100 a.u.), decreased in proportion to the increasing SARS-CoV-2 gRNA concentration (CSARS-CoV-2). Notably, the lowest target concentration (0.5 copies/μL) yielded the threshold fluorescent signal within 30 min for both E and N genes. When the Tt values were plotted against the logarithm (log) of CSARS-CoV-2, excellent linear relationship was obtained for E gene (Tt = - 3.514 log (CSARS-CoV-2) + 27.605, R2 = 0.996) and N gene (Tt = - 4.216 log (CSARS-CoV-2) + 33.805, R2 = 0.99), confirming that the NESBA reaction is quite capable of quantitatively detecting target gRNA in a real-time manner (Fig. 3(b) and (d)). The limit of detection (LOD) for E and N genes was determined to be all 0.5 copies/μL.
Fig. 3.
Sensitivity of the NESBA for E and N genes of SARS-CoV-2 gRNA. Time-dependent fluorescent signals produced from gene-specific MBs during the NESBA reaction for (a) E and (b) N genes of SARS-CoV-2 gRNA at varying concentrations. The linear relationship between Tt and logarithmic CSARS-CoV-2 for (b) E and (d) N genes of SARS-CoV-2 gRNA in the range from 5 × 10−1 to 5 × 105 copies/μL, where Tt is defined as the time when the fluorescent signal reaches the threshold (100 a.u.) and CSARS-CoV-2 is the concentration of SARS-CoV-2 gRNA.
We also conducted the conventional NASBA reactions for the same SARS-CoV-2 gRNA samples and repeated the same procedures to determine the LOD of the NASBA reaction. As a result (Fig. S1), the LOD for E and N genes was determined to be all 50 copies/μL, which is 100-fold higher than that of the NESBA reaction. These results firmly confirm that the proposed NESBA reaction could yield much higher sensitivity than that of the traditional NASBA reaction. Furthermore, the LOD of the NESBA reaction is lower than those of qRT-PCR and other isothermal nucleic acid amplification techniques (Table S2).
3.4. Specificity of the NESBA for SARS-CoV-2
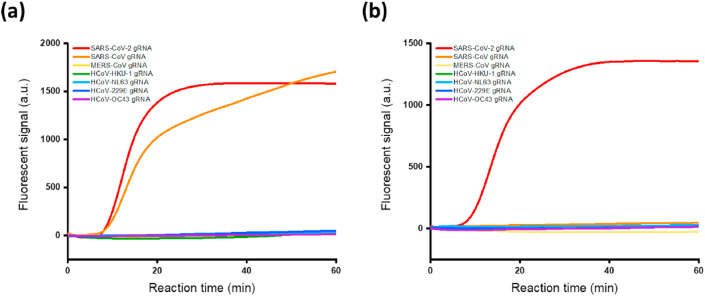
The specificity of the NESBA reaction was next assessed by examining the time-dependent fluorescent signals produced by other types of human coronavirus strains such as SARS-CoV, MERS-CoV, HCoV-229E, HCoV-NL63, HCoV-HKU-1, and HCoV-OC43.
As shown in Fig. 4 , only the sample containing target SARS-CoV-2 gRNA showed highly enhanced fluorescent signal for both E and N genes, while the nontarget samples containing other types of human coronavirus gRNA produced negligible fluorescent signals comparable to that of the blank. In the case of SARS-CoV, however, the fluorescent signal resulting from E gene was quite significant and not negligible. When we analyzed the nucleotide sequence alignment for the six human coronaviruses with respect to the target SARS-CoV-2 (Table S3), the E gene of SARS-CoV was about 94% identical with that of SARS-CoV-2 and this high sequence homology resulted in the nonspecific signal from E gene of SARS-CoV. This result indicates that a single region is not enough and two regions are normally needed for the testing to accurately identify and discriminate SARS-CoV-2 against other human coronaviruses. Except for the E gene of SARS-CoV having abnormally high sequence homology, the NESBA completely discriminated both the E and N genes of target SARS-CoV-2 against all the nontarget human coronaviruses, confirming the excellent specificity of the NESBA.
Fig. 4.
Specificity of the NESBA for E and N genes of SARS-CoV-2 gRNA. Time-dependent fluorescent signals produced from gene-specific MBs during the NESBA reaction for (a) E and (b) N genes of SARS-CoV-2 or other types of human coronaviruses such as SARS-CoV, MERS-CoV, HCoV-HKU-1, HCoV-229E, HCoV-NL63, and HCoV-OC43. The concentration of SARS-CoV-2 gRNA and other viral RNAs is 5 × 105 copies/μL for all seven human coronaviruses. We obtained the representative lines in (a) and (b) from the triplicate experiments and the reproducibility of the experiments were confirmed by the CV values for Tt in Table S6, which were all smaller than 5%. Tt is the time when the fluorescent signal reaches the threshold (100 a.u.) and CV is calculated as (Standard deviation of Tt)/(Mean of Tt) × 100.
3.5. Clinical applicability of the NESBA in COVID-19 diagnosis
To verify its clinical applicability, the NESBA reaction was applied to test a large cohort of clinical samples (nasopharyngeal swab and sputum; n = 98) and the resulting fluorescent signals were measured. The samples that produce fluorescent signals that exceed the threshold, which is defined as background + 5SD, where the background is the mean fluorescent signal of the blank at 30 min and SD is the standard deviation of the blank, were evaluated as positive whereas those that do not exceed the threshold were evaluated as negative (Diao et al., 2020; Fotis et al., 2021; Nessler et al., 2020).
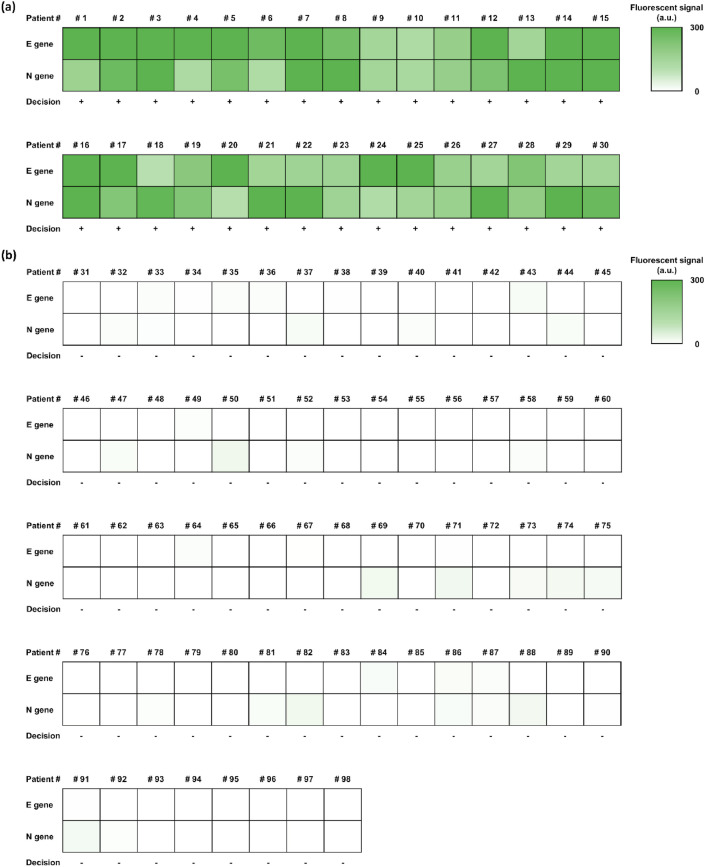
As shown in Fig. S3, the samples that were determined as positive by qRT-PCR lead to higher fluorescent signals than the threshold, while the samples that were determined as negative by qRT-PCR lead to lower fluorescent signals than the threshold and the overall results of the NESBA well agree with those of the qRT-PCR (Table S4). In order to visually compare the diagnosis results of the clinical samples, we display a heatmap based on the fluorescent signal intensities that correspond to the samples (Fig. 5 ). The NESBA was able to correctly identify 30 positive and 68 negative samples with 100% clinical sensitivity (95% CI 88.43%–100.00%) and 100% clinical specificity (95% CI 94.72%–100%) with short turnaround time. These results show that NESBA is a clinically robust technique with great potential for POC application that could serve as a compelling alternative to the current gold standard, qRT-PCR in COVID-19 diagnosis (Table 1 ).
Fig. 5.
The heat map of the clinical sample diagnosis with the NESBA. Fluorescent signals produced from E(I)-MB and N(I)-MB at 30 min in response to gRNA of representative, suspicious COVID-19 patients. The heat map results indicate the fluorescent signals from 0 to 300 a.u., determining the final diagnosis decision as (a) positive and (b) negative based on the threshold (background + 5SD).
Table 1.
Clinical testings for 30 positive and 68 negative cases by qRT-PCR and NESBA.a
| Diagnostic parameter |
qRT-PCRb |
NESBA |
|||
|---|---|---|---|---|---|
| E gene | N gene | E gene | N gene | ||
| Positive | True | 30 | 29 | 30 | 30 |
| False | - | - | - | - | |
| Negative | True | 68 | 68 | 68 | 68 |
| False | - | 1 | - | - | |
| Sensitivity (%) (95% CIc%) | 100 (88.4 to 100) | 96.67 (82.8 to 99.9) | 100 (88.4 to 100) | 100 (88.4 to 100) | |
| Specificity (%) (95% CI%) | 100 (94.7 to 100) | 100 (94.7 to 100) | 100 (94.7 to 100) | 100 (94.7 to 100) | |
The cases were confirmed as positive and negative based on the clinical sample analysis in Table S4.
The qRT-PCR was carried out using AllplexTM 2019-nCoV assay kit (Seegene, Seoul, Korea).
Confidence interval (MedCalc software, version 20.011).
3.6. Multiplex detection of the NESBA for SARS-CoV-2
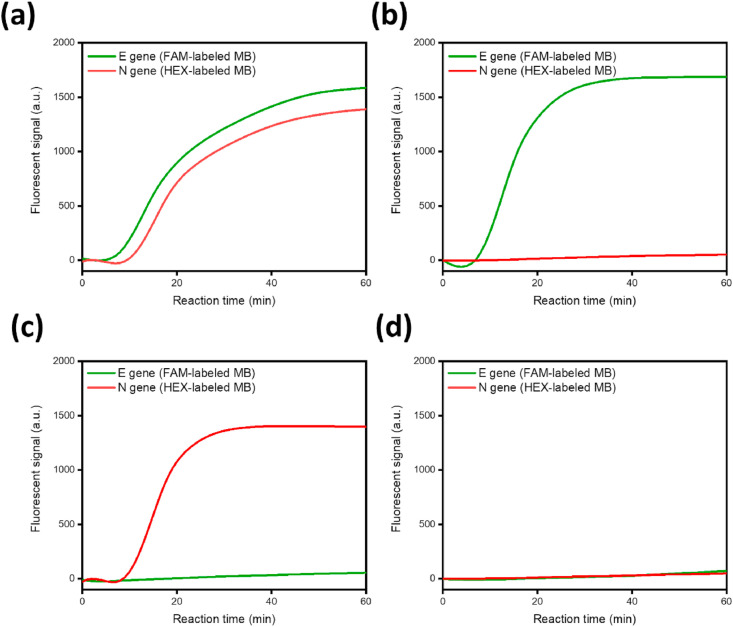
By employing spectrally distinct MBs, we finally validated the multiplex analytical capability of the NESBA for the simultaneous detection of SARS-CoV-2 E and N genes in one-pot. For this purpose, the FAM of N-gene specific MB (N(I)-MB) was replaced with HEX (N(I)-MB-HEX) which does not spectrally overlap with FAM, while the FAM of E(I)-MB was kept. As shown in Fig. 6 , both E and N genes of SARS-CoV-2 gRNA were very successfully identified by producing the intense fluorescent signals in the corresponding fluorescent channels when the NESBA primer sets for E and N genes were applied together or separately while the blank sample did not produce any significant signal in both channels. These results clearly demonstrate that the NESBA is capable of simultaneously identifying several different regions of target nucleic acids in a multiplexed manner.
Fig. 6.
Feasibility of the NESBA multiplex detection. Time-dependent fluorescent signals produced from E(I)-MB (green curve) and N(I)-MB-HEX (red curve), which correspond to the respective E and N genes of SARS-CoV-2 gRNA, were simultaneously measured in FAM and HEX channels, respectively ((a): Both E and N genes of SARS-CoV-2 gRNA, (b): E gene of SARS-CoV-2 gRNA, (c): N gene of SARS-CoV-2 gRNA, (d): Blank). The concentration of SARS-CoV-2 gRNA is 5 × 105 copies/μL. We obtained the representative lines in Fig. 6 from the triplicate experiments and the reproducibility of the experiments were confirmed by the CV values for Tt in Table S7, which were all smaller than 5%. Tt is the time when the fluorescent signal reaches the threshold (100 a.u.) and CV is calculated as (Standard deviation of Tt)/(Mean of Tt) × 100. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusion
In this study, we developed the NESBA system for rapid, accurate COVID-19 diagnosis with simple yet ingenious design of NESBA primer set. The NESBA primer set, which possesses the NRS at the 5′-end of conventional NASBA primers, exerts the nicking enzyme-aided exponential amplification and significantly enhanced the detection sensitivity. With NESBA, we assayed SARS-CoV-2 gRNA within 30 min in a sample-to-detect manner and achieved high detection sensitivity (0.5 copies/μL (= 10 copies/reaction) for E and N genes and specificity.
Through tests of a large cohort of clinical samples (nasopharyngeal swab and sputum; n = 98) and comparison with qRT-PCR results, its clinical validity was also demonstrated with 100% clinical sensitivity (95% CI 88.4%–100.00%) and 100% clinical specificity (95% CI 94.7%–100%). Moreover, we successfully applied the NESBA to multiplex detection of E and N genes of SARS-CoV-2 gRNA in one-pot by employing spectrally distinct MBs. Overall, the developed method is highly convenient and more advantageous compared to previous detection systems for COVID-19 in terms of probe design, assay procedure, and detection performances including clinical accuracy and multiplex capability. Therefore, the NESBA would play a central role in clinical and on-site COVID-19 diagnostics as a compelling alternative to qRT-PCR. Furthermore, it would serve as a universal diagnostic platform to upcoming or existing infectious diseases due to intrinsically flexible probe design.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by BioNano Health-Guard Research Center funded by the Ministry of Science and ICT (MSIT) of Korea as Global Frontier Project (Grant number H-GUARD_2013M3A6B2078964) and Kyung Nam Pharm Co.,Ltd.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113689.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Cui F., Zhou H.S. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wen K., Chen J., Liu Y., Yuan Z., Han C., Chen J., Pan Y., Chen L., Dan Y. 2020. Medrxiv. [Google Scholar]
- Fotis C., Meimetis N., Tsolakos N., Politou M., Akinosoglou K., Pliaka V., Minia A., Terpos E., Trougakos I.P., Mentis A. Sci. Rep. 2021;11:1–11. doi: 10.1038/s41598-021-86035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibzadeh P., Mofatteh M., Silawi M., Ghavami S., Faghihi M.A. Crit. Rev. Clin. Lab Sci. 2021;6:385–398. doi: 10.1080/10408363.2021.1884640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Li Z., Liu C., Cheng Y. Angew. Chem. Int. Ed. 2010;49:5498–5501. doi: 10.1002/anie.201001375. [DOI] [PubMed] [Google Scholar]
- Ju Y., Kim H.Y., Ahn J.K., Park H.G. Nanoscale. 2021;13:10785–10791. doi: 10.1039/d1nr00564b. [DOI] [PubMed] [Google Scholar]
- Jung C., Chung J.W., Kim U.O., Kim M.H., Park H.G. Anal. Chem. 2010;82:5937–5943. doi: 10.1021/ac100606m. [DOI] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.Y.-P., Lin R.T., Renia L., Ng L.F. Front. Immunol. 2020;11:879. doi: 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Jang H., Kim H.Y., Park H.G. Biosens. Bioelectron. 2020;147:111762. doi: 10.1016/j.bios.2019.111762. [DOI] [PubMed] [Google Scholar]
- Liu G., Rusling J.F. ACS sen. 2021;6:593–612. doi: 10.1021/acssensors.0c02621. [DOI] [PubMed] [Google Scholar]
- Lizardi P.M., Huang X., Zhu Z., Bray-Ward P., Thomas D.C., Ward D.C. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J. Emerg. Infect. Dis. 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C., Lam E.T., Chan R.C., Tsang D.N. J. Clin. Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nessler S., Franz J., van der Meer F., Kolotourou K., Venkataramani V., Hasan C., Pollok-Kopp B.B., Zautner A.E., Stadelmann C., Weig M. 2020. medRxiv. [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.12.e63. e63-e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T., Maeng J.-S. J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T. Nat. Biomed. Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Peeling R.W., Olliaro P.L., Boeras D.I., Fongwen N. Lancet Infect. Dis. 2021;9:e290–e295. doi: 10.1016/S1473-3099(21)00048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. Nat. Biotechnol. 2020;38:515–518. doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- Song J., Kim H.Y., Kim S., Jung Y., Park H.G. Biosens. Bioelectron. 2021;178:113051. doi: 10.1016/j.bios.2021.113051. [DOI] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness J., Van Ness L.K., Galas D.J. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4504–4509. doi: 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Xu Y., Kong H. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2021. Coronavirus Disease (COVID-19) Weekly Epidemiological Update and Operational Updates. [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.