Abstract
Integrase-defective lentiviral vectors (IDLVs) represent an attractive platform for vaccine development as a result of the ability to induce persistent humoral- and cellular-mediated immune responses against the encoded transgene. Compared with the parental integrating vector, the main advantages for using IDLV are the reduced hazard of insertional mutagenesis and the decreased risk for vector mobilization by wild-type viruses. Here we report on the development and use in the mouse immunogenicity model of simian immunodeficiency virus (SIV)-based IDLV containing a long deletion in the U3 region and with the 3′ polypurine tract (PPT) removed from the transfer vector for improving safety and/or efficacy. Results show that a safer extended deletion of U3 sequences did not modify integrase-mediated or -independent integration efficiency. Interestingly, 3′ PPT deletion impaired integrase-mediated integration but did not reduce illegitimate, integrase-independent integration efficiency, contrary to what was previously reported in the HIV system. Importantly, although the extended deletion in the U3 did not affect expression or immunogenicity from IDLV, deletion of 3′ PPT considerably reduced both expression and immunogenicity of IDLV.
Keywords: lentiviral vector, integrase, vaccine, immunization, human immunodeficiency virus, simian immunodeficiency virus, HIV, SIV, episomal DNA, LTR
Graphical abstract
Integrase-defective lentiviral vectors (IDLVs) are replication defective and do not integrate in the host genome. We designed a new set of IDLVs to implement the efficiency and safety profile. New modified IDLVs express the immunogen from the vector’s episomal forms and induce persistent humoral and cellular immune responses.
Introduction
Development of safe and effective gene transfer systems is critical to the success of gene therapy and vaccination protocols for human diseases, and an ideal vectored vaccine should maximize immune response (both cellular and humoral) without affecting safety. Virus-based vectors are natural vehicles of genetic information, and among them lentivirus-based vectors are widely exploited for in vivo gene delivery because of their ability to efficiently transduce dividing and non-dividing cells,1 transfer large antigens (up to 8 kb),2 and establish persistent long-lasting immune responses,3,4 along with a low potential for genotoxicity and their absence of preexisting anti-vector immunity.5, 6, 7 The development of self-inactivating (SIN) lentiviral vectors (LVs) reduced the risk for mobilization of the vector genome,8,9 although transcriptional regulatory elements into the 5′ untranslated region may activate cryptic promoters.10,11 The further development of integrase-defective lentiviral vectors (IDLVs) greatly decreased the risk for insertional mutagenesis and improved LV safety, while maintaining a good expression profile, thus making them more appealing, particularly for immunization purposes.12 In the absence of a functional integrase (IN) protein, IDLVs are unable to integrate in the host genome, and they produce unintegrated extrachromosomal DNA (E-DNA), which express viral proteins. Furthermore, the E-DNA persists in vitro and in vivo in non-dividing cells.13,14 IDLVs are currently under evaluation for gene therapy and vaccines in preclinical and clinical studies.15 In this context, we and others showed that IDLVs elicited durable humoral and cellular responses after a single immunization.12,16 In particular, our work is focused on the use of SIN simian immunodeficiency virus (SIV)-based IDLV for immunization. The use of SIV-based IDLV for immunization purposes may be more advantageous because they showed a more favorable expression profile compared with human immunodeficiency virus (HIV)-based IDLVs in primary human and simian dendritic cells, thus increasing their ability to act as functional antigen-presenting cells.17 In addition, SIV-based IDLVs are not based on a human pathogen, and therefore recombination events between SIV and HIV sequences, including cross-packaging, are less likely to occur.
Previous work using HIV-based vectors showed that a large U3 deletion in the long terminal repeat (LTR) of the lentiviral transfer vector increased expression from IDLVs, and that 3′ polypurine tract (PPT)-deleted IDLVs significantly reduced IN-independent illegitimate integration.18, 19, 20 However, these modifications were not tested in the context of SIV-based IDLVs. In this report, we developed SIV-based transfer vectors with the aim of enhancing safety and expression profiles of SIV-based IDLVs by introducing deletions that were previously evaluated in the HIV-based system. Deletions in the SIV-based IDLVs were evaluated separately or were combined in double mutants to assess cumulative effects.
In the present report, we show that SIV-based IDLVs have a high safety profile with low mobilization from both episomal and integrated templates. Also, expression profile and immunogenicity, using codon-optimized HIV-1JR-FL glycoprotein 120 (gp120) envelope as model antigen for immunization in BALB/c mice, were not affected by the removal of additional U3 sequence. Conversely, introduction of 3′ PPT deletion in the IDLVs reduced its expression profile and consequent immunogenicity. Taken together, these results indicate that although extended deletion in the U3 region should be considered for improving SIV-based IDLV design, 3′ PPT deletions are detrimental in the context of SIV-based IDLV expression and immunization.
Results
PPT deletion impairs expression efficiency of SIV-based lentiviral vectors
The basic features of the SIV-based transfer vectors used in this study are shown in Figure S1. Parental pGAE SIN SIV-based transfer vector contains a 149-base pair (bp) deletion in the enhancer/promoter U3 region (deleted from −20 to −169).21 pGAEdU3 transfer vector contains a longer 423-bp deletion in the modulatory U3 region (deleted from –20 to –453). Both pGAE and pGAEdU3 transfer vectors were deleted in the 3′ PPT (21-bp deletion) to produce pGAEdPPT and pGAEdU3dPPT transfer vectors, respectively (Figure S2).
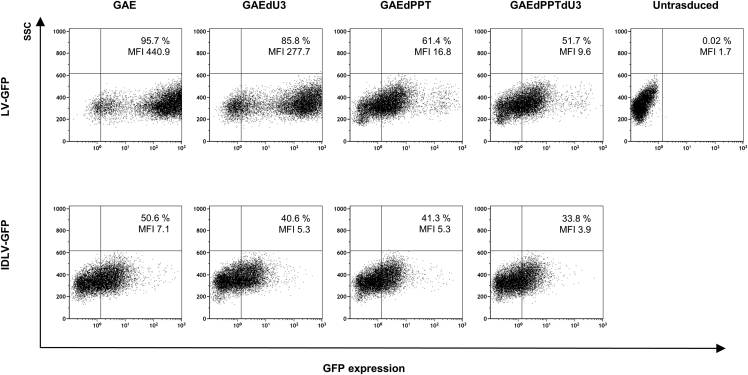
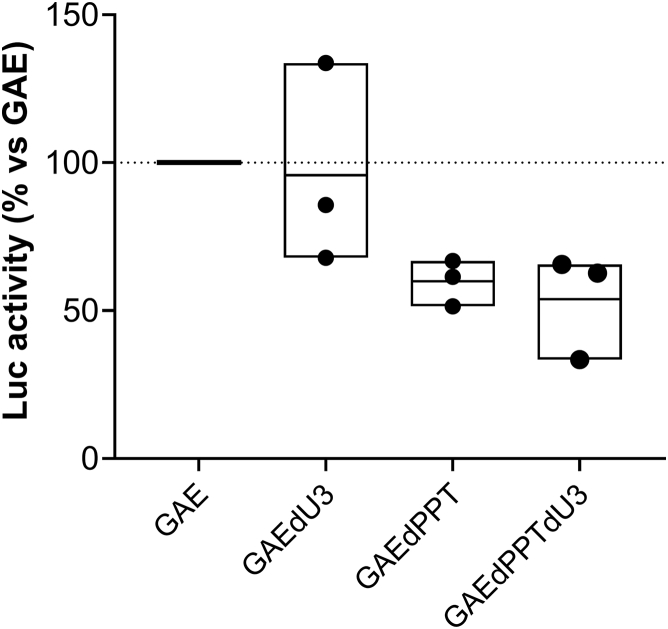
To compare transduction efficiency and gene expression of the different SIV-based vectors, we produced IN-competent LVs and IDLVs delivering reporter green fluorescent protein (GFP) as described in the Materials and methods. All vectors were released at comparable levels, as measured by recovery of reverse transcriptase (RT) activity in the supernatant of 293T Lenti-X packaging cells (data not shown), confirming that the introduced mutations did not significantly alter virus assembly or release. Subsequently, we performed fluorescence-activated cell sorting (FACS) analysis on 293T Lenti-X cells transduced with normalized amounts of GFP-expressing vectors (3 × 105 RT counts/105 cells/well; Figure 1). Concerning the IN-competent LV, a large U3 deletion (LV-GAEdU3-GFP) did not affect transgene expression compared with parental LV-GAE-GFP, consistent with previously reported deletion in HIV-based integrating vectors.18 Conversely, deletion of 3′ PPT greatly decreased the GFP mean fluorescence intensity (MFI) expression profile from both GAEdPPT and GAEdPPTdU3 IN-competent vectors, regardless of U3 deletion (Figure 1). This is similar to what has been shown using the HIV-based system, where the 3′ PPT deletion decreased the levels of linear vector forms, substrate of integration, and enhanced the formation of 1-LTR circles.19 Because the episomal forms of the lentiviral vectors express at lower levels than the integrated form,19 this leads to decreased GFP MFI expression profiles in both 3′ PPT-deleted IN-competent lentiviral vectors compared with the parental vector containing the wild-type 3′ PPT. Concerning parental and mutated IDLVs expressing GFP, we did not detect measurable differences by FACS analyses (Figure 1, bottom). To better assess differences in expression activity from parental and mutated IDLVs, we evaluated luciferase activity on 293T Lenti-X cells transduced with an equal amount of all IDLVs expressing the luciferase transgene (Figure 2). Results indicated that although a large U3 deletion (IDLV-GAEdU3) did not affect luciferase expression compared with parental IDLV-GAE, deletion of 3′ PPT decreased luciferase expression from both IDLV-GAEdPPT and IDLV-GAEdPPTdU3, suggesting that altering the reverse transcription process by 3′ PPT deletion in SIV-based vectors negatively affects production of IDLV templates and consequent expression.
Figure 1.
FACS analysis of GFP expression in 293T Lenti-X cells transduced with SIV-based lentiviral vectors
Cells were transduced with equal amounts (3 × 105 RT counts) of integrating LVs (top) or IDLVs (bottom) delivering GFP. GAE, parental self-inactivating (SIN) SIV-based transfer vector; GAEdPPT, mutant transfer vector with 3′ PPT deleted; GAEdPPTdU3, double mutant with an extended 423-bp U3 deletion and with 3′ PPT deleted; GAEdU3, mutant transfer vector with an extended 423-bp U3 deletion; GFP, green fluorescent protein; MFI, mean fluorescence intensity.
Figure 2.
Luciferase activity from 293T Lenti-X cells transduced with parental and mutated integrase-defective lentiviral vectors delivering luciferase
Cells were transduced with an equal amount (1.5 × 104 RT counts) of IDLVs delivering luciferase. Data are expressed as percentage of luciferase activity compared with the corresponding parental control IDLV-GAE-Luc (100% luciferase activity). Mean with range of three independent experiments is shown. GAE, parental SIN SIV-based transfer vector; GAEdPPT, mutant transfer vector with 3′ PPT deleted; GAEdPPTdU3, double mutant with an extended 423-bp U3 deletion and with 3′ PPT deleted; GAEdU3, mutant transfer vector with an extended 423-bp U3 deletion.
3′ PPT, but not extended U3 deletion, in the SIV-based transfer vector impairs IN-mediated integration
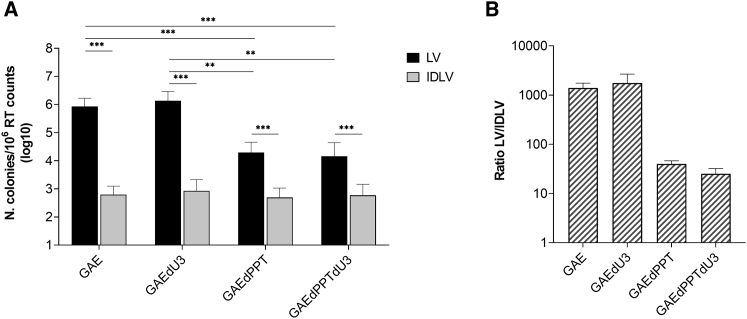
In previous work, we showed that SIV-based IDLVs integrated over 3 logs less frequently than the parental IN-competent vector.22 This is in line with what has been generally reported in the context of HIV-based vectors, where HIV-based IDLVs integrated over 2–3 logs less frequently than the parental IN-competent vector.19,20,23,24 To evaluate the integration activity of the SIV-based U3 and 3′ PPT-deleted mutants, we produced and used recombinant integrative LVs and IDLVs expressing the neomycin resistance (Tn/neoR) gene to transduce human HeLa cells, which were subsequently screened with geneticin in a colony formation assay.
As shown in Figure 3A, the longer U3 deletion did not significantly modify integration frequency of the IN-competent LVs (black bars: GAE versus GAEdU3 and GAEdPPT versus GAEdPPTdU3). Conversely, 3′ PPT-deleted LVs exhibited impaired IN-mediated integration, as shown by a significant reduction in colonies formation with respect to parental LV with wild-type 3′ PPT (Figure 3A, black bars: GAE versus GAEdPPT and GAEdU3 versus GAEdPPTdU3), regardless of the U3 modification. This is in line with reported integration frequencies using HIV-based 3′ PPT-deleted LVs,19,20 but significantly higher than those obtained using the respective non-integrating vectors (Figure 3A, gray bars). Indeed, compared with each parental LV, any tested IDLV showed a sharp decrease in the number of recovered colonies. In particular, as shown in Figure 3B, IDLVs with a long U3 deletion (GAEdU3) showed a reduction in integration frequencies similar to the parental IDLV GAE vector (IDLV fold average reduction versus respective LVs of 1,750 and 1,400, respectively). IDLV GAEdPPT single 3′ PPT mutant and IDLV GAEdPPTdU3 double mutant behaved similarly, integrating an average of 40 and 25 times less frequently than their respective IN-competent LV counterparts (Figure 3B). These data confirm reported data showing impaired integration efficiency of 3′ PPT-deleted IN-competent LVs, while a long U3 deletion had no effect on integration of both LVs and IDLVs.18, 19, 20 Interestingly, the SIV-based 3′ PPT-deleted IDLV GAEdPPT and IDLV GAEdPPTdU3 showed only a marginal 1.5-fold reduction in integration activity with respect to parental IDLV GAE and IDLV GAEdU3, respectively. This is in contrast with published data using HIV-based 3′ PPT-deleted IDLVs, where reported integration frequencies of 3′ PPT-deleted IDLVs were decreased by 3-fold compared with IDLVs containing 3′ PPT.19,20
Figure 3.
DNA recombination frequencies of SIV-based IN-competent and IN-defective lentiviral vectors
(A) Average geneticin-resistant colony formation titer assay results in transduced HeLa cells comparing the frequency of integration among integrase-competent (LV, black bars) and integrase-defective (IDLV, gray bars) lentiviral vector. (B) Ratio between the number of colonies of LV- and IDLV-transduced HeLa cells from (A). GAE, parental lentiviral vectors; GAEdPPT, vectors with a 3′-PPT deletion; GAEdPPTdU3, vectors with a 3′-PPT deletion and a large U3 deletion; GAEdU3, vectors with a large U3 deletion. y axis in (A) represents number of colonies/106 RT counts of vector supernatant, while in (B), it represents the LV/IDLV ratio among colonies. The error bars indicate the standard deviation among n = 4 independent experiments, and p values are shown for intergroup comparisons. ∗∗p < 0.01, ∗∗∗p < 0.001.
Low level of mobilization from vector episomal forms
Previous work has shown that U3 deletions leading to self-inactivation of LV significantly reduced vector mobilization from integrated templates.11,25 Conversely, circular episomal forms of HIV-based SIN-LV supported mobilization of vector templates and subsequent production of high vector titers.25,26
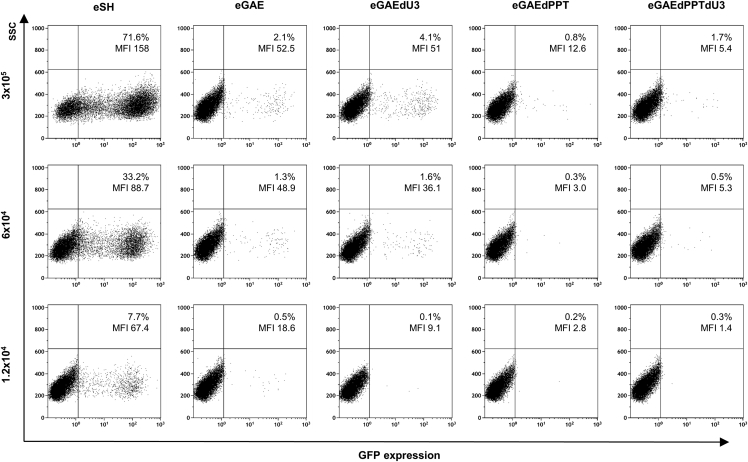
To evaluate mobilization from circular templates resembling the episomal forms of SIV-based IDLV, we produced episomal transfer vectors containing a single LTR and evaluated their ability to support vector production. The basic features of these vectors are shown in Figure S3. Episomal transfer vectors (eGAEs) mimic expression of episomal forms produced after IDLV infection and contain the same mutations described above in the pGAEs vectors, including a short or long U3 deletion (eGAE and eGAEdU3, respectively) or a deletion in the 3′ PPT region (eGAEdPPT) in addition to a long U3 deletion (eGAEdPPTdU3). All episomal transfer vectors include a GFP marker, the Tn/neoR gene (GIN), and elements that allow replication in bacteria (ampicillin resistance [AO] and E. coli replication origin [OriC]). An episomal control vector containing a full-length SIVmac239 LTR and wild-type full-length 3′ PPT (eSH) was used as a positive control of episomal vector mobilization. To evaluate mobilization from episomal templates, we transfected each episomal transfer vector separately together with SIV packaging IN-competent (pADSIV3+) and vesicular stomatitis virus G protein (VSV-G) envelope plasmids into packaging 293T Lenti-X cells. Recovered vector titers were normalized by RT activity and analyzed by FACS using serial dilution on 293T Lenti-X cells and colony formation on HeLa cells.
FACS analysis showed that LV produced with episomal eGAE and eGAEdU3 exhibited a strong decrease in transduction ability with respect to the parental LV produced with eSH episomal transfer vector with full-length LTR, suggesting a sharp reduction of mobilization from episomal templates containing U3 deletions (Figure 4). In addition, deletion of 3′ PPT in both eGAEdPPT and eGAEdPPTdU3 episomal transfer vectors showed an additional reduction in GFP transduction and expression from each respective LV. This is consistent with data shown in Figure 3 using IN-competent GAEdPPT and GAEdPPTdU3 and related to the impairment of integration in the 3′ PPT mutant viruses.19,20
Figure 4.
Mobilization from episomal SIV-based LV
FACS analysis of GFP expression in 293T Lenti-X cells transduced with normalized amounts (RT counts/mL) of LV produced from parental (eGAE), single-mutant (eGAEdU3 and eGAEdPPT), or double-mutant (eGAEdPPTdU3) episomal transfer vectors depicted in Figure S3. Transfer vector eSH containing a full-length SIVmac239 LTR and 3′ PPT was used as a positive control for episomal vector mobilization. All episomal transfer vectors contain a GFP marker and neomycin resistance gene. Mobilization was valuated as % of GFP-expressing cells by FACS analysis.
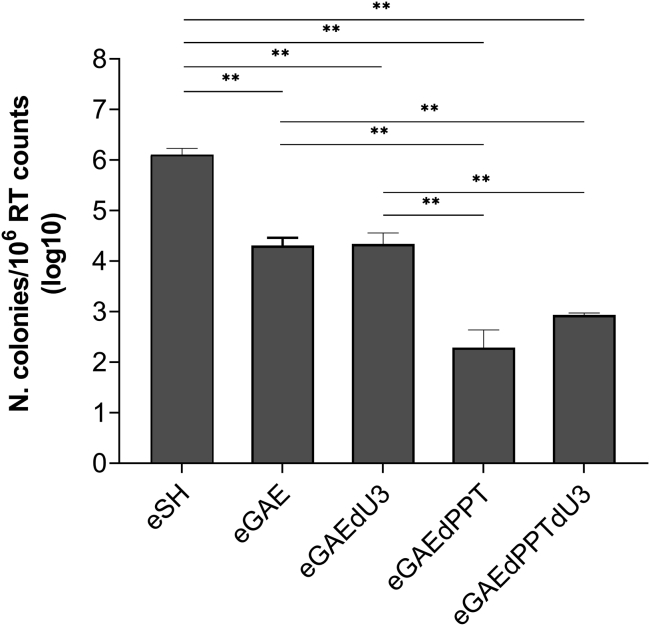
To better quantify episomal transfer vector mobilization, we used supernatants to transduce HeLa cells in a colony formation assay. Results showed a 2-log reduction in colonies formation from eGAE and eGAEdU3 vectors with respect to the full-length LTR (eSH) and an additional 1.5-log reduction in the 3′ PPT-deleted eGAEdPPT and eGAEdPPTdU3 vectors (Figure 5), reproducing the FACS analysis data (Figure 4). These results are in line with integration frequency of 3′ PPT-deleted IN-competent vectors, providing evidence that RNA templates mobilized from 3′ PPT-deleted episomal transfer vectors are impaired in integration and in contrast with published data using HIV-based LV, showing that circular episomal forms of HIV-based SIN-LVs with a large deletion in the U3 region supported mobilization of vector templates and subsequent high vector titers production.25,26
Figure 5.
DNA recombination frequencies of SIV-based episomal transfer vectors
Average geneticin-resistant colony formation titer assay results in transduced HeLa cells comparing the frequency of integration among the episomal lentiviral transfer vectors. y axis represents infectious units/106 RT counts of vector supernatant. The error bars indicate the standard error among n = 4 independent experiments, and p values are shown for intergroup comparisons. ∗∗p < 0.01, ∗∗∗p < 0.001. eGAE, parental episomal lentiviral vector; eGAEdPPT, episomal vector with a 3′ PPT deletion; eGAEdPPTdU3, episomal vector with a 3′-PPT deletion and a large U3 deletion; eGAEdU3, episomal vector with a large U3 deletion; eSH, episomal vector with full-length LTR.
Low level of mobilization from vector episomal forms after cross-packaging
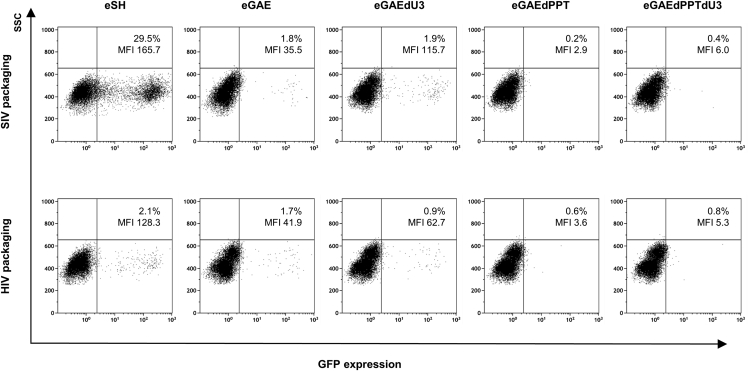
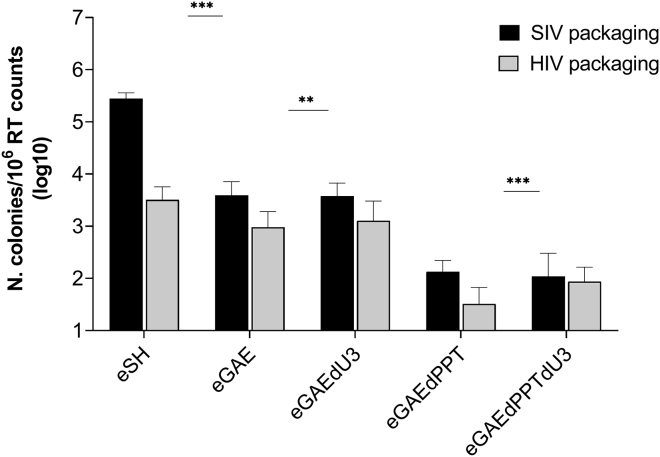
Vectors based on primate lentivirus involve the risk for vector mobilization not only by homologous viruses but also by heterologous viruses. This may have implications in SIV-IDLV recipients who are (or become) infected by HIV. Indeed, the SIV vector RNA genome could be cross-packaged into HIV particle, although at lower levels than the homologous HIV vector, similarly to what was previously described.27, 28, 29 Thus, we investigated the possibility that our episomal SIV-based transfer vectors could be cross-packaged in HIV particles and transferred into recipient cells. To this aim, we produced IN-competent LV with either the SIV-based or the HIV-based packaging system, which were evaluated on 293T Lenti-X and HeLa cells for FACS analysis or colony formation assay, respectively. As shown by FACS analysis (Figure 6), packaging of SIV transfer vector containing the full-length SIV-LTR (eSH) into HIV particles (HIV packaging the SIV genomes) showed over a 10-fold reduction in transduction efficiency with respect to homologous packaging (SIV packaging the SIV genomes). The eGAEs transfer vectors did not show a significant difference with respect to packaging system used, possibly because of the sensitivity of the assay. To better quantify episomal transfer vector mobilization, we used supernatants to transduce HeLa cells in a colony formation assay (Figure 7). The episomal vector with full-length SIV-LTR (eSH) showed a near 2-log decrease in colony formation when cross-packaged into HIV particles, while cross-packaging of episomal eGAEs showed a smaller reduction compared with homologous packaging, possibly because of the lower mobilization ability from the SIN episomal templates. Overall, these data indicated that although SIV episomal genomes can be cross-packaged and mobilized by HIV-based viral particles, this mechanism is less efficient than with homologous SIV viral particles, in line with previously reported data.27, 28, 29
Figure 6.
Cross-packaging of episomal SIV-based LV
FACS analysis of GFP expression in 293T Lenti-X cells transduced with an equal amount (1 × 105 RT counts) of integrase-competent LV produced after SIV homologous or HIV heterologous packaging of SIV-based episomal transfect vectors (eGAEs and eSH). Episomal transfer vector eSH containing a full-length SIVmac239 LTR and 3′ PPT was used as a positive control for episomal vector mobilization. All episomal transfer vectors contain a GFP marker and neomycin resistance gene (GIN). Mobilization was valuated as % of GFP-expressing cells by FACS analysis.
Figure 7.
DNA recombination frequencies of SIV-based episomal transfer vectors after SIV homologous or HIV heterologous packaging
Average geneticin-resistant colony-formation titer assay results in transduced HeLa cells comparing the frequency of integration from the episomal SIV-based lentiviral transfer vectors using SIV homologous (black bars) or HIV heterologous (gray bars) packaging. eGAE, parental episomal lentiviral vector; eGAEdPPT, episomal vector with a 3′ PPT deletion; eGAEdPPTdU3, episomal vector with a 3′ PPT deletion and a large U3 deletion; eGAEdU3, episomal vector with a large U3 deletion; eSH, episomal vector with full-length LTR. y axis represents infectious units/106 RT counts of vector supernatant. The error bars indicate the standard error among n = 4 independent experiments, and p values are shown for intergroup comparisons. ∗∗p < 0.01, ∗∗∗p < 0.001.
Low frequency of mobilization of integrated SIV-based lentiviral vectors
Previous reports showed that mobilization of integrated LV genomes is reduced, but not fully abolished, when in the SIN configuration.11,30 Although we showed a low-level mobilization from episomal templates, we also evaluated mobilization of GAE vectors from the integrated templates. We produced 293 cell lines stably transduced with all SIV-based vectors, including the full-length LTR (293/SH), while a cell line stably transduced with a HIV-based SIN vector (293/LV-GFP-Neo) was used as a control to evaluate mobilization from cross-packaging vectors.17 All cell lines were co-transfected with the pseudotyping VSV-G glycoprotein and either the SIV-based or the HIV-based IN-competent packaging plasmid. Supernatants were collected and used on HeLa cells in a colony formation assay. As shown in Table 1, the SIV-based vector with full-length LTR mobilized with a 3-log higher frequency with respect to all GAE- and HIV-based SIN vector after SIV packaging (Table 1, left columns). Importantly, all vectors, both HIV and SIV based, showed a further reduction of mobilization after cross-packaging with the heterologous packaging system. In particular, the removal of 3′ PPT (in both the single and the double mutants GAEdPPT and GAEdPPTdU3) reduced vector mobilization after cross-packaging with the HIV packaging plasmid compared with the parental SIV packaging plasmid. Of note, homologous mobilization of HIV-based SIN vector was higher than any SIV-based SIN vector, and mobilization of a full-length SIV LTR with the HIV packaging plasmid was 1-log lower than when using the homologous SIV packaging plasmid.
Table 1.
DNA recombination frequencies from integrated lentiviral vectors after SIV homologous or HIV heterologous packaging
| No. of colonies/mL |
||||
|---|---|---|---|---|
| SIV packaging |
HIV packaging |
|||
| Cell type/vector | Average | Rangea | Average | Rangea |
| 293/LV-HIV | 88.25 | 50–135 | 122.75 | 90–160 |
| 293/SH | 5,275.5 | 4,083–6,500 | 382.5 | 180–550 |
| 293/GAE | 10.5 | 8–13 | 0.6 | 0–1.5 |
| 293/GAEdU3 | 4.74 | 4–7 | 0.6 | 0–1.5 |
| 293/GAEdPPT | 4.75 | 3–8 | 0.25 | 0–0.5 |
| 293/GAEdPPTdU3 | 8.25 | 3–13 | 1.25 | 0.5–2 |
n = 4 independent experiments.
3′ PPT deletion in SIV-based IDLV impairs immunogenicity
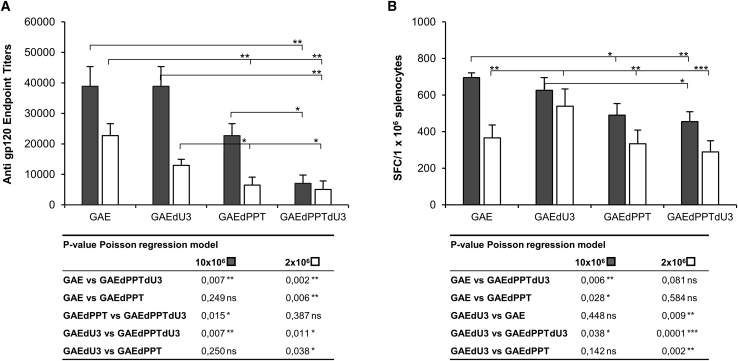
Following in vitro evaluation of gene expression and safety of the modified SIV-based IDLV, we evaluated the impact of these modifications on their ability to induce humoral and cellular responses in vivo. Immune response was assessed following a single intramuscular inoculum of high (10 × 106 RT counts) and low dose (2 × 106 RT counts) of each IDLV expressing the codon-optimized HIV-1JR-FL gp120 envelope protein (JR) as a model antigen in BALB/c mice, as previously described.24,31 Sera samples were collected prior to inoculum and at 5 weeks post-immunization (the day of sacrifice) and subjected to enzyme-linked immunosorbent assay (ELISA) assay to measure anti-Env antibody (Ab) titers (Figure 8A). All groups of mice showed high Ab titers after the single immunization with both doses in a dose-dependent manner. In particular, mice immunized with parental IDLVs (IDLV-GAE-JR) and with IDLVs containing the long U3 deletion (IDLV-GAEdU3-JR) showed the highest anti-Env Ab titers, while introduction of the 3′ PPT deletion in IDLV-GAEdPPT-JR and IDLV-GAEdPPTdU3-JR hindered induction of Ab titers with both vector doses.
Figure 8.
Immune responses in mice immunized with SIV-based IDLVs delivering HIV-1JR-FL gp120 envelope
(A) Anti-gp120 immunoglobulin G (IgG) measured by ELISA in plasma of mice at 5 weeks after immunization with 10 × 106 (gray bars) or 2 × 106 (white bars) RT counts/mouse of each IDLV. Plasma samples from immunized mice were analyzed separately. The error bars indicate the standard error among five mice of the same group, and p values are indicated for intergroup comparisons. (B) T cell responses measured on splenocytes cells at 5 weeks after immunization with 10 × 106 (gray bars) or 2 × 106 (white bars) RT counts/mouse of each IDLV. IFN-γ ELISPOT was performed on cells stimulated overnight with the H-2d-restricted HIV-Env JR 9-mer peptide. IFN-γ-producing T cells are expressed as spot-forming cells (SFCs)/106 splenocytes after background subtraction. The error bars indicate the standard error among five mice of the same group. Comparison among groups was evaluated using the Poisson regression model, as indicated by p values shown in the tables under the graphs. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The cell-mediated immune response was determined by IFNγ enzyme-linked immune absorbent spot (ELISPOT) on ex vivo splenocytes collected at sacrifice (5 weeks after the inoculum) by using a JR-9-mer peptide containing the H-2d-restricted HIV-1 gp120 envelope epitope (IGPGRAFYT).32 All vaccinated animals showed a dose-dependent JR-9-mer-specific immune response (Figure 8B). Mice injected with IDLV containing the 3′ PPT deletion elicited a lower amount of IFNγ-producing cells, which was statistically significant at the highest dose of IDLV-GAE-JR and at the lowest dose of IDLV-GAEdU3-JR. This is in line with a lower induction of anti-Env Ab titers (Figure 8A) and is consistent with the lower transcriptional potential of 3′ PPT-deleted SIV-based IDLV (Figure 2).
Discussion
Soon after infection, IDLVs produce transcriptionally active episomal forms (E-DNA), whereas integrated proviral forms are drastically reduced or virtually absent.33 Although safer than integrated LV, E-DNA may be a target for mobilization from incoming infectious viral particles in the case of superinfection with a replication-competent virus. Vector mobilization may lead to integration events and eventually to insertional mutagenesis or oncogenic activation.34 In this context, we recently reported the absence of recombination/mobilization events in non-human primates (NHP) that were sequentially immunized with SIV-based IDLVs expressing HIV-Env and subsequently challenged with replication-competent SHIV.35 This provided substantial evidence that IDLV is a safe and effective vaccine platform that induces high magnitude and durable immune responses. However, nonintegrating IDLVs express transgenes from E-DNA at a significantly lower level than their integrating counterparts,36 calling for improvements in vector design.
In the present study, we evaluated in cell culture model systems the improvement in expression and safety profile of SIV-based IDLVs after introducing two different deletions (removal of 3′ PPT and extended U3 deletions), alone or in combination, previously characterized in the HIV-based system.18, 19, 20 Previous work showed that the 3′ PPT sequence is required for a correct reverse transcription of the vector genome, and its deletion inhibited strand displacement, thus favoring production of 1-LTR forms of E-DNA and impairing integration of the vector genome.19,20 In addition, Bayer et al.18 reported that a large U3 deletion in the 3′ LTR improved expression from HIV-based IDLVs. Importantly, additional deletions over the current design in the vector genome of IDLVs may further improve bio-safety by decreasing the potential for genetic recombination among components of the vector system, which could potentially lead to the emergence of replication-competent lentivirus (RCL),37,38 although we did not find evidence of RCL in the plasma of IDLV-immunized NHPs.35,39
Our data indicate that deletion of 3′ PPT greatly impaired integration efficiency of IN-competent SIV-based LV, confirming previous reports using HIV-based vectors.19,20 As expected, this also affected the magnitude of expression, which decreased to titers comparable with those of conventional non-integrating vectors. However, 3′ PPT-deleted IDLVs showed only a minor reduction of illegitimate integration compared with parental IDLVs containing the 3′ PPT sequence (IDLV-GAE versus IDLV-GAEdPPT and IDLV-GAEdU3 versus IDLV-GAEdPPTdU3; Figure 1). Interestingly, using a sensitive luciferase assay, expression from 3′ PPT-deleted IDLVs was also impaired, showing titers that were always lower than the parental IDLVs containing 3′ PPT (Figure 2). These data are in contrast with previous reports using HIV-based 3′ PPT-deleted IDLVs, where integration frequencies were 2- to 3-fold lower compared with IDLVs containing 3′ PPT, and expression was maintained at similar levels.19,20
We also showed that a large U3 deletion did not modify expression efficiency in both LVs and IDLVs. These data contrast with previous work showing that HIV-based IDLV expression was improved by an extended U3 deletion,18 but this may not be surprising, because the latter was achieved in the HIV-based system. Although the HIV LTR is similar to the SIV LTR, sequence homology is low and there are some important structural and functional distinctions.40, 41, 42 Indeed, previous work showed that extensive deletions (−384 bp) in the SIV U3 region of the LTR allowed efficient replication of SIVMac239 in vitro in cell culture assays and in vivo in rhesus macaques.43 Therefore, it is plausible that cis elements in the U3 region could inhibit transcription by HIV-based IDLVs as hypothesized by Bayer et al.,18 while in SIV-based vectors these regions may be absent or non-functional in the SIN configuration.
To study mobilization from episomal forms produced after transduction with IDLV, we constructed shuttle transfer vectors containing a single LTR (1-LTR), a Tn/neoR gene, and bacterial DNA sequences providing AmpR and bacterial origin of replication18,26 (Figure S3). These vectors closely mimic the 1-LTR circular forms of E-DNA, which represent the most abundant episomal form produced in the nuclei of IDLV-transduced cells26,33 and can be used conveniently to evaluate mobilization from episomes resembling IDLV genomes by using SIV-based homologous packaging or HIV-based heterologous packaging, the latter to evaluate cross-packaging of vector genomes.
Mobilization of episomal templates from SIN transfer vectors containing the 3′ PPT (eGAE and eGAEdU3 configuration) was 2 logs less efficient compared with mobilization of episomal templates containing a wild-type full-length LTR (eSH). This is in contrast with what was previously reported using HIV-based episomal vectors, which supported mobilization from episomal vector templates and production of high vector titers.25,26 This was due to the presence of an aberrant transcription initiation site located in the SIN-deleted HIV-U3 sequence, upstream of the parental HIV-1 TATA box, which allowed transcription, packaging, and transmission of vector genomic RNA (gRNA),26 suggesting that this site is absent in the SIV-based SIN vectors.
Consistent with our data using SIV-based vectors, 3′ PPT-deleted episomal transfer vectors (eGAEdPPT and eGAEdPPTdU3 configuration) showed a further 2-log reduction in colony formation, compared with PPT-plus vector (Figure 5). This was expected because templates mobilized from 3′ PPT-deleted genomes have a reduced competence for integration, as shown in Figure 3.
We next evaluated heterologous cross-packaging of SIV-based episomal templates with HIV packaging system, because E-DNA produced from SIV-based IDLVs may be a target for mobilization from incoming infectious HIV-1 particles in the case of infection with a replication-competent virus.
Results showed that mobilization of episomal templates from transfer vector containing the wild-type full-length SIV-LTR (eSH) was almost 2 logs less efficient when using the heterologous HIV cross-packaging system (Figure 7). Interestingly, when using the SIN episomal transfer vectors, reduction of mobilization after cross-packaging using the HIV system was less marked. This may be because of the low mobilization capacity from SIV-based SIN episomal templates.
Although integration of IDLV genomes is an unlikely event,22,24 integration into the host genome can occur,44,45 and integrated genomes may be a target for mobilization in the case of infection with a replication-competent virus.10,11 Our data using homologous SIV packaging indicate that mobilization of integrated SIV-based SIN vector templates is very inefficient and was 3 logs lower than mobilization from cells with integrated vector containing the full-length LTR (293/SH), similarly to what was previously described30 and much lower than mobilization of integrated HIV-based SIN vectors templates using homologous HIV packaging (Table 1). Interestingly, mobilization of 3′ PPT-deleted integrated vector templates was negligible, which was expected, given that competence for integration is strongly reduced in the 3′ PPT-deleted mobilized genomes. We also tested the SIV-based vectors using HIV-based heterologous cross-packaging. Results are in line with data we obtained with the episomal templates. Indeed, mobilization of integrated SIV-based SIN vectors using the heterologous HIV cross-packaging system was negligible and close to background levels, whereas mobilization from cells with integrated templates containing the full-length SIV-LTR (293/SH) was over 1 log less efficient when using the heterologous HIV cross-packaging system (Table 1). Overall, these data indicate that heterologous cross-packaging of viral genome is not very efficient, and they highlight that SIV-based SIN IDLVs may be safer for use in humans in the setting of an HIV infection. Additional modifications, such as the one described by Hu et al.,25 may further improve the safety profile of IDLV.
Interestingly, when using the SIN episomal templates (eGAEs configuration), reduction of mobilization after cross-packaging using the HIV packaging system was less marked than from the integrated templates. This may be because of a higher number of episomal templates available for mobilization after transfection with the episomal transfer vectors, compared with a single vector genome/cell in the integrated vector templates. Although 2-LTR circles are also formed following the transduction of cells, we focused our attention on the 1-LTR circular form because it is the most abundant episomal form recovered in HIV-infected or lentiviral vector-transduced cells,18,46 and expression from the 2-LTR circles has been shown to be present at levels comparable with those of the 1-LTR circles in the HIV system.26,47
In our system, mobilization is evaluated by drug-resistant colonies resulting from vector integration (which depends on the design of the transfer vectors) and gRNA availability (which also depends on the number of vector templates). Transcription of gRNA from SIN templates could also be mediated by either internal vector promoter25 or cryptic initiation sequences into/near lentiviral SIN-LTR.19,48 Furthermore, the polyadenylation signal of SIN-LV is leaky and allows gRNA transcription beyond the termination signal.49 Because we forced the system by transfecting episomal transfer vectors or stably selected cell lines, we assume that the gRNA transcribed is greater than that which would be produced from circular forms after IDLV infection or in the case of a rare event of integration; consequently, the mobilization events we described here may be representative of the worst-case scenario.
The elicitation of humoral and cellular immune responses was evaluated in mice by comparing a single injection of parental and modified IDLVs expressing a codon-optimized HIV-1JR-FL gp120.24 All modified IDLVs were able to elicit both humoral and cell-mediated responses after a single intramuscular immunization using two different doses of each vector, although these responses were somewhat weaker in 3′ PPT-deleted vectors. In particular, a 3′ PPT deletion had a negative effect in terms of induction of Ab production and, to a lesser extent, in the induction of antigen-specific IFN-γ-expressing cells. This was not surprising and was consistent with the lower transcriptional activity measured in cell culture using 3′ PPT-deleted IDLVs (Figure 2).
Safety and efficacy are the main characteristics of a successful vaccine. Overall, our data provide evidence that safer SIV-based IDLVs can be constructed with a very low level of mobilization from both episomal and integrated templates. However, improvement of safety after removal of 3′ PPT partly reduced efficacy, measured by a decrease in transcriptional activity and consequent immunogenicity of the safety-modified IDLVs. Conversely, inclusion of an extended U3 deletion did not improve expression and efficacy of IDLV but improved the design of the transfer vector genome by decreasing the potential for genetic recombination during IDLV production. Thus, combining the high efficiency of gene transfer mediated by these modified IDLVs with a higher safety profile is highly attractive for clinical application of this delivery system for future vaccines.
Materials and methods
Cell lines
293T Lenti-X human embryonic kidney (HEK) (Clontech, Mountain View, CA, USA), HEK293 (CRL-1573; American Type Culture Collection, LGC Standards, Italy), and HeLa (CCL-2; American Type Culture Collection) cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO, Life Technologies Italia, Monza, Italy) supplemented with 10% fetal bovine serum (FBS) (Corning, Mediatech, Manassas, VA, USA) and 100cU/mL penicillin/streptomycin/glutamine (PSG) (GIBCO).
Transfer vector plasmids
A schematic depiction of the vectors used in this study is provided in Figures S1 and S3. Details on vectors construction are provided in supplemental experimental procedures. In brief, the SIV-based SIN lentiviral transfer vector pGAE-CMV-GFPW (pGAE-GFP in this report) and pGAE-GFP-IRES-Neo (pGAE-GIN in this report) have been described previously.22 All parental transfer vectors with pGAE/eGAE configuration maintain the 3′ PPT and contain a 149-bp deletion in the U3 region of the LTR (deleted from –20 to –169 from the R region).21 All transfer vectors with pGAEdPPTdU3/eGAEdPPTdU3 configuration are deleted in the 3′ PPT and contain a longer 433-bp deletion in the modulatory U3 region (deleted from –20 to –453 from the R region). All transfer vectors with pGAEdPPT/eGAEdPPT configuration are deleted in the 3′ PPT and contain a short 149-bp deletion in the modulatory U3 region (deleted from –20 to –169 from the R region). All transfer vectors with pGAEdU3/eGAEdU3 configuration maintain the 3′ PPT and contain a longer 433-bp deletion in the modulatory U3 region (deleted from –20 to –453 from the R region). Alignment of 3′ PPT-LTR regions of all vectors is shown in Figure S2. The HIV- and SIV-based IN-defective packaging plasmids (pcHelp/IN and pADSIVD64V, respectively), the HIV- and SIV-based IN-competent packaging plasmids (pCMVdR8.2 and pADSIV3+, respectively), and the phCMV-VSV-G plasmid producing the pseudotyping VSV-G envelope have already been described.22,24
Production and titration of lentiviral vectors
293T Lenti-X cells were seeded on 10-cm Petri dishes (Corning Incorporated-Life Sciences, Oneonta, NY, USA) and transiently transfected with transfer vector plasmid, IN-competent or IN-defective packaging plasmid, and VSV-G envelope plasmid using the Profection Mammalian Transfection System (Clontech) as previously described24 using a total of 15 μg of plasmid DNA for each plate in a ratio 4:8:3 (transfer vector/packaging plasmid/VSV-G plasmid). After 48 h, cell culture supernatants were recovered, cleared from cellular debris, and passed through a 0.45-μM pore-size filter (Millipore Corporation, Billerica, MA, USA). For in vivo animal studies, supernatants containing IDLV particles were concentrated by ultracentrifugation (Beckman Coulter, Fullerton, CA, USA) for 2 h at 27,000 rpm on a 20% sucrose cushion (Sigma Chemicals, St. Louis, MO, USA). Finally, the viral pellets were resuspended in 1 × PBS and stored at −80°C until use. Vector transducing units (TUs/mL) were determined on 293T Lenti-X cells using serial dilution of GFP-expressing vector, and titers were calculated by scoring the percentage of GFP-positive cells measured with a FACSCalibur analytical flow cytometer with CellQuest software (BD Biosciences Immunocytometry Systems, San Jose, CA, USA). Each vector stock was titered by the RT activity assay,50 and the corresponding TUs were calculated by comparing the RT activity with the one of IDLV-GFP virions with known infectious titers, thus allowing for the determination of their infectious titer units.51
Luciferase assay
293T cells Lenti-X (1 × 104/well) were seeded in a flat-bottom 96-well plate and transduced with vectors expressing luciferase (1.5 × 104 RT counts/well). Seventy-two hours post-transduction, luciferase activity was evaluated using the Britelite Ultra-High Sensitivity Luminescence Reporter Gene Assay System (Perkin-Elmer, Groningen, the Netherlands) with a microplate reader Wallac luminometer (Perkin-Elmer), as already reported.52 Each experiment was performed and calculated in quadruplicate, and results were recorded as relative light units (RLUs) per well.
Colony formation assay
HeLa cells (5 × 104/well) were seeded in six-well plates. After 24 h from seeding, cells were transduced in duplicate with serial dilutions of IN-competent or IN-defective lentiviral vectors expressing the Tn/neoR coding sequence (range 1 × 105 to 1 × 101 RT counts for the IN-competent vectors and 1 × 106 to 1 × 102 RT counts for the IN-defective vectors). After 24 h, the medium was replaced with fresh medium supplemented with 800 μg of geneticin (GIBCO) and replaced every 3 days. Cells were grown for 2 weeks, and developed colonies were fixed with methanol and stained with Giemsa solution (Sigma Chemicals). Colonies on each well were counted and expressed as the number of colonies/106 RT counts as described.17,22,24 293 cells transduced with each IN-competent vector were allowed to grow under geneticin pressure for production of 293/SH, 293/GAE, 293/GAEdU3, 293/GAEdPPT, and 293/GAEdPPTdU3 cell lines, used to evaluate mobilization of integrated vector sequences. The 293/LV-HIV cell line containing an integrated copy of a HIV-based SIN vector expressing GFP-Neo has been previously described.17
Mobilization from integrated vector
The earlier cell lines stably transduced with lentiviral vectors were used for evaluating mobilization of integrated genomes. Cells were transfected using the Profection Mammalian Transfection System (Clontech) with IN-competent packaging plasmid (pADSIV3+ or pCMVdR8.2) and VSV-G envelope plasmid. Supernatants were collected 48 h later and used to transduce 5 × 104 HeLa cells in six-well plates in a colony formation assay (see above). The next day, the medium was replaced with fresh medium supplemented with 800 μg/mL geneticin (GIBCO) and replaced every 3 days. After selection, developed clones were counted and expressed as number of colonies per milliliter of supernatant.
Mouse immunization
Six- to eight-week-old female BALB/c mice were purchased from Harlan (Harlan Laboratory, San Pietro al Natisone, Italy) and housed under specific pathogen-free conditions in the animal facility of the Istituto Superiore di Sanità (ISS, Rome, Italy). All animal procedures have been performed in accordance with European Union guidelines and Italian legislation for animal care. Animal studies were authorized by the Italian Ministry of Health and reviewed by the Service for Animal Welfare at ISS (Authorization no. 314/2015-PR of 30/04/2015). One week after arrival, five mice per group were injected once intramuscularly with 10 × 106/RT U/mouse or 2 × 106/RT U/mouse of (1) IDLV-GAE, (2) IDLV-GAEdU3, (3) IDLV-GAEdPPT, and (4) IDLV-GAEdPPTdU3, expressing the codon-optimized HIV-1JR-FL gp120 envelope protein (JR). A control group of naive, non-immunized mice were kept for parallel analysis. Abs were measured in plasma at the day of immunization (day 0) and at sacrifice (5 weeks). The cellular immune responses were analyzed at sacrifice (5 weeks) in splenocytes. Heparin-treated glass Pasteur pipettes were used to collect plasma samples and stored at −20°C until assayed. Splenocytes were prepared by mechanical disruption and passage through cell strainers (BD Biosciences), counted, and resuspended in complete RPMI 1640 (GIBCO) medium containing 10% FBS (Lonza, Treviglio, Milan, Italy), 100 U/mL PSG (GIBCO), non-essential amino acids (GIBCO), sodium pyruvate 1 mM (GIBCO), HEPES buffer solution 25 mM (GIBCO), and 50 mM 2-mercaptoethanol (Sigma Chemicals) or were frozen in liquid nitrogen for storage.
IFN-γ ELISPOT assay
The IFNγ ELISPOT assay was carried out using the BD ELISPOT kit reagents and protocol (BD Biosciences). In brief, 2.5 × 105 splenocytes/well were seeded in flat-bottom 96-well plates previously coated with 5 mg/mL anti-mouse IFNγ and stimulated overnight with 2 μg/mL of the H-2Kd-restricted HIV-1 gp120 V3 loop epitope (IGPGRAFYT) (UFPeptides, Ferrara, Italy), or using medium alone and concanavalin A (ConA; 5 mg/mL; Sigma Chemicals) as negative and positive controls, respectively. Spot-forming cells (SFC) were counted with an ELISPOT reader (A.EL.VIS, Hannover, Germany), and results were expressed as number of IFNγ-secreting cells (SFCs)/106 cells. Samples positive score was attributed when a minimum of 50 spots per 106 cells were present and 2-fold higher than unstimulated cells.
Measurement of Ab titers
Plasma was collected from each mouse by retro-orbital bleeding at pre-immunization and at 5 weeks post-IDLV immunization and frozen at −20°C. Specific Ab titers against gp120 were measured by ELISA. In brief, Microlon High binding, flat-bottom 96-well polystyrene microplates (Greiner Bio-One) were coated with 3 μg/ml HIV-1 JRFL gp140 Recombinant Protein (B.JRFL gp140CF; NIH AIDS Reagent Program Catalog [Cat.] No. 12573) in PBS and incubated overnight at 4°C. Plates were blocked at room temperature for 2 h using PBS with 1% BSA (Sigma Chemicals). Following three washes with PBS supplemented with 0.05% Tween 20, mouse plasma was serially diluted in dilution buffer (0.1% BSA and 0.025% Tween 20 in PBS), transferred into the JR-coated plate (100 μL), and incubated at room temperature for 2 h. Plates were washed again, and biotin-conjugated goat anti-mouse IgG secondary Ab (Southern Biotechnology, Birmingham, AL, USA) diluted 1:20,000 was added into wells. After 2 h of incubation at room temperature, plates were washed, and 100 μL of 1:20,000 streptavidin horseradish peroxidase-conjugated (HRP) substrate (AnaSpec) was added to each well for 30 min. Plates were washed and incubated in the dark for 30 min with 100 μL of TMB substrate (SurModics). Enzymatic reaction was stopped with H2SO4 1M (VWR), and OD was measured at 450 nm with a microplate reader Wallac (Perkin-Elmer). The starting dilution was 1:100 for all plasma samples, with serial 2-fold dilutions thereafter. Sera with no activity at 1:100 were assigned titers of 1:100. Endpoint titers were defined as the reciprocal of the highest dilution that gave an OD reading three standard deviations above the mean of the pre-immunization plasma.
Statistical analysis
Comparisons among number of colonies were statistically analyzed by paired two-tailed t test using Prism software (GraphPad); nsp > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, as specified in the figure legends. ELISA titers and ELISPOT results were jointly modeled by Poisson regressions within a generalized structural equation modeling (GSEM) framework.53,54 ELISA titers (at two different dosages: 1 × 106 and 2 × 106) at 5 weeks and ELISPOT results (at two different dosages: 1 × 106 and 2 × 106) at 5 weeks were used as the dependent variables, while type of IDLV was used as the independent variable. Dependent variables were allowed to be correlated. All analyses were carried out in Stata 16.
Acknowledgments
The authors are grateful to Stefania Donnini for secretarial assistance and Ferdinando Costa and Patrizia Cocco for technical support. This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (NIAID) (1P01AI110485-01A1), Italian Ministry of Health Ricerca Finalizzata (PE-2011-02347035 to A. Cara), and Italian Ministry of Health Ricerca Finalizzata (PE-2016-02364927). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 681137 (EAVI2020) and from the European Union’s Seventh Programme for Research, Technological Development and Demonstration under grant agreement no. 280873 (ADITEC) (to A. Cara and D.N.). We thank Fondation Dormeur, Vaduz for the donation of laboratory instruments relevant to this project. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: B.JRFL gp140CF.
Author contributions
R.B. and Z.M. participated in study design, performed the majority of the experiments, analyzed the data, and wrote the manuscript. M.B. performed the RT assay and contributed to performing the ELISPOT assays. C.M., A. Canitano, A.G., and M.F.P. contributed to construction and preparation of vectors. M.F.V. performed statistical analysis. A.D.V. performed the animal experiments. M.E.K. contributed to data analysis and editing of the manuscript. D.N. contributed to study design, data analysis, and editing of the manuscript and oversaw the animal experiments. A. Cara oversaw the planning and direction of the project, including analysis and interpretation of the data and editing of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2021.09.011.
Supplemental information
References
- 1.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 2.Hu B., Tai A., Wang P. Immunization delivered by lentiviral vectors for cancer and infectious diseases. Immunol. Rev. 2011;239:45–61. doi: 10.1111/j.1600-065X.2010.00967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iglesias M.C., Frenkiel M.P., Mollier K., Souque P., Despres P., Charneau P. A single immunization with a minute dose of a lentiviral vector-based vaccine is highly effective at eliciting protective humoral immunity against West Nile virus. J. Gene Med. 2006;8:265–274. doi: 10.1002/jgm.837. [DOI] [PubMed] [Google Scholar]
- 4.Buffa V., Negri D.R.M., Leone P., Bona R., Borghi M., Bacigalupo I., Carlei D., Sgadari C., Ensoli B., Cara A. A single administration of lentiviral vectors expressing either full-length human immunodeficiency virus 1 (HIV-1)(HXB2) Rev/Env or codon-optimized HIV-1(JR-FL) gp120 generates durable immune responses in mice. J. Gen. Virol. 2006;87:1625–1634. doi: 10.1099/vir.0.81706-0. [DOI] [PubMed] [Google Scholar]
- 5.Montini E., Cesana D., Schmidt M., Sanvito F., Bartholomae C.C., Ranzani M., Benedicenti F., Sergi L.S., Ambrosi A., Ponzoni M. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry C., Hannenhalli S., Leipzig J., Bushman F.D. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biffi A., Bartolomae C.C., Cesana D., Cartier N., Aubourg P., Ranzani M., Cesani M., Benedicenti F., Plati T., Rubagotti E. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- 8.Iwakuma T., Cui Y., Chang L.J. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology. 1999;261:120–132. doi: 10.1006/viro.1999.9850. [DOI] [PubMed] [Google Scholar]
- 9.Déglon N., Tseng J.L., Bensadoun J.C., Zurn A.D., Arsenijevic Y., Pereira de Almeida L., Zufferey R., Trono D., Aebischer P. Self-inactivating lentiviral vectors with enhanced transgene expression as potential gene transfer system in Parkinson’s disease. Hum. Gene Ther. 2000;11:179–190. doi: 10.1089/10430340050016256. [DOI] [PubMed] [Google Scholar]
- 10.Logan A.C., Haas D.L., Kafri T., Kohn D.B. Integrated self-inactivating lentiviral vectors produce full-length genomic transcripts competent for encapsidation and integration. J. Virol. 2004;78:8421–8436. doi: 10.1128/JVI.78.16.8421-8436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanawa H., Persons D.A., Nienhuis A.W. Mobilization and mechanism of transcription of integrated self-inactivating lentiviral vectors. J. Virol. 2005;79:8410–8421. doi: 10.1128/JVI.79.13.8410-8421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negri D.R.M., Michelini Z., Bona R., Blasi M., Filati P., Leone P., Rossi A., Franco M., Cara A. Integrase-defective lentiviral-vector-based vaccine: a new vector for induction of T cell immunity. Expert Opin. Biol. Ther. 2011;11:739–750. doi: 10.1517/14712598.2011.571670. [DOI] [PubMed] [Google Scholar]
- 13.Cara A., Klotman M.E. Retroviral E-DNA: persistence and gene expression in nondividing immune cells. J. Leukoc. Biol. 2006;80:1013–1017. doi: 10.1189/jlb.0306151. [DOI] [PubMed] [Google Scholar]
- 14.Lin Y.Y., Belle I., Blasi M., Huang M.N., Buckley A.F., Rountree W., Klotman M.E., Cara A., Negri D. Skeletal Muscle Is an Antigen Reservoir in Integrase-Defective Lentiviral Vector-Induced Long-Term Immunity. Mol. Ther. Methods Clin. Dev. 2020;17:532–544. doi: 10.1016/j.omtm.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luis A. The Old and the New: Prospects for Non-Integrating Lentiviral Vector Technology. Viruses. 2020;12:1103. doi: 10.3390/v12101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Nunzio F., Félix T., Arhel N.J., Nisole S., Charneau P., Beignon A.S. HIV-derived vectors for therapy and vaccination against HIV. Vaccine. 2012;30:2499–2509. doi: 10.1016/j.vaccine.2012.01.089. [DOI] [PubMed] [Google Scholar]
- 17.Negri D.R.M., Rossi A., Blasi M., Michelini Z., Leone P., Chiantore M.V., Baroncelli S., Perretta G., Cimarelli A., Klotman M.E., Cara A. Simian immunodeficiency virus-Vpx for improving integrase defective lentiviral vector-based vaccines. Retrovirology. 2012;9:69. doi: 10.1186/1742-4690-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayer M., Kantor B., Cockrell A., Ma H., Zeithaml B., Li X., McCown T., Kafri T. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol. Ther. 2008;16:1968–1976. doi: 10.1038/mt.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kantor B., Bayer M., Ma H., Samulski J., Li C., McCown T., Kafri T. Notable reduction in illegitimate integration mediated by a PPT-deleted, nonintegrating lentiviral vector. Mol. Ther. 2011;19:547–556. doi: 10.1038/mt.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw A.M., Joseph G.L., Jasti A.C., Sastry-Dent L., Witting S., Cornetta K. Differences in vector-genome processing and illegitimate integration of non-integrating lentiviral vectors. Gene Ther. 2017;24:12–20. doi: 10.1038/gt.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangeot P.E., Duperrier K., Nègre D., Boson B., Rigal D., Cosset F.L., Darlix J.L. High levels of transduction of human dendritic cells with optimized SIV vectors. Mol. Ther. 2002;5:283–290. doi: 10.1006/mthe.2002.0541. [DOI] [PubMed] [Google Scholar]
- 22.Michelini Z., Negri D.R., Baroncelli S., Spada M., Leone P., Bona R., Klotman M.E., Cara A. Development and use of SIV-based Integrase defective lentiviral vector for immunization. Vaccine. 2009;27:4622–4629. doi: 10.1016/j.vaccine.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornu T.I., Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- 24.Negri D.R., Michelini Z., Baroncelli S., Spada M., Vendetti S., Buffa V., Bona R., Leone P., Klotman M.E., Cara A. Successful immunization with a single injection of non-integrating lentiviral vector. Mol. Ther. 2007;15:1716–1723. doi: 10.1038/sj.mt.6300241. [DOI] [PubMed] [Google Scholar]
- 25.Hu P., Bi Y., Ma H., Suwanmanee T., Zeithaml B., Fry N.J., Kohn D.B., Kafri T. Superior lentiviral vectors designed for BSL-0 environment abolish vector mobilization. Gene Ther. 2018;25:454–472. doi: 10.1038/s41434-018-0039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma H., Kafri T. A single-LTR HIV-1 vector optimized for functional genomics applications. Mol. Ther. 2004;10:139–149. doi: 10.1016/j.ymthe.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Strappe P.M., Hampton D.W., Brown D., Cachon-Gonzalez B., Caldwell M., Fawcett J.W., Lever A.M. Identification of unique reciprocal and non reciprocal cross packaging relationships between HIV-1, HIV-2 and SIV reveals an efficient SIV/HIV-2 lentiviral vector system with highly favourable features for in vivo testing and clinical usage. Retrovirology. 2005;2:55. doi: 10.1186/1742-4690-2-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al Shamsi I.R., Al Dhaheri N.S., Phillip P.S., Mustafa F., Rizvi T.A. Reciprocal cross-packaging of primate lentiviral (HIV-1 and SIV) RNAs by heterologous non-lentiviral MPMV proteins. Virus Res. 2011;155:352–357. doi: 10.1016/j.virusres.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Ali L.M., Rizvi T.A., Mustafa F. Cross- and Co-Packaging of Retroviral RNAs and Their Consequences. Viruses. 2016;8:276. doi: 10.3390/v8100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunwald T., Pedersen F.S., Wagner R., Uberla K. Reducing mobilization of simian immunodeficiency virus based vectors by primer complementation. J. Gene Med. 2004;6:147–154. doi: 10.1002/jgm.479. [DOI] [PubMed] [Google Scholar]
- 31.Negri D.R.M., Michelini Z., Baroncelli S., Spada M., Vendetti S., Bona R., Leone P., Klotman M.E., Cara A. Nonintegrating Lentiviral Vector-Based Vaccine Efficiently Induces Functional and Persistent CD8+ T Cell Responses in Mice. J. Biomed. Biotechnol. 2010;2010:534501. doi: 10.1155/2010/534501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexander-Miller M.A., Parker K.C., Tsukui T., Pendleton C.D., Coligan J.E., Berzofsky J.A. Molecular analysis of presentation by HLA-A2.1 of a promiscuously binding V3 loop peptide from the HIV-envelope protein to human cytotoxic T lymphocytes. Int. Immunol. 1996;8:641–649. doi: 10.1093/intimm/8.5.641. [DOI] [PubMed] [Google Scholar]
- 33.Wanisch K., Yáñez-Muñoz R.J. Integration-deficient lentiviral vectors: a slow coming of age. Mol. Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cesana D., Ranzani M., Volpin M., Bartholomae C., Duros C., Artus A., Merella S., Benedicenti F., Sergi Sergi L., Sanvito F. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol. Ther. 2014;22:774–785. doi: 10.1038/mt.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blasi M., Negri D., Saunders K.O., Baker E.J., Stadtler H., LaBranche C., Mildenberg B., Morton G., Ciarla A., Shen X. Immunogenicity, safety, and efficacy of sequential immunizations with an SIV-based IDLV expressing CH505 Envs. NPJ Vaccines. 2020;5:107. doi: 10.1038/s41541-020-00252-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Negri D.R., Bona R., Michelini Z., Leone P., Macchia I., Klotman M.E., Salvatore M., Cara A. Transduction of human antigen-presenting cells with integrase-defective lentiviral vector enables functional expansion of primed antigen-specific CD8(+) T cells. Hum. Gene Ther. 2010;21:1029–1035. doi: 10.1089/hum.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pauwels K., Gijsbers R., Toelen J., Schambach A., Willard-Gallo K., Verheust C., Debyser Z., Herman P. State-of-the-art lentiviral vectors for research use: risk assessment and biosafety recommendations. Curr. Gene Ther. 2009;9:459–474. doi: 10.2174/156652309790031120. [DOI] [PubMed] [Google Scholar]
- 38.Mátrai J., Chuah M.K., VandenDriessche T. Recent advances in lentiviral vector development and applications. Mol. Ther. 2010;18:477–490. doi: 10.1038/mt.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negri D., Blasi M., LaBranche C., Parks R., Balachandran H., Lifton M., Shen X., Denny T., Ferrari G., Vescio M.F. Immunization with an SIV-based IDLV expressing HIV-1 Env 1086 clade C elicits durable humoral and cellular responses in rhesus macaques. Mol. Ther. 2016;24:2021–2032. doi: 10.1038/mt.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renjifo B., Speck N.A., Winandy S., Hopkins N., Li Y. cis-acting elements in the U3 region of a simian immunodeficiency virus. J. Virol. 1990;64:3130–3134. doi: 10.1128/jvi.64.6.3130-3134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pöhlmann S., Flöss S., Ilyinskii P.O., Stamminger T., Kirchhoff F. Sequences just upstream of the simian immunodeficiency virus core enhancer allow efficient replication in the absence of NF-kappaB and Sp1 binding elements. J. Virol. 1998;72:5589–5598. doi: 10.1128/jvi.72.7.5589-5598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Münch J., Rajan D., Rücker E., Wildum S., Adam N., Kirchhoff F. The role of upstream U3 sequences in HIV-1 replication and CD4+ T cell depletion in human lymphoid tissue ex vivo. Virology. 2005;341:313–320. doi: 10.1016/j.virol.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 43.Münch J., Adam N., Finze N., Stolte N., Stahl-Hennig C., Fuchs D., Ten Haaft P., Heeney J.L., Kirchhoff F. Simian immunodeficiency virus in which nef and U3 sequences do not overlap replicates efficiently in vitro and in vivo in rhesus macaques. J. Virol. 2001;75:8137–8146. doi: 10.1128/JVI.75.17.8137-8146.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yáñez-Muñoz R.J., Balaggan K.S., MacNeil A., Howe S.J., Schmidt M., Smith A.J., Buch P., MacLaren R.E., Anderson P.N., Barker S.E. Effective gene therapy with nonintegrating lentiviral vectors. Nat. Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- 45.Blasi M., Negri D., LaBranche C., Alam S.M., Baker E.J., Brunner E.C., Gladden M.A., Michelini Z., Vandergrift N.A., Wiehe K.J. IDLV-HIV-1 Env vaccination in non-human primates induces affinity maturation of antigen-specific memory B cells. Commun. Biol. 2018;1:134. doi: 10.1038/s42003-018-0131-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler S.L., Hansen M.S., Bushman F.D. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 47.Cara A., Cereseto A., Lori F., Reitz M.S., Jr. HIV-1 protein expression from synthetic circles of DNA mimicking the extrachromosomal forms of viral DNA. J. Biol. Chem. 1996;271:5393–5397. doi: 10.1074/jbc.271.10.5393. [DOI] [PubMed] [Google Scholar]
- 48.Cockrell A.S., Kafri T. Gene delivery by lentivirus vectors. Mol. Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- 49.Zaiss A.K., Son S., Chang L.J. RNA 3′ readthrough of oncoretrovirus and lentivirus: implications for vector safety and efficacy. J. Virol. 2002;76:7209–7219. doi: 10.1128/JVI.76.14.7209-7219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss R., Teich N., Varmus H., Coffin J. Second Edition. Cold Spring Harbor Laboratory Press; 1992. RNA Tumor Viruses; pp. 1205–1218. [Google Scholar]
- 51.Berger G., Durand S., Goujon C., Nguyen X.N., Cordeil S., Darlix J.L., Cimarelli A. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat. Protoc. 2011;6:806–816. doi: 10.1038/nprot.2011.327. [DOI] [PubMed] [Google Scholar]
- 52.Bona R., Andreotti M., Buffa V., Leone P., Galluzzo C.M., Amici R., Palmisano L., Mancini M.G., Michelini Z., Di Santo R. Development of a human immunodeficiency virus vector-based, single-cycle assay for evaluation of anti-integrase compounds. Antimicrob. Agents Chemother. 2006;50:3407–3417. doi: 10.1128/AAC.00517-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acock A.C. Stata Press; 2013. Discovering Structural Equation Modeling Using Stata. [Google Scholar]
- 54.STATA . Stata Press; 2015. Structural Equation Modelling Reference Manual. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.