Abstract
In vivo cell fate conversions have emerged as potential regeneration-based therapeutics for injury and disease. Recent studies reported that ectopic expression or knockdown of certain factors can convert resident astrocytes into functional neurons with high efficiency, region-specificity and precise connectivity. However, using stringent lineage tracing in the mouse brain, we show that the presumed astrocyte-converted neurons are actually endogenous neurons. AAV-mediated co-expression of NEUROD1 and a reporter specifically and efficiently induces reporter-labeled neurons. However, these neurons cannot be traced retrospectively to quiescent or reactive astrocytes using lineage-mapping strategies. Instead, through a retrograde labeling approach, our results reveal that endogenous neurons are the source for these viral reporter-labeled neurons. Similarly, despite efficient knockdown of PTBP1 in vivo genetically traced resident astrocytes were not converted into neurons. Together, our results highlight the requirement of lineage-tracing strategies, which should be broadly applied to studies of cell fate conversions in vivo.
Keywords: in vivo reprogramming, astrocyte-to-neuron conversion, lineage tracing, AAV, NEUROD1, PTBP1, DLX2, PAX6, CRISPR-CasRx, shRNA
Graphical Abstract
In Brief
Stringent lineage tracings reveal that the presumed astrocyte-converted neurons are not originated from the resident astrocytes but from the AAV-infected endogenous neurons.
INTRODUCTION
Neural injury or degeneration often leads to neuron loss and impaired neurocircuits. Regeneration of neurons could constitute an ideal therapeutic strategy since they will be immunocompatible with the host. Except for a few neurogenic niches (Zhao et al., 2008), however, the adult mammalian brain or spinal cord lacks intrinsic capacity to produce new neurons.
In vivo conversion of resident glial cells to functional new neurons emerges as a promising strategy for neural regeneration (Barker et al., 2018; Chen et al., 2015; Tai et al., 2020; Wang and Zhang, 2018). This is largely accomplished by ectopic expression of a single or a combination of fate-determining factors or through knockdown of a single gene (Grande et al., 2013; Guo et al., 2014; Liu et al., 2015; Matsuda et al., 2019; Mattugini et al., 2019; Niu et al., 2013; Qian et al., 2020; Tai et al., 2021; Torper et al., 2015; Zhou et al., 2020). Resident glial cells are ideal cell sources for fate-conversions since they become reactive and can proliferate to replenish themselves in response to injury or degeneration. While SOX2-mediated reprogramming passes through an expandable progenitor state resembling endogenous neurogenesis (Niu et al., 2015; Niu et al., 2013; Su et al., 2014a; Tai et al., 2021; Wang et al., 2016), the glia-to-neuron conversions by other strategies are direct without a proliferative intermediate (Liu et al., 2020; Liu et al., 2015; Matsuda et al., 2019; Mattugini et al., 2019; Pereira et al., 2017; Qian et al., 2020; Torper et al., 2015; Wu et al., 2020; Zhou et al., 2020). Employing the AAV-mediated gene delivery, recent studies further show that resident glia can be directly and efficiently converted into mature neurons with brain region-specificity and precise connectivity (Liu et al., 2020; Liu et al., 2015; Matsuda et al., 2019; Mattugini et al., 2019; Pereira et al., 2017; Qian et al., 2020; Torper et al., 2015; Wu et al., 2020; Zhou et al., 2020). Importantly, direct conversion of astrocytes shows therapeutic benefits under pathological conditions (Chen et al., 2020; Qian et al., 2020; Wu et al., 2020; Zhou et al., 2020). Such results, if confirmed, will revolutionize regenerative medicine.
During the past decade, our laboratory has conducted a series of in vivo screens for glia reprogramming (Islam et al., 2015; Niu et al., 2015; Niu et al., 2018; Niu et al., 2013; Su et al., 2014a; Tai et al., 2021; Wang et al., 2016). We noticed that endogenous neurons could have been misidentified as the glia-converted if not verified by stringent lineage-tracing methods. In this study, we reexamined direct astrocyte-to-neuron (AtN) conversion by employing multiple lineage-tracing strategies. Our results do not support those phenomenal claims that resident astrocytes can be directly and efficiently converted into mature neurons.
RESULTS
Targeting brain astrocytes through AAVs.
We compared astrocyte-specificity of the commonly used AAV serotypes: AAV2, AAV5, AAV8, AAV9, and PHP.eB (Brulet et al., 2017; Liu et al., 2020; Liu et al., 2015; Pereira et al., 2017; Qian et al., 2020; Torper et al., 2015; Zhou et al., 2020). To drive gene expression, we used the synthetic 681-bp gfaABC1D human GFAP promoter (hGFAP) (Lee et al., 2008), which exhibits astrocyte-restricted activity as the 2.2-kb gfa2 promoter. Cell type-specificity was examined by the GFP reporter after intracerebral AAV injections. When examined at 4 days post virus injection (dpv), GFP was predominantly detected in GFAP+ cortical astrocytes for most of these AAVs (Fig. S1A-C; 97.0 ±1.2 % for AAV2, 92.1 ± 1.4% for AAV5, 97.6 ± 1.2% for AAV8, and 94.1 ± 0.9% for AAV9). The number of GFP+ neurons was minimal (Fig. S1A-C). However, cell type-specificity was dramatically different at 14 dpv (Fig. S1C-E). Only 62.4 ± 8.0% and 69.2 ± 8.0% of the virus-transduced cells were GFAP+ astrocytes for AAV8 and AAV9, respectively, whereas the neuronal fractions were correspondingly increased. Furthermore, both AAV8 and AAV9 showed spatial preferences with predominant neurons or astrocytes in different cortical regions (Fig. S1E). AAV.PHP.eB, on the other hand, showed a similar targeting efficiency for neurons (34.7 ± 1.5% at 4 dpv and 51.8 ± 1.7% at 14 dpv) and astrocytes (64.8 ± 1.4% for 4 dpv and 47.9 ± 1.6% for 14 dpv) (Fig. S1B, C, E). In contrast, the cell type-specificity of AAV2 and AAV5 remained rather stable with 98.6 ± 1.3% and 90.4 ± 8.0% of the GFP reporter in astrocytes, respectively (Fig. S1B-D). Since AAV2 infected fewer cells when compared to AAV5 with the same virus titer, AAV5 was selected for most of the subsequent experiments.
Highly efficient in vivo AtN conversion by NEUROD1?
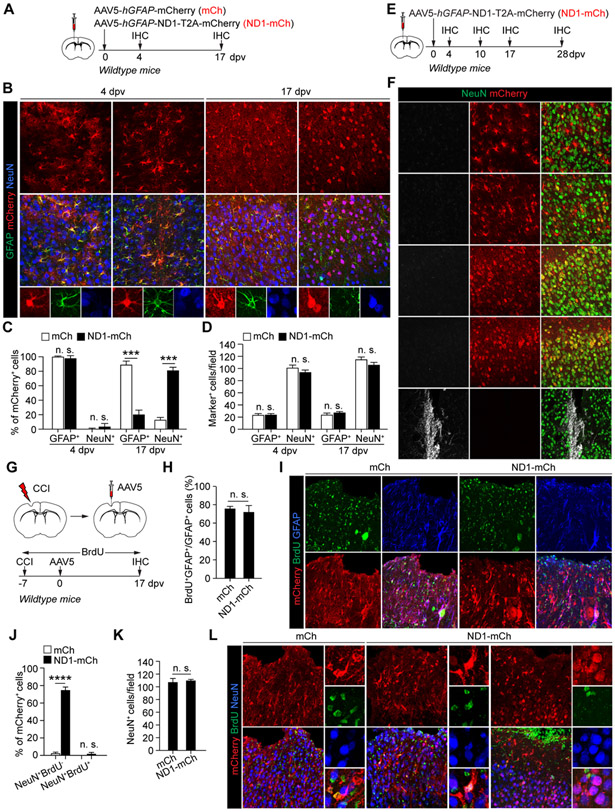
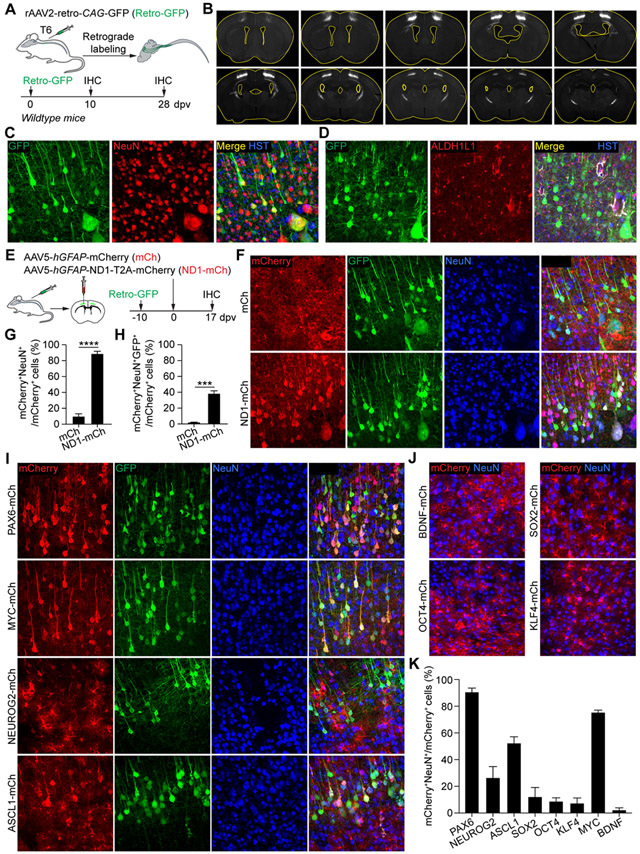
We injected hGFAP-mCherry (mCh) or hGFAP-NEUROD1-T2A-mCherry (ND1-mCh) AAV5 virus into the adult mouse cortex (Fig. 1A). At 4 dpv, the reporter mCherry was highly restricted to GFAP+ astrocytes (Fig. 1B, C; 99.9 ± 0.1% for mCh and 96.4 ± 3.6% for ND1-mCh). Remarkably, at 17 dpv, 79.8 ± 4.4% of mCherry+ cells were NeuN+ neurons in the cortex injected with the ND1-mCh virus (Fig. 1B, C). This was in sharp contrast to the control mCh virus-injected mice, in which 87.7 ± 5.0% of mCherry+ cells were still astrocytes. Such a result was largely consistent with what was reported on NEUROD1 (Chen et al., 2020; Liu et al., 2020; Wu et al., 2020). Notwithstanding such abundant mCherry+ neurons in the ND1-mCh group, we failed to observe significant changes on the density of either neurons or astrocytes in the virus-injected cortex (Fig. 1D). However, it should be noted that proliferative astrocytes might have replenished those lost. On the other hand, a time-course analysis of the ND1-mCh virus-injected brains failed to show cells expressing doublecortin (DCX; Fig. 1E, F), a well-established marker for immature neurons (Couillard-Despres et al., 2005). Together, these results indicate that, despite robust induction of reporter-positive neurons by ND1-mCh, these neurons do not pass through an immature stage and do not lead to an overall increase of neuronal density in the virus-injected brain areas.
Figure 1. NEUROD1-induced reporter+ neurons neither pass through an immature stage nor come from reactive astrocytes.
(A) Study design to examine AtN conversion. IHC, immunohistochemistry; dpv, days post virus-injection.
(B) Confocal images of the indicated markers. Scales, 50 μm.
(C) Efficient induction of mCherry+ neurons by NEUROD1 (n = 3-4 mice per group per time-point; mean ± SEM; ***p<0.001; n.s., not significant).
(D) Cell numbers in virus-injected cortex (n = 3-4 mice per group per time-point; mean ± SEM; n.s., not significant).
(E) Study design to examine immature neurons.
(F) Confocal images showing a lack of DCX+ cells in virus-injected cortex. Cells in the lateral ventricle (LV) are used as controls. Scales, 50 μm.
(G) Study design to trace reactive cells. BrdU was delivered through drinking water. CCI, controlled cortical impact.
(H) Robust BrdU-labeling of reactive astrocytes (n = 4 mice per group; mean ± SEM; n.s., not significant).
(I) Confocal images of BrdU-labeled cells. Scales, 50 μm.
(J) Quantification of mCherry+ cells (n = 4 mice per group; mean ± SEM; ****p < 0.0001; n.s., not significant).
(K) Quantification of neurons in virus-injected cortex (n = 4 mice per group; mean ± SEM; n.s., not significant).
(L) Confocal images of the indicated markers. A BrdU+ astrocyte-like NeuN+ cell is shown in ND1-mCh group. Scales, 50 μm.
Reactive astrocytes are not an origin for NEUROD1-induced reporter-positive neurons.
A therapeutic promise for in vivo reprogramming is to convert reactive astrocytes into neurons under pathological conditions. We subjected adult mice to brain injury through controlled cortical impact (CCI) (Chen et al., 2019). Immediately following CCI, these mice were continuously administered with BrdU in drinking water to label proliferating cells including reactive astrocytes. Seven days later, we injected either mCh or ND1-mCh virus into the penumbra of the injured cortex and performed analyses at 17 dpv (Fig. 1G). Over 72% of GFAP+ cells were BrdU+, indicating robust labeling of reactive astrocytes (Fig. 1H, I). Under this injury condition, 74.0 ± 3.9% of mCherry+ cells in the ND1-mCh group were NeuN+, comparing to 2.3 ± 1.7% in the control mCh group (Fig. 1J, L). However, only 2.0 ± 1.5% of mCherry+ cells were BrdU+NeuN+ in the ND1-mCh group (Fig. 1J, L), indicating a very low BrdU-labeling efficiency. Of note, many of these BrdU+NeuN+ cells exhibited a glia-like morphology with small nuclei, implicating induction of NeuN expression in glial cells (Fig. 1L). Despite such a high ratio of mCherry+ cells being NeuN+ in the ND1-mCh group, we failed to observe a significant increase of neuronal density surrounding the injury penumbra (Fig. 1K). These results indicate that reactive astrocytes do not contribute to NEUROD1-induced mCherry+ neurons.
Genetically traced astrocytes are not an origin for NEUROD1-induced reporter-positive neurons.
To follow the fate of astrocytes, we employed the tamoxifen-inducible Aldh1l1-CreERT2 mouse (Srinivasan et al., 2016). We confirmed cell type-specificity and labeling efficiency by crossing this line to the R26R-YFP reporter (Fig. S2A). After tamoxifen treatments, 95.7 ± 2.6% and 99.9 ± 0% of YFP+ cells expressed the astrocyte marker ALDH1L1 and ALDOC, respectively (Fig. S2B-E). Only 4.3 ± 2.6% of YFP+ cells were NeuN+ (Srinivasan et al., 2016). Conversely, 96.5 ± 1.8% of ALDH1L1+ cells and 94.1 ± 2.2% of ALDOC+ cells could be traced by YFP (Fig. S2C-E), showing highly efficient tracing of resident astrocytes.
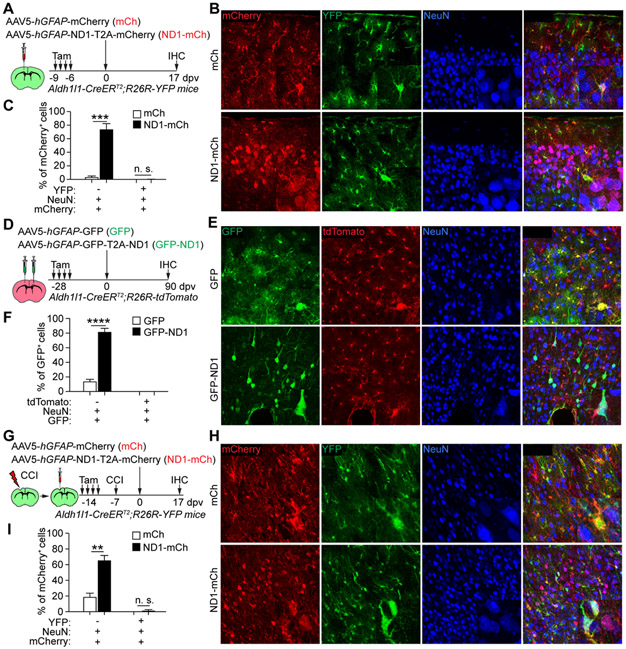
We then injected either mCh or ND1-mCh AAV5 virus into the cortex of tamoxifen-treated adult Aldh1l1-CreERT2;R26R-YFP mouse (Fig. 2A). At 17 dpv, 73.3 ± 8.9% of mCherry+ cells were NeuN+ neurons in the ND1-mCh group (Fig. 2B, C), consistent with the results in wildtype mice (Fig. 1B, C). However, these mCherry+NeuN+ cells were not labeled with YFP indicating a non-astrocyte origin (Fig. 2C).
Figure 2. Genetically traced astrocytes are not an origin for NEUROD1-induced reporter+ neurons.
(A) Study design to trace resident astrocytes with the YFP reporter. Tam, tamoxifen.
(B) Confocal images of YFP-traced cells. Scales, 50 μm.
(C) Quantifications of mCherry+ cells (n = 3-4 mice per group; mean ± SEM; ***p = 0.0003; n.s., not significant).
(D) Study design to trace resident astrocytes with the tdTomato reporter.
(E) Confocal images of tdTomato-traced cells. Scales, 50 μm.
(F) Quantifications of GFP+ cells (n = 4 mice per group; mean ± SEM; ****p < 0.0001; n.s., not significant).
(G) Study design to examine the effect of brain injury on AtN conversion.
(H) Confocal images of YFP-traced cells. Scales, 50 μm.
(I) Quantifications of mCherry+ cells after CCI (n = 4 mice per group; mean ± SEM; **p = 0.0022; n.s., not significant).
See also Figure S2.
As controls, we also examined AtN conversion in adult Aldh1l1-CreERT2;R26R-tdTomato mice (Xiang et al., 2021). Tamoxifen treatments induced efficient labeling of resident astrocytes by tdTomato (Fig. S2J-N). Nonetheless, it should be noted that in rare cases endogenous neurons could also be traced in various brain regions (Fig. S2O, P). These rare mice could be excluded by examining tdTomato+ neurons in regions that were not virus-injected during immunohistological analyses. Tamoxifen-treated Aldh1l1-CreERT2;R26R-tdTomato mice were intracerebrally injected with hGFAP-GFP (GFP) or hGFAP-GFP-T2A-NEUROD1 (GFP-ND1) virus (Fig. 2D). At 90 dpv, GFP+ cells were predominantly NeuN+ neurons in the GFP-ND1 group (81.0 ± 5.2% for GFP-ND1 vs. 13.4 ± 3.8% for GFP control; Fig. 2E, F). However, these GFP+NeuN+ neurons were not traced with tdTomato indicating a non-astrocyte origin (Fig. 2E, F).
Brain injury fails to facilitate NEUROD1 to convert traced astrocytes.
The environmental milieu may account for the failure of detecting AtN conversion by NEUROD1 in Aldh1l1-CreERT2;R26R-YFP mice, since injury may precondition resident glial cells for fate reprogramming (Grande et al., 2013; Heinrich et al., 2014). We examined this possibility by subjecting tamoxifen-treated tracing mice to CCI, followed by injections of either mCh or ND1-mCh virus near the injury penumbra (Fig. 2G). mCherry+NeuN+ cells could be efficiently induced in the ND1-mCh group (Fig. 2H, I; 64.9 ± 7.2% for ND1-mCh vs. 18.5 ± 5.4% for mCh); however, very few of them were YFP+ (Fig. 2H, I; 1.4 ± 1.4% for ND1-mCh). These mCherry+NeuN+YFP+ cells might be a result of variable labeling background, since 4.3 ± 2.6% of endogenous neurons could be genetically traced in Aldh1l1-CreERT2 mice (Srinivasan et al., 2016) (Fig. S2C, D).
The AAV-based Cre-FLEX system fails to restrict gene expression in astrocytes.
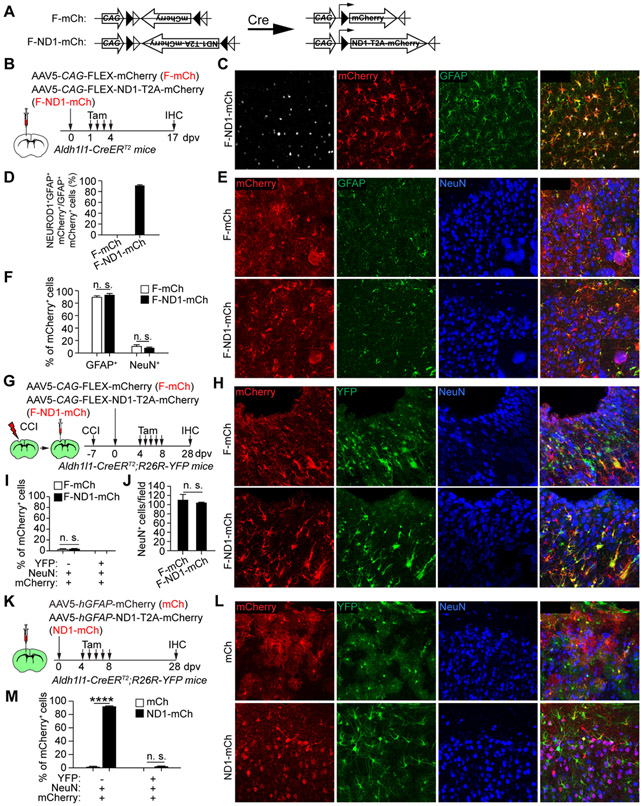
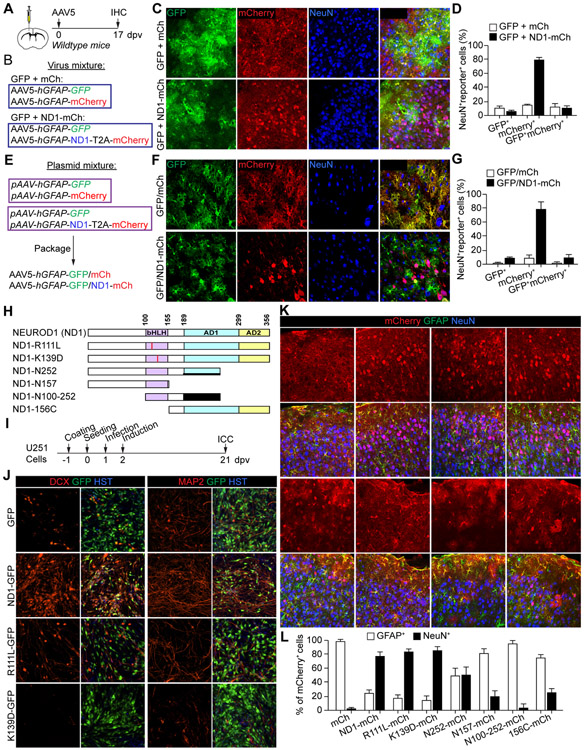
The AAV-based Cre-FLEX system was previously used for AtN conversion (Chen et al., 2020; Wu et al., 2020). It employed two AAV vectors (Fig. 3A, S3A). One used hGFAP-Cre to target astrocytes (Fig. S3A); and the other, CAG-FLEX-mCherry (F-mCh) or CAG-FLEX-NEUROD1-T2A-mCherry (F-ND1-mCh), expressed the genes in a Cre-dependent manner under the constitutively active CAG promoter (Fig. 3A). We examined cell type specificity of the hGFAP-Cre with the commonly used serotypes. When examined at 4 or 14 dpv, both AAV8 and AAV9 serotypes showed Cre expression in NeuN+ neurons (Fig. S3C-E). The AAV5-hGFAP-Cre showed astrocyte-restricted expression at 4 dpv; however, neurons became the predominant cell type at 14 dpv (Fig. S3C-E). We also examined AAV5.GFAP.Cre.WPRE.hGH (referred as AAV5-hGFAP*-Cre), in which Cre was driven by the 2.2 kb gfa2 hGFAP promoter (Fig. S3B). It showed a similar pattern with astrocytes and neurons as the predominant cells at 4 and 14 dpv, respectively (Fig. S3C-E). Despite these results, we co-injected hGFAP-Cre with either F-mCh or F-ND1-mCh AAV5 virus into the adult cortex. mCherry+ neurons were observed at 17 dpv in all groups, though the F-ND1-mCh group showed less within-group variations (Fig. S3F-H; 88.6 ± 3.5% for F-ND1-mCh vs. 61.8 ± 12.0% for F-mCh). Thus, these results indicate that the dual AAV-based Cre-FLEX system lacks the required cell type-specificity to trace AtN conversions.
Figure 3. Astrocyte-restricted NEUROD1 cannot rapidly induce reporter+ neurons.
(A) A schematic illustration of the Cre-FLEX system.
(B) Study design to examine AtN conversion through the Cre-FLEX system.
(C) Confocal images of the indicated markers. Scale, 50 μm.
(D) Quantifications showing robust NEUROD1 expression in astrocytes (n = 4 mice per group; mean ± SEM).
(E) Confocal images of the indicated markers. Scales, 50 μm.
(F) Quantifications showing a basal number of mCherry+ neurons (n = 4 mice per group; mean ± SEM; n.s., not significant).
(G) Study design to examine AtN conversion from reactive cells.
(H) Confocal images of the indicated markers. Scales, 50 μm.
(I) Quantifications showing a basal number of mCherry+ neurons (n = 4 mice per group; mean ± SEM; n.s., not significant).
(J) Quantification of neurons in virus-injected cortex (n = 3-4 mice per group; mean ± SEM; n.s., not significant).
(K) Study design to examine the effect of tamoxifen on induction of mCherry+ neurons.
(L) Confocal images of the indicated markers. Scales, 50 μm.
(M) Quantifications of mCherry+ cells (n = 4 mice per group; mean ± SEM; ****p < 0.0001; n.s., not significant).
See also Figures S3 and S4.
Astrocyte-restricted NEUROD1 fails to induce AtN conversion.
To restrict NEUROD1 in astrocytes, we employed the tamoxifen-inducible Aldh1l1-CreERT2 mice. They were injected with F-mCh or F-ND1-mCh AAV5 virus, treated with tamoxifen, and analyzed at 17 dpv (Fig. 3A, B). Immunostaining showed robust NEUROD1 expression in the F-ND1-mCh group (Fig. 3C, D). Unexpectedly, the reporter mCherry was mainly detected in GFAP+ astrocytes (Fig. 3E, F; 92.8 ± 6.1% for F-ND1-mCh vs. 87.8 ± 6.4% for F-mCh), indicating that mCherry+ neurons cannot be rapidly induced by NEUROD1 when its expression is restricted to adult astrocytes.
To examine whether injury might facilitate astrocyte-restricted NEUROD1 to induce AtN conversion, adult Aldh1l1-CreERT2;R26R-YFP mice were subjected to CCI, injected with F-mCh or F-ND1-mCh virus, and treated with tamoxifen (Fig. 3G). A few mCherry+ NeuN+ cells were detected at 28 dpv in both groups and none of them were YFP+ (Fig. 3H, I). Neuronal density was also similar between groups (Fig. 3J). These results indicate that astrocyte-restricted NEUROD1 is incapable of inducing AtN conversion even after brain injury.
To determine whether a longer time was required for NEUROD1 to induce AtN conversion, adult Aldh1l1-CreERT2;R26R-YFP mice were injected with virus, treated with tamoxifen, and examined at 98 dpv (Fig. S4A, B). mCherry+ cells in the control F-mCh group were largely NeuN− (Fig. S4C). Interestingly, the phenotypes of mCherry+ cells in the F-ND1-mCh group were variable: in some areas they were predominantly NeuN− whereas in other areas they were largely NeuN+ (Fig. S4C). Notwithstanding, these mCherry+NeuN+ cells were not traced with YFP indicating a non-astrocyte origin.
Neither tamoxifen nor genetic background affects NEUROD1’s ability to induce reporter-positive neurons.
An alternative explanation for these above results was that tamoxifen might have abolished the reprogramming ability of NEUROD1. However, this was unlikely the case. As shown in Fig. 2, tamoxifen treatment prior to virus injection had no effect on the induction of mCherry+ neurons by NEUROD1 in Aldh1l1-CreERT2;R26R-YFP mice, although these neurons were not traced with YFP (Fig. 2A-C, G-I). We also examined a delayed treatment scheme, in which tamoxifen was administered following the virus injections (Fig. 3K). 92.0 ± 1.1% of mCherry+ cells were NeuN+ neurons at 28 dpv in mice injected with the ND1-mCh AAV5 virus; nonetheless, only a low background level of YFP+ neurons was observed (Fig. 3L, M). Together, these results show that neither tamoxifen-treatment nor the transgenic mouse background blocks the ability of NEUROD1 to induce mCherry+NeuN+ neurons.
NEUROD1 and DLX2 fail to reprogram genetically traced striatal astrocytes.
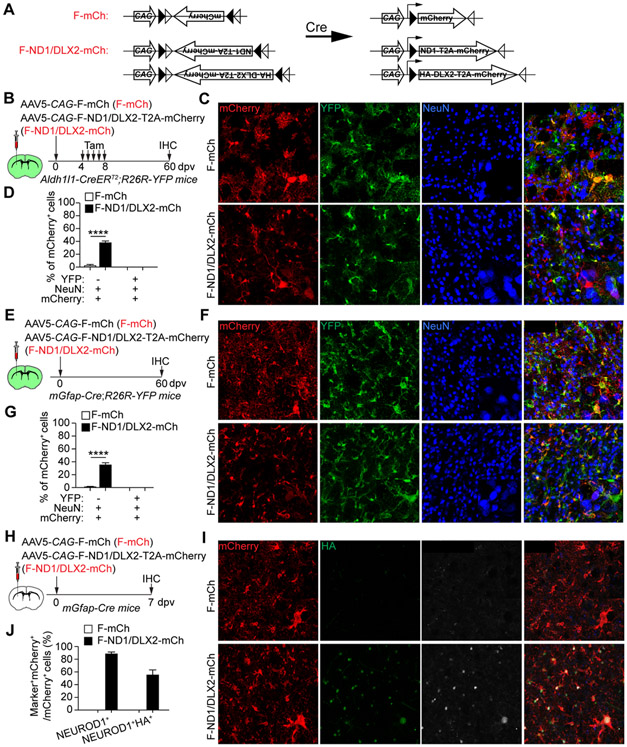
A recent study showed that co-expression of NEUROD1 and DLX2 can efficiently convert astrocytes into local neurons in the adult mouse striatum (Wu et al., 2020), suggesting that the reprogramming ability of NEUROD1 might be enhanced by DLX2. To test such a possibility, we injected the Cre-dependent F-mCh AAV5 virus or a combination of F-ND1-mCh and CAG-FLEX-HA-DLX2-mCh (F-ND1/DLX2-mCh) into the striatum of adult Aldh1l1-CreERT2;R26R-YFP mouse. These mice were then treated with tamoxifen at 4 dpv and examined at 60 dpv (Fig. 4B). A larger number of mCherry+NeuN+ neurons was indeed observed in the F-ND1/DLX2-mCh group (38.3 ± 2.7 % for F-ND1/DLX2-mCh vs. 2.7 ± 1.2% for F-mCh; Fig. 4C, D); however, they were not traced with YFP indicating a non-astrocyte origin.
Figure 4. A combination of NEUROD1 and DLX2 fails to convert striatal astrocytes.
(A) A schematic illustration of the Cre-FLEX system.
(B) Study design to examine AtN conversion in the striatum. Astrocytes were traced with YFP after Tamoxifen treatments.
(C) Confocal images of the indicated markers. Scales, 50 μm.
(D) Quantifications showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; ****p < 0.0001).
(E) Study design to examine AtN conversion in the striatum. Astrocytes were constitutively traced with YFP.
(F) Confocal images of the indicated markers. Scales, 50 μm.
(G) Quantifications showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; ****p < 0.0001).
(H) Study design to examine NEUROD1 and DLX2 expression.
(I) Confocal images of the indicated markers. DLX2 expression is indicated by the HA staining. Scales, 50 μm.
(J) Quantifications showing robust expression of NEUROD1 and DLX2 (indicated by HA) in mCherry+ cells (n = 3 mice per group; mean ± SEM).
See also Figure S5.
To verify these above results, we also employed mGfap-Cre;R26R-YFP mice (Gregorian et al., 2009). Under normal condition, YFP+NeuN+ neurons were observed in various brain regions including the cortex and midbrain (Fig. S5A, B). In contrasts, striatal astrocytes were specifically and highly efficiently traced with the reporter YFP (Fig. S5C-F). 95.8 ± 1.2% and 95.9 ± 1.5% of YFP+ cells were ALDH1L1+ and ALDOC+ astrocytes, respectively (Fig. S5D, E). Conversely, 88.5 ± 3.9% of ALDH1L1+ and 87.0 ± 4.1% of ALDOC+ striatal astrocytes were traced with YFP indicating highly efficient labeling (Fig. S5D, F). We then injected F-mCh or F-ND1/DLX2-mCh AAV5 viruses into the striatum of mGfap-Cre;R26R-YFP mouse (Fig. 4E). mCherry+NeuN+ neurons were observed at 60 dpv in the F-ND1/DLX2-mCh group (35.7 ± 2.4% for F-ND1/DLX2-mCh vs. 1.7 ± 0.9% for F-mCh; Fig, 4F, G). Once again, however, these neurons were not traced with YFP. The co-expression of Cre-dependent NEUROD1 and DLX2 (indicated by the HA-tag) was verified (Fig. 4H-J). Together, these above results clearly show that a combination of NEUROD1 and DLX2 fails to convert striatal astrocytes into neurons.
NEUROD1-induced reporter-positive neurons are endogenous neurons.
Our results with genetic lineage tracing and BrdU-labeling show that resident astrocytes, reactive or quiescent, are not a cell source for NEUROD1-induced mCherry+ neurons. We finally examined whether they were actually endogenous neurons. We first labeled cortical neurons through the rAAV2-retro virus, an engineered AAV2 variant enabling retrograde access to projection neurons (Tervo et al., 2016). We injected the rAAV2-retro-CAG-GFP (Retro-GFP) virus to target the cortical spinal tract (CST) at the 6th thoracic spinal cord level (T6) (Fig. 5A). When examined at 10 or 28 dpv (Fig. 5A), a group of cortical motor neurons were retrogradely labeled with GFP (Fig. 5B, C). Importantly, GFP was not detected in any of the ALDH1L1+ or GFAP+ cortical astrocytes (Fig. 5D), showing stringent cell type-specificity of the retrograde-labeling method. Adult wildtype mice were then first injected with the Retro-GFP virus into the T6 CST, followed by cortical injections of mCh or ND1-mCh AAV5 virus (Fig. 5E). Many mCherry+ neurons were detected at 17 dpv in the ND1-mCh group (Fig. 5F, G). Of these, 37.3 ± 4.3% were also GFP+ surrounding the AAV5-injected cortex (Fig. 5H). This number was a huge underestimate of the contribution of endogenous neurons to NEUROD1-induced mCherry+NeuN+ cells, since only a fraction of endogenous motor neurons were traced by the retrograde GFP reporter. Together, these results show that endogenous neurons are the cell source for NEUROD1-induced mCherry+ neurons.
Figure 5. Virus-induced reporter+ neurons are endogenous neurons.
(A) Study design to label corticomotor neurons. T6, the 6th thoracic spinal cord level.
(B) Series of brain sections showing retrograde labeling of the motor cortex. Ventricles are outlined. Scale, 1 mm.
(C) Confocal images of the motor cortex showing GFP-traced neurons. Scales, 50 μm.
(D) Confocal images showing a complete lack of GFP-traced astrocytes in the motor cortex. Scale, 50 μm.
(E) Study design to determine the contribution of endogenous neurons to virus-induced mCherry+ neurons.
(F) Confocal images of the motor cortex showing reporter-labeled neurons. Arrowheads show examples of neurons with dual reporters. Scales, 50 μm.
(G) Quantifications showing robust induction of mCherry+ neurons by NEUROD1 in the cortex (n = 4-5 mice per group; mean ± SEM; ****p < 0.0001).
(H) Quantifications showing endogenous neurons as the cell source for NEUROD1-induced mCherry+ neurons (n = 4-5 mice per group; mean ± SEM; ***p = 0.0002).
(I) Confocal images showing reporter-labeled cortical neurons using study designs as shown in panel E. Arrowheads show examples of neurons with dual reporters. Scales, 50 μm.
(J) Confocal images showing examples of factors with minimal induction of mCherry+ neurons. Scales, 50 μm.
(K) Quantifications of the indicated factors for their ability to induce mCherry+ neurons (n = 4 mice per group; mean ± SEM).
Reporter-positive neurons can be induced by selective AAV-expressed factors.
Our results indicate that the cell type-specificity of AAV5 virus under the hGFAP promoter can be altered by the downstream NEUROD1. To determine how broadly applicable this phenomenon was, we examined additional factors, including PAX6, NEUROG2, ASCL1, SOX2, OCT4, KLF4, MYC, and BDNF. They were individually cloned into the pAAV-hGFAP-xxx-T2A-mCherry vector and packaged into AAV5 virus. As shown in Fig. 5E, adult mice were first injected with the Retro-GFP virus into the T6 CST, followed by intracerebral injections of hGFAP-xxx-T2A-mCherry AAV5 virus into the motor cortex. At 17 dpv, we observed 90.7 ± 3.6% and 76.2 ± 1.9% of mCherry+ neurons in brains injected with PAX6 and MYC viruses, respectively (Fig. 5I, K). ASCL1 and NEUROG2 also induced approximately 52.7 ± 5.4% and 27.3 ± 8.5% of mCherry+ neurons (Fig. 5I, K). Importantly, many of these mCherry+ neurons were also labeled with GFP indicating an origin of endogenous neurons (Fig. 5I, K). In contrast, only a background level of mCherry+ neurons were observed in brains injected with virus expressing SOX2, OCT4, KLF4, or BDNF (Fig. 5J, K). These results indicate that the cell type-specificity of AAV5 virus under the hGFAP promoter can be selectively altered by the downstream factors.
Promoter specificity is likely cis-regulated by NEUROD1.
How NEUROD1 induces reporter expression in endogenous neurons? One possibility is that NEUROD1 broadly changes the specificity of hGFAP promoter. We examined this through virus co-injections. We mixed and co-injected hGFAP-GFP AAV5 virus (GFP) with mCh or ND1-mCh AAV5 virus (Fig. 6A, B). If NEUROD1 ectopic expression can broadly upregulate hGFAP promoter, GFP expression from the co-injected hGFAP-GFP virus should accumulate in neurons. As previously shown, many mCherry+ neurons were specifically observed in the ND1-mCh group at 17 dpv (Fig. 6C, D). Unexpectedly, GFP+ neurons were rarely observed in brains co-injected with either mCh or ND1-mCh virus (Fig. 6C, D), suggesting that NEUROD1 might modulate in cis the specificity of hGFAP promoter.
Figure 6. NeuroD1 cis-regulates the viral promoter and does not require its neurogenic activity to induce reporter+ neurons.
(A) Study design to examine NEUROD1 on viral promoter activity.
(B) Study design to examine a mixture of individually packaged AAV5 viruses.
(C) Confocal images showing cell type-specificity of the indicated viral reporters. Scales, 50 μm.
(D) Quantifications showing neuronal expression of the cis-expressed mCherry but not the trans-expressed GFP (n = 3 mice per group; mean ± SEM).
(E) Study design to examine co-packaged AAV5 viruses.
(F) Confocal images showing cell type-specificity of the indicated viral reporters. Scales, 50 μm.
(G) Quantifications showing neuronal expression of the cis-expressed mCherry but not the trans-expressed GFP (n = 3 mice per group; mean ± SEM).
(H) A schematic of NEUROD1 and its mutants. bHLH, basic helix-loop-helix; AD1, activation domain 1; AD2, activation domain 2.
(I) Study design to examine neurogenic activity in U251 cells. ICC, immunocytochemistry.
(J) Confocal images showing a loss of neurogenic ability of NEUROD1 point-mutants (n = 4 biological replicates). DCX and MAP2 were used as markers of neurons. Scales, 50 μm.
(K) Confocal images of the indicated markers in the cortex at 17 dpv. Scales, 50 μm.
(L) Quantifications showing robust induction of mCherry+ neurons by NEUROD1 point-mutants (n = 3-4 mice per group; mean ± SEM).
Alternatively, the AAV5 serotype could be altered by NEUROD1 expression during virus packaging, such that the ND1-mCh AAV5 virus was prone to infecting neurons when compared to the control mCh AAV5 virus. To examine this possibility, we co-transfected an equal amount of AAV-hGFAP-GFP plasmid with either AAV-hGFAP-mCherry or AAV-hGFAP-ND1-mCherry plasmid during AAV5 packaging (Fig. 6E). If NEUROD1 expression somehow altered viral capsid during production, the co-packaged hGFAP-GFP should be similarly impacted. These co-packaged AAV5 viruses (GFP/mCh or GFP/ND1-mCh) were then injected into the brains of adult mice and examined at 17 dpv. Again, GFP expression was rarely detected in neurons in both groups, unlike mCherry which was predominantly in neurons in the GFP/ND1-mCh group (Fig. 6F, G). Together, these results indicate that NEUROD1 likely cis-regulates the specificity of hGFAP promoter for its expression in neurons.
Neurogenic activity of NEUROD1 is not required for induction of reporter-positive neurons.
To further examine the molecular mechanism, we made a few NEUROD1 mutants (Fig. 6H). The point mutations, R111L and K139D, were made in the basic helix-loop-helix (bHLH) domain to abolish DNA-binding (Malecki et al., 1999; Voronova and Baltimore, 1990). NEUROD1 and its point-mutants were cloned into the lentiviral CSC-IRES-GFP vector and their neurogenic activity was then determined in human U251 cells (Fig. 6I, J), as previously described (Liu et al., 2013; Su et al., 2014b; Wang et al., 2021). As expected, wildtype NEUROD1 efficiently induced U251 cells to become DCX+ or MAP2+ neurons by 21 dpv, whereas the point mutants lost such neurogenic activity (Fig. 6J). Of note, the K139D mutant behaved like a dominant-negative since it blocked basal neuronal induction observed in the GFP control. We then cloned all NEUROD1 mutants into the pAAV-hGFAP-xxx-T2A-mCherry vector and packaged them into AAV5 virus. These viruses were individually injected into the adult cortex and examined at 17 dpv. Whereas the truncation mutants showed a variable ratio of mCherry expression in NeuN+ neurons, NEUROD1 and its point mutants induced many mCherry+NeuN+ neurons (76.2± 5.5% for ND1, 82.2 ± 4.5% for ND1-R111L, and 84.2 ± 5.4% for ND1-K139D; Fig. 6K, L). These results indicate that efficient induction of mCherry+ neurons by NEUROD1 does not require its neurogenic activity.
AAV-GFAP-CasRx-Ptbp1 neither efficiently downregulates PTBP1 nor converts striatal astrocytes in vivo.
Our above results show that resident astrocytes cannot be converted to neurons by NEUROD1. How about the other factors? We examined PTBP1 since its downregulation was reported to efficiently convert striatal astrocytes into either dopaminergic (Zhou et al., 2020) or striatal neurons (Qian et al., 2020). We first employed the CRISPR-CasRx system (Zhou et al., 2020). AAV-GFAP-CasRx or AAV-GFAP-CasRx-Ptbp1 was packaged with PHP.eB serotype, mixed with hGFAP-mCherry AAV5 virus, and injected into the adult mouse striatum (Fig. S6A, B). Instead of using wildtype mice, we used mGfap-Cre;R26R-YFP mice such that striatal astrocytes could be lineage-traced. At 60 dpv, 22.7± 3.2% of mCherry+ cells were NeuN+ in the CasRx-Ptbp1 group; however, they were not traced with YFP indicating a non-astrocyte origin (Fig. S6C, D). We next examined PTBP1 expression by using an antibody that was verified by both western blots and immunostaining (Qian et al., 2020). Surprisingly, PTBP1 expression was only mildly reduced in cells infected with CasRx-Ptbp1 virus at 60 dpv (Fig. S6E-G). For this above analysis, the FLAG-tagged CasRx was indicated by the co-expressed mCherry reporter due to the incompatibility of FLAG antibody and PTBP1 antibody for co-staining. On the other hand, co-staining of mCherry and FLAG-tagged CasRx showed that over 95% of mCherry+ cells were also FLAG+ (Fig. S6H, I), indicating that mCherry could be used as a valid indicator for CasRx or CasRx-Ptbp1 virus-transduced cells. Together, these results show that AAV-GFAP-CasRx-Ptbp1 neither efficiently downregulates PTBP1 nor converts striatal astrocytes in vivo, in sharp contrast to what was reported (Zhou et al., 2020).
PTBP1 knockdown fails to convert striatal astrocytes in vivo.
We then employed the shRNA-based approach (Qian et al., 2020). The reported Ptbp1-targeting sequence was cloned into a miR30-based vector to generate hGFAP-mCherry-shPtbp1. A luciferase-targeting sequence (shLuc) was used as a control. Western blots showed that endogenous Ptbp1 was efficiently knocked down in cultured primary mouse brain astrocytes (Fig. S7A). hGFAP-mCherry-shPtbp1 (mCh-shPtbp1) or hGFAP-mCherry-shLuc (mCh-shLuc) AAV5 virus was then injected into the striatum of tamoxifen-treated young (P35-40) Aldh1l1-CreERT2;R26R-YFP mouse (Fig. S7B). PTBP1 expression was reduced to the background level when examined at 60 dpv (Fig. S7C, D). Unexpectedly, only a small number of mCherry+ cells were NeuN+ (2.8 ± 0.9% for shPtbp1 vs. 3.7 ± 1.7% for shLuc; Fig. S7E, F). Furthermore, these mCherry+NeuN+ neurons were not traced with YFP indicating a non-astrocyte origin. To rule out a potential effect of mouse background or tamoxifen treatments, we injected mCh-shPtbp1 or mCh-shLuc virus into the striatum of young mGfap-Cre;R26R-YFP mouse (Fig. S7G). The majority of mCherry+ cells were NeuN− at 60 dpv (Fig. S7H, I; 97.2 ± 0.7% for shLuc and 98.1 ± 1.5% for shPtbp1), indicating no AtN conversion by PTBP1 knockdown.
We also made the Cre-dependent AAV-CAG-LSL-mCherry-shPtbp1 and the control AAV-CAG-LSL-mCherry-shLuc AAV5 virus. Young Aldh1l1-CreERT2;R26R-YFP mice were injected with AAV5 virus into the striatum, treated with tamoxifen, and examined at 60 dpv (Fig. S7J). A larger number of mCherry+NeuN+ neurons were observed in the striatum injected with the shPtbp1 virus (31.1 ± 1.2% for shPtbp1 vs. 6.7 ± 2.9% for shLuc); however, they were not traced with YFP (Fig. S7K, L). To verify such a result, we injected these above viruses into the striatum of young mGfap-Cre;R26R-YFP mouse (Fig. S7M). Despite a relatively larger number of mCherry+NeuN+ neurons were observed in the shPtbp1 group at 60 dpv (34.3± 1.9% for shPtbp1 vs. 7.3 ± 1.5% for shLuc), they were not traced with YFP indicating a non-astrocyte origin (Fig. S7N, O).
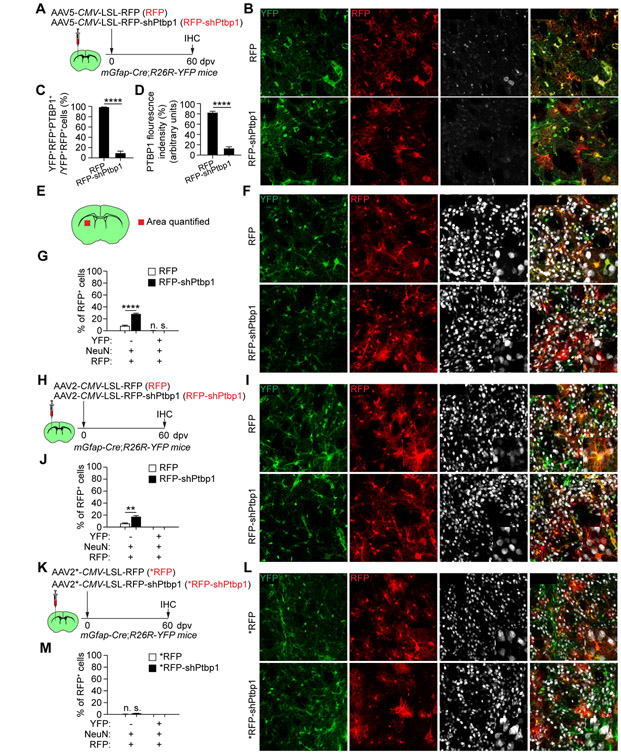
To further examine PTBP1 knockdown, we obtained AAV-CMV-LSL-RFP (RFP) and AAV-CMV-LSL-RFP-shPtbp1 (RFP-shPtbp1) vectors (Qian et al., 2020) and packaged them into AAV5 virus. They were injected into the striatum of young mGfap-Cre;R26R-YFP mouse and examined at 60 dpv (Fig. 7A). We confirmed efficient knockdown of endogenous PTBP1 (Fig. 7B-D). Despite RFP+NeuN+ neurons were observed in the RFP-shPtbp1 group (32.1 ± 0.8% for RFP-shPtbp1 vs. 7.8 ± 1.1% for RFP), they were not traced with YFP (Fig. 7E-G). To rule out a potential effect of AAV serotype, we also packaged these vectors into AAV2 (Qian et al., 2020) and examined them in young mGfap-Cre;R26R-YFP mouse at 60 dpv (Fig. 7H). Once again, YFP-traced RFP+NeuN+ neurons were not observed (Fig. 7I, J).
Figure 7. PTBP1 knockdown fails to convert striatal astrocytes in vivo.
(A) Study design. Striatal astrocytes were constitutively traced with YFP.
(B) Confocal images of the indicated markers. Scales, 50 μm.
(C, D) Quantifications showing robust and efficient PTBP1 knockdown (n = 4 mice per group; mean ± SEM; ****p < 0.0001).
(E) A schematic showing the striatal region for analysis
(F) Confocal images of the indicated markers. Scales, 50 μm.
(G) Quantifications showing a lack of YFP-traced RFP+ neurons (n = 4 mice per group; mean ± SEM; ****p < 0.0001).
(H) Study design. Striatal astrocytes were constitutively traced with YFP.
(I) Confocal images of the indicated markers. Scales, 50 μm.
(J) Quantifications showing a lack of YFP-traced RFP+ neurons (n= 4 mice per group; mean ± SEM; **p = 0.0056).
(K) Study design. AAV2 viruses were from Fu laboratory. Striatal astrocytes were constitutively traced with YFP.
(L) Confocal images of the indicated markers. Scales, 50 μm.
(M) Quantifications showing a lack of YFP-traced RFP+ neurons (n = 4 mice per group; mean ± SEM; n.s., not significant).
See also Figures S6 and S7.
As a final attempt, we obtained the RFP and RFP-shPtbp1 AAV2 viruses (Qian et al., 2020) and injected them into the striatum of young mGfap-Cre;R26R-YFP mouse (Fig. 7K). Again, YFP-traced RFP+NeuN+ neurons were not detected at 60 dpv (Fig. 7L, M). All together, these above results clearly indicate that, despite efficient knockdown of PTBP1, resident striatal astrocytes cannot be converted into neurons in vivo.
DISCUSSION
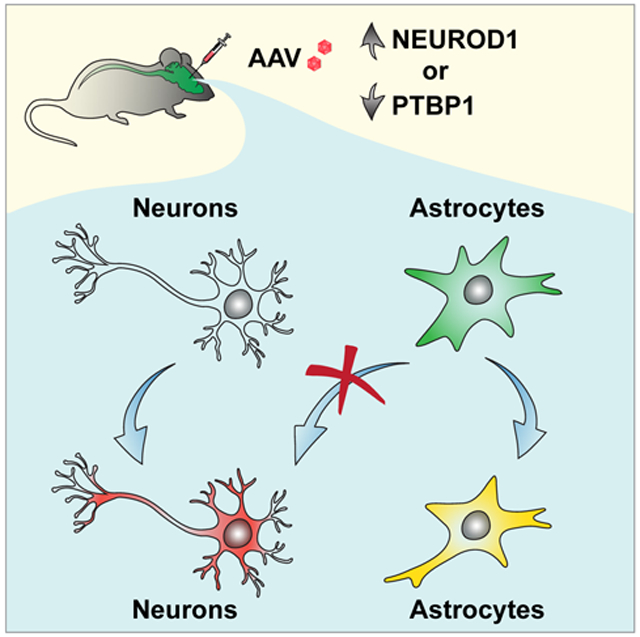
By employing multiple stringent lineage-tracing methods and examining many experimental conditions, our results do not support some of the recent extraordinary claims that resident astrocytes can be directly and efficiently converted into mature neurons with local identity and connectivity (Chen et al., 2020; Liu et al., 2020; Qian et al., 2020; Wu et al., 2020; Xiang et al., 2021; Zhou et al., 2020). Rather, our results indicate that those reporter-labeled neurons are endogenous neurons.
Using AAV-mediated co-expression of NEUROD1 and the mCherry reporter (Chen et al., 2020; Liu et al., 2020), our results indeed confirm that NEUROD1 can specifically, rapidly, and highly efficiently induce mCherry+ neurons (Chen et al., 2020; Liu et al., 2020). Nonetheless, these viral reporter-positive neurons are not converted from resident astrocytes.
First, an immature stage is expected if astrocytes are converted into neurons. However, DCX, a well-established marker for immature neurons (Couillard-Despres et al., 2005), cannot be detected in any time points post injection of NEUROD1 virus. Secondly, continuous BrdU-labeling after brain injury fails to show a contribution of reactive glial cells to the mCherry+ neurons. A caveat might be that BrdU is toxic to the newly converted neurons; however, this is unlikely since new neurons from reprogrammed glia can be robustly traced by such a BrdU-labeling strategy (Niu et al., 2015; Niu et al., 2013; Su et al., 2014a; Tai et al., 2021; Wang et al., 2016). Thirdly, NEUROD1-induced mCherry+ neurons cannot be traced by the astrocyte lineage reporter YFP, despite an approximate 95% YFP-labeling efficiency. One might argue that tamoxifen-treatments and the transgenic mouse background impede the reprogramming ability of NEUROD1; however, this is not the case since tamoxifen-independent NEUROD1 expression can induce numerous mCherry+ neurons in tamoxifen-treated Aldh1l1-CreERT2;R26R-YFP mice. By switching the viral reporter to GFP and the lineage tracing reporter to tdTomato, we also fail to detect astrocytes-converted neurons in Aldh1l1-CreERT2;R26R-tdTomato mice at 90 dpv. Of note, some endogenous neurons could be labeled in variable brain regions of rare Aldh1l1-CreERT2;R26R-tdTomato mice, which might account for what was observed in a recent rebuttal article to our preprint (Wang et al., 2020; Xiang et al., 2021). Fourthly, brain injury fails to precondition genetically traced astrocytes to become neurons by NEUROD1. Since injury can promote fate reprogramming (Grande et al., 2013; Guo et al., 2014; Heinrich et al., 2014), we examined such condition but failed to show AtN conversion. Fifthly, despite numerous mCherry+ neurons, the overall neuronal density remains unchanged in the virus-injected regions, arguing against a net increase of neurons due to presumed robust AtN conversion. If any neurons were converted from astrocytes, the number would be extremely small, consistent with a previous report (Brulet et al., 2017). Sixthly, viral reporter-positive neurons could still be induced by NEUROD1 mutants lacking any neurogenic activity, arguing against a potential function in cell fate conversion. And finally, retrograde labeling clearly reveals that endogenous neurons are the cell source for NEUROD1-induced reporter+ neurons.
Our results, however, do not argue against a neurogenic activity of NEUROD1 under certain conditions, such as in cell culture or retrovirus-mediated expression after injury (Guo et al., 2014; Pataskar et al., 2016; Wang et al., 2021; Zhang et al., 2015). Regarding retrovirus-mediated conversion (Guo et al., 2014), nonetheless, the cell origin requires further delineation. Brain injury can induce migration of endogenous neural stem cells, which normally differentiate into astrocytes that are molecularly distinct from those in the local cortex (Benner et al., 2013). These migrating and differentiating neural stem cells, but not local cortical astrocytes, might well be the cell origin for the neurons that were induced by retroviral NEUROD1 (Guo et al., 2014). Such a possibility could have been excluded through genetic lineage tracing of endogenous neural stem cells, as we have previously shown (Niu et al., 2013; Tai et al., 2021). Alternatively, local astrocytes could be genetically traced and examined for fate changes during the conversion process. Furthermore, retroviral infection could lead to fusion of microglia and endogenous neurons (Ackman et al., 2006), which might be the cell origin for the beautiful neurons observed in regions with retroviral NEUROD1 (Xiang et al., 2021). Because of these above caveats, stringent lineage tracings are also essential even for retrovirus-mediated fate conversion (Guo et al., 2014; Xiang et al., 2021).
An unresolved question is how NEUROD1 specifically induce reporter expression in endogenous neurons. Obviously, viral toxicity is not the underlying mechanism as recently proposed (Xiang et al., 2021). First, if any virus-induced cell toxicity, neurons would be more prone than astrocytes to toxicity-induced death and therefore, the viral reporter would be more frequently detected in astrocytes but not neurons. Secondly, viral reporter-positive neurons are also observed in brain regions far away from the injection core, in which the virus concentration would be lower. Thirdly, neuronal expression of the viral reporter is dependent on the factors expressed. The reporter is predominantly in astrocytes for the control virus (mCherry or GFP) and a few others (such as SOX2, OCT4, KLF4, and BDNF), whereas it is largely in neurons of brains injected with AAVs expressing NEUROD1, PAX6, ASCL1, or MYC. Viral toxicity would not confer such gene-specificity. And fourthly, injection of mixed viruses or co-packaged viruses shows a clear separation of the viral reporter with expression in astrocytes for the control virus and in neurons for the NEUROD1 virus. Such a phenomenon could not be explained by viral toxicity. These latter results rather suggest that the hGFAP promoter might be cis-regulated by the downstream factors. This conclusion would be consistent with a prior observation that cell type-specificity of the GFAP promoter can be altered by the expressed genes (Su et al., 2004). Because of the detection limits of the virus-expressed fluorescent reporters, however, we cannot exclude that NEUROD1 might also regulate in trans the co-injected hGFAP-Cre virus. The residual NEUROD1-induced Cre expression might form a positive feedforward loop with the AAV FLEX system to induce robust viral reporter expression in neurons (Chen et al., 2020; Wu et al., 2020; Xiang et al., 2021). Regarding the AAV FLEX system, a recent report showed Cre-independent leaky expression in neurons (Fischer et al., 2019). Such leaky neuronal expression may be further exacerbated by the downstream genes such as NEUROD1 and DLX2. Interestingly, AAVs can form circular concatamers through intermolecular recombination (Yang et al., 1999). Through concatamerization, NEUROD1 may cooperate with the co-transduced AAVs to further increase reporter expression in neurons. Clearly, future studies are required to understand how the promoter in virus can be influenced by the downstream genes.
As another test case, our results also do not support the phenomenal claims that PTBP1 knockdown induces AtN conversion in vivo (Qian et al., 2020; Zhou et al., 2020). It was extremely surprising that PTBP1 knockdown converts striatal astrocytes into either dopaminergic (Zhou et al., 2020) or striatal neurons (Qian et al., 2020). By using the AAV-GFAP-CasRx-Ptbp1 virus (Zhou et al., 2020), we not only failed to obtain neurons from genetically traced striatal astrocytes but also failed to detect efficient downregulation of PTBP1. Upon closer examination of the Cell report (Zhou et al., 2020), the image quality is too poor to tell whether new striatal dopaminergic neurons were ever generated. Similarly, PTBP1 staining in the Cell report seems nonspecific, since endogenous PTBP1 should be mainly localized in the nucleus (Qian et al., 2020). On the other hand, we confirmed shRNA-mediated efficient knockdown of PTBP1 in vitro and in vivo. However, we failed to detect AtN conversion in two mouse lines (Aldh1l1-CreERT2;R26R-YFP and mGfap-Cre;R26R-YFP) and under several conditions: 1) AAV5 hGFAP-mCherry-shPtbp1, 2) AAV5 CAG-LSL-mCherry-shPtbp1 3) AAV5 CMV-LSL-RFP-shPtbp1, 4) AAV2 CMV-LSL-RFP-shPtbp1, and 5) AAV2 CMV-LSL-RFP-shPtbp1 from Fu laboratory. Of note, a variable number of viral reporter-labeled neurons could be detected under certain conditions indicating leaky expression. The underlying cause is not clear for the discrepancies between our results and Fu lab’s, although young (P35-40) mice were also used in our shPtbp1 study. One possibility is the mGfap-Cre line, which was reported to exhibit Cre expression in neural stem cells and in the male germline (https://www.jax.org/strain/024098). As recommended, only hemizygous females were used for breeding in our study. Even under such a condition, we also noticed leaky reporter expression in neurons of multiple brain regions including the cortex and midbrain.
Without in vivo AtN conversion as the cellular basis for function, how could mouse behaviors be improved under pathological conditions (Chen et al., 2020; Qian et al., 2020; Wu et al., 2020; Zhou et al., 2020)? If those behavioral improvements could be independently reproduced, neuronal protection might be an underlying mechanism. NEUROD1 is known to be cell-autonomously required for neuronal survival (Gao et al., 2009; Jahan et al., 2010), whereas PTBP1 might contribute to neuronal vulnerability by controlling tau exon 10 splicing (Roussarie et al., 2020). Future experiments are needed to examine such scenarios.
In conclusion, our results show that virus expressed reporters and Cre are prone to leakage and that stringent lineage-tracing strategies are indispensable when studying in vivo cell fate conversions. Unlike cultured cells the fate of which can be directly observed, in vivo conversion occurs in a complex microenvironment often filled with endogenous neurons. It is crucial to use well-controlled lineage mappings to confirm the cell origin. For tracing the lineage of astrocytes, the tamoxifen-inducible Aldh1l1-CreERT2;R26R-YFP line seems superior with minimal labeling of endogenous neurons. The Aldh1l1-CreERT2;R26R-tdTomato line also is generally specific; however, some leaky expression of tdTomato in neurons of variable brain regions is observed in rare mice. These rare mice should be excluded by broadly examining brain regions not injected with the reprogramming factors. The constitutive mGfap-Cre;R26R-YFP line exhibits specific labeling of striatal astrocytes but leaky expression in multiple brain regions. Since AtN conversion should transit through an immature stage, these immature neurons can be identified by the marker DCX (Couillard-Despres et al., 2005). A promise of AtN conversion is to generate new neurons from disease-responsive reactive astrocytes. These reactive astrocytes can be prelabeled with BrdU and examined for BrdU-traced neurons after AtN conversion. A superior method for examining AtN conversion is in vivo time-lapse imaging, as elegantly demonstrated for adult neurogenesis (Pilz et al., 2018); however, such a method will be technically and costly prohibitive for most research labs. Single cell RNA-seq may be considered; nonetheless, the trajectory analysis should be cautiously interpreted since the reprogramming factors can indeed regulate a plethora of developmental genes. Overall, stringent genetic lineage tracings, prelabeling of reactive cells with BrdU, and examination of immature neuronal stages can be readily conducted in most research labs and should be broadly applied in the studies of cell fate conversion in vivo.
Limitations of the Study
Future experiments are needed to tease out the molecular mechanisms by which NEUROD1 or others alter the cell type specificity of the viral promoter. For example, NEUROD1 and PAX6 share no sequence similarity at either the DNA or protein levels, yet they both robustly induce the viral reporter in endogenous neurons. Truncation analyses of NEUROD1 also failed to identify a region responsible for the altered cell type specificity. Furthermore, it is not clear how NEUROD1 or others (alone or in combinations) exacerbate leaky neuronal expression of the AAV-based FLEX system (Fischer et al., 2019), although we suspect that concatamerization of AAVs through intermolecular recombination (Yang et al., 1999) may play a critical role.
STAR ★ METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Chun-Li Zhang (chun-li.zhang@utsouthwestern.edu).
Materials availability
All materials generated in this study are available from the lead contact.
Data and code availability
Microscopy data reported in this paper will be shared by the lead contact upon request.
All software were commercially or freely available and are listed in the STAR methods description and Key resources table.
Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-ALDH1L1 | NeuroMab | Cat# 75-140; RRID: AB_10673448 |
| Goat polyclonal anti-ALDOC (Aldolase C) | Santa Cruz Biotechnology | Cat# sc-12065; RRID: AB_2242641 |
| Rat monoclonal anti-BrdU | BIO-RAD | Cat# OBT0030; RRID: AB_609568 |
| Rabbit polyclonal anti-Cre | Covance | Cat# PRB-1061C-200; RRID: AB_11220031 |
| Goat polyclonal anti-DCX | Santa Cruz Biotechnology | Cat# sc-8066; RRID: AB_2088494 |
| Rabbit polyclonal anti-FLAG | Sigma-Aldrich | Cat# F7425; RRID: AB_439687 |
| Mouse monoclonal anti-GFAP | Sigma-Aldrich | Cat# G3893; RRID: AB_477010 |
| Chicken polyclonal anti-GFAP | Abcam | Cat# ab4674; RRID: AB_304558 |
| Chicken polyclonal anti-GFP | Aves Labs | Cat# GFP-1020; RRID: AB_10000240 |
| Chicken polyclonal anti-HA | Aves Labs | ET-HA100; RRID: AB_2313511 |
| Mouse monoclonal anti-MAP2 | Proteintech | Cat# 67015-1-Ig; RRID: AB_2882331 |
| Goat polyclonal anti-tdTomato (mCherry) | MyBioSource | Cat# MBS448092; RRID: AB_2827808 |
| Rabbit monoclonal anti-NeuN | Abcam | Cat# ab177487; RRID: AB_2532109 |
| Rabbit recombinant monoclonal anti-NEUROD1 | Abcam | Cat# ab109224; RRID: AB_10861489 |
| Rabbit polyclonal anti-PTBP1 | ABclonal | Cat# A6107; RRID: AB_2766748 |
| Mouse monoclonal anti-ACTB | Proteintech | Cat# 66009-1; RRID: AB_2687938 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202; RRID: AB_141607 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | Cat# 703-545-155; RRID: AB_2340375 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11006; RRID: AB_2534074 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21432; RRID: AB_2535853 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID: AB_2536183 |
| Hoechst 33342 | Thermo Fisher Scientific | REF H3570 |
| HRP-conjugated anti-rabbit IgG | Jackson Laboratory | Cat# 111-035-144; RRID: AB_2307391 |
| HRP-conjugated anti-mouse IgG | Jackson Laboratory | Cat# 715-035-150; RRID: AB_2340770 |
| Bacterial and virus strains | ||
| AAV5-hGFAP*-Cre virus | Addgene | Cat# 105550-AAV5; RRID: Addgene_105550 |
| AAV2*-CMV-LSL-RFP | Fu Laboratory | N/A |
| AAV2*-CMV-LSL-RFP-shPtbp1 | Fu Laboratory | N/A |
| Biological samples | ||
| Mouse brain tissue | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Cayman Chemical | CAS: 10540-29-1 |
| BrdU | Alfa Aesar Chemical TFS | CAS: 59-14-3 |
| N-2 | Gibco | Cat# 17502-048 |
| B-27 | Gibco | Cat# 17504-044 |
| Forskolin | Selleckchem | Cat# S2449 |
| Dorsomorphin | Sigma-Aldrich | Cat# 171260 |
| Polybrene (Hexadimethrime bromide) | Sigma-Aldrich | Cat# H9268 |
| Matrigel | Corning | Cat# CB-40230 |
| Gelatin | BIO-RAD | Cat# 1706537 |
| Papain | Worthington Biochemical Corporation | Cat# LS003126 |
| DNase | Invitrogene | Cat# 18068-015 |
| Nuclease (Benzonase) | Chem Cruz | Cat# sc-202391A |
| Poly ethylene glycol (PEG) | Sigma | Cat# 81268 |
| Iodixanol gradient | Alere Technologies | Prod. No. 1114542 |
| Protease inhibitor cocktail | Thermo Scientific Pierce | Cat# A32965 |
| Western Chemiluminescent HRP substrate | Millipore | Cat# WBKLS0500 |
| Critical commercial assays | ||
| none | ||
| Deposited data | ||
| none | ||
| Experimental models: Cell lines | ||
| U251 glioma cells | Sigma | Cat# 09063001 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Mouse: R26R-YFP: B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | The Jackson Laboratory | JAX: 006148; RRID: IMSR_JAX:006148 |
| Mouse: R26R-tdTomato: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007914; RRID: IMSR_JAX:007914 |
| Mouse: Aldh1l1-CreERT2: B6;FVB-Tg(Aldh1l1-cre/ERT2)1Khakh/J | The Jackson Laboratory | JAX: 029655; RRID: IMSR_JAX: 029655 |
| Mouse: mGfap-Cre line 77.6: B6.Cg-Tg(Gfap-cre)77.6Mvs/2J | The Jackson Laboratory | JAX: 024098; RRID: IMSR_JAX: 024098 |
| Oligonucleotides | ||
| ITR primer: forward: 5-GGAACCCCTAGTGATGGAGTT-3 | Sigma-Aldrich | Customized |
| ITR primer: reverse: 5-CGGCCTCAGTGAGCGA-3 | Sigma-Aldrich | Customized |
| Recombinant DNA | ||
| pAAV-hGFAP-GFP | This paper | N/A |
| pAAV-hGFAP-Cre | This paper | N/A |
| pAAV-hGFAP-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-PAX6-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-MYC-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROG2-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-ASCL1-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-BDNF-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-SOX2-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-OCT4-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-KLF4-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-R111L-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-K139D-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-N252-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-N157-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-N100-252-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-156C-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-mCherry-shLuc | This paper | N/A |
| pAAV-hGFAP-mCherry-shPtbp1 | This paper | N/A |
| pAAV-CAG-GFP | Addgene | Cat# 37825; RRID:Addgene_37825 |
| pAAV-CAG-FLEX-GFP | Addgene | Cat# 28304; RRID:Addgene_28304 |
| pAAV-CAG-FLEX-mCherry | This paper | N/A |
| pAAV-CAG-FLEX-NEUROD1-T2A-mCherry | This paper | N/A |
| pAAV-CAG-FLEX-HA-DLX2-T2A-mCherry | This paper | N/A |
| Ai9 (to clone LSL) | Addgene | Cat# 22799; RRID:Addgene_22799 |
| pAAV-CAG-LSL-mCherry-shLuc | This paper | N/A |
| pAAV-CAG-LSL-mCherry-shPtbp1 | This paper | N/A |
| pAAV-GFAP-CasRx | Addgene | Cat# 154000; RRID:Addgene_154000 |
| pAAV-GFAP-CasRx-Ptbp1 | Addgene | Cat# 154001; RRID:Addgene_154001 |
| pAAV-CMV-LSL-RFP | Fu Laboratory | N/A |
| pAAV-CMV-LSL-RFP-shPtbp1 | Fu Laboratory | N/A |
| pAd-deltaF6 | Addgene | Cat# 112867; RRID:Addgene_112867 |
| helper pAAV2/2 | Addgene | Cat# 104963; RRID:Addgene_104963 |
| helper pAAV2/5 | Addgene | Cat# 104964; RRID:Addgene_104964 |
| helper pAAV2/8 | Addgene | Cat# 112864; RRID:Addgene_112864 |
| helper pAAV2/9 | Addgene | Cat# 112865; RRID:Addgene_112865 |
| pUCmini-iCAP-PHP.eB | Addgene | Cat# 103005; RRID:Addgene_103005 |
| rAAV2-retro | Addgene | Cat# 81070; RRID:Addgene_81070 |
| pCSC-IRES-GFP | This paper | N/A |
| pCSC-NEUROD1-IRES-GFP | This paper | N/A |
| pCSC-NEUROD1-R111L-IRES-GFP | This paper | N/A |
| pCSC-NEUROD1-K139D-IRES-GFP | This paper | N/A |
| pLV-hGFAP-mCherry-shPtbp1 | This paper | N/A |
| pLV-hGFAP-GFP-shLuc | This paper | N/A |
| pLV-hGFAP-GFP | This paper | N/A |
| pMDL (pMDLg-pRRE) | Addgene | Cat# 12251; RRID:Addgene_12251 |
| VSV-G (pMD2.G) | Addgene | Cat# 12259; RRID:Addgene_12259 |
| pREV (pRSV-Rev) | Addgene | Cat# 12253; RRID:Addgene_12253 |
| Software and algorithms | ||
| ImageJ | NIH | RRID:SCR_003070, https://imagej.net/Fiji |
| Graphpad Prism | Graphpad | RRID:SCR_002798), https://www.graphpad.com |
| ZEN | Zeiss | RRID:SCR_013672, https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| ChemiDoc iMP Imaging System | BioRad | RRID:SCR_019037, https://www.bio-rad.com/en-in/product/chemidoc-mp-imaging-system?ID=NINJ8ZE8Z |
| NIS-Elements C | Nikon | RRID:SCR_020317, https://www.microscope.healthcare.nikon.com/about/news/nikon-instruments-a1-confocal-laser-microscope-series-with-nis-elements-c-software-delivers-fully-integrated-comprehensive-confocal-imaging-capabilities |
| Adobe Photoshop | Adobe | RRID:SCR_014199, https://www.adobe.com/products/photoshop.html |
| Adobe Illustrator | Adobe | RRID:SCR_010279, https://www.adobe.com/products/illustrator.html |
| Other | ||
| 33 gauge, 45-degree-beveled needle | Hamilton, Reno, NV | Cat# 22033 (Customized) |
| Impact One™ Stereotaxic CCI Instrument | Leica | Model 39463920 |
| 100K PES concentrator (Pierce™) | Thermo Fisher | Cat# 88524 |
| 40-μm nylon strainer | Falcon | REF: 352340 |
| Sliding microtome | Leica | Model SM200R |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
The following wildtype or transgenic mouse lines were obtained from the Jackson Laboratory: C57BL/6J (stock #000664), Aldh1l1-CreERT2 (stock #029655) (Srinivasan et al., 2016), mGfap-Cre line 77.6 (stock #024098) (Gregorian et al., 2009), R26R-YFP (stock #006148) (Srinivas et al., 2001), and R26R-tdTomato (stock #007914) (Madisen et al., 2010). Hemizygous Aldh1l1-CreERT2 males or females were used for breeding, whereas only hemizygous females of mGfap-Cre were used for breeding. After breeding to the reporter mouse lines, cell type-specificity of the traced cells was extensively examined by immunohistochemistry in different brain regions. Of note, except for the core striatum the mGfap-Cre line shows variable leaky expression in neurons of different brain regions including the cortex and midbrain. Both adult male and female mice at 8 weeks or older were used for most of the experiments, whereas young mice at postnatal day (P) 35 to 40 days were used for the shRNA experiments as described (Qian et al., 2020). All mice were housed under controlled temperature and a 12-h light/dark cycle with free access to water and food in an animal barrier facility at UT Southwestern. Animal procedures were approved by the Institutional Animal Care and Use Committee at UT Southwestern.
Primary astrocytes culture
Primary mouse cortical astrocytes were isolated from wildtype mice at postnatal day 2 as previous described (Ma et al., 2020). Briefly, cortices were dissociated with a solution containing papain (10 U/mL, with 1 mM Ca2+ and 0.5 mM EDTA) and 1% DNase for 20 min at 37 °C. Tissues were pelleted through brief centrifugation and further dissociated using a pipette in FBS-containing medium. Cells were passed through a 40-μm nylon strainer, and the cell mixture was spun at 400 g for 3 min and re-suspended in growth media consisting of DMEM supplemented with 10% FBS and plated into 0.1% gelatin-coated 75 cm2 flasks. Media was exchanged every 3 days. To test PTBP1 knock down efficiency in vitro, astrocytes were seeded on matrigel-coated culture vessels. 24 hours later, cells were transduced with lentivirus in the presence of 6 μg/mL polybrene. After overnight incubation, culture media were refreshed. Medium was half-changed every other day. Cell lysates were collected at 7 dpv for analysis.
U251 culture
Human U251 glioma cells (Sigma) were cultured in DMEM supplemented with 10% fetal bovine serum. Cells were seeded on Matrigel-coated culture vessels. The following day, they were infected with the indicated lentivirus in the presence of 6 μg/mL polybrene. After overnight incubation, culture media were refreshed. One day later, the cells were then switched to neuronal induction medium, consisting of DMEM:F12:Neurobasal (2:2:1), 0.8% N-2 (Gibco), and 0.4% B-27 (Gibco). In addition, forskolin (10 μM) and dorsomorphin (1 μM) were added to the above induction medium. Induction medium was half-changed every other day. Immuno-fluorescence staining for neuronal markers was performed on 21 days post viral infection.
METHOD DETAILS
AAV vectors and virus production
The AAV vectors driven by hGFAP promoter listed in KEY RESOURCES TABLE were constructed through PCR-based subcloning. Unless indicated otherwise, the hGFAP promoter used in this study is the synthetic 681-bp gfaABC1D derived from the 2.2-kb gfa2 promoter (Lee et al., 2008). Both promoters exhibit identical astrocyte-restricted expression patterns through transgenic analyses (Lee et al., 2008). The GAG promoter was derived from the pAAV-CAG-GFP vector (Addgene #37825), whereas the FLEX system was based on the pAAV-CAG-FLEX-GFP vector (Addgene #28304). The Cre-dependent loxP-STOP-loxP (LSL) cassette was subcloned from the Ai9 plasmid (Addgene #22799). pAAV-GFAP-CasRx and pAAV-GFAP-CasRx-Ptbp1 were obtained from Addgene (#154000 and #154001), whereas pAAV-CMV-LSL-RFP and pAAV-CMV-LSL-RFP-shPtbp1 were provided by Fu Laboratory (Qian et al., 2020). All vectors were verified through restriction enzyme digestions and DNA sequencing. AAV viruses were packaged with pAd-deltaF6 (Addgene #112867) and the helper pAAV2/2 (Addgene #104963), pAAV2/5 (Addgene #104964), pAAV2/8 (Addgene #112864), pAAV2/9 (Addgene #112865), pUCmini-iCAP-PHP.eB (Addgene #103005), or rAAV2-retro (Addgene #81070) in HEK293T cells. Briefly, HEK293T cells were transfected with the packaging plasmids and a vector plasmid. Three days later, virus was collected from the cell lysates and culture media. Virus was purified through iodixanol gradient ultracentrifugation, washed with PBS, and concentrated with 100K PES concentrator (Pierce™, Thermo Scientific). Viral titers were determined by quantitative PCR with ITR primers (forward: 5-GGAACCCCTAGTGATGGAGTT-3; reverse: 5-CGGCCTCAGTGAGCGA-3). The AAV5-hGFAP*-Cre virus was obtained from Addgene (#105550), in which Cre is driven by the 2.2-kb gfa2 promoter. Based on publications, virus with a titer range from 1e12 - 4e13 GC/mL was used for experiments (details in Table S1).
Brain injury and intracerebral injection
The controlled cortical impact (CCI) model of TBI was employed as previous described (Chen et al., 2019). Under anesthesia, a skin incision was made in the mouse forehead to expose the skull. A craniotomy was performed over the right hemisphere and the bone flap (2.2 mm in diameter) was carefully removed. A cortical injury (0 bregma, 1 mm lateral to the sagittal suture line) was introduced at an impact depth of 0.5 mm with a 2-mm diameter round impact tip (3.0 m/s speed and 100 ms dwell time) by using an electromagnetically driven CCI device (Impact One™ Stereotaxic CCI Instrument, Leica). After injury, skin was sutured and further secured with liquid adhesive. Intracerebral viral injections (1~4 μL per injection, details in Table S1) were performed on a stereotaxic frame. The injection coordinates were as the following: +1.0 mm anterior/posterior (AP), ±1.5 mm medial/lateral (ML), and −0.8 mm dorsal/ventral from the skull (DV) for uninjured cortex; 0 mm AP, ±1.5 mm ML, and −0.8 mm DV for injured cortex; 1.0 mm AP, ±2.0 mm ML, and −3.0 mm DV for adult striatum; 0.8 mm AP, ±1.8 mm ML, and −2.7 mm DV for young striatum.
Retrograde labeling and intracerebral injection
rAAV2-retro-CAG-GFP virus (2 μL per mouse) was injected into the T6 level of dorsal spinal cord to target the corticospinal tract. 3-10 days post rAAV2-retro injection, we stereotaxically injected AAV5 virus into the cortex with the following coordinates: −0.5 mm AP, ±1.5 mm ML, and −1.0 mm DV.
Tamoxifen and BrdU administration
Tamoxifen (Cayman Chemical) was dissolved in a mixture of ethanol and sesame oil (1:9 by volume) at a concentration of 40 mg/mL. Tamoxifen was administered through intraperitoneal injections at a daily dose of 1 mg/10g body weight for 3-9 days. BrdU (Alfa Aesar Chemical; 0.5 g/L) was supplied in drinking water for durations as indicated in the text.
Lentivirus preparation
The following lentivirus vectors were constructed through PCR-based subcloning into the pCSC-IRES-GFP vector (Ding et al., 2021; Liu et al., 2016; Liu et al., 2013; Su et al., 2014b): pCSC-NEUROD1-IRES-GFP, pCSC-NEUROD1-R111L-IRES-GFP, and pCSC-NEUROD1-K139D-IRES-GFP.pLV-hGFAP-mCherry-shPtbp and pLV-hGFAP-GFP-shLuc were based on the pLV-hGFAP-GFP vector (Niu et al., 2013; Wang et al., 2016) and an enhanced miR30 backbone for shRNA (Fellmann et al., 2013). The third generation, replication-deficient lentivirus was generated in HEK293T cells by transient transfections with lentiviral vectors and the packaging plasmids (pMDL, VSV-G and pREV).
Immunohistochemistry
Mice were euthanized and perfused with intracardial injection of 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS). Brains were isolated and post-fixed overnight with 4% (w/v) paraformaldehyde at 4 °C. After cryoprotection with 30% sucrose in PBS for 72 h at 4 °C, 40-μm brain slices were collected through a sliding microtome (Leica). For immunostaining, brain sections were treated with 50% formamide in 1 X SSC buffer for 2 h at 65 °C. For BrdU staining, the sections were pretreated with 2N HCl for 30 min at 37 °C. The following primary antibodies were used: GFP (Chicken; 1:2,000; Aves Labs), mCherry (Goat; 1:2,000; Mybiosource), NeuN (Rabbit; 1:10,000; Abcam), GFAP (Mouse; 1:2,000; Sigma), Cre (Rabbit; 1:500; Covance), ALDH1L1 (Mouse; 1:200; Neuromab), ALDOC (Goat; 1:100; Santa Cruz), BrdU (Rat; 1:500; Bio-Rad), NEUROD1 (Rabbit; 1:1,000; Abcam), PTBP1 (Rabbit, 1:1,000, ABclonal), FLAG (Rabbit; 1:300; Sigma), HA (Chicken; 1:300; Aves), DCX (Goat; 1:500; Santa Cruz), and MAP2 (Mouse; 1:1,000; Proteintech). Alexa Fluor 488-, 555-, or 647-conjugated corresponding secondary antibodies from Jackson ImmunoResearch were used for indirect fluorescence (1:2,000). Nuclei were counterstained with Hoechst 33342 (Hst). Images were captured using a Zeiss LSM700 or Nikon A1R confocal microscope for analysis.
Western blot
Whole-cell lysates were obtained through direct lysis of cells in 50 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl, 1% NP40, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, and protease inhibitor cocktail (Roche). Equal amounts of these lysates (10 μg per lane) were used for SDS-PAGE and western blot analysis. Samples were immunoblotted against PTBP1 (Rabbit; 1:1,000; ABclonal) or the loading control ACTB (Mouse; 1:20,000; Proteintech), followed by their corresponding HRP-conjugated secondary antibodies (HRP conjugated anti rabbit or anti mouse IgG 1:10,000, Jackson Laboratory). Blots were developed with Immobilon Western Chemiluminescent HRP substrate (Millipore) and visualized using ChemiDoc imaging system (BioRad).
QUANTIFICATION AND STATISTICAL ANALYSIS
Four representative images from the top and middle layer of the cortex near the injection site were collected and quantified for each brain sample to confirm cell identity or conversion rate. To determine the local density of neurons or astrocytes, NeuN+ or GFAP+ cells were quantified from a 1-μm thick confocal image with an area of 0.102977 mm2. Four to six random confocal images surrounding the viral injection area with evenly distributed reporter+ cells were analyzed for each animal. Quantification data are presented as mean ± SEM from 3-5 mice per group. Statistical analysis was performed by homoscedastic two-tailed Student’s t-test using the GraphPad Prism software v. 6.0. A p value < 0.05 was considered significant. Significant differences are indicated by *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Supplementary Material
Figure S1. Targeting brain astrocytes through hGFAP promoter and AAV serotypes, related to Figure 1. (A) Study design to analyze cell type-specificity of AAVs. IHC, immunohistochemistry; dpv, days post virus-injection. (B) Confocal images showing marker expression in the virus-injected cortex. Samples were examined at 4 dpv. Scales, 50 μm. (C) Quantifications showing cell type-specificity of the examined AAV serotypes in the cortex at 4 and 14 dpv (n = 3-4 mice per serotype per time-point). (D) Confocal images showing astrocytes-restricted reporter expression mediated by AAV2 or AAV5 at 14 dpv. Scales, 50 μm. (E) Confocal images showing variable neuronal expression of the viral reporter mediated by AAV8, AAV9, or PHP.eB in the cortex at 14 dpv. Scales, 50 μm.
Figure S2. Cell type- and region-specificity of genetically traced cells in Aldh1l1-CreERT2 mice, related to Figure 2. (A) Study design to determine Tam-induced YFP+ cells in adult mice. (B) A schematic showing the cortical region analyzed for YFP expression. (C) Confocal images of the indicated markers in the cortex. Scales, 50 μm. (D) Quantifications showing astrocyte-restricted YFP expression (n = 4 mice; mean ± SEM). (E) Quantifications showing efficient YFP-tracing of resident cortical astrocytes (n = 4 mice; mean ± SEM). (F) A schematic showing the striatal region analyzed for YFP expression. (G) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (H) Quantifications showing astrocyte-restricted YFP expression in the striatum (n = 4 mice; mean ± SEM). (I) Quantifications showing efficient YFP-tracing of resident striatal astrocytes (n = 4 mice; mean ± SEM). (J) Study design to determine Tam-induced tdTomato (tdT)+ cells in adult mice. (K) A schematic showing the cortical region analyzed for tdT expression for results on panels L-N. (L) Confocal images of the indicated markers. Scales, 50 μm. (M) Quantifications showing astrocyte-restricted tdT expression (n = 3 mice; mean ± SEM). (N) Quantifications showing efficient tdT-tracing of resident astrocytes (n = 3 mice; mean ± SEM). (O) Neuronal expression of the tdT reporter in variable cortical regions of rare Tam-treated Aldh1l1-CreERT2;R26R-tdT mice. Enlarged views of the boxed regions are shown on the bottom panels. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views. (P) Neuronal expression of the tdT reporter in variable cortical and striatal regions of rare Tam-treated Aldh1l1-CreERT2;R26R-tdT mice. Enlarged views of the boxed regions are also shown. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views.
Figure S3. AAV-mediated Cre-FLEX system fails to restrict gene expression in astrocytes, related to Figure 3. (A) Study design to determine cell type-specificity of hGFAP-Cre AAV virus. (B) A schematic representation of the GFAP promoters used in this study. (C) Confocal images of the indicated markers in the virus-injected cortex at 4 dpv. AAV5* was obtained from Addgene. Scales, 50 μm. (D) Quantifications showing Cre expression in neurons mediated by the indicated AAV serotypes (n = 4 mice per serotype per time-point). (E) Confocal images of the indicated markers in the virus-injected cortex at 14 dpv. Scales, 50 μm. (F) Study design to examine AtN conversion by the AAV-based Cre-FLEX system. (G) Confocal images of the indicated markers at 17 dpv. Scales, 50 μm. (H) Quantifications showing an unsuitability of the AAV-based Cre-FLEX system for examining AtN conversion (n = 4 mice per group; mean ± SEM; n.s., not significant).
Figure S4. Leaky expression of the AAV FLEX system, related to Figure 3. (A) The AAV5 viruses used in the experiment. (B) Study design. (C) Confocal images of the indicated markers in the cortex injected with F-mCherry or F-ND1-mCherry AAV5 virus at 98 dpv. Higher magnification views of the boxed regions are shown on the right panels. Leaky Cre-independent mCherry+ cells were detected in cortical regions. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views.
Figure S5. Cell type- and region-specificity of genetically traced cells in mGfap-Cre;R26R-YFP mice, related to Figure 4. (A) Confocal images of coronal brain sections at the core striatal level. Higher magnification views of the boxed regions are shown on the right panels. YFP is restricted to striatal astrocytes but also in neuron of variable cortical regions. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views. (B) Confocal images of coronal brain sections at the midbrain level. Higher magnification views of the boxed regions are shown on the right panels. YFP is also expressed in neurons of multiple midbrain regions. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views. (C) A schematic showing the striatal region analyzed for YFP expression. (D) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (E) Quantifications showing astrocyte-restricted YFP expression (n = 4 mice; mean ± SEM). (F) Quantifications showing highly efficient YFP-tracing of striatal astrocytes (n = 4 mice; mean ± SEM).
Figure S6. AAV-GFAP-CasRx-Ptbp1 neither efficiently downregulates PTBP1 nor converts striatal astrocytes in vivo, related to Figure 7. (A) AAVs used in the experiments. hGFAP-mCherry virus was co-injected to trace CasRx-expressing cells. (B) Study design to examine AtN conversion by PTBP1 knockdown through the CRISPR-CasRx system. Striatal astrocytes were constitutively traced with YFP. (C) Quantifications showing a lack of YFP-traced mCherry+ neurons at 60 dpv (n = 4 mice per group; mean ± SEM; *p = 0.032). (D) Confocal images of the indicated markers at 60 dpv. Scales, 50 μm. (E) Quantifications of virus-transduced cells showing few of them with knockdown of PTBP1 at 60 dpv (n = 4 mice per group; mean ± SEM; *p = 0.0332). (F) Quantifications of PTBP1 fluorescence intensity in virus-transduced cells showing low knockdown efficiency (n = 4 mice per group; mean ± SEM; **p = 0.0012). (G) Confocal images of the indicated markers showing less efficient knockdown of PTBP1 by the CRISPR-CasRx system at 60 dpv. Scales, 50 μm. (H) Quantification of the indicated markers showing that mCherry+ cells indeed expressed CasRx (indicated by the FLAG tag; n = 4 mice per group; mean ± SEM). (I) Confocal images of the indicated markers showing co-expression of mCherry and FLAG-tagged CasRx at 60 dpv. Scales, 50 μm.
Figure S7. Cre-independent and -dependent PTBP1 knockdown fails to convert striatal astrocytes in vivo, related to Figure 7. (A) Western blot analysis of PTBP1 knockdown in cultured astrocytes that were transduced with shRNA-expressing lentiviruses at 7 dpv. shRNA against luciferase was used a control. Expression of β-actin (ACTB) was used as a loading control. (B) Study design to trace AtN conversion by shRNA-mediated knockdown of PTBP1 in the striatum. Striatal astrocytes were genetically traced with YFP after Tam treatments. (C) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (D) Quantifications showing efficient shRNA-mediated PTBP1 knockdown (n= 4 mice per group; mean ± SEM; ****p < 0.0001). (E) Quantifications of shRNA-expressing cells showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; n.s., not significant). (F) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (G) Study design to trace AtN conversion by shRNA-mediated knockdown of PTBP1 in the striatum. Striatal astrocytes were constitutively traced with YFP. (H) Quantifications of shRNA-expressing cells showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; n.s., not significant). (I) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (J) Study design to trace AtN conversion by Cre-dependent shRNA-mediated knockdown of PTBP1 in the striatum. Striatal astrocytes were traced with YFP after Tam treatments. (K) Quantifications of shRNA-expressing cells showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; ****p < 0.0001). (L) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (M) Study design to trace AtN conversion by Cre-dependent shRNA-mediated knockdown of PTBP1 in the striatum. Striatal astrocytes were constitutively traced with YFP. (N) Quantifications of shRNA-expressing cells showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; ****p < 0.0001). (O) Confocal images of the indicated markers in the striatum. Scales, 50 μm.
Highlights.
NEUROD1 specifically and efficiently induces viral reporter-labeled neurons in vivo
Lineage-traced resident astrocytes are not converted into neurons by NEUROD1
NEUROD1-induced viral reporter-labeled neurons are actually endogenous neurons
Knockdown of PTBP1 fails to convert brain astrocytes into neurons in vivo
Acknowledgements
We thank Xiang-Dong Fu, Hao Qian, and members of the Zhang laboratory for discussions and reagents. C.L.Z. is a W. W. Caruth, Jr. Scholar in Biomedical Research and supported by the Welch Foundation (I-1724), the Decherd Foundation, Kent Waldrep Foundation Center for Basic Research on Nerve Growth and Regeneration, the Texas Alzheimer’s Research and Care Consortium (TARCC2020), and the NIH (NS099073, NS092616, NS111776, NS117065, and NS088095).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
REFERENCES
- Ackman JB, Siddiqi F, Walikonis RS, and LoTurco JJ (2006). Fusion of microglia with pyramidal neurons after retroviral infection. J Neurosci 26, 11413–11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker RA, Gotz M, and Parmar M (2018). New approaches for brain repair-from rescue to reprogramming. Nature 557, 329–334. [DOI] [PubMed] [Google Scholar]
- Benner EJ, Luciano D, Jo R, Abdi K, Paez-Gonzalez P, Sheng H, Warner DS, Liu C, Eroglu C, and Kuo CT (2013). Protective astrogenesis from the SVZ niche after injury is controlled by Notch modulator Thbs4. Nature 497, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulet R, Matsuda T, Zhang L, Miranda C, Giacca M, Kaspar BK, Nakashima K, and Hsieh J (2017). NEUROD1 Instructs Neuronal Conversion in Non-Reactive Astrocytes. Stem cell reports 8, 1506–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhong X, Smith DK, Tai W, Yang J, Zou Y, Wang LL, Sun J, Qin S, and Zhang CL (2019). Astrocyte-Specific Deletion of Sox2 Promotes Functional Recovery After Traumatic Brain Injury. Cereb Cortex 29, 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wernig M, Berninger B, Nakafuku M, Parmar M, and Zhang CL (2015). In Vivo Reprogramming for Brain and Spinal Cord Repair. eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Ma NX, Pei ZF, Wu Z, Do-Monte FH, Keefe S, Yellin E, Chen MS, Yin JC, Lee G, et al. (2020). A NeuroD1 AAV-Based Gene Therapy for Functional Brain Repair after Ischemic Injury through In Vivo Astrocyte-to-Neuron Conversion. Mol Ther 28, 217–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard-Despres S, Winner B, Schaubeck S, Aigner R, Vroemen M, Weidner N, Bogdahn U, Winkler J, Kuhn HG, and Aigner L (2005). Doublecortin expression levels in adult brain reflect neurogenesis. Eur J Neurosci 21, 1–14. [DOI] [PubMed] [Google Scholar]
- Ding B, Tang Y, Ma S, Akter M, Liu ML, Zang T, and Zhang CL (2021). Disease Modeling with Human Neurons Reveals LMNB1 Dysregulation Underlying DYT1 Dystonia. J Neurosci 41, 2024–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al. (2013). An optimized microRNA backbone for effective single-copy RNAi. Cell reports 5, 1704–1713. [DOI] [PubMed] [Google Scholar]
- Fischer KB, Collins HK, and Callaway EM (2019). Sources of off-target expression from recombinase-dependent AAV vectors and mitigation with cross-over insensitive ATG-out vectors. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbels S, Eisch AJ, and Hsieh J (2009). Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci 12, 1090–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande A, Sumiyoshi K, Lopez-Juarez A, Howard J, Sakthivel B, Aronow B, Campbell K, and Nakafuku M (2013). Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat Commun 4, 2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorian C, Nakashima J, Le Belle J, Ohab J, Kim R, Liu A, Smith KB, Groszer M, Garcia AD, Sofroniew MV, et al. (2009). Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J Neurosci 29, 1874–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zhang L, Wu Z, Chen Y, Wang F, and Chen G (2014). In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell stem cell 14, 188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Bergami M, Gascon S, Lepier A, Vigano F, Dimou L, Sutor B, Berninger B, and Gotz M (2014). Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem cell reports 3, 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Smith DK, Niu W, Fang S, Iqbal N, Sun G, Shi Y, and Zhang CL (2015). Enhancer Analysis Unveils Genetic Interactions between TLX and SOX2 in Neural Stem Cells and In Vivo Reprogramming. Stem cell reports 5, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Kersigo J, Pan N, and Fritzsch B (2010). Neurod1 regulates survival and formation of connections in mouse ear and brain. Cell and tissue research 341, 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Messing A, Su M, and Brenner M (2008). GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia 56, 481–493. [DOI] [PubMed] [Google Scholar]
- Liu MH, Li W, Zheng JJ, Xu YG, He Q, and Chen G (2020). Differential neuronal reprogramming induced by NeuroD1 from astrocytes in grey matter versus white matter. Neural Regen Res 15, 342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ML, Zang T, and Zhang CL (2016). Direct Lineage Reprogramming Reveals Disease-Specific Phenotypes of Motor Neurons from Human ALS Patients. Cell reports 14, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ML, Zang T, Zou Y, Chang JC, Gibson JR, Huber KM, and Zhang CL (2013). Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun 4, 2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Miao Q, Yuan J, Han S, Zhang P, Li S, Rao Z, Zhao W, Ye Q, Geng J, et al. (2015). Ascl1 Converts Dorsal Midbrain Astrocytes into Functional Neurons In Vivo. J Neurosci 35, 9336–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Zang T, Liu ML, and Zhang CL (2020). Aging-relevant human basal forebrain cholinergic neurons as a cell model for Alzheimer's disease. Mol Neurodegener 15, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki MT, Jhala US, Antonellis A, Fields L, Doria A, Orban T, Saad M, Warram JH, Montminy M, and Krolewski AS (1999). Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet 23, 323–328. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Irie T, Katsurabayashi S, Hayashi Y, Nagai T, Hamazaki N, Adefuin AMD, Miura F, Ito T, Kimura H, et al. (2019). Pioneer Factor NeuroD1 Rearranges Transcriptional and Epigenetic Profiles to Execute Microglia-Neuron Conversion. Neuron 101, 472–485 e477. [DOI] [PubMed] [Google Scholar]
- Mattugini N, Bocchi R, Scheuss V, Russo GL, Torper O, Lao CL, and Gotz M (2019). Inducing Different Neuronal Subtypes from Astrocytes in the Injured Mouse Cerebral Cortex. Neuron 103, 1086–1095 e1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Smith DK, Vue TY, Zou Y, Bachoo R, Johnson JE, and Zhang CL (2015). SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem cell reports 4, 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Wang LL, Zou Y, and Zhang CL (2018). Phenotypic Reprogramming of Striatal Neurons into Dopaminergic Neuron-like Cells in the Adult Mouse Brain. Stem cell reports 11, 1156–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, and Zhang CL (2013). In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nature cell biology 15, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, and Tiwari VK (2016). NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J 35, 24–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M, Birtele M, Shrigley S, Benitez JA, Hedlund E, Parmar M, and Ottosson DR (2017). Direct Reprogramming of Resident NG2 Glia into Neurons with Properties of Fast-Spiking Parvalbumin-Containing Interneurons. Stem cell reports 9, 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz GA, Bottes S, Betizeau M, Jorg DJ, Carta S, Simons BD, Helmchen F, and Jessberger S (2018). Live imaging of neurogenesis in the adult mouse hippocampus. Science 359, 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Kang X, Hu J, Zhang D, Liang Z, Meng F, Zhang X, Xue Y, Maimon R, Dowdy SF, et al. (2020). Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature 582, 550–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussarie JP, Yao V, Rodriguez-Rodriguez P, Oughtred R, Rust J, Plautz Z, Kasturia S, Albornoz C, Wang W, Schmidt EF, et al. (2020). Selective Neuronal Vulnerability in Alzheimer's Disease: A Network-Based Analysis. Neuron 107, 821–835 e812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, and Khakh BS (2016). New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo. Neuron 92, 1181–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Hu H, Lee Y, d'Azzo A, Messing A, and Brenner M (2004). Expression specificity of GFAP transgenes. Neurochem Res 29, 2075–2093. [DOI] [PubMed] [Google Scholar]
- Su Z, Niu W, Liu ML, Zou Y, and Zhang CL (2014a). In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat Commun 5, 3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zang T, Liu ML, Wang LL, Niu W, and Zhang CL (2014b). Reprogramming the fate of human glioma cells to impede brain tumor development. Cell Death Dis 5, e1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W, Wu W, Wang LL, Ni H, Chen C, Yang J, Zang T, Zou Y, Xu XM, and Zhang CL (2021). In vivo reprogramming of NG2 glia enables adult neurogenesis and functional recovery following spinal cord injury. Cell stem cell 28, 923–937 e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W, Xu XM, and Zhang CL (2020). Regeneration Through in vivo Cell Fate Reprogramming for Neural Repair. Front Cell Neurosci 14, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DG, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. (2016). A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O, Ottosson DR, Pereira M, Lau S, Cardoso T, Grealish S, and Parmar M (2015). In Vivo Reprogramming of Striatal NG2 Glia into Functional Neurons that Integrate into Local Host Circuitry. Cell reports 12, 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronova A, and Baltimore D (1990). Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci U S A 87, 4722–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-L, Garcia CS, Zhong X, Ma S, and Zhang C-L (2020). Rapid and efficient in vivo astrocyte-to-neuron conversion with regional identity and connectivity? bioRxiv, 2020.2008.2016.253195. [Google Scholar]
- Wang LL, Su Z, Tai W, Zou Y, Xu XM, and Zhang CL (2016). The p53 Pathway Controls SOX2-Mediated Reprogramming in the Adult Mouse Spinal Cord. Cell reports 17, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, and Zhang CL (2018). Engineering new neurons: in vivo reprogramming in mammalian brain and spinal cord. Cell and tissue research 371, 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pei Z, Hossain A, Bai Y, and Chen G (2021). Transcription factor-based gene therapy to treat glioblastoma through direct neuronal conversion. Cancer Biol Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Parry M, Hou XY, Liu MH, Wang H, Cain R, Pei ZF, Chen YC, Guo ZY, Abhijeet S, et al. (2020). Gene therapy conversion of striatal astrocytes into GABAergic neurons in mouse models of Huntington's disease. Nat Commun 11, 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Xu L, Liu M, Wang Q, Li W, Lei W, and Chen G (2021). Lineage tracing of direct astrocyte-to-neuron conversion in the mouse cortex. Neural Regen Res 16, 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhou W, Zhang Y, Zidon T, Ritchie T, and Engelhardt JF (1999). Concatamerization of adeno-associated virus circular genomes occurs through intermolecular recombination. J Virol 73, 9468–9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yin JC, Yeh H, Ma NX, Lee G, Chen XA, Wang Y, Lin L, Chen L, Jin P, et al. (2015). Small Molecules Efficiently Reprogram Human Astroglial Cells into Functional Neurons. Cell stem cell 17, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, and Gage FH (2008). Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660. [DOI] [PubMed] [Google Scholar]
- Zhou H, Su J, Hu X, Zhou C, Li H, Chen Z, Xiao Q, Wang B, Wu W, Sun Y, et al. (2020). Glia-to-Neuron Conversion by CRISPR-CasRx Alleviates Symptoms of Neurological Disease in Mice. Cell 181, 590–603 e516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Targeting brain astrocytes through hGFAP promoter and AAV serotypes, related to Figure 1. (A) Study design to analyze cell type-specificity of AAVs. IHC, immunohistochemistry; dpv, days post virus-injection. (B) Confocal images showing marker expression in the virus-injected cortex. Samples were examined at 4 dpv. Scales, 50 μm. (C) Quantifications showing cell type-specificity of the examined AAV serotypes in the cortex at 4 and 14 dpv (n = 3-4 mice per serotype per time-point). (D) Confocal images showing astrocytes-restricted reporter expression mediated by AAV2 or AAV5 at 14 dpv. Scales, 50 μm. (E) Confocal images showing variable neuronal expression of the viral reporter mediated by AAV8, AAV9, or PHP.eB in the cortex at 14 dpv. Scales, 50 μm.
Figure S2. Cell type- and region-specificity of genetically traced cells in Aldh1l1-CreERT2 mice, related to Figure 2. (A) Study design to determine Tam-induced YFP+ cells in adult mice. (B) A schematic showing the cortical region analyzed for YFP expression. (C) Confocal images of the indicated markers in the cortex. Scales, 50 μm. (D) Quantifications showing astrocyte-restricted YFP expression (n = 4 mice; mean ± SEM). (E) Quantifications showing efficient YFP-tracing of resident cortical astrocytes (n = 4 mice; mean ± SEM). (F) A schematic showing the striatal region analyzed for YFP expression. (G) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (H) Quantifications showing astrocyte-restricted YFP expression in the striatum (n = 4 mice; mean ± SEM). (I) Quantifications showing efficient YFP-tracing of resident striatal astrocytes (n = 4 mice; mean ± SEM). (J) Study design to determine Tam-induced tdTomato (tdT)+ cells in adult mice. (K) A schematic showing the cortical region analyzed for tdT expression for results on panels L-N. (L) Confocal images of the indicated markers. Scales, 50 μm. (M) Quantifications showing astrocyte-restricted tdT expression (n = 3 mice; mean ± SEM). (N) Quantifications showing efficient tdT-tracing of resident astrocytes (n = 3 mice; mean ± SEM). (O) Neuronal expression of the tdT reporter in variable cortical regions of rare Tam-treated Aldh1l1-CreERT2;R26R-tdT mice. Enlarged views of the boxed regions are shown on the bottom panels. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views. (P) Neuronal expression of the tdT reporter in variable cortical and striatal regions of rare Tam-treated Aldh1l1-CreERT2;R26R-tdT mice. Enlarged views of the boxed regions are also shown. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views.
Figure S3. AAV-mediated Cre-FLEX system fails to restrict gene expression in astrocytes, related to Figure 3. (A) Study design to determine cell type-specificity of hGFAP-Cre AAV virus. (B) A schematic representation of the GFAP promoters used in this study. (C) Confocal images of the indicated markers in the virus-injected cortex at 4 dpv. AAV5* was obtained from Addgene. Scales, 50 μm. (D) Quantifications showing Cre expression in neurons mediated by the indicated AAV serotypes (n = 4 mice per serotype per time-point). (E) Confocal images of the indicated markers in the virus-injected cortex at 14 dpv. Scales, 50 μm. (F) Study design to examine AtN conversion by the AAV-based Cre-FLEX system. (G) Confocal images of the indicated markers at 17 dpv. Scales, 50 μm. (H) Quantifications showing an unsuitability of the AAV-based Cre-FLEX system for examining AtN conversion (n = 4 mice per group; mean ± SEM; n.s., not significant).
Figure S4. Leaky expression of the AAV FLEX system, related to Figure 3. (A) The AAV5 viruses used in the experiment. (B) Study design. (C) Confocal images of the indicated markers in the cortex injected with F-mCherry or F-ND1-mCherry AAV5 virus at 98 dpv. Higher magnification views of the boxed regions are shown on the right panels. Leaky Cre-independent mCherry+ cells were detected in cortical regions. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views.
Figure S5. Cell type- and region-specificity of genetically traced cells in mGfap-Cre;R26R-YFP mice, related to Figure 4. (A) Confocal images of coronal brain sections at the core striatal level. Higher magnification views of the boxed regions are shown on the right panels. YFP is restricted to striatal astrocytes but also in neuron of variable cortical regions. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views. (B) Confocal images of coronal brain sections at the midbrain level. Higher magnification views of the boxed regions are shown on the right panels. YFP is also expressed in neurons of multiple midbrain regions. Scales, 500 μm for lower magnification views and 50 μm for higher magnification views. (C) A schematic showing the striatal region analyzed for YFP expression. (D) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (E) Quantifications showing astrocyte-restricted YFP expression (n = 4 mice; mean ± SEM). (F) Quantifications showing highly efficient YFP-tracing of striatal astrocytes (n = 4 mice; mean ± SEM).
Figure S6. AAV-GFAP-CasRx-Ptbp1 neither efficiently downregulates PTBP1 nor converts striatal astrocytes in vivo, related to Figure 7. (A) AAVs used in the experiments. hGFAP-mCherry virus was co-injected to trace CasRx-expressing cells. (B) Study design to examine AtN conversion by PTBP1 knockdown through the CRISPR-CasRx system. Striatal astrocytes were constitutively traced with YFP. (C) Quantifications showing a lack of YFP-traced mCherry+ neurons at 60 dpv (n = 4 mice per group; mean ± SEM; *p = 0.032). (D) Confocal images of the indicated markers at 60 dpv. Scales, 50 μm. (E) Quantifications of virus-transduced cells showing few of them with knockdown of PTBP1 at 60 dpv (n = 4 mice per group; mean ± SEM; *p = 0.0332). (F) Quantifications of PTBP1 fluorescence intensity in virus-transduced cells showing low knockdown efficiency (n = 4 mice per group; mean ± SEM; **p = 0.0012). (G) Confocal images of the indicated markers showing less efficient knockdown of PTBP1 by the CRISPR-CasRx system at 60 dpv. Scales, 50 μm. (H) Quantification of the indicated markers showing that mCherry+ cells indeed expressed CasRx (indicated by the FLAG tag; n = 4 mice per group; mean ± SEM). (I) Confocal images of the indicated markers showing co-expression of mCherry and FLAG-tagged CasRx at 60 dpv. Scales, 50 μm.
Figure S7. Cre-independent and -dependent PTBP1 knockdown fails to convert striatal astrocytes in vivo, related to Figure 7. (A) Western blot analysis of PTBP1 knockdown in cultured astrocytes that were transduced with shRNA-expressing lentiviruses at 7 dpv. shRNA against luciferase was used a control. Expression of β-actin (ACTB) was used as a loading control. (B) Study design to trace AtN conversion by shRNA-mediated knockdown of PTBP1 in the striatum. Striatal astrocytes were genetically traced with YFP after Tam treatments. (C) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (D) Quantifications showing efficient shRNA-mediated PTBP1 knockdown (n= 4 mice per group; mean ± SEM; ****p < 0.0001). (E) Quantifications of shRNA-expressing cells showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; n.s., not significant). (F) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (G) Study design to trace AtN conversion by shRNA-mediated knockdown of PTBP1 in the striatum. Striatal astrocytes were constitutively traced with YFP. (H) Quantifications of shRNA-expressing cells showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; n.s., not significant). (I) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (J) Study design to trace AtN conversion by Cre-dependent shRNA-mediated knockdown of PTBP1 in the striatum. Striatal astrocytes were traced with YFP after Tam treatments. (K) Quantifications of shRNA-expressing cells showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; ****p < 0.0001). (L) Confocal images of the indicated markers in the striatum. Scales, 50 μm. (M) Study design to trace AtN conversion by Cre-dependent shRNA-mediated knockdown of PTBP1 in the striatum. Striatal astrocytes were constitutively traced with YFP. (N) Quantifications of shRNA-expressing cells showing a lack of YFP-traced mCherry+ neurons (n = 4 mice per group; mean ± SEM; ****p < 0.0001). (O) Confocal images of the indicated markers in the striatum. Scales, 50 μm.
Data Availability Statement
Microscopy data reported in this paper will be shared by the lead contact upon request.
All software were commercially or freely available and are listed in the STAR methods description and Key resources table.
Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-ALDH1L1 | NeuroMab | Cat# 75-140; RRID: AB_10673448 |
| Goat polyclonal anti-ALDOC (Aldolase C) | Santa Cruz Biotechnology | Cat# sc-12065; RRID: AB_2242641 |
| Rat monoclonal anti-BrdU | BIO-RAD | Cat# OBT0030; RRID: AB_609568 |
| Rabbit polyclonal anti-Cre | Covance | Cat# PRB-1061C-200; RRID: AB_11220031 |
| Goat polyclonal anti-DCX | Santa Cruz Biotechnology | Cat# sc-8066; RRID: AB_2088494 |
| Rabbit polyclonal anti-FLAG | Sigma-Aldrich | Cat# F7425; RRID: AB_439687 |
| Mouse monoclonal anti-GFAP | Sigma-Aldrich | Cat# G3893; RRID: AB_477010 |
| Chicken polyclonal anti-GFAP | Abcam | Cat# ab4674; RRID: AB_304558 |
| Chicken polyclonal anti-GFP | Aves Labs | Cat# GFP-1020; RRID: AB_10000240 |
| Chicken polyclonal anti-HA | Aves Labs | ET-HA100; RRID: AB_2313511 |
| Mouse monoclonal anti-MAP2 | Proteintech | Cat# 67015-1-Ig; RRID: AB_2882331 |
| Goat polyclonal anti-tdTomato (mCherry) | MyBioSource | Cat# MBS448092; RRID: AB_2827808 |
| Rabbit monoclonal anti-NeuN | Abcam | Cat# ab177487; RRID: AB_2532109 |
| Rabbit recombinant monoclonal anti-NEUROD1 | Abcam | Cat# ab109224; RRID: AB_10861489 |
| Rabbit polyclonal anti-PTBP1 | ABclonal | Cat# A6107; RRID: AB_2766748 |
| Mouse monoclonal anti-ACTB | Proteintech | Cat# 66009-1; RRID: AB_2687938 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202; RRID: AB_141607 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | Cat# 703-545-155; RRID: AB_2340375 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11006; RRID: AB_2534074 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21432; RRID: AB_2535853 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID: AB_2536183 |
| Hoechst 33342 | Thermo Fisher Scientific | REF H3570 |
| HRP-conjugated anti-rabbit IgG | Jackson Laboratory | Cat# 111-035-144; RRID: AB_2307391 |
| HRP-conjugated anti-mouse IgG | Jackson Laboratory | Cat# 715-035-150; RRID: AB_2340770 |
| Bacterial and virus strains | ||
| AAV5-hGFAP*-Cre virus | Addgene | Cat# 105550-AAV5; RRID: Addgene_105550 |
| AAV2*-CMV-LSL-RFP | Fu Laboratory | N/A |
| AAV2*-CMV-LSL-RFP-shPtbp1 | Fu Laboratory | N/A |
| Biological samples | ||
| Mouse brain tissue | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Cayman Chemical | CAS: 10540-29-1 |
| BrdU | Alfa Aesar Chemical TFS | CAS: 59-14-3 |
| N-2 | Gibco | Cat# 17502-048 |
| B-27 | Gibco | Cat# 17504-044 |
| Forskolin | Selleckchem | Cat# S2449 |
| Dorsomorphin | Sigma-Aldrich | Cat# 171260 |
| Polybrene (Hexadimethrime bromide) | Sigma-Aldrich | Cat# H9268 |
| Matrigel | Corning | Cat# CB-40230 |
| Gelatin | BIO-RAD | Cat# 1706537 |
| Papain | Worthington Biochemical Corporation | Cat# LS003126 |
| DNase | Invitrogene | Cat# 18068-015 |
| Nuclease (Benzonase) | Chem Cruz | Cat# sc-202391A |
| Poly ethylene glycol (PEG) | Sigma | Cat# 81268 |
| Iodixanol gradient | Alere Technologies | Prod. No. 1114542 |
| Protease inhibitor cocktail | Thermo Scientific Pierce | Cat# A32965 |
| Western Chemiluminescent HRP substrate | Millipore | Cat# WBKLS0500 |
| Critical commercial assays | ||
| none | ||
| Deposited data | ||
| none | ||
| Experimental models: Cell lines | ||
| U251 glioma cells | Sigma | Cat# 09063001 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Mouse: R26R-YFP: B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | The Jackson Laboratory | JAX: 006148; RRID: IMSR_JAX:006148 |
| Mouse: R26R-tdTomato: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007914; RRID: IMSR_JAX:007914 |
| Mouse: Aldh1l1-CreERT2: B6;FVB-Tg(Aldh1l1-cre/ERT2)1Khakh/J | The Jackson Laboratory | JAX: 029655; RRID: IMSR_JAX: 029655 |
| Mouse: mGfap-Cre line 77.6: B6.Cg-Tg(Gfap-cre)77.6Mvs/2J | The Jackson Laboratory | JAX: 024098; RRID: IMSR_JAX: 024098 |
| Oligonucleotides | ||
| ITR primer: forward: 5-GGAACCCCTAGTGATGGAGTT-3 | Sigma-Aldrich | Customized |
| ITR primer: reverse: 5-CGGCCTCAGTGAGCGA-3 | Sigma-Aldrich | Customized |
| Recombinant DNA | ||
| pAAV-hGFAP-GFP | This paper | N/A |
| pAAV-hGFAP-Cre | This paper | N/A |
| pAAV-hGFAP-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-PAX6-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-MYC-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROG2-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-ASCL1-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-BDNF-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-SOX2-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-OCT4-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-KLF4-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-R111L-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-K139D-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-N252-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-N157-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-N100-252-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-NEUROD1-156C-T2A-mCherry | This paper | N/A |
| pAAV-hGFAP-mCherry-shLuc | This paper | N/A |
| pAAV-hGFAP-mCherry-shPtbp1 | This paper | N/A |
| pAAV-CAG-GFP | Addgene | Cat# 37825; RRID:Addgene_37825 |
| pAAV-CAG-FLEX-GFP | Addgene | Cat# 28304; RRID:Addgene_28304 |
| pAAV-CAG-FLEX-mCherry | This paper | N/A |
| pAAV-CAG-FLEX-NEUROD1-T2A-mCherry | This paper | N/A |
| pAAV-CAG-FLEX-HA-DLX2-T2A-mCherry | This paper | N/A |
| Ai9 (to clone LSL) | Addgene | Cat# 22799; RRID:Addgene_22799 |
| pAAV-CAG-LSL-mCherry-shLuc | This paper | N/A |
| pAAV-CAG-LSL-mCherry-shPtbp1 | This paper | N/A |
| pAAV-GFAP-CasRx | Addgene | Cat# 154000; RRID:Addgene_154000 |
| pAAV-GFAP-CasRx-Ptbp1 | Addgene | Cat# 154001; RRID:Addgene_154001 |
| pAAV-CMV-LSL-RFP | Fu Laboratory | N/A |
| pAAV-CMV-LSL-RFP-shPtbp1 | Fu Laboratory | N/A |
| pAd-deltaF6 | Addgene | Cat# 112867; RRID:Addgene_112867 |
| helper pAAV2/2 | Addgene | Cat# 104963; RRID:Addgene_104963 |
| helper pAAV2/5 | Addgene | Cat# 104964; RRID:Addgene_104964 |
| helper pAAV2/8 | Addgene | Cat# 112864; RRID:Addgene_112864 |
| helper pAAV2/9 | Addgene | Cat# 112865; RRID:Addgene_112865 |
| pUCmini-iCAP-PHP.eB | Addgene | Cat# 103005; RRID:Addgene_103005 |
| rAAV2-retro | Addgene | Cat# 81070; RRID:Addgene_81070 |
| pCSC-IRES-GFP | This paper | N/A |
| pCSC-NEUROD1-IRES-GFP | This paper | N/A |
| pCSC-NEUROD1-R111L-IRES-GFP | This paper | N/A |
| pCSC-NEUROD1-K139D-IRES-GFP | This paper | N/A |
| pLV-hGFAP-mCherry-shPtbp1 | This paper | N/A |
| pLV-hGFAP-GFP-shLuc | This paper | N/A |
| pLV-hGFAP-GFP | This paper | N/A |
| pMDL (pMDLg-pRRE) | Addgene | Cat# 12251; RRID:Addgene_12251 |
| VSV-G (pMD2.G) | Addgene | Cat# 12259; RRID:Addgene_12259 |
| pREV (pRSV-Rev) | Addgene | Cat# 12253; RRID:Addgene_12253 |
| Software and algorithms | ||
| ImageJ | NIH | RRID:SCR_003070, https://imagej.net/Fiji |
| Graphpad Prism | Graphpad | RRID:SCR_002798), https://www.graphpad.com |
| ZEN | Zeiss | RRID:SCR_013672, https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| ChemiDoc iMP Imaging System | BioRad | RRID:SCR_019037, https://www.bio-rad.com/en-in/product/chemidoc-mp-imaging-system?ID=NINJ8ZE8Z |
| NIS-Elements C | Nikon | RRID:SCR_020317, https://www.microscope.healthcare.nikon.com/about/news/nikon-instruments-a1-confocal-laser-microscope-series-with-nis-elements-c-software-delivers-fully-integrated-comprehensive-confocal-imaging-capabilities |
| Adobe Photoshop | Adobe | RRID:SCR_014199, https://www.adobe.com/products/photoshop.html |
| Adobe Illustrator | Adobe | RRID:SCR_010279, https://www.adobe.com/products/illustrator.html |
| Other | ||
| 33 gauge, 45-degree-beveled needle | Hamilton, Reno, NV | Cat# 22033 (Customized) |
| Impact One™ Stereotaxic CCI Instrument | Leica | Model 39463920 |
| 100K PES concentrator (Pierce™) | Thermo Fisher | Cat# 88524 |
| 40-μm nylon strainer | Falcon | REF: 352340 |
| Sliding microtome | Leica | Model SM200R |