Abstract
A cell line was generated that expresses the poliovirus 2A protease in an inducible manner. Tightly controlled expression was achieved by utilizing the muristerone A-regulated expression system. Upon induction, cleavage of the eukaryotic translation initiation factor 4GI (eIF4GI) and eIF4GII is observed, with the latter being cleaved in a somewhat slower kinetics. eIF4G cleavage was accompanied by a severe inhibition of protein synthesis activity. Upon induction of the poliovirus 2A protease, the cells displayed fragmented nuclei, chromatin condensation, oligonucleosome-size DNA ladder, and positive TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) staining; hence, their death can be characterized as apoptosis. These results indicate that the expression of the 2A protease in mammalian cells is sufficient to induce apoptosis. We suggest that the poliovirus 2A protease induces apoptosis either by arresting cap-dependent translation of some cellular mRNAs that encode proteins required for cell viability, by preferential cap-independent translation of cellular mRNAs encoding apoptosis inducing proteins, or by cleaving other, yet unidentified cellular target proteins.
Infection with poliovirus results in a dramatic shutoff of host protein synthesis that is followed by a selective and efficient translation of the viral mRNA (9). Cellular mRNAs contain a 5′-terminal cap structure which plays a pivotal role in the process of initiation of their translation (40). In contrast, poliovirus mRNA is uncapped (15, 32), and its translation is initiated by an alternative mechanism that involves direct landing of the ribosomes at an internal site termed internal ribosome entry site (IRES) (19, 36). An early event occurring during poliovirus infection is cleavage of the eukaryotic translation initiation factor 4G (eIF4G) by the viral 2A protease (10). Since eIF4G acts as an important mediator that bridges eIF3 (which is complexed to the 40S ribosomal subunit) and the cap binding protein eIF4E (18, 23), the translation of cap-dependent mRNAs is selectively inhibited (10, 34). Picornavirus RNAs utilize the C-terminal fragment of eIF4G for translation (35, 38). A recent study suggested that the translational inhibitory effect of poliovirus infection may under certain conditions trigger apoptotic cell death (44). Since infection with the entire virus is likely to complicate any interpretation concerning the contribution of individual viral proteins to cellular effects, it was important to express individual viral genes in cells. In this respect, the viral 2A protease is of particular interest. Since expression of the 2A protease is likely to be toxic to cells, all previous attempts to express it were by means of transient transfection (2, 33, 39). However, transient transfections are subjected to variations in the proportion of successfully transfected cells, and therefore the effects are measured in a mixed cell population. We have therefore efficiently expressed the poliovirus 2A protease in an inducible manner in stably transformed human 293 cells, using the ecdysone-inducible system (31). Induction of the 2A protease results in cleavage of eIF4Gs, strong inhibition of protein synthesis activity, and apoptotic cell death.
MATERIALS AND METHODS
Cells and cell culture conditions.
The human embryonic kidney epithelial 293 cells were stably transfected with the pVgRXR vector (Invitrogen), which encodes a Drosophila ecdysone receptor modified to contain the VP16 transactivation domain and the mammalian homologue of its heterodimeric partner. Fifty of the resulting clones were then transiently transfected with plasmid pIND-LacZ and screened for those that express minimal LacZ activity under basal conditions. Twenty such clones were selected, transfected with plasmid pIND-LacZ, and treated with muristerone A (MurA). One of these clones in which maximal induction of LacZ activity was selected for further studies and termed 293-EcR. The 293-EcR cells were stably transfected with pIND plasmids containing different forms of the poliovirus 2A protease (see below). Some of the resulting clones were treated with MurA, and a clone in which 2A protease activity is efficiently induced was selected as described in Results; this clone was designated 293-EcR-2AI2A.
Structure of the pIND vector encoding the poliovirus type 1 (Mahoney) 2A protease.
The following constructs encoding the poliovirus 2A protease were prepared using the pIND plasmid as a template. The first construct contains two copies of DNA encoding the poliovirus 2A protease spaced by the encephalomyocarditis virus IRES (pIND-2AI2A). The second construct produces a monocistronic mRNA encoding a VP1-2A fusion protein from which the 2A protease is released by autocleavage (pIND-2A). Two other constructs contain DNA encoding wild-type 2A protease or an inactive variant of the protease designated D2A in which cysteine 109 was replaced by alanine by site-directed mutagenesis. In these two constructs, the 2A sequence is preceded by an in-frame hemagglutinin (HA)-tagged segment.
Antibody.
Polyclonal antibody against the poliovirus 2A protease and against the N- and C-terminal fragments of eIF4G, with respect to the 2A protease cleavage site (24, 45) (amino acids 173 to 457 and 934 to 1139 respectively), were prepared in New Zealand White rabbits. These proteins were expressed in Escherichia coli, using the pRSET expression vector (Invitrogen). The antibodies used against the N-terminal fragment of eIF4GII are as described previously (11). The peptide EQRREMLETVKQLTGGVDVERNSTEAE (Sheldon Biotechnology Center, McGill University, Montreal, Quebec, Canada) from the eIF4GII C-terminal region (amino acids 1168 to 1194) conjugated with keyhole limpet hemacyanin (Inject activated immunogen conjugation kit; Pierce, Rockford, Ill.) was used to raise antipeptide antibodies in rabbits.
Western blot analysis.
293-EcR and 293-EcR-2AI2A cells were harvested at various times following the addition of MurA to the growth medium. At the indicated times, the cells were trypsinized or scraped, washed with cold phosphate-buffered saline, and lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.2], 0.5% NP-40, 1% Triton X-100, 1% sodium deoxycholate). Protein concentration was determined in the cellular extracts by the Bradford method. Samples containing equal amounts of protein were denatured in Laemmli buffer, fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and blotted onto nitrocellulose. The resulting blots were probed with the indicated antibodies.
Assays for the determination of cell viability and apoptosis.
Cells were treated as described in the relevant experiments. Cell viability was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, using an assay kit from Boehringer Mannheim as instructed by the manufacturer. Percentage viability was determined by comparing the number of viable cells in treated cultures to the number of cells in a parallel control culture (treated with ethanol for equivalent time). The appearance of apoptotic cells was determined at the indicated times by inspecting May-Grünwald-Giemsa- or 4′,6-diamidino-2-phenylindole (DAPI)-stained cytocentrifuge preparations. Apoptotic cells were smaller and contained condensed chromatin and fragmented nuclei. DNA fragmentation was determined by isolating DNA from treated and untreated cells using the G NOME DNA isolation kit (Bio 101, La Jolla, Calif.). Equal portions of DNA were fractionated by electrophoresis in a 1.5% agarose gel, stained with ethidium bromide, and visualized under UV light. For the TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assay, 293-EcR-2AI2A cells were grown on chamber slides (Nunc) covered with poly-d-Lysine (Sigma). Cells were then fixed with 4% paraformaldehyde, the chambers were removed, and the cells were permeabilized and stained with an in situ cell death detection kit (Boehringer Mannheim TUNEL/FITC kit) according to the manufacturer's instructions.
Monitoring protein synthesis.
Cells were treated with MurA (2.5 μM). Following the indicated times of treatment, the cells were labeled for 1 h with [35S]methionine (200 μCi/ml, 1,000 Ci/mmol; Amersham). The cells were then lysed in lysis buffer (100 mM Tris-HCl [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.1% Triton X-100, 0.1% SDS) by three cycles of freezing and thawing. Rate of protein synthesis was measured by determination of radioactivity in the trichloroacetic acid (TCA)-insoluble material.
RESULTS
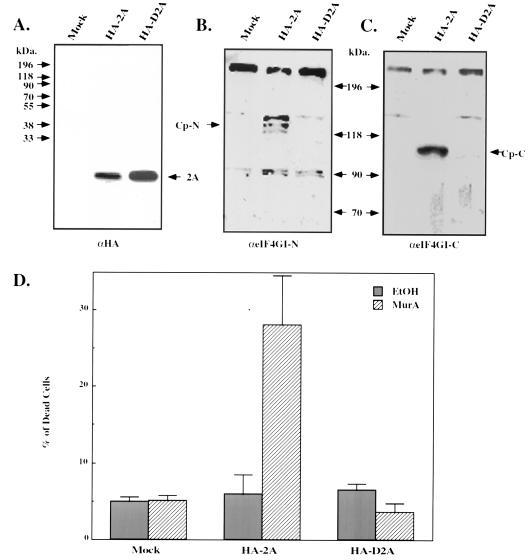
The ecdysone-inducible system (31), was used to conditionally express the poliovirus 2A protease in mammalian cells. DNA encoding wild-type 2A protease and an inactive 2A protease mutant (14), both tagged by an HA epitope, were cloned into the mammalian expression vector pIND (Invitrogen), which contains five modified ecdysone response elements located upstream to a minimal heat shock promoter. Human 293-EcR cells, engineered to express a modified Drosophila ecdysone receptor (see Materials and Methods), were transiently transfected with each of the two 2A protease constructs together with green fluorescent protein (GFP)-expressing vector. Cellular extracts were prepared before and after induction with muristerone, and the level of the expressed 2A protease as well as the status of eIF4GI was examined by Western blotting (using anti-HA and anti-eIF4GI antibodies, respectively). While induction with MurA resulted in efficient expression of the two 2A variants (Fig. 1A), only expression of the wild-type enzyme resulted in the cleavage of eIF4GI (Fig. 1B and C) and in induction of cell death. The percentage of dead cells was determined by microscopic inspection of transfected cells expressing GFP from the cotransfected vector (Fig. 1D). However, transient transfections are subjected to variations, as not all cells are successfully transfected, and therefore the effects measured are obtained from a mixed cell population. We therefore wished to produce a cell line which stably expresses the poliovirus 2A protease in a tightly regulated manner. For this purpose, DNA encoding an untagged wild-type 2A protease was cloned into the pIND vector and transfected into the 293-EcR cells that express a modified Drosophila ecdysone receptor. Two constructs were used. In the first, one copy of the 2A protease was cloned; in the second, two copies of the DNA were cloned to produce a bicistronic mRNA that has the potential to encode twice of the amount of 2A protease. In the first construct (pIND-2A), the 2A protease is encoded from a monocistronic mRNA that is translated in a cap-dependent manner. In the second construct (pIND2AI2A), the first copy of 2A is translated in a cap-dependent manner, while the second copy, which is located downstream to the encephalomyocarditis virus IRES, is translated in a cap-independent manner. We inferred that by using a bicistronic mRNA we will assure continues expression of the 2A protease even under conditions that prevent cap-dependent translation. The resulting constructs were transfected into the 293-EcR cells. Since expression of the viral 2A protease may be harmful to the cells, as was indeed noted during the transient transfections (Fig. 1D), we chose viability as our screening parameter. To this end, large number of individual clones were cultured in duplicates in wells of a 96-well microtiter plate, and MurA was added to one well of each pair. Several clones were identified in which cell death was noted shortly after stimulation. We next wished to examine the status of eIF4GI, whose cleavage during poliovirus infection is considered to be manifested by the action of the 2A protease (10). For this purpose, cellular extracts were prepared at various times following the addition of MurA, and the integrity of eIF4GI was tested by Western blotting analysis. Significant cleavage of eIF4GI was noted in cell lines expressing the bicistronic (293-EcR-2AI2A) or the monocistronic (293-EcR-2A) mRNA (Fig. 2A). The cleavage products noted following the activation of the 2A protease were identical to those observed in cells infected with poliovirus at a multiplicity of infection (MOI) of 100 PFU/cell (Fig. 2B). Although eIF4GI was efficiently cleaved in our 293-EcR-2AI2A cells following their stimulation with MurA, this cleavage was significantly slower than that noted in poliovirus-infected cells (Fig. 2B). This observation is compatible with the significantly lower levels of the 2A protease in our cells than in poliovirus-infected cells (not shown). Since the cleavage of eIF4GI was significantly more efficient in the clones expressing the 2A protease from the bicistronic mRNA, one of these clones was selected for further studies.
FIG. 1.
Transient expression of the poliovirus 2A protease results in the cleavage of eIF4GI and in induction of cell death. 293-EcR cells were transiently transfected with pEGFP-N1 (Clontech) together with a pIND plasmid encoding an HA-tagged 2A protease (HA-2A), with an inactive variant of this protease in which cysteine 109 was replaced by alanine (HA-D2A), or with a pIND plasmid encoding LacZ (Mock). MurA was added to the growth medium 6 h posttransfection, and cellular extracts were prepared after additional 18 h. Cellular extracts were fractionated by electrophoresis and subjected to Western blot analysis with an anti-HA antibody (A) and anti-eIF4GI antibodies that recognize specifically the N-terminal fragment (B) or the C-terminal fragment (C). The N (Cp-N)- and C (Cp-C)-terminal cleavage products of eIF4GI are indicated. The percentage of dead cells out of all GFP-expressing cells was determined by microscopic inspection (D). EtOH, ethanol.
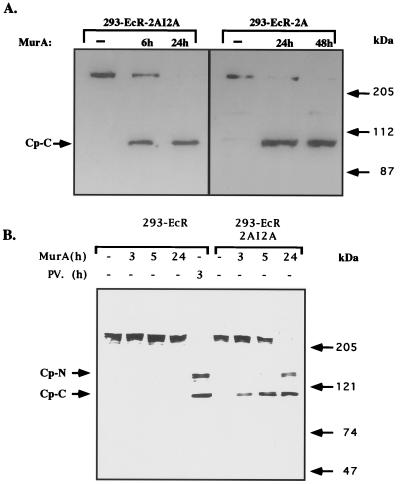
FIG. 2.
Expression of poliovirus 2A protease in 293-ECR-2AI2A cells promote cleavage of eIF4GI to fragments identical to those observed in poliovirus-infected cells. (A) 293-ECR-2A and 293-ECR-2AI2A cells were treated with MurA (2.5 μM). At the indicated times, the cells were harvested and portions of cellular extracts containing 60 μg of protein were fractionated by electrophoresis on an SDS–8% polyacrylamide gel. The fractionated material was transferred to a nitrocellulose membrane which was then probed with an antibody raised against the C-terminal fragment of eIF4GI. (B) 293-EcR and 293-ECR-2AI2A were treated with MurA (2 μM) or infected with poliovirus (PV) at an MOI of 100 PFU/cell as previously described (13). At the indicated times, cells were harvested and cellular extracts were fractionated by electrophoresis on an SDS–6% polyacrylamide gel and probed with an antibody raised against the N- and C-terminal fragments of eIF4GI. The N (Cp-N)- and C (Cp-C)-terminal cleavage products of eIF4GI are indicated.
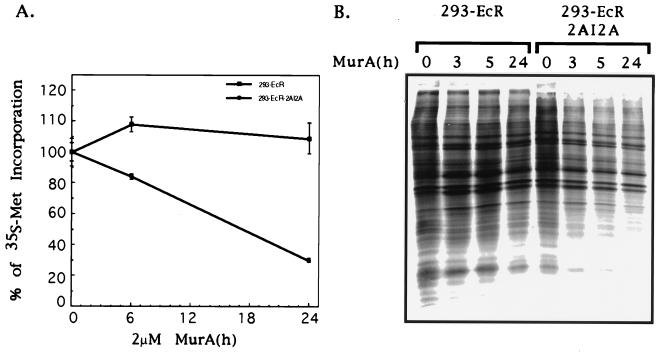
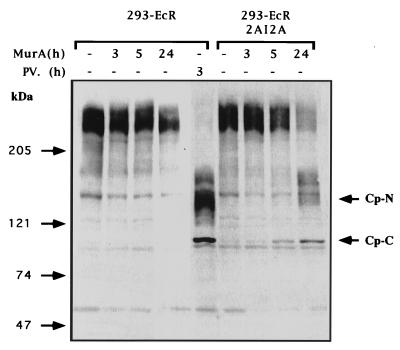
Since eIF4G is a key player in the cellular protein synthesis machinery, we monitored the effect of induced 2A protease on protein synthesis activity by measuring the incorporation of [35S]methionine into proteins. 293-EcR-2AI2A and the control 293-EcR cells were stimulated with MurA and pulse-labeled with [35S]methionine at various time points. Incorporation of [35S]methionine was determined either by TCA precipitation (Fig. 3A) or by fractionation of equal portions of cellular extracts in SDS-polyacrylamide gels (Fig. 3B). As expected, protein synthesis activity was severely inhibited (Fig. 3). However, this inhibition was not complete, which may suggest that translation is partially maintained by another cellular translation factor. Recent studies have reported the existence of a functional homologue of eIF4GI termed eIF4GII (11). In poliovirus- or rhinovirus-infected cells, eIF4GII was less sensitive to proteolysis than eIF4GI (12, 43). We demonstrate here that eIF4GII is also cleaved in the 293-EcR-2AI2A cells which express the 2A protease as the sole poliovirus protein with a slower kinetics of cleavage compared to eIF4GI (Fig. 4). This increased resistance of eIF4GII may explain the residual protein synthesis activity noted in our cells following induction of the 2A protease.
FIG. 3.
Expression of 2A protease inhibits host cell protein synthesis activity. (A) 293-EcR and 293-ECR-2AI2A cells were plated in a 24-well plate. Triplicate wells were treated with MurA (2 μM) for the indicated times. One hour before harvesting, [35S]methionine was added to a final concentration of 200 μCi/ml. At the indicated times cells were collected, washed with phosphate-buffered saline, and lysed by three cycles of freeze-thaw in lysis buffer containing protease inhibitors (Sigma). Aliquots containing equal amount of cellular proteins were subjected to TCA precipitation, and incorporated radioactivity was determined. The ratio of radioactivity (in counts per minute) to cellular protein (in micrograms) was calculated for each time point. (B) 293-EcR and 293-ECR-2AI2A cells were treated with MurA for the indicated times and pulse-labeled for 1 h with [35S]methionine; then equal portions of cellular extracts fractionated by SDS-PAGE as previously described (13).
FIG. 4.
Poliovirus 2A protease in 293-ECR-2AI2A cells promotes cleavage of eIF4GII to fragments identical to those observed in poliovirus-infected cells. 293-EcR and 293-ECR-2AI2A were treated with MurA (2.5 μM) or infected with poliovirus (PV) at MOI of 100 PFU/cell as previously described (13). At indicated times, cells were harvested and portions of cellular extracts containing 100 μg of protein were fractionated by SDS-PAGE on a 6% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with antibodies raised against the N- and C-terminal fragments of eIF4GII. The N (Cp-N)- and C (Cp-C)-terminal cleavage products of eIF4GII are indicated.
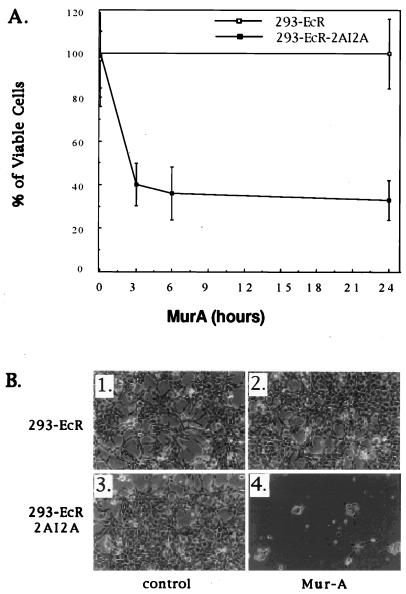
As demonstrated in Fig. 1, death is provoked in cells transiently transfected with 2A protease-expressing constructs. Since death was the parameter used for the identification of stable transfectants that express the 2A protease in an inducible manner, we wished to characterize the mode of death. To this end, we treated 293-EcR-2AI2A and control cells with MurA and determined their viability by the MTT assay (Fig. 5A) and microscopic examination (Fig. 5B). As shown by the MTT assay, cell death was rapidly induced. To exclude the possibility that the observed cell death was not merely a result of growth arrest, cells were left for a longer period (72 h) in the presence of MurA. At this time point, the MTT assay is inapplicable because of the small number of remaining cells. However, microscopic inspection demonstrates the irreversibility of the MurA effect.
FIG. 5.
Poliovirus 2A protease induces cell death. (A) 293-EcR and 293-ECR-2AI2A cells were cultured for 24 h in a 96-well microtiter plate. At the indicated times, 16 wells of each cell line were treated with 2 μM MurA. MTT reagent (Boehringer Mannheim) was added to each well 2.5 h prior to viability determination, which was performed as recommended by the manufacturer. (B) 293-EcR cells (1, 2) and 293-ECR-2AI2A cells (3, 4) were cultured for 72 h in the presence of ethanol (1 and 3) or 2 μM MurA (2 and 4) and were observed by phase-contrast microscopy.
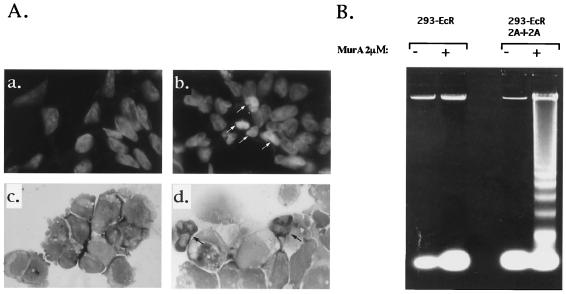
Cells that undergo apoptosis are characterized by a distinct morphology and by a typical pattern of DNA fragmentation (DNA ladder) that results from cleavage of chromosomal DNA in internucleosomal segments (8). To determine whether the 2A protease-provoked death can be characterized as apoptosis, these features were investigated in 293-EcR-2AI2A cells and in the control 293-EcR cells following stimulation with MurA. At 6 h poststimulation, cells were spread on a microscope slide using a cytocentrifuge and then stained with DAPI (Fig. 6A, a and b) or Giemsa (Fig. 6A, c and d), which demonstrated typical cell shrinkage, nuclear fragmentation, and chromatin condensation. Genomic DNA isolated from cells 24 h poststimulation was fractionated by agarose gel electrophoresis and clearly demonstrated a characteristic DNA ladder (Fig. 6B). Apoptotic death of 293-EcR-2AI2A cells following 2A protease induction was also tested by the TUNEL assay (4). For this purpose, 293-ECR-2AI2A cells were treated with MurA and apoptotic cells were revealed by staining with a TUNEL reagent. Figure 7 demonstrates clear and significant staining of 293-EcR-2AI2A cells, with almost no staining observed in the control 293-EcR cells. Thus, induction of the 2A protease as the only poliovirus protein in mammalian cells provokes apoptotic cell death.
FIG. 6.
Apoptotic markers are provoked by the expressed poliovirus 2A protease. (A) 293-ECR-2AI2A cells were treated with ethanol (a and c) or MurA (2 μM) (b and d). Six hours poststimulation, the cells were cytocentrifuged and the resulting preparations were fixed and stained either with DAPI (a and b) or with May-Grünwald-Giemsa stain (c and d). Arrows show apoptotic cells. (B) 293-ECR and 293-ECR-2AI2A cells were treated with 2 μM MurA (+) or with ethanol (−); 24 h poststimulation, the cells were harvested, and genomic DNA was extracted and resolved (65 μg/lane) on a 1.5% agarose gel.
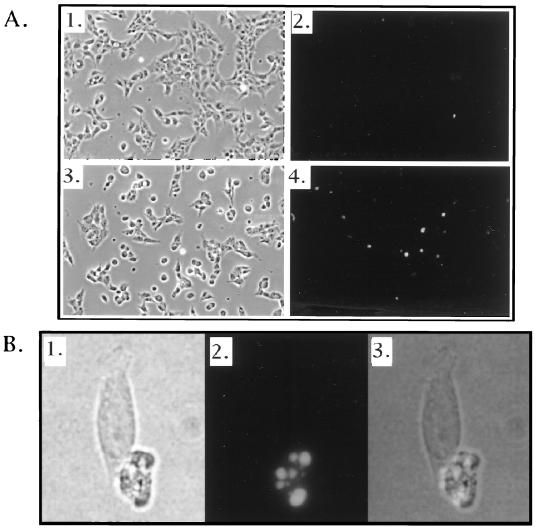
FIG. 7.
TUNEL staining of 293-ECR-2AI2A following the induction of 2A protease expression. (A) 293-ECR-2AI2A cells were treated for 18 h with ethanol (1 and 2) or 2 μM MurA (3 and 4). Cells were fixed and stained with a Boehringer Mannheim TUNEL/FITC kit according to the manufacturer's instructions. Cells were photographed at a magnification of ×20. Fields 1 and 2 and fields 3 and 4 represent the same fields visualized by visible light (1 and 3) or fluorescent light (2 and 4). (B) Sixtyfold magnification of field 4 from panel A. 1, visible light; 2, fluorescent light; 3, superimposition of fields 1 and 2.
It should be noted, however, that the proportion of individual cells demonstrating apoptotic markers revealed by DAPI and Giemsa staining or by the TUNEL assay is lower than what could be expected based on the efficiency of killing of these cells by the activated 2A protease. This discrepancy is probably a result of the observation that the late stages of apoptosis in which the characteristic features are noted are rather short. Therefore, even in a population in which the majority of cells are undergoing apoptosis within 24 h, at any given time point only a minor proportion of the cells exhibit the characteristic apoptotic markers. Therefore, the true apoptotic index should be obtained by comparing the number of cells displaying apoptotic markers in the treated culture to the control culture that display an insignificantly small proportion of apoptotic cells (Fig. 6A and 7).
DISCUSSION
In this paper we describe the construction of an experimental system that enables the evaluation of the effect of poliovirus 2A protease on cellular functions. Determination of the net effect of an individual viral gene during viral infections is complicated by the expression of other viral genes products. A way to overcome this problem is to express individual viral genes that are cloned in expression vectors. However, since the poliovirus 2A protease is likely to be toxic to mammalian cells, all previous attempts to express it were by means of transient transfection, a method that suffers from problems of lack of uniformity and reproducibility. We have therefore utilized the recently developed ecdysone expression system and constructed a stable cell line that conditionally expresses the poliovirus 2A protease. We demonstrate here that expression of 2A protease as the only poliovirus component results in complete cleavage of eIF4GI, somewhat slower cleavage of eIF4GII, severe inhibition of protein synthesis, and apoptotic cell death.
The molecular basis for the host protein synthesis shutoff during poliovirus infection has been investigated intensively. Although the correlation between eIF4GI cleavage and the shutoff of host protein synthesis was questioned (37), it is clear that eIF4GI is not absolutely required for the translation of capped cellular mRNAs, because of the existence of a functional eIF4GI homologue, eIF4GII (11), which is more resistant to poliovirus infection than is eIF4GI (13). As we demonstrate here also in our 2A protease-producing cells, eIF4GII is more resistant to cleavage by the 2A protease. It is therefore likely that residual eIF4GII supports some protein synthesis activity even though eIF4GI is completely cleaved. Alternatively, it is possible that some cellular capped mRNAs are capable of cap-independent translation. Some cellular mRNAs were demonstrated to contain potential IRES element as determined in a bicistronic mRNA (1, 5, 17, 26, 28, 30, 41, 42, 46). It will be of interest to determine whether such cellular mRNAs are selectively translated after induction of the 2A protease in our 293-EcR-2AI2A cells.
As demonstrated here, the induction of the viral 2A protease provoked apoptotic cell death. Although only partial inhibition of host protein synthesis was noted, this inhibition may account for the observed death due, for instance, to the disappearance of some survival proteins. Alternatively, the apoptotic death may be provoked by the appearance of death-inducing proteins that are encoded by cellular mRNAs which are selectively translated under conditions that prefer cap-independent translation. If this is the case, then mRNAs that are associated with heavy polysomes may be enriched in cellular mRNAs that are capable of cap-independent initiation. The cells we describe here, in which expression of the poliovirus 2A protease can be efficiently manipulated, can be used for the selective cloning of such mRNAs as was recently done with a mutant poliovirus (21).
Recent studies have demonstrated cleavage of eIF4GI by caspase 3 in cells that undergo apoptotic cell death (7, 27, 29). It should be noted that the cleavage products observed in poliovirus-infected cells are different from those observed after activation of caspases. It is well documented that there are caspase-independent apoptotic processes (6). In this respect, it should be mentioned that our preliminary studies suggest that the 2A protease-induced apoptotic process is caspase independent. Although the cleavage products generated by the activity of caspase 3 are different from those generated by the 2A protease, our present study provides evidence that the targeted cleavage of eIF4GI and -II by the poliovirus 2A protease results in severe inhibition of translation and in apoptotic cell death. Such cleavage of eIF4GI and -II can lead to apoptotic death either by inhibiting the cap-dependent translation of the cellular mRNAs encoding proteins that are required for maintaining cellular viability or by allowing preferential cap-independent translation of cellular mRNAs that encode proteins that actively induce the apoptotic state. In support of this possibility are recent findings for IRES elements in mRNAs encoding proteins that are involved in regulating apoptosis (16, 21, 25, 30). Although it is highly likely that the primary target of activity of the 2A protease is the translational machinery, we cannot exclude the possibility that the 2A protease induces apoptosis by cleaving other cellular proteins which may be crucial for other cellular functions. Indeed, recent studies have demonstrated that the poly(A)-binding protein (20, 22) and the dystrophin protein (3) are cleaved by the 2A protease. As for the case of eIF4Gs, cleavage of the poly(A)-binding protein may also cause apoptosis via a translational mechanism. In contrast, cleavage of the dystrophin protein may induce apoptosis due to cytoskeleton disruption. Additional studies are required to differentiate between these possibilities.
ACKNOWLEDGMENTS
This study was supported by QBI Enterprises, Nes Ziona, Israel, to C.K. and by grant from the Medical Research Council of Canada to N.S.
REFERENCES
- 1.Akiri G, Nahari D, Finkelstein Y, Le S Y, Elroy-Stein O, Levi B Z. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe R, Feduchi E, Novoa I, Carrasco L. Expression of poliovirus 2Apro in mammalian cells: effects on translation. FEBS Lett. 1995;377:1–5. doi: 10.1016/0014-5793(95)01269-9. [DOI] [PubMed] [Google Scholar]
- 3.Badorff C, Lee G H, Lamphear B J, Martone M E, Campbell K P, Rhoads R E, Knowlton K U. Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nat Med. 1999;5:320–326. doi: 10.1038/6543. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Sasson S A, Sherman Y, Gavrieli Y. Identification of dying cells—in situ staining. Methods Cell Biol. 1995;46:29–39. [PubMed] [Google Scholar]
- 5.Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 6.Borner C, Monney L. Apoptosis without caspases: an inefficient molecular guillotine? Cell Death Differ. 1999;6:497–507. doi: 10.1038/sj.cdd.4400525. [DOI] [PubMed] [Google Scholar]
- 7.Clemens M J, Bushell M, Morley S J. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- 8.Eastman A. Assays for DNA fragmentation, endonucleases, and intracellular pH and Ca2+ associated with apoptosis. Methods Cell Biol. 1995;46:41–55. doi: 10.1016/s0091-679x(08)61923-8. [DOI] [PubMed] [Google Scholar]
- 9.Ehrenfeld E. Initiation of translation by picornavirus mRNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–575. [Google Scholar]
- 10.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 11.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradi A, Svitkin Y, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellen C U, Fäcke M, Kräusslich H G, Lee C K, Wimmer E. Characterization of poliovirus 2A proteinase by mutational analysis: residues required for autocatalytic activity are essential for induction of cleavage of eukaryotic initiation factor 4F polypeptide p220. J Virol. 1991;65:4226–4231. doi: 10.1128/jvi.65.8.4226-4231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewlett M, Rose J, Baltimore D. 5′-terminal structure of poliovirus polyribosomal RNA is pUp. Proc Natl Acad Sci USA. 1976;73:327–330. doi: 10.1073/pnas.73.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holcik M, Lefebvret C, Yeh C, Chow T, Korneluk R G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 17.Huez I, Creancier L, Audigier S, Gensac M C, Prats A C, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joachims M, Van Breugel P C, Lloyd R E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerekatte V, Keiper B D, Badorff C, Cai A, Knowlton K U, Rhoads R E. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 24.Lamphear B J, Yan R, Yang F, Waters D, Liebig H D, Klump H, Kuechler E, Skern T, Rhoads R E. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human coxsackievirus and rhinovirus. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 25.Lazarus P, Parkin N, Sonenberg N. Developmental regulation of translation by the 5′ noncoding region of murine c-myc mRNA in Xenopus laevis. Oncogene. 1988;3:517–521. [PubMed] [Google Scholar]
- 26.Le S Y, Maizel J V., Jr A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marissen W E, Lloyd R E. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol. 1998;18:7565–7674. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller D L, Dibbens J A, Damert A, Risau W, Vadas M A, Goodall G J. The vascular endothelial growth factor mRNA contains an internal ribosome entry site. FEBS Lett. 1998;434:417–420. doi: 10.1016/s0014-5793(98)01025-4. [DOI] [PubMed] [Google Scholar]
- 29.Morley S J, McKendrick L, Bushell M. Cleavage of translation initiation factor 4G (eIF4G) during anti-Fas IgM-induced apoptosis does not require signalling through the p38 mitogen-activated protein (MAP) kinase. FEBS Lett. 1998;438:41–48. doi: 10.1016/s0014-5793(98)01269-1. [DOI] [PubMed] [Google Scholar]
- 30.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 31.No D, Yao T P, Evans R M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomoto A, Lee Y F, Wimmer E. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc Natl Acad Sci USA. 1976;73:375–380. doi: 10.1073/pnas.73.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novoa I, Martinez A F, Fortes P, Ortin J, Carrasco L. Cleavage of p220 by purified poliovirus 2A(pro) in cell-free systems: effects on translation of capped and uncapped mRNAs. Biochemistry. 1997;36:7802–7809. doi: 10.1021/bi9631172. [DOI] [PubMed] [Google Scholar]
- 34.Nuss D L, Oppermann H, Koch G. Selective blockage of initiation of host protein synthesis in RNA-virus-infected cells. Proc Natl Acad Sci USA. 1975;72:1258–1262. doi: 10.1073/pnas.72.4.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohlmann T, Pain V M, Wood W, Rau M, Morley S J. The proteolytic cleavage of eukaryotic initiation factor (eIF) 4G is prevented by eIF4E binding protein (PHAS-I; 4E-BP1) in the reticulocyte lysate. EMBO J. 1997;16:844–855. doi: 10.1093/emboj/16.4.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 37.Perez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology. 1992;189:178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 38.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts L O, Seamons R A, Belsham G J. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA. 1998;4:520–529. doi: 10.1017/s1355838298971989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonenberg N, Gingras A C. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 41.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoneley M, Paulin F E, Le Quesne J P, Chappell S A, Willis A E. C-myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 43.Svitkin Y V, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolskaya E A, Romanova L I, Kolesnikova M S, Ivannikova T A, Smirnova E A, Raikhlin N T, Agol V I. Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J Virol. 1995;69:1181–1189. doi: 10.1128/jvi.69.2.1181-1189.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan R, Rychlik W, Etchison D, Rhoads R E. Amino acid sequence of the human protein synthesis initiation factor eIF-4 gamma. J Biol Chem. 1992;267:23226–23231. [PubMed] [Google Scholar]
- 46.Yang Q, Sarnow P. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]