Abstract
Background
It is well established that obesity is a major health risk in diabetes and associated diseases. Epigenetic changes, specially DNA methylation, play an important role in regulation of adipokines. The objective of the present study was to evaluate the DNA methylation status at the promoter region of the leptin gene in obese individuals and its association with metabolic risk factors.
Methods
The study included obese (n=100) and non-obese (n=75) individuals aged 25-45 years, and measured their physical, biochemical parameters (glucose, insulin, and lipid profiles) and leptin, DNA methyltransferase 1 (DNMT1), and DNA methyltransferase 3 beta (DNMT3b) mRNA expressions with real-time reverse transcription-polymerase chain reaction (qRT-PCR). DNA methylation of the leptin gene at the promoter region was analyzed by methyl-specific qPCR .
Results
The study found that the DNA methylation level at the promoter area of the leptin gene was negatively associated with weight in obese subjects. Furthermore, study findings showed that the DNA methylation level was negatively associated with fasting insulin, glucose, homeostatic model assessment for insulin resistance, and total cholesterol. There was also a higher expression of DNMT1 and DNMT-3b in obese subjects as compared with non-obese subjects.
Conclusion
The leptin epigenetic profile may be associated with obesity and its associated metabolic risk factors.
Keywords: DNA methylation, DNA methyltransferase, Leptin, Obesity
INTRODUCTION
Obesity is a long-standing condition1 that “results from the interaction between genetic factors with environmental influences”.2 An overflow of triglycerides (TGs) in adipose tissue due to an imbalance between energy intake and expenditure causes obesity.3 The World Health Organization estimates that 39% of the world adult population is overweight and 13% is obese. Around 3.0 million individuals die each year from obesity-associated complications. Obesity is now considered a key risk factor for a broad range of diseases such as type 2 diabetes, cardiovascular disease, and cancer.4 In 2015, an estimated 1.6 million deaths were caused by diabetes.5
Adipose tissue expresses leptin, a 16 kDa protein6 that works like a hormone and changes whole body energy homeostasis by controlling appetite. The arcuate nucleus in the hypothalamus has a leptin receptor where leptin acts like a hormone and maintains energy homeostasis by regulating appetite.7,8 Leptin has a key association between leptinaemia and the chronic sub inflammatory state in obesity, which suggests other possible, peripheral biological effects associated with its cytokine-like structure.9 Accordingly, supplementation of leptin in generalized lipodystrophy (absence of adipose tissue) polish up insulin sensitivity and metabolic parameters.10 Leptin expression11 and its serum levels are directly associated with obesity. Increased adipose tissue size (total fat mass) is significantly associated with increase in leptin, hence leptin is a signature of total fat mass.12 Leptin is a signaling molecule from peripheral adipose tissue to the brain. Leptin gene-lacking mice were obese and had more complicated type 2 diabetes.6 These opinions enlighten further work to uncover the mechanism of regulation for leptin expression in adipocytes.
Gene expression in physiologic and pathologic states is regulated by DNA methylation, which signals major epigenetic changes. Methylation of cytosine in CpG sites is one of the important epigenetic modifications that suppresses gene expression. Methylation usually prevents transcriptional factors from binding to promoters, and recruits transcriptional repressors such as methyl CpG binding protein 2.13,14 As a result, genes associated with the CpG-rich promoter and their expression level of genes have a tendency to rely on the methylation grade. In fact, the reason for down-regulated activity of leptin is methylated CpG rich promoter.15
Previous studies have reported that DNA methylation levels at candidate gene loci associated to obesity and metabolic abnormalities are impaired in peripheral blood and fat tissue of obese patients16,17 and in low weight loss responder to diet and exercise interventions.18-21 In total, these findings suggest that leptin DNA methylation profiles might be involved in the pathology of obesity and metabolic disorder. Thus, the aim of the present study was to explore the role of DNA methylation in leptin gene expression in obese subjects and its link with metabolic risk factors in a northwest Indian Population.
METHODS
Study design and participants
The researchers designed a hospital based, cross-sectional study. The study design was approved by the Institutional Human Ethics Committee (IHEC-LOP/2016/EF0026 dated 7th Mar 2016) and a written informed consent was obtained from all participants before inclusion. Individuals of both sexes between the age of 25–45 years and who presented to the outpatient department of All India Institute of Medical Sciences, Jodhpur (a tertiary care hospital in northwestern India) for a routine health-check-up were recruited. A questionnaire was administered to all participants to determine their known morbidities, and anthropometry was performed using standard means to classify them as obese (body mass index [BMI] ≥5 kg/m2) or non-obese (BMI, 18.5–24.9 kg/m2) on the basis of BMI. A fasting blood sample was obtained for biochemical and molecular assays.
The obese subjects were further divided into two sub-groups, those with metabolic syndrome and those without metabolic syndrome, according to the National Cholesterol Education Program’s Adult Treatment Panel III, which requires at least three of the following: central obesity (waist circumference [WC] of ≥102 cm in men and ≥88 cm in women); dyslipidemia (TG levels ≥150 mg/dL; decreased high-density lipoprotein [HDL] cholesterol levels <40 mg/dL [males] and <50 mg/dL [female]); blood pressure of 130/85 mmHg or higher or drug treatment for hypertension; and fasting plasma glucose ≥110 mg/dL.22
Biochemical estimation
Serum insulin concentrations were determined using the ADVIA Centaur Insulin assay kit (Tarrytown, NY, USA). Plasma glucose and lipid profile (Beckman Coulter kit) concentrations were determined using an autoanalyzer (Beckman Coulter 480).
Nucleic acid (DNA and RNA) extraction
DNA was purified from whole blood samples with the Relia Prep Blood gDNA Miniprep (Promega Corp., Madison, WI, USA). Total RNA was isolated using Trizol reagent (Thermo Fischer Life Science, Carlsbad, CA ,USA). Complementary DNA (cDNA) was synthesized from RNA using high-capacity cDNA RT kit (Thermo Fischer Life Science) according to manufacturer instruction.
Sodium bisulfite modification
The isolated DNA was modified with sodium bisulfite, which transforms unmethylated cytosine into uracil without changing methylated cytosine by using the EpiTect Bisufite Kit (Qiagen) according to the manufacturer’s instructions.
Quantitative methylation analysis
Real time PCR was carried out using Methylamp MS-qPCR (Epigenetek). PCR amplification was carried out in Bio-Rad CFX96 Real time PCR (Bio-Rad). A 96-well PCR plate using a temperature profile of 95°C, 15 minutes (initial denaturation) followed by 40 cycles of 94°C, 15 seconds, 59°C, 30 seconds and 72°C, 30 seconds for denaturation, annealing, and extension steps, respectively. The primers were designed by using Meth Primer (Li Lab) software. The PCR primers were synthesized by (Eurofins Scientific). Primer sequence for leptin methylated are forward: 5´-TAGGATTAACGAGGGCGTAGTC-3´ and reverse: 5´-AACCCCTTAAAAAAATACTTCGAA-3´ and for unmethylated are forward: 5´-TTAGGATTAATGAGGGTGTAGTTGT-3´ and reverse: 5´-CAACCCCTTAAAAAAATACTTCAAA-3´.
Real time PCR measurement of leptin mRNA, DNMT1, and DNMT3b
Real time-PCR was carried out using the iTaq Universal SYBR Green Supermix kit (Bio-Rad). PCR amplification was conducted in Bio-Rad CFX96 Realtime PCR (Bio-Rad). A 96 well PCR plate using the temperature profile of 95°C, 15 minutes, (initial denaturation) was followed by 40 cycles of 94°C, 15 seconds, 59°C, 30 seconds and 72°C, 30 seconds for denaturation, annealing, and extension steps, respectively. Primer sequence of human leptin, DNA methyltransferase 1 (DNMT1) and DNA methyltransferase 3 beta (DNMT3b) were 5´-GCTGTGCCCATCCAAAAAGT-3´ (forward) 5´-ACTGCCAGTGTCTGGTCCAT-3´ (reverse), 5´-TACCTGGACCCTGACCTC-3´ (forward), 5´-CGTTGGCATCAAAGATGGACA-3´ (reverse) and 5´-GGCAAGTTCTCCGAGGTC TCTG-3´ (forward) 5´-TGGTACATGGCTTTTCGATAGGA-3´ (reverse). The primer sequence of GAPDH as an internal control with the following sequence was 5′-AGGGCTGCTTTTAACTCTGGT-3′ (forward) and 5′-CCCCACTTGATTTTGGAGGGA-3′ (reverse). The PCR primers were synthesized by Eurofins Scientific (Munich, Germany).
Calculation
Insulin resistance
The homeostatic model assessment for insulin resistance (HOMA-IR) was calculated using the HOMA-IR [HOMA-IR=fasting insulin (μU/L)×fasting glucose (mM)/22.5].23
Relative gene expression
The relative gene expression was calculated using [(1/2)ΔCt].
Statistical analysis
Data were summarized as the mean±standard error of the mean. Two independent groups (non-obese and obese; and obese with non-metabolic and metabolic syndrome) were compared using the Student t-test. A Pearson correlation analysis was performed to assess the association of methylation (leptin) with gene expression (DNMT1, DNMT3b and leptin) and metabolic risk factors (demographic, biochemical, and clinical variables). A two-tailed P<0.05 was considered statistically significant. Analyses were performed with STATISTICA software version 6.0 (StatSoft, Tulsa, OK, USA).
RESULTS
Demographic, biochemical, and clinical characteristics
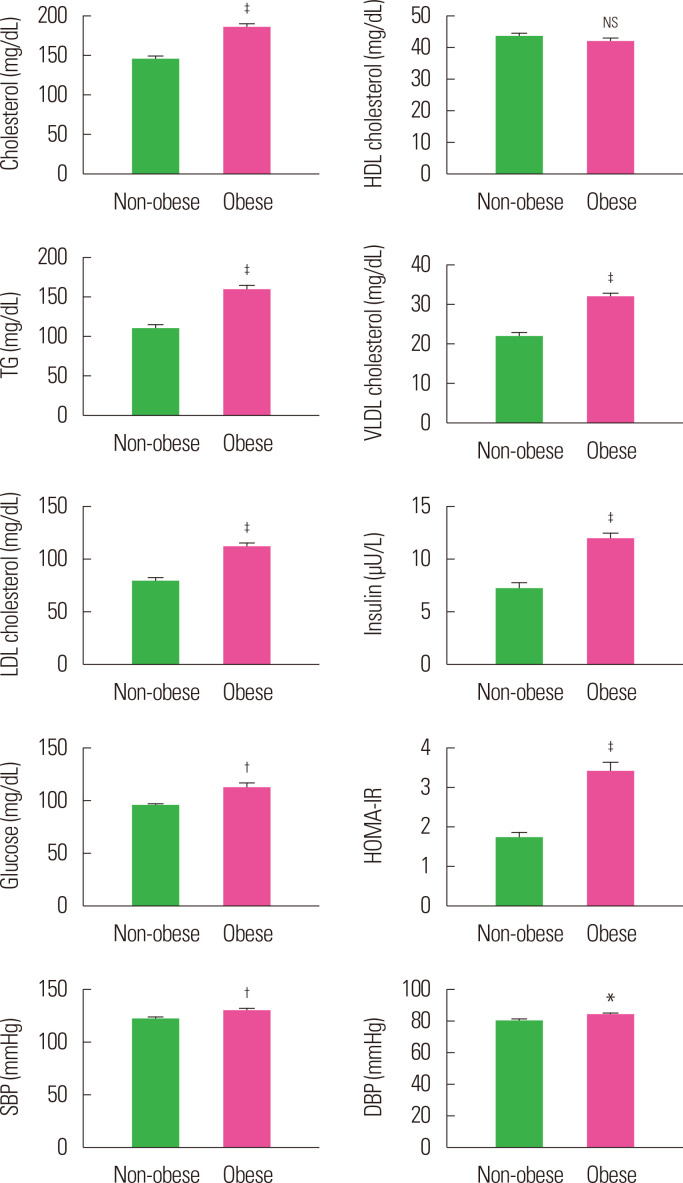
A total of 175 individuals were included in the study; 100 of them were obese and 75 were non-obese. The demographic (age, sex, height, weight, BMI, WC, biochemical (cholesterol, HDL cholesterol, TG, very low density lipoprotein (VLDL) cholesterol, low-density lipoprotein (LDL) cholesterol, and clinical (insulin, glucose, HOMA-IR, systolic blood pressure [SBP], and diastolic blood pressure [DBP]) characteristics of these two groups are summarized in Table 1 and Fig. 1. Age and sex distribution of the participants in the two groups was similar (P>0.05). However, the mean weight, BMI, WC, cholesterol, TG, VLDL, LDL, insulin, HOMA-IR, SBP, and DBP were found to be significantly (P<0.05, P<0.01, or P<0.001) different and higher in obese individuals when compared to non-obese individuals.
Table 1.
Demographic, biochemical, and clinical characteristics of non-obese and obese groups
| Variable | Non-obese group (n = 75) | Obese group (n = 100) | t-value | P |
|---|---|---|---|---|
| Age (yr) | 36.04 ± 0.82 | 37.56 ± 0.63 | 1.50 | 0.136 |
| Sex | 0.09 | 0.760 | ||
| Female | 38 (50.7) | 53 (53.0) | ||
| Male | 37 (49.3) | 47 (47.0) | ||
| Height (cm) | 161.93 ± 0.98 | 160.92 ± 1.07 | 0.68 | 0.500 |
| Weight (kg) | 58.06 ± 0.93 | 76.53 ± 1.14 | 11.94 | < 0.001 |
| BMI (kg/m2) | 22.10 ± 0.24 | 29.58 ± 0.38 | 15.52 | < 0.001 |
| WC (cm) | 83.47 ± 0.90 | 99.06 ± 0.89 | 12.13 | < 0.001 |
| Cholesterol (mg/dL) | 144.34 ± 3.75 | 185.03 ± 4.57 | 6.57 | < 0.001 |
| HDL cholesterol (mg/dL) | 43.53 ± 0.90 | 41.69 ± 1.02 | 1.31 | 0.193 |
| TG (mg/dL) | 109.43 ± 4.86 | 159.00 ± 6.28 | 5.91 | < 0.001 |
| VLDL cholesterol (mg/dL) | 21.89 ± 0.97 | 31.80 ± 1.26 | 5.91 | < 0.001 |
| LDL cholesterol (mg/dL) | 78.92 ± 3.33 | 111.54 ± 4.10 | 5.88 | < 0.001 |
| Insulin (μU/L) | 7.21 ± 0.57 | 11.87 ± 0.64 | 5.22 | < 0.001 |
| Glucose (mg/dL) | 94.81 ± 1.52 | 111.86 ± 4.62 | 3.10 | 0.002 |
| HOMA-IR | 1.70 ± 0.15 | 3.39 ± 0.24 | 5.47 | < 0.001 |
| SBP (mmHg) | 121.67 ± 1.88 | 129.84 ± 1.62 | 3.30 | 0.001 |
| DBP (mmHg) | 80.11 ± 1.16 | 84.05 ± 1.04 | 2.51 | 0.013 |
Values are presented as mean± standard error of the mean or number (%).
BMI, body mass index; WC, waist circumference; HDL, high-density lipoprotein; TG, triglyceride; VLDL, very low density lipoprotein; LDL, low-density lipoprotein; HOMA-IR, homeostatic model assessment for insulin resistance; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Figure. 1.
Mean biochemical risk factor levels of non-obese and obese subjects. Values are presented as mean± standard error of the mean. *P< 0.05, †P< 0.01, ‡P< 0.001, and NSP> 0.05, as compared to non-obese. HDL, high-density lipoprotein; TG, triglyceride; VLDL, very low density lipoprotein; LDL, low-density lipoprotein; SE, standard error of the mean; SBP, systolic blood pressure; DBP, diastolic blood pressure; NS, not significant.
Relative gene expression
The relative gene expression of markers (DNMT1, DNMT3b, and leptin) of the two groups (obese and non-obese) is summarized in Table 2. The relative mean gene expression of markers was found to be similar (P>0.05) between the two groups though it was 63.3%, 78.4%, and 96.4% higher, respectively, in obese as compared to non-obese individuals.
Table 2.
Relative gene expression of markers of the two groups
| Variable | Non-obese group | Obese group | t-value | P | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| n | Mean± SE | n | Mean± SE | |||
| DNMT1 | 29 | 0.169 ± 0.027 | 52 | 0.462 ± 0.127 | 1.70 | 0.092 |
| DNMT3b | 29 | 0.033 ± 0.005 | 52 | 0.154 ± 0.054 | 1.66 | 0.101 |
| Leptin | 29 | 0.012 ± 0.005 | 52 | 0.321 ± 0.178 | 1.29 | 0.200 |
SE, standard error of the mean; DNMT, DNA methyltransferase.
Methylation and unmethylation
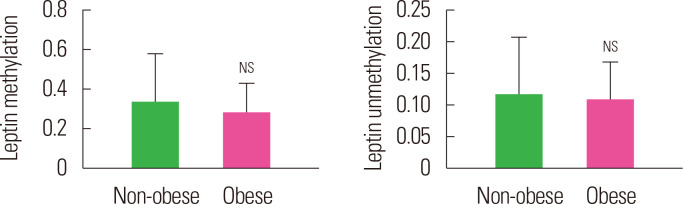
The methylation and unmethylation levels of leptin in the two groups (obese and non-obese) is summarized in Table 3 and Fig. 2. The mean methylation and unmethylation levels of leptin were also found to be similar (P>0.05) between the two groups though it was lower, 16.5% and 7.3% respectively, in obese as compared to non-obese individuals.
Table 3.
Leptin methylation and unmethylation levels of non-obese and obese groups
| Variable | Non-obese | Obese | t-value | P | |||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| n | Mean ± SE | n | Mean ± SE | ||||
| Leptin M | 29 | 0.328 ± 0.248 | 33 | 0.274 ± 0.154 | 0.19 | 0.849 | |
| Leptin U | 20 | 0.116 ± 0.093 | 28 | 0.107 ± 0.062 | 0.08 | 0.938 | |
SE, standard error of the mean; M, methylation; U, unmethylation.
Figure. 2.
Mean methylation and unmethylation leptin levels of non-obese and obese subjects. Values are presented as mean ± standard error of the mean. NSP> 0.05 as compared to non-obese. NS, not significant.
Correlation
The correlation of leptin methylation and unmethylation with metabolic risk factors (demographic, biochemical, and clinical) and relative markers (DNMT1, DNMT3b, and leptin) gene expression in the obese is summarized in Table 4. Leptin methylation and unmethylation showed an insignificant (P>0.05) correlation with metabolic risk factors and relative markers gene expression except leptin. Leptin showed a significant and positive (direct) correlation with age (r=0.560, P<0.001) and a significant and negative (inverse) correlation with weight (r=–0.449, P<0.01).
Table 4.
Correlation of methylation with metabolic risk factors and relative markers gene expression in obese subjects
| Variable | Leptin methylation | Leptin unmethylation | |||
|---|---|---|---|---|---|
|
|
|
||||
| Correlation (r) | P | Correlation (r) | P | ||
| Age | 0.560 | < 0.001† | 0.171 | 0.357 | |
| Sex | –0.267 | 0.213 | –0.097 | 0.584 | |
| Height | –0.287 | 0.128 | 0.169 | 0.506 | |
| Weight | –0.449 | 0.001* | 0.059 | 0.933 | |
| BMI | –0.329 | 0.194 | –0.067 | 0.665 | |
| WC | –0.360 | 0.148 | –0.025 | 0.855 | |
| Cholesterol | –0.092 | 0.548 | 0.012 | 0.952 | |
| HDL cholesterol | 0.033 | 0.996 | 0.136 | 0.457 | |
| TG | 0.001 | 0.987 | –0.210 | 0.279 | |
| VLDL cholesterol | 0.001 | 0.987 | –0.210 | 0.279 | |
| LDL cholesterol | –0.119 | 0.482 | 0.029 | 0.897 | |
| Insulin | –0.063 | 0.927 | –0.160 | 0.390 | |
| Glucose | –0.174 | 0.358 | 0.046 | 0.813 | |
| HOMA-IR | –0.178 | 0.467 | –0.113 | 0.539 | |
| SBP | –0.273 | 0.170 | –0.177 | 0.403 | |
| DBP | –0.187 | 0.292 | –0.023 | 0.865 | |
| DNMT1 | 0.022 | 0.918 | 0.168 | 0.413 | |
| DNMT3b | 0.022 | 0.919 | 0.269 | 0.183 | |
| Leptin | –0.034 | 0.872 | –0.089 | 0.664 | |
*P< 0.01; †P< 0.001.
BMI, body mass index; WC, waist circumference; HDL, high-density lipoprotein; TG, triglyceride; VLDL, very low density lipoprotein; LDL, low-density lipoprotein; HOMA-IR, homeostatic model assessment for insulin resistance; SBP, systolic blood pressure; DBP, diastolic blood pressure; DNMT, DNA methyltransferase.
Methylation and unmethylation in obese groups
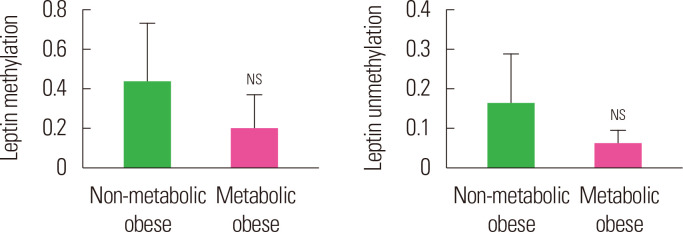
The methylation and unmethylation levels of leptin in two obese groups (non-metabolic and metabolic) is summarized in Table 5 and Fig. 3. The mean methylation and unmethylation levels of leptin were also found to be similar (P>0.05) between the two obese groups though it was lower, 55.0% and 62.1% respectively, in the metabolic obese as compared to the non-metabolic obese group.
Table 5.
Leptin methylation and unmethylation levels of two obese groups
| Variable | Non-metabolic obese group | Metabolic obese group | t-value | P-value | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| n | Mean± SE | n | Mean± SE | |||
| Leptin M | 11 | 0.433 ± 0.302 | 22 | 0.195 ± 0.177 | 0.73 | 0.474 |
| Leptin U | 13 | 0.160 ± 0.128 | 15 | 0.061 ± 0.035 | 0.80 | 0.432 |
SE, standard error of the mean; M, methylation; U, unmethylation.
Figure. 3.
Mean methylation and unmethylation leptin levels of non-metabolic obese and metabolic obese subjects. NSP> 0.05 as compared to non-obese. NS, not significant.
DISCUSSION
In obesity and obesity-related metabolic disorders, leptin regulation is profoundly altered,24 and some of these changes are regulated by DNA methylation. The present study reports that leptin DNA methylation levels at the promoter region, investigated in peripheral blood specimens, are associated with obesity-related anthropometric variables, biochemical parameters, and leptin mRNA expression.
Study findings report that the hypomethylation (reduced methylation) of the leptin promoter in the peripheral blood of the obese was like that of the non-obese. Further, it demonstrated reduced DNA methylation in the metabolic obese as compared to the non-metabolic obese subjects. Various authors have reported reduced levels of leptin methylation in the blood cells of children with higher birth weight and BMI,25 in obese and insulin resistant adolescents,26 and obese women.27 Another report stipulates that genetic variation may have a substantial impact on local methylation patterns,28,29 but as to the extent that methylation is not affected by genetic variation, the mechanisms remain unclear.
This study also found higher leptin gene expression in obese subjects. Interestingly, leptin was also higher in non-metabolic obese subjects. This finding is supported by Tiwari et al.24 who reported higher gene expression in the adipose tissue of obese subjects. Another study involving human subjects showed that the developmental increase in leptin mRNA expression in adipose tissue during childhood, reaching maximal capacity in adulthood.30 Moreover, a study also supported associating leptin with a direct supervision of body fat (adipose tissue) metabolism through inhibition of TG synthesis and stimulation of lipid breakdown.31 Reduced leptin methylation could account for obesity-related leptin upregulation. These findings indicate that methylation at the promoter region regulates leptin gene expression. A possible mechanism, the in silico analysis, is in line with a study that noted that methylation surrounding the transcription start site is tightly associated to transcriptional silencing while methylation of more downstream regions is not associated with the magnitude of gene expression.32,33
Researchers in the current study also observed the significant negative association between methylation levels of leptin promoter and body weight. This finding is consistent with the recent finding of García-Cardona et al.26 who reported a negative correlation between methylation frequency and BMI in obese adolescents.
In addition, the study also found a negative association of methylation levels of leptin promoter with BMI, cholesterol, insulin, HOMA-IR and glucose. However, it was not significant. These insignificant results may be due to small sample size. This finding is also consistent with previous findings.26,34 Moreover, these results also showed higher expression of DNMT1 and DNMT3b in obese rather than non-obese groups, but there was a lower methylation level at the promoter of leptin. This may be due to changes in DNMT protein levels. Moreover, the study found a higher expression of DNMT1 and DNMT3b in the non-metabolic obese than in the metabolic obese. Thus, these findings support that lower methylation might be the cause of the development of obesity and its associated metabolic risks.
The strength of the current study includes analysis of the importance of adipokine central in energy metabolism regulation in blood cells. Blood is the most clinically within-reach tissue. Limitations of the current study are its small sample size and the fact that it did not include analysis in adipose tissue. In conclusion, the present findings add evidence that leptin DNA methylation levels in blood cells are associated with obesity-related anthropometric measures.
ACKNOWLEDGMENTS
This study was funded by Science and Engineering Research Board (SERB), New Delhi, India (Grant No. YSS/2015/000054).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Study concept and design: S; acquisition of data: AM and MK; analysis and interpretation of data: PS; drafting of the manuscript: SSM; critical revision of the manuscript: RJ; statistical analysis: VVB and MPSN; obtained funding: S; administrative, technical, or material support: RB; and study supervision: ST and PKS.
REFERENCES
- 1.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity and trends in body mass index among US children and adolescents, 1999-2010. JAMA. 2012;307:483–90. doi: 10.1001/jama.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462:307–14. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 3.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–53. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pi-Sunyer X. The medical risks of obesity. Postgrad Med. 2009;121:21–33. doi: 10.3810/pgm.2009.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization, author. World health report 2012 fact sheet 311. World Health Organization; Geneva: 2016. [Google Scholar]
- 6.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–32. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 7.Minokoshi Y, Kim YB, Peroni OD, Fryer LG, Müller C, Carling D, et al. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature. 2002;415:339–43. doi: 10.1038/415339a. [DOI] [PubMed] [Google Scholar]
- 8.Muoio DM, Dohm GL, Fiedorek FT, Jr, Tapscott EB, Coleman RA. Leptin directly alters lipid partitioning in skeletal muscle. Diabetes. 1997;46:1360–3. doi: 10.2337/diab.46.8.1360. [DOI] [PubMed] [Google Scholar]
- 9.Ahima RS, Flier JS. Leptin. Annu Rev Physiol. 2000;62:413–37. doi: 10.1146/annurev.physiol.62.1.413. [DOI] [PubMed] [Google Scholar]
- 10.Oral EA, Simha V, Ruiz E, Andewelt A, Premkumar A, Snell P, et al. Leptin-replacement therapy for lipodystrophy. N Engl J Med. 2002;346:570–8. doi: 10.1056/NEJMoa012437. [DOI] [PubMed] [Google Scholar]
- 11.Vidal H, Auboeuf D, De Vos P, Staels B, Riou JP, Auwerx J, et al. The expression of ob gene is not acutely regulated by insulin and fasting in human abdominal subcutaneous adipose tissue. J Clin Invest. 1996;98:251–5. doi: 10.1172/JCI118786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Considine RV, Sinha MK, Heiman ML, Kriauciunas A, Stephens TW, Nyce MR, et al. Serum immunoreactive-leptin concentrations in normal-weight and obese humans. N Engl J Med. 1996;334:292–5. doi: 10.1056/NEJM199602013340503. [DOI] [PubMed] [Google Scholar]
- 13.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–43. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 14.Bird AP, Wolffe AP. Methylation-induced repression: belts, braces, and chromatin. Cell. 1999;99:451–4. doi: 10.1016/S0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 15.Komori N, Tawata M, Onaya T. DNA demethylation modulates mouse leptin promoter activity during the differentiation of 3T3-L1 cells. Diabetologia. 2002;45:140–8. doi: 10.1007/s125-002-8255-4. [DOI] [PubMed] [Google Scholar]
- 16.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aïssi D, Wahl S, et al. DNA methylation and body-mass index: a genomewide analysis. Lancet. 2014;383:1990–8. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 17.Milagro FI, Gómez-Abellán P, Campión J, Martínez JA, Ordovás JM, Garaulet M. CLOCK, PER2 and BMAL1 DNA methylation: association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chronobiol Int. 2012;29:1180–94. doi: 10.3109/07420528.2012.719967. [DOI] [PubMed] [Google Scholar]
- 18.Jacobsen SC, Gillberg L, Bork-Jensen J, Ribel-Madsen R, Lara E, Calvanese V, et al. Young men with low birthweight exhibit decreased plasticity of genome-wide muscle DNA methylation by high-fat overfeeding. Diabetologia. 2014;57:1154–8. doi: 10.1007/s00125-014-3198-8. [DOI] [PubMed] [Google Scholar]
- 19.Bouchard L, Rabasa-Lhoret R, Faraj M, Lavoie ME, Mill J, Pérusse L, et al. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr. 2010;91:309–20. doi: 10.3945/ajcn.2009.28085. [DOI] [PubMed] [Google Scholar]
- 20.Rönn T, Volkov P, Davegårdh C, Dayeh T, Hall E, Olsson AH, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moleres A, Campión J, Milagro FI, Marcos A, Campoy C, Garagorri JM, et al. Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON study. FASEB J. 2013;27:2504–12. doi: 10.1096/fj.12-215566. [DOI] [PubMed] [Google Scholar]
- 22.Oh JE. Relationship between weekly physical activity frequency and metabolic syndrome. Korean J Obes. 2016;25:77–83. doi: 10.7570/kjo.2016.25.2.77. [DOI] [Google Scholar]
- 23.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–9. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 24.Tiwari S, Sadashiv, Paul BN, Kumar S, Chandra A, Dhananjai S, et al. TNF-α gene expression in subcutaneous adipose tissue associated with HOMA in Asian Indian postmenopausal women. Horm Metab Res. 2014;46:94–9. doi: 10.1055/s-0033-1358706. [DOI] [PubMed] [Google Scholar]
- 25.Obermann-Borst SA, Eilers PH, Tobi EW, de Jong FH, Slagboom PE, Heijmans BT, et al. Duration of breastfeeding and gender are associated with methylation of the LEPTIN gene in very young children. Pediatr Res. 2013;74:344–9. doi: 10.1038/pr.2013.95. [DOI] [PubMed] [Google Scholar]
- 26.García-Cardona MC, Huang F, García-Vivas JM, López-Camarillo C, Del Río Navarro BE, Navarro Olivos E, et al. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. Int J Obes (Lond) 2014;38:1457–65. doi: 10.1038/ijo.2014.30. [DOI] [PubMed] [Google Scholar]
- 27.Lesseur C, Armstrong DA, Paquette AG, Koestler DC, Padbury JF, Marsit CJ. Tissue-specific Leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Mol Cell Endocrinol. 2013;381:160–7. doi: 10.1016/j.mce.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoof E, Stuppy A, Harig F, Carbon R, Horbach T, Stöhr W, et al. Comparison of leptin gene expression in different adipose tissues in children and adults. Eur J Endocrinol. 2004;150:579–84. doi: 10.1530/eje.0.1500579. [DOI] [PubMed] [Google Scholar]
- 31.Yang WS, Lee WJ, Funahashi T, Tanaka S, Matsuzawa Y, Chao CL, et al. Weight reduction increases plasma levels of an adipose-derived anti-inflammatory protein, adiponectin. J Clin Endocrinol Metab. 2001;86:3815–9. doi: 10.1210/jcem.86.8.7741. [DOI] [PubMed] [Google Scholar]
- 32.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–3. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–94. doi: 10.1128/MCB.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houde AA, Légaré C, Biron S, Lescelleur O, Biertho L, Marceau S, et al. Leptin and adiponectin DNA methylation levels in adipose tissues and blood cells are associated with BMI, waist girth and LDL-cholesterol levels in severely obese men and women. BMC Med Genet. 2015;16:29. doi: 10.1186/s12881-015-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]