Abstract
Erythrovirus (formerly parvovirus) B19 causes a wide range of diseases in humans, including anemia due to aplastic crisis. Diagnosis of B19 infection relies on serology and the detection of viral DNA by PCR. These techniques are usually thought to detect all erythrovirus field isolates, since the B19 genome is known to undergo few genetic variations. We have detected an erythrovirus (V9) markedly different from B19 in the serum and bone marrow of a child with transient aplastic anemia. The B19 PCR assay yielded a product that hybridized only very weakly to the B19-specific probe and whose sequence diverged more from those of 24 B19 viruses (11 to 14%) than the divergence found within the B19 group (≤6.65%). Restriction enzyme analysis of the V9 genome revealed that this genetic divergence extended beyond the amplified region. Interestingly, serological tests failed to demonstrate a response characteristic of acute B19 infection. V9 could be a new erythrovirus, and new diagnostic tests are needed for its detection.
Erythrovirus B19, called parvovirus before the revision of the taxonomy in 1995 (17), causes erythema infectiosum, usually in children and young adults. In most patients B19 infection causes an acute illness from which patients recover spontaneously and confers protective, lifelong immunity (12, 27). Complications can occur when the viral infection arises in patients with particular backgrounds: chronic thrombocytopenia or anemia in immunocompromised patients and fetal infection in pregnant women. Transient aplastic crisis (TAC), a frequent complication of acute B19 infection, was originally described as the abrupt onset of severe anemia with reticulopenia in patients suffering from chronic hemolysis due to cessation of erythrocyte production in the bone marrow secondary to the tropism of the virus for erythroid progenitor cells. TAC can also occur under conditions of erythroid stress, such as hemorrhage or iron deficiency (5, 27). Erythrovirus B19 is a common infectious agent in humans: B19 seroprevalence is approximately 50% by the age of 15 and rises further among elderly people because infection occurs throughout adult life (5). Although it is generally accepted that B19 infection is transmitted by the respiratory route, it can also be transmitted by blood or blood products, even those treated with heat or a solvent-detergent to inactivate viruses (5). Genotypes were previously established on the basis of the restriction enzyme polymorphism of the viral genome (15, 16). However, until now, the single-stranded DNA of erythrovirus has been known to undergo little genetic variation (<1% of the entire genome [4, 11, 22]), and there is only one species in the genus Erythrovirus, which is referred to as B19 (17).
MATERIALS AND METHODS
B19 serological assays.
Testing for B19-specific antibodies was performed with commercial assays (Parvovirus B19 IgG Enzyme Immunoassay or Parvovirus B19 IgM Enzyme Immunoassay; Biotrin, Dublin, Ireland), according to the manufacturer’s recommendations.
Viral DNA extraction.
To isolate leukocytes, the bone marrow sample was layered onto Histopaque 1119 (Sigma Diagnostics, Saint Quentin-Fallavier, France) according to the manufacturer’s instructions. After two washes in 150 mM NaCl, the leukocytes were pelleted by centrifugation and stored at −80°C. The cells were lysed in 250 μl of buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 0.5% Tween 20, 0.5% Nonidet P-40) with 15 μg of proteinase K at 56°C for 90 min. After inactivation of the proteinase K by heating at 100°C for 10 min, the solution was extracted with 250 μl of phenol and then 250 μl of chloroform-isoamyl alcohol (25/1). The DNA was precipitated at −80°C for 1 h with 20 μg of glycogen (as the carrier), 25 μl of 3 M sodium acetate (pH 5.2), and 500 μl of ethanol. After washing with 70% ethanol, the DNA pellet was dried at 56°C for 10 min, resuspended in 100 μl of H2O, and stored at −20°C.
Viral DNA was extracted from serum samples with the QIAamp Blood Kit (Qiagen, Courtabœuf, France), according to the manufacturer’s instructions, and was stored at −20°C.
Restriction map.
NaCl was added to the (single-stranded) viral DNA extracted from the serum sample to a final concentration of 50 mM. The solution was heated at 95°C for 2 min and annealed at 55°C for 16 h. The double-stranded DNA was digested with a restriction enzyme (BamHI, HindIII, or PvuII).
PCR for B19 detection.
A standard PCR procedure was performed with 10 μl of either the extracted DNA or a 1/10 dilution of it with the following program: 1 cycle of 95°C for 7 min; 40 cycles of 94°C for 1 min, 48°C for 1 min, and 72°C for 1 min; and 1 cycle of 72°C for 7 min (on a thermocycler 480, Perkin-Elmer, Courtabœuf, France). The PCR products were separated by electrophoresis on a 1.5% agarose gel and Southern transferred by capillary blotting (0.4 M NaOH, 0.6 M NaCl) onto a charged nylon membrane. The blot was hybridized at 42°C for 16 h in a buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2% blocking reagent [Boehringer, Mannheim, Germany], 0.1% N-laurylsarcosine, 0.02% sodium dodecyl sulfate) with a B19-specific probe, P2560, labeled with the Dig Oligonucleotide Tailing Kit (Boehringer) according to the manufacturer’s instructions. Posthybridization washes were done at 60°C, and detection was performed with alkaline phosphatase conjugate and CSPD (DIG Luminescent Detection Kit; Boehringer). Sequences corresponding to the primers and probe are located in the unique region of the VP1 gene (VP1u) of the B19 sequence (22): primer 376 from positions 2408 to 2428, primer 377 from positions 2790 to 2809 (in the reverse orientation), and probe P2560 from positions 2560 to 2600 (13).
Sequencing of PCR products.
Both strands of the PCR products from the bone marrow and serum extracts were sequenced with primers 376 and 377 by using the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer Applied Biosystems, Courtabœuf, France) and the ABI Prism 377 DNA Sequencer (Perkin-Elmer Applied Biosystems).
Sequence analysis and phylogeny.
The sequence of the PCR product was used as the query sequence in FASTA (19) and BLAST (1) searches of the GenBank and EMBL data banks with the GCG package (Genetics Computer Group, Madison, Wis.).
The 24 nonidentical data bank B19 sequences covering 346 bp of the amplified region were aligned with the V9 sequence by using CLUSTAL W or CLUSTAL X (25). Phylogenetic analyses were performed with the PHYLIP package (9): Nucleotide distances were estimated with DNADIST, and the phylogenetic tree was then calculated with NEIGHBOR. The phylogenetic tree was plotted by using TREE TOOL (14).
Nucleotide sequence accession numbers.
The EMBL nucleotide sequence accession no. of the sequences reported here are AJ223617 and AJ242810.
RESULTS
Clinical and biological findings.
A 6-year-old male child who had lived in France since his birth was admitted to a pediatric unit with headache and dysuria in May 1995. Clinical examination noted only paleness of the conjunctiva and a mild fever (temperature, 38.6°C). Initial blood analyses (Table 1) revealed severe microcytic anemia with a sharp drop in reticulocytes associated with lymphopenia and neutropenia (5 May 1995). The myelogram (12 May 1995) showed a relative richness in erythroblasts and few abnormalities: insufficient hemoglobin in erythroblasts and negative Perls’ staining for iron. No giant pronormoblasts were found. Complementary analyses indicated that numerous factors could have contributed to this anemia. Gastrofibroscopy conducted because of initially low serum iron levels (5.7 μmol/liter; normal range, 11 to 24 μmol/liter) revealed mild interstitial gastritis associated with Helicobacter pylori in biopsy specimens of the antrum. Three months after the initial crisis, investigation of the chronic hemolysis (haptoglobin concentration, <0.09 g/liter) revealed a glucose-6-phosphate dehydrogenase (G6PD) defect (on 8 August 1995 tests for G6PD revealed 0 U per g of hemoglobin; normal range, 5.3 to 7.9 U per g of hemoglobin). A familial inquiry found a G6PD defect in one of his four siblings.
TABLE 1.
Hematological and biological data
| Date | Hemoglobin concn (g/dl) | Erythrocyte count (106/μl) | Mean cell vol (fl) | Reticulocyte count (no./μl) | Total leukocyte count (no./μl) | Neutrophil count (no./μl) | Lymphocyte count (no./μl) | Monocyte count (no./μl) | Platelet count (no./μl) | Haptoglobin concn (g/liter) |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 May 1995 | 6.3 | 3.21 | 65.3 | 80,300 | 4,500 | 2,070 | 1,760 | 585 | 320,000 | NDa |
| 5 May 1995 | 6.7 | 2.88 | 65 | 23,000 | 3,100 | 983 | 1,550 | 459 | 303,000 | <0.09 |
| 13 May 1995 | 6.4 | 2.75 | 65.3 | 138,000 | 13,600 | 8,120 | 3,160 | 1,700 | 707,000 | ND |
| 22 May 1995 | 7.2 | 3.27 | 74.9 | 526,000 | 11,500 | 4,720 | 5,060 | 920 | 481,000 | ND |
| Normal range | 12–14 | 4.1–5.5 | 73–89 | >50,000 | 5,000–16,000 | 1,500–8,500 | 2,000–8,000 | 0–800 | 200,000–400,000 | 1–3 |
ND, not done.
The anemia evolved favorably over a few weeks with treatment of the sideropenia and the gastritis (ferrous fumarate, folic acid, omeprazol, amoxicillin, and clarithromycin). No transfusion or gamma globulin injection was given.
Virological findings.
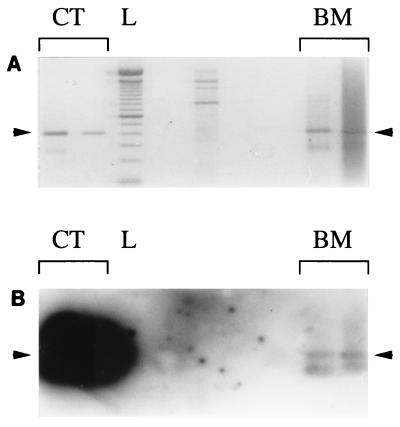
A search for a B19 infection was conducted. B19 serology was negative for immunoglobulin M (IgM) and positive for IgG both initially (on 2 May 1995 the optical density and cutoff values were 0.030 and 0.090 respectively, for IgM and 0.473 and 0.376, respectively, for IgG) and 3 months later (on 7 August 1995 the optical density and cutoff values were 0.012 and 0.082 respectively, for IgM and 1.130 and 0.279, respectively, for IgG). Despite the serological results which suggested a past B19 infection, a bone marrow sample was taken on 12 May 1995 for a PCR search for the B19 viral genome. This B19 PCR was inconclusive (Fig. 1), giving contrasting results: a PCR band that migrated the same distance as the B19-positive control band was clearly visible on the electrophoresis gel stained with ethidium bromide, but this band gave only a very faint signal after hybridization of the Southern blot with our B19-specific probe (Fig. 1). The intensity of the hybridization signal was enhanced when less stringent washing conditions were used (data not shown). This finding strongly suggests a mispairing of the B19-specific probe with an erythrovirus PCR product but could also denote nonspecific hybridization of the probe to a PCR product unrelated to B19 (false-positive result). Similar results were obtained when the B19 PCR was done with the first serum (drawn on 2 May). Conversely, the PCR assay was negative with the serum sample drawn 3 months later (data not shown).
FIG. 1.
B19 PCR with bone marrow extract. (A) Agarose gel stained with ethidium bromide. (B) Southern blot hybridized to a B19-specific probe. The arrowheads indicate the positions of the expected 402-bp PCR product. CT, B19 positive control; L, 100-bp DNA molecular size ladder; BM, bone marrow extract. The B19 PCR assay performed with the bone marrow extract gave a band which migrated the same distance as that of the B19-positive control (A), but this PCR product hybridized very poorly to the B19-specific probe (B). Exposure times were identical for both sides of the figure.
Sequence analysis and phylogeny.
To determine whether this weak hybridization signal could be due to mutations in the probe recognition sequence, the PCR products obtained from both the bone marrow and the serum DNA extracts were sequenced. Both sequences were identical, with six mutations mismatching the 41-bp B19-specific probe, and this weak similarity (85%) could explain the very faint hybridization signal that was observed (Fig. 1).
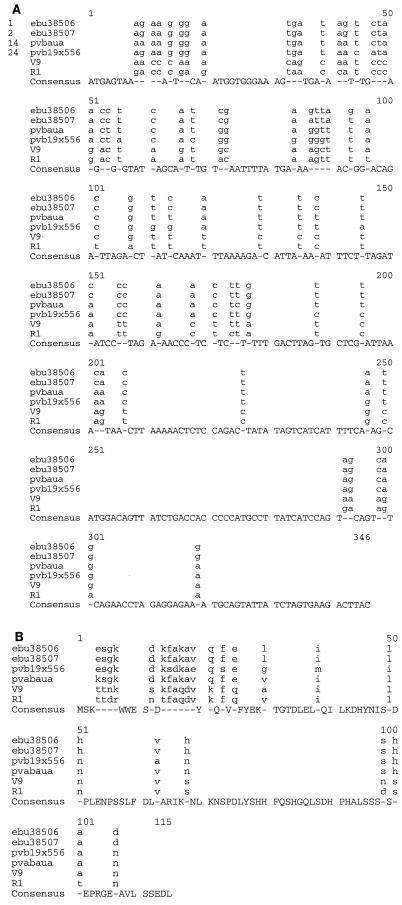
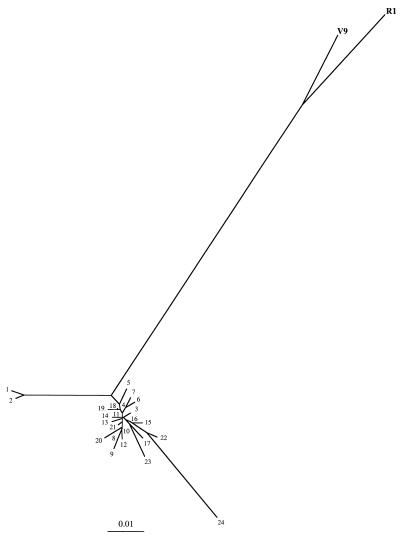
When compared to sequences stored in GenBank and EMBL databases, this 346-bp sequence resembled B19 sequences of the VP1u gene by FASTA analysis (data not shown). However, the sequence from this novel erythrovirus (which we called V9) was unexpectedly more divergent from the sequences of 24 B19 isolates (≥11.07% divergence) (Fig. 2) than these B19 sequences are among themselves (≤6.65% divergence). The unrooted phylogenetic tree based on these data showed that the V9 sequence was outside the B19 group (Fig. 3). Figure 3 would have been even more striking and the B19 cluster would have been even more condensed if we had excluded sequence 24 (pvb19x556), which was obtained from a patient with a persistent B19 infection (10), which is quite unusual in a nonimmunocompromised patient (5, 7, 27).
FIG. 2.
Multiple alignments of partial VP1u sequences from V9, R1, and the most divergent B19 sequences. (a) DNA sequences (346 bp) Modified from reference 18 with permission of the publisher. The genetic divergence between the sequences of V9 and R1 strains and the sequences of B19 strains resides in the 5′ part of the VP1 gene, which corresponds to one of the two major neutralization epitopes localized in the first 80 amino acids of VP1 (2, 21).
FIG. 3.
Unrooted phylogenetic tree of partial VP1u gene sequences from V9, R1, and B19 genomes. Branch lengths are proportional to genetic distances, expressed as percent divergence (a scale bar is shown at the bottom of the figure). Correspondence of the numbers used in the figure and the GenBank mnemonics for B19 sequences are as follows: 1, ebu38506; 2, ebu38507; 3, pvb19x572; 4, bvu31358; 5, ebu38510; 6, ebu38511; 7, ebu38514; 8, pvb19x583; 9, pvb19x591; 10, ebu38508; 11, pvb19x560; 12, ebu38512; 13, ebu38546; 14, pvbaua; 15, pvb19x528; 16, ebu38515; 17, pvbpro; 18, ebu38513; 19, pvb19nsvp; 20, pvb19x599; 21, e09420; 22, pvb19x541; 23, ebu38509; 24, pvb19x556. Erythrovirus B19 sequences 1 (ebu38506) and 2 (ebu38507) were isolated from Chinese patients, and B19 sequence 24 (pvb19x556) came from a patient suffering from persistent B19 infection. V9 and R1 are clearly outside the B19 group.
To determine whether V9 could be derived from an animal parvovirus, all known animal parvovirus sequences were subjected to phylogenetic analysis. The V9 sequence was situated very far from the animal parvovirus sequences, as is the B19 sequence (6). Among animal parvovirus sequences, the simian parvovirus was the closest to both V9 and B19, with divergences of 47 and 44%, respectively.
In light of these results, we rescreened samples for which, like the V9 sample, the results of our routine B19 PCR test were initially found to be inconclusive (work on this is in progress). From the first eight samples drawn from patients in France in early 1998, one, named R1, gave a PCR product whose sequence was similar to that of V9 (5.20% divergence) and even more distant from B19 (≥12.14% divergence) than V9 is (Fig. 2 and 3). This patient suffered from macrocytic anemia in a background of chronic renal insufficiency.
Restriction fragment length analysis.
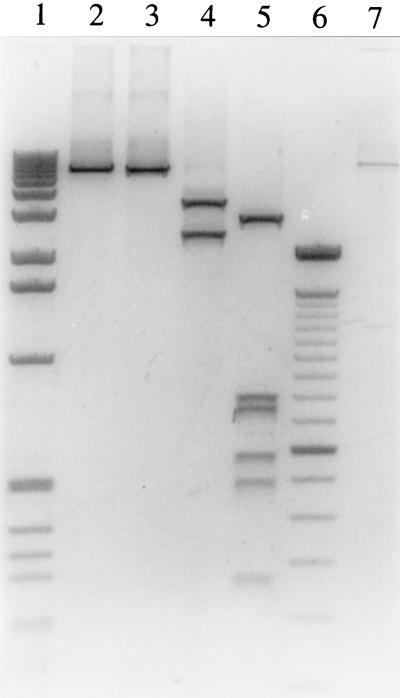
One should point out that the amplified VP1u region was recently found to be the most variable region of the B19 genome (8, 10), especially for B19 strains isolated during persistent infections. In this region, pvb19x556 shows 4% divergence with pvbaua, according to Hemauer et al. (10), and up to 6.65% divergence when its sequence is compared to those of the Chinese isolates. To determine whether the sequence divergence that we observed between V9 and B19 extended beyond the VP1u region, the entire V9 genome isolated from serum was subjected to restriction enzyme analysis (Fig. 4). Undigested V9 DNA had an apparent molecular size similar to that of B19 DNA. The V9 DNA restriction patterns obtained with the enzymes BamHI, HindIII, and PvuII differed markedly from those of 65 B19 strains from different geographic origins reported in the literature (15, 16, 26). V9 DNA had no BamHI site and one HindIII site, while all the B19 strains have one and two to three sites for these enzymes, respectively. Five PvuII sites were found for V9, whereas one to three PvuII sites were found for B19 strains. On the basis of its restriction map, V9 cannot be assigned to one of the five B19 genotypes described by Mori et al. (15), nor does it seem to be related to any of the B19 strains unassigned to a given genotype. Thus, it is quite likely that the sequence divergence between V9 and B19 is not restricted to the VP1u region but extends along the whole genome.
FIG. 4.
Restriction map of V9 genome. Restriction enzymes were used to cut V9 DNA. Lane 2, uncut DNA; lane 3, BamHI-digested DNA; lane 4, HindIII-digested DNA; lane 5, PvuII-digested DNA. DNA molecular size markers were as follows: lane 1, 1-kb ladder; lane 6, 100-bp ladder (Life Technologies, Cergy Pontoise, France). Lane 7, uncut B19 DNA.
DISCUSSION
This case report demonstrates that human erythrovirus genomic sequences may be much more divergent than has so far been accepted. Sequence divergence in the VP1u region can be due to antigenic drift within a patient during persistent B19 infection due to selective pressure by the immune system over a long period of viral replication (10). However, with regard to the V9 virus, such a mechanism is unlikely for two reasons: (i) V9 was found during an acute infection and (ii) when compared to the B19 sequence (22), the DNA mutations did not systematically result in nonconservative amino acid substitutions, as has been shown for persistent B19 infections (10). Furthermore, contrary to what was described for persistent B19 infections (10), the genetic divergence between V9 and B19 seems to extend beyond the VP1u region, as indicated by the V9 restriction map. Such a high level of divergence raises numerous questions which must be addressed.
First, this high level of divergence can affect the diagnosis of erythrovirus infection either by B19 PCR and DNA hybridization assays or by B19 serological assays. The results of the B19 PCR test that we used were indeed inconclusive, and the results of B19 IgM serological assay were negative, while this patient’s clinical and biological presentations suggested an acute human erythrovirus infection, for which both tests are usually positive (7, 12). Diagnostic tests specific for V9 DNA and antibody detection are being developed to address this point. Using these tools, large-scale epidemiological studies will be conducted in order to evaluate the medical importance of V9-related erythroviruses. The finding that such viruses are widespread would require the virological diagnosis of erythrovirus infection to be changed, thereby extending the etiological role of the erythroviruses in human pathology.
Second, the initial B19 IgG serological assay positivity could indicate a prior B19 infection or, alternatively, an (early) cross-reactivity to the B19 serological assay. If the former hypothesis is true, it appears (at least for this patient) that the acquired immunity against B19 could not prevent the occurrence of an acute infection with variant erythrovirus V9. Because a previous B19 infection usually confers strong and durable protective immunity against reinfection with B19 (12), this case questions whether anti-B19 antibodies ensure cross-protective immunity against V9. Measurements of VP1 IgG avidity (23) or epitope type-specific IgG reactivity to VP2 (24) should indicate whether this patient had preexisting B19 immunity. Should large epidemiological studies confirm this supposition, it will be necessary to include V9 proteins in the development of any candidate human erythrovirus vaccine (3, 12, 20). It should be noted that in the VP1u region most of the divergence between V9 and B19 resides in the 5′ portion (Fig. 2), which corresponds to one of the two major neutralization epitopes (2, 21).
Third, the taxonomic position of erythrovirus variant V9 as a new genotype in the B19 species or a new species in the genus Erythrovirus remains to be established. We favor the latter hypothesis for two reasons: the V9 genome restriction pattern, which differs from those of unassigned and established B19 genotypes reported worldwide (15, 16, 26), and the genetic divergence between V9 and B19s. The results of phylogenetic analysis with V9, R1, and B19 sequences reinforce the hypothesis that V9 and R1 could belong to another species besides the species B19 in the genus Erythrovirus. All B19 sequences group in a compact cluster (≤6.65% divergence), while the V9 and R1 sequences clearly segregate from this group (with divergences of ≥11.07 and ≥12.14%, respectively) and seem to belong to a second cluster (5.20% divergence between V9 and R1). This hypothesis must be confirmed by phylogenetic analysis with both a wider portion of erythrovirus genome and a larger number of variant erythrovirus sequences. Cloning and sequencing of the whole V9 viral genome, which is already in progress, may help to fully elucidate its taxonomic position.
Fourth, because erythrovirus variant V9 was found during a TAC in a child and this clinical presentation is typical of a B19 infection, some questions arise: Why was V9 not found earlier? Is it a new variant (or a new virus) that segregated very recently from B19 or, more simply, is it an ancient human virus that has not been detected until now because diagnostic tests (serology, DNA hybridization, PCR) were designed for B19 and are not suitable for the detection of V9? Epidemiological studies with V9-specific assays may help to provide an answer. If the prevalence of V9 is initially low and rises over the next few years, this may be a clue that V9 is an emerging human virus.
ACKNOWLEDGMENTS
We thank Eric Osika, Charles Roth, and Janet Jacobson for critical reading of the manuscript.
This work was supported in part by FRM grant 4001524-01 and DRC AP-HP grant TBI 97029.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Astell C R, Luo W, Brunstein J, St. Amand J. B19 parvovirus: biochemical and molecular features. In: Anderson L J, Young N S, editors. Human parvovirus B19. New York, N.Y: Karger; 1997. pp. 16–41. [Google Scholar]
- 3.Bansal G P, Hatfield J A, Dunn F E, Kramer A A, Brady F, Riggin C H, Collett M S, Yoshimoto K, Kajigaya S, Young N S. Candidate recombinant vaccine for human B19 parvovirus. J Infect Dis. 1993;167:1034–1044. doi: 10.1093/infdis/167.5.1034. [DOI] [PubMed] [Google Scholar]
- 4.Blundell M C, Beard C, Astell C R. In vitro identification of a B19 parvovirus promoter. Virology. 1987;157:534–538. doi: 10.1016/0042-6822(87)90296-0. [DOI] [PubMed] [Google Scholar]
- 5.Brown K E. Human parvovirus B19 epidemiology and clinical manifestations. In: Anderson L J, Young N S, editors. Human parvovirus B19. New York, N.Y: Karger; 1997. pp. 42–59. [Google Scholar]
- 6.Brown K E, Green S W, O’Sullivan M G, Young N S. Cloning and sequencing of the simian parvovirus genome. Virology. 1995;210:314–322. doi: 10.1006/viro.1995.1348. [DOI] [PubMed] [Google Scholar]
- 7.Brown K E, Young N S. Human parvovirus B19: pathogenesis of disease. In: Anderson L J, Young N S, editors. Human parvovirus B19. New York, N.Y: Karger; 1997. pp. 105–119. [Google Scholar]
- 8.Erdman D D, Durigon E L, Wang Q Y, Anderson L J. Genetic diversity of human parvovirus B19: sequence analysis of the VP1/VP2 gene from multiple isolates. J Gen Virol. 1996;77:2767–2774. doi: 10.1099/0022-1317-77-11-2767. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 10.Hemauer A, von Poblotzki A, Gigler A, Cassinotti P, Siegl G, Wolf H, Modrow S. Sequence variability among different parvovirus B19 isolates. J Gen Virol. 1996;77:1781–1785. doi: 10.1099/0022-1317-77-8-1781. [DOI] [PubMed] [Google Scholar]
- 11.Hicks K E, Cubel R C, Cohen B J, Clewley J P. Sequence analysis of a parvovirus B19 isolate and baculovirus expression of the non-structural protein. Arch Virol. 1996;141:1319–1327. doi: 10.1007/BF01718833. [DOI] [PubMed] [Google Scholar]
- 12.Kajigaya S, Momoeda M. Immune response to B19 infection. In: Anderson L J, Young N S, editors. Human parvovirus B19. New York, N.Y: Karger; 1997. pp. 120–136. [Google Scholar]
- 13.Lefrere J J, Mariotti M, De la Croix I, Lerable J, Thauvin M, Burnouf T, Follea G. Albumin batches and B19 parvovirus DNA. Transfusion. 1995;35:389–391. doi: 10.1046/j.1537-2995.1995.35595259148.x. [DOI] [PubMed] [Google Scholar]
- 14.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori J, Beattie P, Melton D W, Cohen B J, Clewley J P. Structure and mapping of the DNA of human parvovirus B19. J Gen Virol. 1987;68:2797–2806. doi: 10.1099/0022-1317-68-11-2797. [DOI] [PubMed] [Google Scholar]
- 16.Morinet F, Tratschin J D, Perol Y, Siegl G. Comparison of 17 isolates of the human parvovirus B19 by restriction enzyme analysis. Arch Virol. 1986;90:165–172. doi: 10.1007/BF01314155. [DOI] [PubMed] [Google Scholar]
- 17.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Virus taxonomy—classification and nomenclature of viruses. Arch Virol. 1995;10(Suppl.):1–530. [Google Scholar]
- 18.Nguyen Q T, Sifer C, Schneider V, Bernaudin F, Auguste V, Garbarg-Chenon A. Detection of an erythrovirus sequence distinct from B19 in a child with acute anemia. Lancet. 1998;352:1524. doi: 10.1016/S0140-6736(05)60330-3. [DOI] [PubMed] [Google Scholar]
- 19.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenfeld S J, Yoshimoto K, Kajigaya S, Anderson S, Young N S, Field A, Warrener P, Bansal G, Collett M S. Unique region of the minor capsid protein of human parvovirus B19 is exposed on the virion surface. J Clin Invest. 1992;89:2023–2029. doi: 10.1172/JCI115812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saikawa T, Anderson S, Momoeda M, Kajigaya S, Young N S. Neutralizing linear epitopes of B19 parvovirus cluster in the VP1 unique and VP1-VP2 junction regions. J Virol. 1993;67:3004–3009. doi: 10.1128/jvi.67.6.3004-3009.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shade R O, Blundell M C, Cotmore S F, Tattersall P, Astell C R. Nucleotide sequence and genome organization of human parvovirus B19 isolated from the serum of a child during aplastic crisis. J Virol. 1986;58:921–936. doi: 10.1128/jvi.58.3.921-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soderlund M, Brown C S, Cohen B J, Hedman K. Accurate serodiagnosis of B19 parvovirus infections by measurement of IgG avidity. J Infect Dis. 1995;171:710–713. doi: 10.1093/infdis/171.3.710. [DOI] [PubMed] [Google Scholar]
- 24.Soderlund M, Brown C S, Spaan W J, Hedman L, Hedman K. Epitope type-specific IgG responses to capsid proteins VP1 and VP2 of human parvovirus B19. J Infect Dis. 1995;172:1431–1436. doi: 10.1093/infdis/172.6.1431. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalities and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umene K, Nunoue T. The genome type of human parvovirus B19 strains isolated in Japan during 1981 differs from types detected in 1986 to 1987: a correlation between genome type and prevalence. J Gen Virol. 1990;71:983–986. doi: 10.1099/0022-1317-71-4-983. [DOI] [PubMed] [Google Scholar]
- 27.Young N S. Parvoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2190–2220. [Google Scholar]