Abstract
Yes-associated protein 1 (YAP1), a key player in the Hippo pathway, has been shown to play a critical role in tumor progression. However, the role of YAP1 in prostate cancer cell invasion, migration, and metastasis is not well defined. Through functional, transcriptomic, epigenomic, and proteomic analyses, we showed that prolyl hydroxylation of YAP1 plays a critical role in the suppression of cell migration, invasion, and metastasis in prostate cancer. Knockdown (KD) or knockout (KO) of YAP1 led to an increase in cell migration, invasion, and metastasis in prostate cancer cells. Microarray analysis showed that the EMT pathway was activated in Yap1-KD cells. ChIP-seq analysis showed that YAP1 target genes are enriched in pathways regulating cell migration. Mass spectrometry analysis identified P4H prolyl hydroxylase in the YAP1 complex and YAP1 was hydroxylated at multiple proline residues. Proline-to-alanine mutations of YAP1 isoform 3 identified proline 174 as a critical residue, and its hydroxylation suppressed cell migration, invasion, and metastasis. KO of P4ha2 led to an increase in cell migration and invasion, which was reversed upon Yap1 KD. Our study identified a novel regulatory mechanism of YAP1 by which P4HA2-dependent prolyl hydroxylation of YAP1 determines its transcriptional activities and its function in prostate cancer metastasis.
Keywords: YAP1, prolyl hydroxylation, P4A2, prostate cancer, cell migration, invasion, metastasis
INTRODUCTION
Yes-associated protein 1 (YAP1), a key transcriptional coactivator in the Hippo pathway, is an important driver in cancer development and progression (1). Although YAP1 plays an oncogenic role in various cancer types, multiple studies also support a tumor-suppressive function for YAP1 in head and neck (2), breast (3–5), hematological (6), and colorectal (7, 8) cancers. Thus, the functions of YAP1 are likely context-dependent (9). YAP1 was shown to be overexpressed in prostate adenocarcinoma (PCa) and associated with cell proliferation and invasiveness in castration-resistant prostate cancer (CRPC) (10–12). However, YAP1 was found to be downregulated in the highly aggressive NEPC subset (10). Importantly, YAP1 deletion (heterozygous and homozygous) and mutation were observed in ~3.6% of prostate cancers and was strongly associated with metastasis (Supplementary Fig. 1A, B & Supplementary Tables 1–3). On the contrary, deletion/mutation of TAZ, another transcriptional coactivator in the Hippo pathway, was not significantly different between the primary and metastatic PCa (Supplementary Fig. 1B). On the cellular level, YAP1 promotes prostate cancer cell proliferation through cell-autonomous and non-autonomous mechanisms (11–14), but its role in prostate cancer metastasis is not clearly defined.
Post-translational modification of YAP1, such as phosphorylation, has also been shown to regulate YAP1 cellular localization, stability, and activities (15, 16). Interestingly, we found that YAP1 proteins in prostate cancer cells are modified by proline hydroxylation, an important post-translational modification that modulates protein folding and stability in mammalian cells (17, 18). Proline hydroxylation is induced by prolyl hydroxylases, such as prolyl hydroxylase domain proteins (PHD) and collagen prolyl 4-hydroxylase (P4H). Whether YAP1 is subjected to proline hydroxylation was previously unknown, as were the effects of such modification on YAP1 function. In this study, we identified a surprising role for YAP1 in the suppression of cell migration, invasion, and metastasis in prostate, pancreatic, and breast cancers. We found that YAP1 interacts with the P4H complex and is hydroxylated at multiple proline residues. The status of proline hydroxylation of YAP1 determines its oncogenic activity in regulating cell migration, invasion, and metastasis in prostate cancer and possibly in other cancer cell types.
RESULTS
YAP1 suppresses cancer cell migration, invasion, and metastasis
Previously, we showed that YAP1 was highly expressed in primary tumors from the metastatic Pten/Smad4 prostate conditional knockout (KO) model (12). We first examined the effect of Yap1 knockdown (KD) on cell migration, invasion, and metastasis in a highly metastatic Pten/Smad4-deficient CRPC cell line (referred to as PS cells hereafter) (19). Surprisingly, Yap1 KD and Yap1 KO led to a significant increase in cell migration and invasion (Fig. 1A–C and Supplementary Fig. 2A). Of note, Yap1 KD or KO did not have a significant effect on cell proliferation as measured by the total number of cells at the end of the assays (data not shown). Furthermore, re-expression of human YAP1 in Yap1-KO cells suppressed cell migration and invasion (Fig. 1D and Supplementary Fig. 2B). YAP1-KD in C4-2b, IGR-CaP1, and PC3 cells similarly increased migration and invasion (Fig. 1E–G and Supplementary Fig. 2C). However, YAP1 KD in DU145 led to a decrease in cell migration (Supplementary Fig. 2D). Also, we examined whether YAP1 also suppressed cell migration in other metastatic cancers in which YAP1 has been implicated to play an important role in tumor progression (20–24). We found that Yap1 KO or KD led to increased cell migration in iKPC mouse pancreatic cancer cells (25) and MDA-MB-231 human breast cancer cells (Fig. 1H, I) but not in SYO-1 synovial sarcoma cells (Supplementary Fig. 2E). Moreover, Yap1 KD in PS cells promoted lung metastasis (Fig. 1J). Taken together, our data suggest that YAP1 suppresses cell migration, invasion, and metastasis in multiple cancer types.
Figure 1. YAP1 suppresses cell migration, invasion, and metastasis.
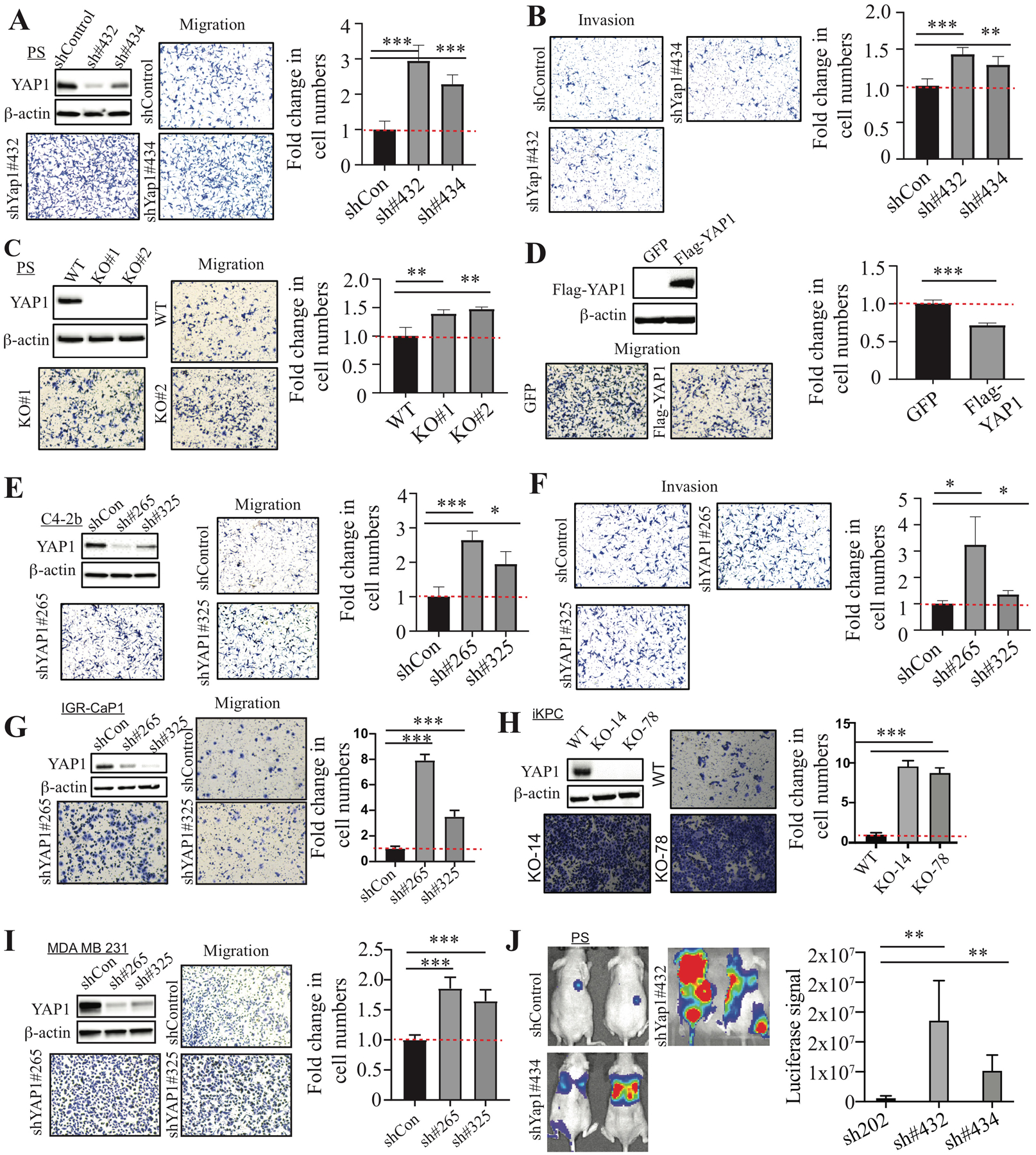
(A, B) Cell migration and invasion assay using PS cells transduced with control shRNA and Yap1 shRNAs. The Yap1 KD efficiency was confirmed by WB analysis. (C) Cell migration assay using Yap1-WT PS cells and Yap1-KO cells. WB analysis confirmed the KO of YAP1 expression. (D) Cell migration assay using Yap1-KO cells with GFP overexpression and YAP1 overexpression. WB analysis confirmed the overexpression of YAP1 in Yap1-KO PS cells. (E, F) Cell migration and invasion assay using C4-2b cells transduced with control shRNA and YAP1 shRNAs. The YAP1 KD efficiency was confirmed by WB analysis. (G–I) Cell migration assay using IGR-CaP1 (G), iKPC (H), and MDA-MB-231 (I) with YAP1 KD or KO compared to control cells. (J) Luciferase imaging in mice injected with PS cells transduced with control shRNA and Yap1 shRNAs through the tail vein.
YAP1 interacts with the prolyl 4-hydroxylase complex, and its proline residues are hydroxylated
To understand the mechanisms by which YAP1 suppresses cell migration, invasion, and metastasis, we performed microarray analysis of RNA isolated from Yap1-KD and control PS cells (Supplementary Table 4). As expected, gene set enrichment analysis (GSEA) (26) identified epithelial to mesenchymal transition (EMT) as the top pathway activated in Yap1-KD cells (Fig. 2A). We also confirmed that the expression of several EMT genes, including Postn, Cdh11, Acta2, Rgs4, and Mgp, were upregulated upon Yap1 KD (Supplementary Fig. 3A).
Figure 2. Microarray, ChIP-seq, and immunoprecipitation-mass spectrometry analyses.
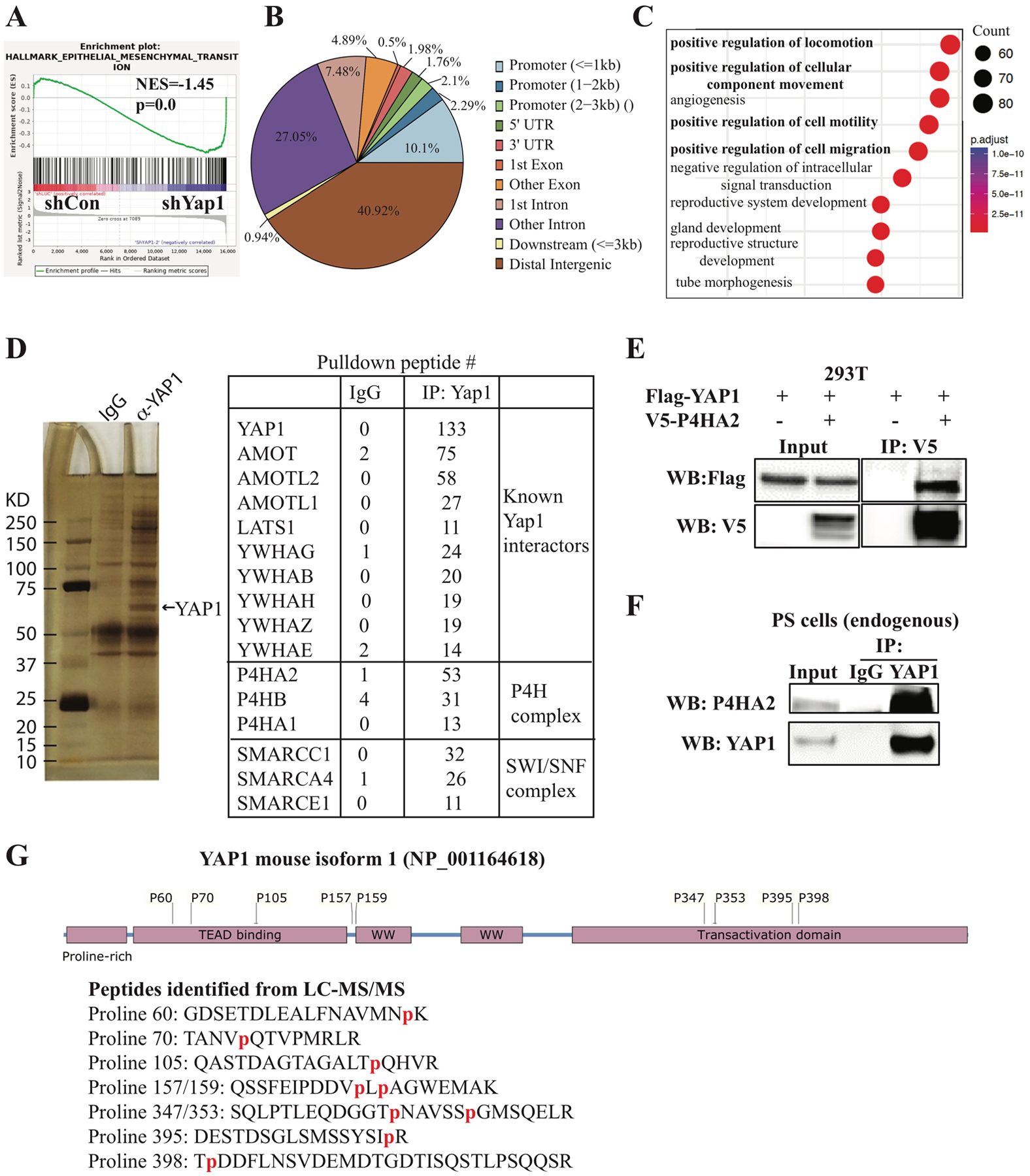
(A) GSEA analysis of microarray data from PS cells transduced with doxycycline-inducible Yap1 shRNA identified EMT as the top pathway activated in Yap1-KD cells. (B, C) ChIP-seq analysis identified YAP1 binding sites and YAP1-regulated pathways. (D) Immunoprecipitation-mass spectrometry analysis identified known YAP1-interacting proteins and novel YAP1-interacting proteins. (E) Exogenous YAP1 interacts with exogenous P4HA2 when overexpressed in 293T cells by transfection of the indicated plasmids for co-immunoprecipitation experiments. (F) Endogenous YAP1 interacts with endogenous P4HA2 in PS cells. (G) Multiple prolyl hydroxylation sites were identified in peptides of YAP1 isoform 3 from the LC- MS/MS analysis.
Since YAP1 acts as a transcriptional coactivator, we sought to determine whether these upregulated EMT genes are direct target genes of YAP1 using ChIP-seq in PS cells. We found that YAP1 binds mostly to the distant intergenic region and other introns (Fig. 2B), which is consistent with previous reports (27). Pathway analyses of the top 2000 YAP1 binding sites using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes identified multiple pathways related to cell migration as top pathways and the expected “Hippo signaling pathway” (Fig. 2C and Supplementary Fig. 3B). Motif analysis using Homer (28) identified TEAD1 and AP1 motifs (Supplementary Fig. 3C and data not shown), which is consistent with the known physical and functional interaction between YAP1, TEAD, and AP1 (27). Importantly, we found that 194 upregulated genes and 247 downregulated genes in Yap1 KD cells were among the top 6000 YAP1-target genes predicted by Cistrome-GO (29) (Supplementary Tables 5 and 7). Among these genes, YAP1 binds to the distant intergenic region upstream or downstream of several genes upregulated in Yap1-KD cells (e.g., Postn, Cdh11, Acta2, Rgs4, and Mgp) (Supplementary Fig. 3D and data not shown), suggesting that YAP1 directly represses their expression. In addition, we confirmed the binding of YAP1 to its known target genes, including Cxcl5 and Ccnd1 (Supplementary Fig. 3E).
Since the functions of YAP1 are regulated through its interacting partners, as well as by post-translational modifications (15, 16), we performed immunoprecipitation (IP) of YAP1 in PS cells followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify YAP1-interacting proteins and novel post-translational modifications (Fig. 2D). As expected, we identified multiple proteins previously shown to interact with YAP1, such as AMOT and the SWI/SNF complex (Fig. 2D). Interestingly, proteins of P4H complex (P4HA1, P4HA2, and P4HB) were identified among the top YAP1-interacting proteins (Fig. 2D). On the contrary, prolyl 3-hydroxylase was not identified in the IP-MS (data not shown). Collagen P4H, an α2β2 tetrameric complex, specifically catalyzes 4-hydroxylation of proline (17, 18) through its catalytic α subunit (P4HA). Because P4HA2 was the most abundant protein of the P4H complex pulled down by YAP1, we focused on its interaction with YAP1. We showed that overexpressed Flag-YAP1 efficiently pulled down overexpressed P4HA2 in 293T cells (Fig. 2E). Also, endogenous YAP1 was found to interact with P4HA2 in PS cells (Fig. 2F).
Give the proline hydroxylase activity of the P4H complex (17, 18), we examined whether proline residues in YAP1 were hydroxylated. We identified nine hydroxylated proline residues (proline 60, 70, 105, 157, 159, 347, 353, 395, 398) in mouse YAP1, eight of which were evolutionarily conserved between mouse and human (Fig. 2G and Supplementary Fig. 4A, B), suggesting that they might play a role in regulating YAP1 functions.
Hydroxylation at proline 174 of YAP1 plays a critical role in suppressing cell migration, invasion, and metastasis
Given the critical regulatory roles of proline hydroxylation in proteins (17, 18), we decided to examine whether proline hydroxylation modulates YAP1 functions. We first generated hydroxylation-defective human YAP1 mutants by mutating proline to alanine (Mut1-3: P75/85/120A; Mut4-5: P172/174A; Mut6-9: P348/352/394/397A) (Fig. 3A and Supplementary Fig. 4B). These YAP1 mutants were overexpressed in Yap1-KO PS cells to avoid the possible interference of the endogenous wild type (WT) YAP1. The expression of all the YAP1 mutants was similar but higher than the WT (Fig. 3B). We did not observe any significant difference in cell growth in vitro and in vivo (Supplementary Fig. 5A, B) between YAP1 WT and mutants. However, we found that there is an increase in cell migration and invasion in Mut4-5–overexpressing cells compared to WT-overexpressing cells (Fig. 3C, D), suggesting that mutations of proline 172 and 174 to alanine abolished the activity of YAP1 in suppressing cell migration and invasion. Consistent with the in vitro findings, PS cells with YAP1 Mut4-5 overexpression increased the colonization of cancer cells in the lung compared to WT, Mut1-3, and Mut6-9 (Fig. 3E and Supplementary Fig. 5C).
Figure 3. Prolyl hydroxylation of YAP1 suppressed cell migration, invasion, and metastases.
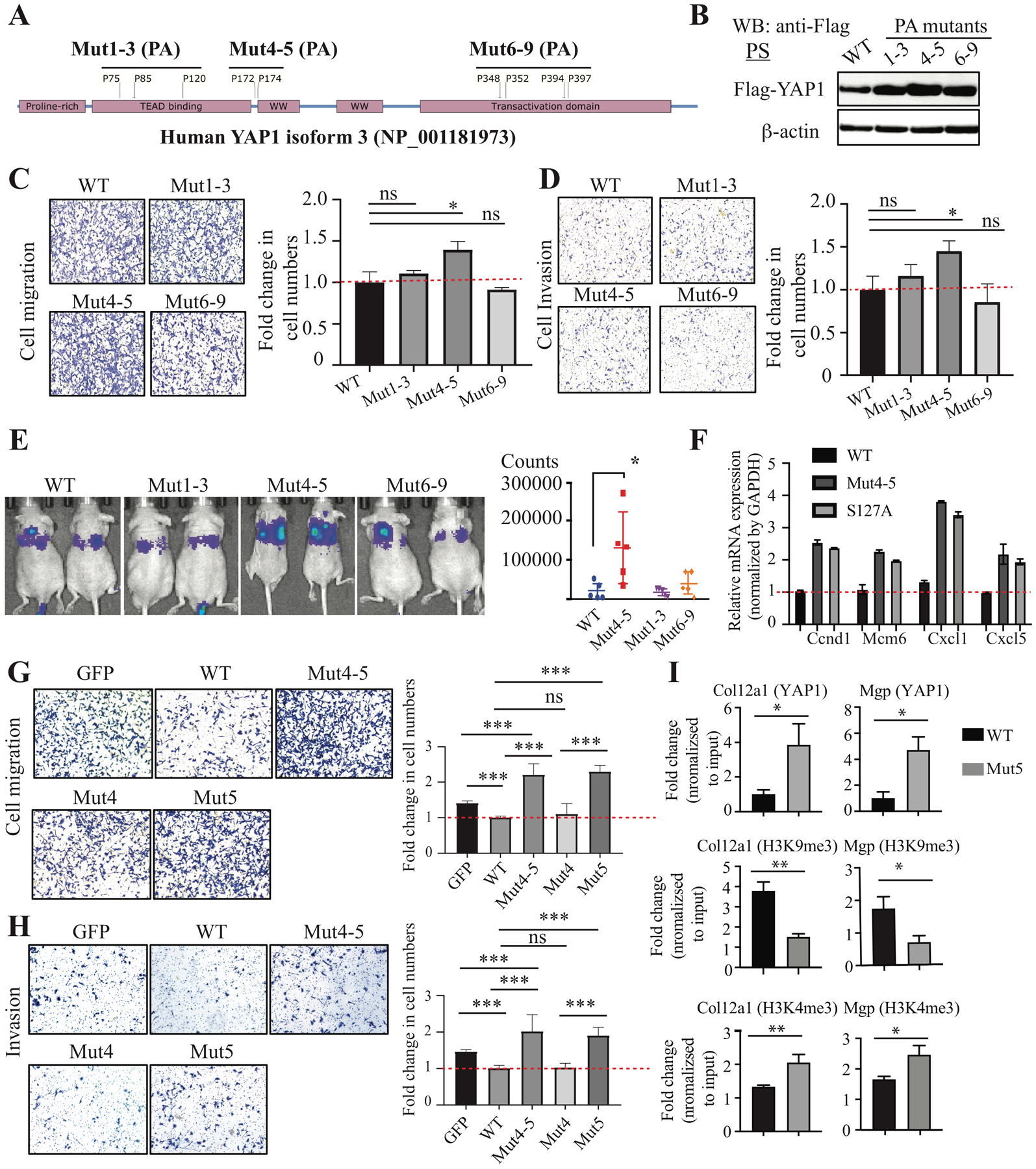
(A) Scheme showing the strategy to generate prolyl hydroxylation–defective YAP1 mutants by mutating proline to alanine (PA): Mut1-3 (P75/85/120A), Mut4-5 (P172/174A), and Mut6-9 (P348/352/394/397). Human YAP1 isoform 3 was used. (B) Expression of YAP1 WT and PA mutants in Yap1-KO PS cells. (C, D) Cell migration and invasion assay in Yap1-KO PS cells with overexpression of YAP1 WT and PA mutants. (E) Tail vein injection of Yap1-KO cells with overexpression of YAP1 WT and PA mutants. (F) qPCR analysis of YAP1 target genes in Yap1-KO PS cells with overexpression of YAP1 WT, Mut4-5, and the constitutively active S127A mutant. (G, H) Cell migration and invasion assay using Yap1-KO PS cells with overexpression of GFP, YAP1 WT, Mut4-5, Mut4, and Mut5. (I) ChIP-qPCR analysis of YAP1, H3K4me3, and H3K9me4 binding sites in Col12a1 and Mgp.
To determine whether prolyl hydroxylation controls the transcriptional activities of YAP1, we examined the expression of YAP1 target genes Ccnd1, Mcm6, Cxcl1, and Cxcl5 in Yap1-KO cells that overexpressed YAP1 WT, Mut4-5, or the constitutively active S127A mutant (30). Both Mut4-5 and the S127A mutant dramatically increased the expression of these genes compared to the YAP1 WT (Fig. 3F and Supplementary Fig. 5D), suggesting that the non-hydroxylated YAP1 is more transcriptionally active than the hydroxylated YAP1.
To further pinpoint which proline residue of YAP1 is critical for its function in suppressing cell migration and invasion, we generated site-specific proline-to-alanine mutants of human YAP1 (P172A and P174A mutants), which corresponded to proline 157 and 159 in mouse YAP1. We overexpressed GFP control, YAP1 WT, and YAP1 mutants (Mut4-5: P172/174A; Mut4: P172A; Mut5: P174A) in Yap1-KO cells (Supplementary Fig. 5E) and examined their effects on cell migration and invasion. We found that both Mut4-5 and Mut5, but not Mut4, dramatically increased cell migration and invasion compared to the GFP control (Fig. 3G, H), suggesting proline 174 is the critical hydroxylation site that regulates YAP1 activity in cell migration and invasion. Importantly, overexpression of Mut4-5 and Mut5 similarly increased the cell migration of PC3 and TRAMPC2 cells (Supplementary Fig. 5F). We then performed ChIP-qPCR to determine whether the increased expression of YAP1 target genes in Mut4-5 expressing cells is due to increased YAP1 binding to chromatin. We found that Mut5 binding to the promoter/distal enhancers of its target genes was significantly increased compared to WT (Fig. 3I and Supplementary Fig. 5G). To examine the transcriptional activation and repression of YAP1 target genes, we examined H3K9me3, a mark associated with transcriptional repression (31), and H3K4me3, a hallmark of active chromatin enriched at active promoters and correlates with transcriptional activity (32), in the promoters/enhancers of YAP1 target genes. We found that H3K9me3 was significantly decreased and H3K4me4 was significantly increased in the regulatory region of several YAP1 target genes (e.g., Col12a1, Mgp, Postn, Cxcl12) (Fig. 3I and Supplementary Fig. 5G). Taken together, our data suggest that hydroxylation at proline 174 of YAP1 plays a critical role in regulating cell migration, invasion, and metastasis by repressing the expression of a subset of its target genes.
Loss of P4HA2 promotes cell migration and invasion through YAP1
Given the observed hydroxylation of YAP1 in PS cells (Fig. 2G) and the effect of hydroxylation-defective YAP1 mutants on migration and invasion (Fig. 3C–H and Supplementary Fig. 5F), the ability of YAP1 to suppress cell migration and invasion appeared to be regulated by the P4H complex. We examined the effect of P4ha2 KO on cell migration and invasion and found that P4ha2 KO significantly increased cell migration and invasion compared to WT cells (Fig. 4A–C), which was not due to an increase in cell proliferation (data not shown). Interestingly, we found that YAP1 target genes Postn, Col12a1, Mgp, Ccl5, and Cxcl12 were significantly upregulated in P4ha2-KO cells compared to control cells (Fig. 4D and data not shown), suggesting YAP1 is transcriptionally more active upon loss of P4HA2. Importantly, KD of Yap1 in P4ha2-KO cells abolished the effect of P4ha2 KO on cell migration and invasion (Fig. 4E, F and Supplementary Fig. 5H). Taken together, our data indicate that P4HA2 suppresses cell migration and invasion through proline hydroxylation of YAP1.
Figure 4. P4HA2 suppresses cell migration and invasion through Yap1.
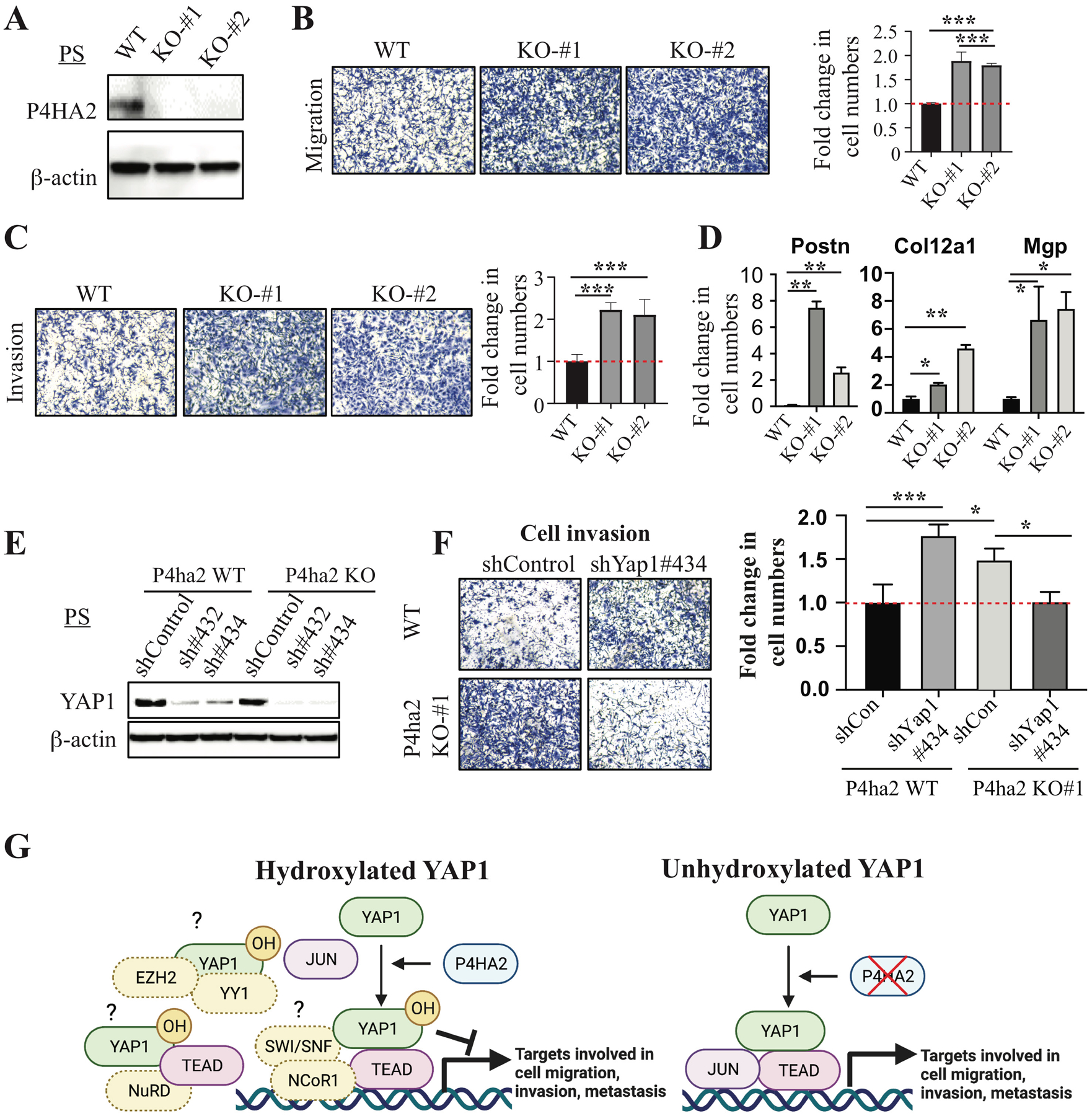
(A) WB analysis of P4HA2 in P4ha2-WT and P4ha2-KO PS cells. (B, C) Cell migration and invasion using P4ha2-WT and P4ha2-KO PS cells. (D) qPCR analysis of YAP1 target genes (Postn, Col12a1, and Mgp) in P4ha2 KO and WT cells. (E) WB analysis of YAP1 in P4ha2-WT and P4ha2-KO PS cells transduced with Yap1 shRNAs. (F) Cell invasion assay in P4ha2-WT and P4ha2-KO PS cells transduced with shYap1#434. (G) A model for hydroxylation-dependent YAP1 function in cell migration, invasion, and metastasis (Created with BioRender.com). Left: P4HA2-mediated hydroxylation of YAP1 may impair its interactions with transcription factors such as JUN or enhance the recruitment of corepressor, such as SWI/SNF-NCoR1, NuRD, and EZH2/YY1, which results in a decrease in the expression of genes involved in cell migration, invasion, and metastasis. Right: In the absence of P4HA2, non-hydroxylated YAP1 may efficiently interact with transcription factors such as JUN to activate genes involved in cell migration, invasion, and metastasis.
DISCUSSION
In contrary to previous findings that KD of YAP1 in LNCaP-C4–2 cells impaired cell migration and invasion, we demonstrated that KD or KO of Yap1 in mouse (PS and TRAMPC2) and human (C4-2b, IGR-CaP1, and PC3) prostate cancer cells led to enhanced cell migration, invasion, and metastasis. The discrepancy between our study and the previous one is not clear. Our data also showed that YAP1 KD in DU145 cells suppressed cell migration whereas YAP1 KD/KO promoted cell migration in iKPC cells and MDA-MB-231 cells. These findings strongly suggest that YAP1 plays a context-dependent function in cell migration, invasion, and metastasis. Further studies are necessary to define molecular basis underlying the context-dependent functions of YAP1 in cell migration, invasion, and metastasis. Importantly, the clinical significance of YAP1 loss in PCa patients was supported by the strong association of YAP1 deletion with metastatic PCa and the loss of YAP1 protein in advanced PCa (10, 12). Loss of YAP1 function via post-translational modification by proline hydroxylation will be also an important mechanism of clinical significance. Furthermore, our unpublished data showed that TAZ similarly suppresses cell migration and regulates a common set of genes as YAP1, suggesting functional redundancy between YAP1 and TAZ.
Mechanistically, our data suggest that prolyl hydroxylation plays an important role in the regulation of YAP1 activities, which can both activate and repress transcription (Fig. 4G). In P4HA2 WT cells, YAP1 may suppress gene expression through its interaction with SWI/SNF complex, which was identified as YAP1-interating proteins in our study and has been shown to regulate both activation and repression of the same promoters (33, 34), in part through corepressor NCoR1 (35). Also, YAP1 may recruit YY1 and EZH2 (36) or recruit the NuRD complex to suppress the expression of its target genes (37). Interestingly, proline 174 is within the first WW domain of YAP1, which is crucial for the transcriptional activities of YAP1 through its interaction with transcription factors that contain PPxY motifs (38). Thus, our findings suggest that prolyl hydroxylation at P174 of YAP1 may impair its interaction with key transcription [e.g., c-JUN (27), TEAD], resulting in a decrease in both binding to its target gene and reduced transcriptional activation. This notion is supported by our findings that YAP1-Mut5 binds more efficiently to its target genes. YAP1 hydroxylation may also be required for the efficient recruitment of the transcription corepressor complexes (e.g., NCoR1, NuRD), as our data showed that YAP1 P174A OE increased H3K4me3 and reduced H3K9me3 for a subset of YAP1 target genes. Thus, our data suggest that P174A YAP1 mutant not only has increased binding to its target genes, but also induces a switch from repressive transcription to active transcription for a subset of genes. Further studies are needed to delineate the effect of prolyl hydroxylation of YAP1 on the dynamics of the epigenomic landscape. YAP1 is known to be regulated by phosphorylation at multiple serine residues (39). Our studies shed light on a new aspect of YAP1 regulation, which may also be involved in many YAP1-mediated cellular activities yet to be identified.
The P4H α subunit (P4HA) has three isoforms (P4HA1–3) in mammalian cells (18). Since P4HA1 and P4HA2 were both identified as YAP1-interacting proteins in our MS analysis, P4HA1 may also suppress cell migration, invasion, and metastasis through prolyl hydroxylation of YAP1. However, P4HA1 was previously shown to promote prostate cancer progression (40), and further studies are needed to clarify the role of P4HA1 in regulating YAP1 functions and cell migration, invasion, and metastasis. Since LS-MS mass spectrometry analysis cannot distinguish 3-prolyl hydroxylation from 4-prolyl hydroxylation, we cannot rule out the presence of 3-hydroxyl proline in YAP1. Given that P3H1 was not identified as YAP1-interacting proteins, the high specificity of the P4H and P3H towards prolyl hydroxylation strongly indicates that the hydroxyproline identified in YAP1 is 4-hydroxyproline. Future experiments combining liquid chromatography retention time differences with mass spectrometry using ETD-HCD fragmentation, complemented by ab initio calculations, are needed to address this issue (41).
Although collagen deposition is generally associated with tumor progression and invasive behavior (42), it also plays a tumor-suppressive role (43). P4HA1 is the major isoenzyme in most cells, and P4ha1−/− leads to embryonic lethality in mice due to abnormal deposition of collagen IV (44). In contrast, P4ha2−/− mice had no apparent abnormalities (45). Given that we did not observe any significant difference in the expression of P4HA1 between P4ha2 WT and KO cells (Supplementary Fig. 5I), collagen deposition in P4ha2 KO/KD cells may not be significantly impacted. Given the role of P4HA2 in promoting secretion and deposition of collagen and cell invasion in breast cancer (46, 47), the seemly contradictory findings on the differential effect of P4ha2 KO/KD and collagen deposition on cell migration may warrant further studies. Furthermore, due to the lack of an antibody that can specifically recognize the hydroxylated proline 174 of YAP1, we cannot assess the clinical relevance of prolyl hydroxylation of YAP1 in tumor samples from prostate cancer patients. The development of such antibodies is warranted and would allow us to examine the association of prolyl hydroxylation of YAP1 and P4HA2 expression in prostate cancer specimens.
In summary, our findings support a model in which P4HA2-mediated prolyl hydroxylation serves as a molecular switch that controls the activities of YAP1 in cell migration, invasion, and metastasis (Fig. 4G).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ronald DePinho and Haoqiang Ying for helpful discussions, Sarah E. Townsend for editing, and Dr Timothy C. Thompson for providing RM-1 cells. GW is supported by the funding from MDACC (Moon Shot, IRG, and PCRP), UT STARs Award, NIH R00 CA194289, and P50 CA140388. SHL is supported by grants from the NIH R01 CA174798 (S.-H. Lin, L.-y. Yu-Lee), NIH 5 P50 CA140388 (C. Logothetis, S.-H. Lin), and Cancer Prevention Research Institute of Texas grants RP150179 & RP190252 (S.-H. Lin and L.-Y. Yu-Lee). This study is supported by NIH P30 CA016672 for the use of Research Animal Support Facility, Flow Cytometry and Cellular Imaging Core Facility, and Functional Genomics Core.
Footnotes
Conflict of interest: C. J. Logothetis reports receiving commercial research grants from Bayer, Sanofi, Janssen, Astellas Pharma, Pfizer; and honoraria from Bayer, Janssen, Sanofi, Astellas Pharma. No potential conflicts of interest were disclosed by the other authors.
MATERIALS AND METHODS
The reagents and assays as well as bioinformatic/statistical analyses were described in Supplementary Information.
REFERENCES
- 1.Calses PC, Crawford JJ, Lill JR, Dey A. Hippo Pathway in Cancer: Aberrant Regulation and Therapeutic Opportunities. Trends Cancer. 2019;5(5):297–307. [DOI] [PubMed] [Google Scholar]
- 2.Ehsanian R, Brown M, Lu H, Yang XP, Pattatheyil A, Yan B, et al. YAP dysregulation by phosphorylation or DeltaNp63-mediated gene repression promotes proliferation, survival and migration in head and neck cancer subsets. Oncogene. 2010;29(46):6160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008;15(11):1752–9. [DOI] [PubMed] [Google Scholar]
- 4.Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di GH, et al. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res. 2013;19(6):1389–99. [DOI] [PubMed] [Google Scholar]
- 5.Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27(6):962–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014;20(6):599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 2007;14(4):743–51. [DOI] [PubMed] [Google Scholar]
- 8.Cheung P, Xiol J, Dill MT, Yuan WC, Panero R, Roper J, et al. Regenerative Reprogramming of the Intestinal Stem Cell State via Hippo Signaling Suppresses Metastatic Colorectal Cancer. Cell Stem Cell. 2020;27(4):590–604 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou C, Sun Z, Li S, Li G, Li X, Ma J. Dual roles of yes-associated protein (YAP) in colorectal cancer. Oncotarget. 2017;8(43):75727–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng S, Prieto-Dominguez N, Yang S, Connelly ZM, StPierre S, Rushing B, et al. The expression of YAP1 is increased in high-grade prostatic adenocarcinoma but is reduced in neuroendocrine prostate cancer. Prostate Cancer Prostatic Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen LT, Tretiakova MS, Silvis MR, Lucas J, Klezovitch O, Coleman I, et al. ERG Activates the YAP1 Transcriptional Program and Induces the Development of Age-Related Prostate Tumors. Cancer Cell. 2015;27(6):797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, et al. Targeting YAP-Dependent MDSC Infiltration Impairs Tumor Progression. Cancer Discov. 2016;6(1):80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Yang S, Chen X, Stauffer S, Yu F, Lele SM, et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol Cell Biol. 2015;35(8):1350–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuser-Abali G, Alptekin A, Lewis M, Garraway IP, Cinar B. YAP1 and AR interactions contribute to the switch from androgen-dependent to castration-resistant growth in prostate cancer. Nat Commun. 2015;6:8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YA, Lu CY, Cheng TY, Pan SH, Chen HF, Chang NS. WW Domain-Containing Proteins YAP and TAZ in the Hippo Pathway as Key Regulators in Stemness Maintenance, Tissue Homeostasis, and Tumorigenesis. Front Oncol. 2019;9:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan F, Qian M, He Q, Zhu H, Yang B. The posttranslational modifications of Hippo-YAP pathway in cancer. Biochim Biophys Acta Gen Subj. 2020;1864(1):129397. [DOI] [PubMed] [Google Scholar]
- 17.Zurlo G, Guo J, Takada M, Wei W, Zhang Q. New Insights into Protein Hydroxylation and Its Important Role in Human Diseases. Biochim Biophys Acta. 2016;1866(2):208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorres KL, Raines RT. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol. 2010;45(2):106–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su W, Han HH, Wang Y, Zhang B, Zhou B, Cheng Y, et al. The Polycomb Repressor Complex 1 Drives Double-Negative Prostate Cancer Metastasis by Coordinating Stemness and Immune Suppression. Cancer Cell. 2019;36(2):139–55 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158(1):185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158(1):171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiemer SE, Szymaniak AD, Varelas X. The transcriptional regulators TAZ and YAP direct transforming growth factor beta-induced tumorigenic phenotypes in breast cancer cells. J Biol Chem. 2014;289(19):13461–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim T, Yang SJ, Hwang D, Song J, Kim M, Kyum Kim S, et al. A basal-like breast cancer-specific role for SRF-IL6 in YAP-induced cancer stemness. Nat Commun. 2015;6:10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isfort I, Cyra M, Elges S, Kailayangiri S, Altvater B, Rossig C, et al. SS18-SSX-Dependent YAP/TAZ Signaling in Synovial Sarcoma. Clin Cancer Res. 2019;25(12):3718–31. [DOI] [PubMed] [Google Scholar]
- 25.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17(9):1218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Wan C, Zheng R, Fan J, Dong X, Meyer CA, et al. Cistrome-GO: a web server for functional enrichment analysis of transcription factor ChIP-seq peaks. Nucleic Acids Res. 2019;47(W1):W206–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker JS, Nicetto D, Zaret KS. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet. 2016;32(1):29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30(20):4198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, Chambers KJ, Faller DV, Wang S. Reprogramming of the SWI/SNF complex for co-activation or co-repression in prohibitin-mediated estrogen receptor regulation. Oncogene. 2007;26(50):7153–7. [DOI] [PubMed] [Google Scholar]
- 34.Nagl NG Jr., Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26(3):752–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275(51):40463–70. [DOI] [PubMed] [Google Scholar]
- 36.Hoxha S, Shepard A, Troutman S, Diao H, Doherty JR, Janiszewska M, et al. YAP-Mediated Recruitment of YY1 and EZH2 Represses Transcription of Key Cell-Cycle Regulators. Cancer Res. 2020;80(12):2512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M, Kim T, Johnson RL, Lim DS. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 2015;11(2):270–82. [DOI] [PubMed] [Google Scholar]
- 38.Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69(3):1089–98. [DOI] [PubMed] [Google Scholar]
- 39.Ma S, Meng Z, Chen R, Guan KL. The Hippo Pathway: Biology and Pathophysiology. Annu Rev Biochem. 2019;88:577–604. [DOI] [PubMed] [Google Scholar]
- 40.Chakravarthi BV, Pathi SS, Goswami MT, Cieslik M, Zheng H, Nallasivam S, et al. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5(16):6654–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Huizen NA, Burgers PC, Saintmont F, Brocorens P, Gerbaux P, Stingl C, et al. Identification of 4-Hydroxyproline at the Xaa Position in Collagen by Mass Spectrometry. J Proteome Res. 2019;18(5):2045–51. [DOI] [PubMed] [Google Scholar]
- 42.Discher DE, Smith L, Cho S, Colasurdo M, Garcia AJ, Safran S. Matrix Mechanosensing: From Scaling Concepts in ‘Omics Data to Mechanisms in the Nucleus, Regeneration, and Cancer. Annu Rev Biophys. 2017;46:295–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivirikko KI, et al. Loss of assembly of the main basement membrane collagen, type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J Biol Chem. 2007;282(4):2512–9. [DOI] [PubMed] [Google Scholar]
- 45.Aro E, Salo AM, Khatri R, Finnila M, Miinalainen I, Sormunen R, et al. Severe Extracellular Matrix Abnormalities and Chondrodysplasia in Mice Lacking Collagen Prolyl 4-Hydroxylase Isoenzyme II in Combination with a Reduced Amount of Isoenzyme I. J Biol Chem. 2015;290(27):16964–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong G, Deng L, Zhu J, Rychahou PG, Xu R. Prolyl-4-hydroxylase alpha subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer. 2014;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilkes DM, Chaturvedi P, Bajpai S, Wong CC, Wei H, Pitcairn S, et al. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73(11):3285–96. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


