Abstract
The Coronavirus Disease 2019 (COVID-19) outbreak has a devastating impact on health and the economy globally, that’s why it is critical to diagnose positive cases rapidly. Currently, the most effective test to detect COVID-19 is Reverse Transcription-polymerase chain reaction (RT-PCR) which is time-consuming, expensive and sometimes not accurate. It is found in many studies that, radiology seems promising by extracting features from X-rays. COVID-19 motivates the researchers to undergo the deep learning process to detect the COVID- 19 patient rapidly. This paper has classified the X-rays images into COVID- 19 and normal by using multi-model classification process. This multi-model classification incorporates Support Vector Machine (SVM) in the last layer of VGG16 Convolution network. For synchronization among VGG16 and SVM we have added one more layer of convolution, pool, and dense between VGG16 and SVM. Further, for transformations and discovering the best result, we have used the Radial Basis function. CovXmlc is compared with five existing models using different parameters and metrics. The result shows that our proposed CovXmlc with minimal dataset reached accuracy up to 95% which is significantly higher than the existing ones. Similarly, it also performs better on other metrics such as recall, precision and f-score.
Keywords: Coronavirus, COVID-19, SARS Cov-2, Deep learning, Convolutional neural network, Chest X-ray images
1. Introduction
The Coronavirus is still on a surge in many countries. The World Health Organisation (WHO) named 2019-nCOV epidemic as COVID-19 in February 2020[1]. The typical symptoms of the disease are; headache, fever, cough, fatigue, muscular pain and respiratory distress [[2], [3], [4], [5]]. The most prominently used test process called Reverse Transcription-polymerase chain reaction (RT-PCR) is less accurate, particularly in the early stage of the infection and, also it is a time-consuming procedure [6]. Rapid antigen testing can be done to quickly to correlate any symptoms of COVID-19 by collecting sample from the nose using swab[7]. But antigen testing is considered to be less accurate than RT-PCR. Also, repeated testing is required to confirm the status from time to time. In the due course, the patient has to be quarantined until the result is not processed. Negative RT-PCR using nasopharyngeal (NP) or oropharyngeal (OP) swab test does not rule our COVID-19 diagnosis as NP/OP sensitivity in the course of disease lies from 42 % to 71 % [8].
These issues bring the requirement of a more accurate and rapid testing process. The reduced accuracy in early detection of disease in RT-PCR process can be overcome by supplementing radiological chest images such as X-rays and Computed Tomography CT [9]. The radiological images seem promising to detect the disease at the onset, or even before, of any symptoms encountered [10]. Nevertheless, radio imaging and RT-PCR both can be combinedly applied to support more sensitivity for detecting and predicting COVID-19[[11], [12], [13], [14], [15]]. The rapid and more accurate results of radio images persuade researchers to evaluate these images using different deep learning techniques and models. This will help automate a major part of detection process while adding more accuracy. Moreover, these methodologies can be applied to various other tests of infections.
As of now, datasets of X-ray and CT-scan of normal, COVID-19 and pneumonia patients are being made available to support the learning process; Even then, the governments are still facing a huge scarcity of testing kits. The deep learning model is a branch of Artificial Intelligence that applies feature extraction techniques to detect the required outcome. The features extraction of deep learning was applied for face detection initially [[16], [17]]. Later on, it was implemented for the detection of breast cancer, skin cancer and pneumonia [[18], [19]]. Currently, many diseases can be detected using the models of deep earning, such disease can be diabetic retinopathy, detection of tumors in different body organs such as breast, head, lungs and brain [[20], [21], [22]]. This motivates researchers to apply machine learning practices to the data and images available on the network. In the recent past, various work has been proposed to detect COVID-19 with the help of radio imaging [[23], [24]]. Many researchers have shown interest in detecting COVID-19 using medical imagining.
We can throw some light on the similar work using deep learning for COVID-19 diagnosing. In [25] authors proposed a deep-learning model by modifying inception transfer learning to achieve accuracy up to 92.4. A review is presented [26] by authors that cater to an analysis of various proposed models for diagnosing COVID-19 using radiological images. For an unbalanced dataset between COVID and other images, [27] purposes a model based on RestNetV20 and Xception to detect COVID-19 and pneumonia using chest X-rays. For classification [28] DarkNet model is used as the base model with seventeen layers of convolution. It uses an equal number of normal and pneumonia images and uses LeayReLU operations for binary classification. Image augmentation is used to increase the number of infected images [29], it classifies images into three different categories COVID, normal and pneumonia. It is using 66 COVID X-ray images. A metalerner[30] architecture is introduced to process different shapes of input images. It uses ReLU as an activation function to classify images among normal, bacterial pneumonia and viral pneumonia. A quick summery is shown in Table 1 .
Table 1.
Feature summary of surveyed models.
| Model | Base-Model | Classification | COVID images | Activation Function | Input Feature |
|---|---|---|---|---|---|
| [25] | Xception and RestNet | Binary | 149 | ReLU and batch normalization | [300,300,1] |
| [26] | DarkCovidNet | Binary | 127 | LeakyReLUU | [256,256,1] |
| [27] | SqueezNet | Binary | 66 | ReLU | [227,227,3] |
| [28] | CovXNet | Terary | 305 | LeakyReLU | Multiple input zone |
Here we propose COVID-19 detection model using deep learning. We have proposed a model named CovXmlc. That is an abbreviation for COVID-19 X- rays multi-model Classification. CovXmlc takes X-ray image as an input and classifies it into normal or COVID-19. CovXmlc model is based on the deep learning architecture known as VGG16[31] and Support Vector Machine SVM. We have taken VGG16 as a base CNN model and on the top as the model is fully trained on a diversified imagenet dataset. We will use the evaluated weight of VGG16 instead of starting from scratch.
The SVM model is added to the top of VGG16 to get better results. The SVM model utilizes the deep-feature evaluated from the CNN model, to give multi-model classification. Hence, this model is concatenation of VGG16 model and SVM model. Further SVM uses different transformations such as radial bases function, linear function and sigmoid. The dataset is preprocessed to give unified pixel size for CNN i.e. 224 × 224 × 3.
To ready VGG16 model for binary prediction, it is also modified by adding a new convolution layer including max-pooling, dense and dropout layer. The proposed model CovXmlc is compared with existing models where it performed significantly better considering various metrics.
The remaining sections of this article are section 2 that describes the methodology showing the proposed model using VGG16 and SVM with brief elaboration of each one of them. Section 3 evaluates, visualizes and analyzes the results of proposed model considering different performance metrics. Finally, section 4 includes concluding remarks.
2. Methodology
While addressing machine vision problems, use of deep convolution network
[32] is quite extensive. Deep convolution network has a multilayered architecture which is quite helpful in feature extraction for various image classification problems.
The machine vision solutions are applied on images like X-ray, Sonography, MRI in medical diagnosis as well as many other industrial and agricultural solutions. We propose that the CNN can also be applied to X-ray images for various medical diagnoses including COVID-19. This work focuses to classify patients through X-ray images to suggest diagnostics in terms of COVID-19 or normal. The process involving iterative computations to extract features of X-ray images to detect COVID-19 images with good amount of accuracy.
In the recent development of Deep CNN various effective and efficient deep convolutional networks were proposed and examined these includes are VGG16; ResNet [33]; DenseNet [34]; Inception [35]; Xception [36]; VGG16. VGG16 is trained in large classes of ImageNet dataset. Instead of, starting from the scratch the processed weights from the VGG16 can be used for image classification. However, for detecting COVID-19 cases more robust and dependable models are required. For this, our experiment uses multi-model combinations of these machine learning models.
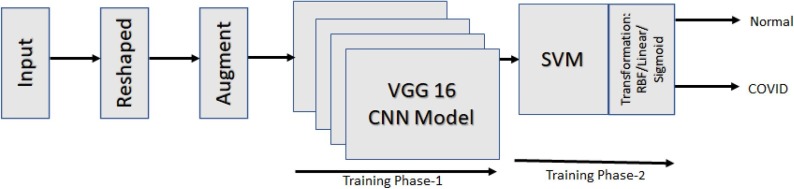
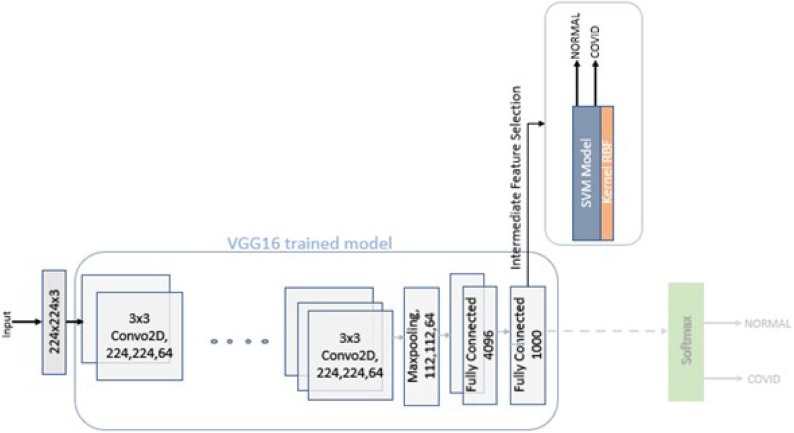
The block diagram of proposed model is depicted in Fig. 1 . The figure shows VGG16 model clubbed with State Vector Machine (SVM) model. In this model instead of depending fully on VGG16, the SVM is also introduced to perform additional processing on the intermediate feature extraction by VGG16. The final output of the model (that is, based on the X-ray image, whether the patient can be diagnosed normal or COVID-19) will be given by the SVM. The VGG16 model is referred in Fig. 3 . The model represents its architecture, the model takes an input of 224*224 RGB images. The input is passed through various layers of Convolution Neural Network it includes Convolution, ReLU, Max-Pooling, fully-connected layer and Softmax.
Fig. 1.
Block model for proposed model.
Fig. 3.
Proposed Model CovXmlc Architecture.
VGG16 model consists of various convolution layers with different numbers of filters. As shown in figure the input image is passed through two convolution layers each of 64 kernels each of size 3x3, it is then followed by Max-Pooling. This gives an output of dimensions 112x112x64. The process will go on with several convolutions and Max-Pooling layers to finally give an output of 7x7x512 dimensions. The output is then further flattened and transferred to the three layers of fully connected network. Finally, a sigmoid function is applied to get the final output.
In our proposed model, convolution channels of 64, 128, 256, 512, and 512 are used to extract features which are then followed by Max-Pooling and ReLU activation function in CNN. The features Y are extracted as follows:
| (1) |
Where, and
2.1. Dataset
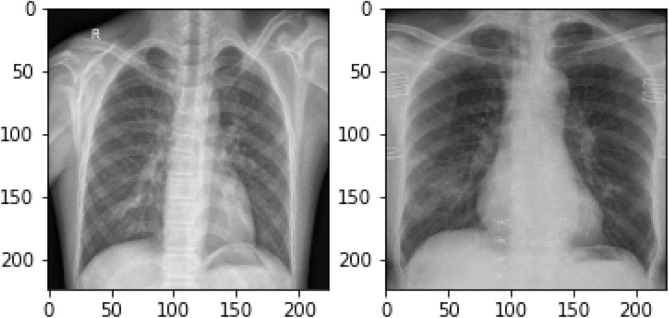
For training our model we are using a total 220 X-ray images from an open dataset COVID-19 Image Data Collection: Prospective Predictions are the Future. Out of these 220 images, 118 are X-ray images of a Normal person while 102 are COVID-19 patient’s X-ray images. The Fig. 2 shows Normal (left) and COVID-19(right) patient X-ray images. These images are randomly picked from the Normal set and COVID-19 images set of the dataset. These images are resized to 224x224 pixel resolution ignoring aspect ratio. The dataset [37] is in the public domain, it is a collection of radiological images collected from various public sources. It consists of COVID-19, bacterial and viral pneumonia. In our experiment COVID-19 and normal images are used with RGB channels. To make images ready for further processing, minimal preprocessing is done to streamline the process of training and testing. These images are modified to have a uniform resolution to make them ready for convolution. The images of different sizes are resized into acceptable dimensions for our model as our model accepts images as input in 224x224 pixels only.
Fig. 2.
(a) Normal (left) and (b) COVID-19(right) X-ray.
3. Proposed CovXmlc
The proposed CovXmlc architecture is shown in Fig. 3. CovXmlc model incorporates the SVM (Support Vector Machine) as the output layer on the intermediate features that are computed at the CNN level. The SVM is widely used for classification and pattern recognition tasks [38]. It can be used for linear as well as non-linear classification problems [39] also.
The transformation of data into required form is done by different kernels available in SVM. These kernels are linear kernel, sigmoid kernel, Radial Basis Kernal (RBF) and polynomial kernel.
| (2) |
| (3) |
| (5) |
The proposed model is shown in Fig. 3. Instead of beginning the CNN from scratch, proposed model exploits the available weight by VGG16 model explored on vast images of ImageNet. Now the model has been trained on the available data set by using the training data and validating it. Once our model is ready with the CNN after processing it with specific counts of epochs, the intermediate feature of a fully connected network is given to SVM as an input. As shown in Fig. 3 the VGG16 model is entirely trained using the whole structure of the basic VGG16 model. However, the features of this fully connected layer are preserved. These features obtained from fully connected layers are given to SVM to provide results. The results finally obtained from SVM models are considered to be CovXmlc results and then systematically compared with other existing models.
3.1. HYPERLINK “SPS:id::Sec3” Evaluation metrics
To evaluate the performance of the proposed model various metrics are utilized are Precision, Sensitivity, Specificity, Accuracy, False Positive Rate and F-Score These can be defined as follows:
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
Here, NTP is number of correct classifications for COVID-19 images, NTN is the number of correctly classified normal images, NFP is incorrectly classified COVID-19 image and NFN is number of misclassified normal images. For a good classification model, the result of all the above-mentioned metrics must be equivalent or near to the best-expected values.
3.2. Experimental setup and result analysis
To train our model we have implemented it on Google Collaborator Notebooks having 25 GB RAM and Tesla P100 GPU. The implementation is done in python using Keras module. The Table 2 shows experimental setup.
Table 2.
Observation table.
| Epochs | Batchsize | Learning Rate | Model |
|---|---|---|---|
| 50/75/100/125 | 20 | 1e−3 | VGG16/VGG19/CovXmlc ResNet50/ InceptionV3/DenseNet121 |
Performance of our proposed model was observed by computation with different epochs at CNN level. The experiment has considered total 60 % random images for training the model and remaining 40 % is used for testing the model. For evaluating all the metrics, the confusion matrix is computed first. Following cases are hereby discussed:
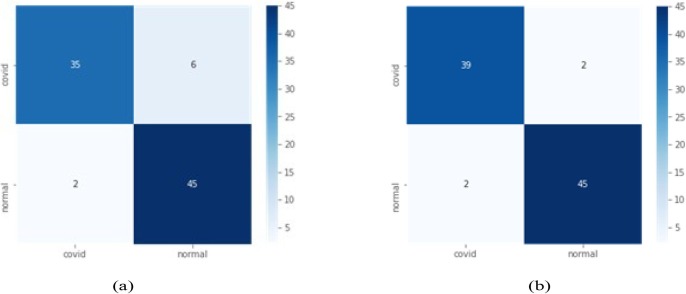
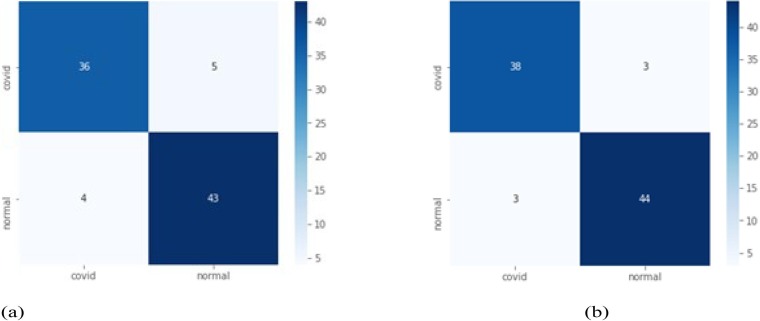
Case 1: In this, we have performed the experiment using total 50 epochs in CNN. The performance of VGG16, VGG19, RestNet50, InceptionV3 and DenseNet121 are compared with proposed CovXmlc, shown in Table 3 . It can be noted that the proposed model’s precision, recall and accuracy are 0.05 better than existing while specificity and F1-score are also better significantly. From figure Fig. 4 it can be seen False Positive in the proposed model is three less than False Positive of VGG16 while True Positive is also three higher than the existing one.
Table 3.
Case 1 with 50 epochs.
| Model | Accuracy | Recall | Specificity | F1-Score | Precision |
|---|---|---|---|---|---|
| VGG16 | 0.90 | 0.90 | 0.95 | 0.91 | 0.91 |
| CovXmlc | 0.95 | 0.95 | 0.96 | 0.95 | 0.96 |
| VGG19 | 0.77 | 0.77 | 0.74 | 0.77 | 0.78 |
| RestNet50 | 0.82 | 0.81 | 0.8 | 0.81 | 0.81 |
| InceptionV3 | 0.9 | 0.91 | 0.91 | 0.91 | 0.91 |
| DenseNet121 | 0.93 | 0.93 | 0.96 | 0.93 | 0.94 |
Fig. 4.
Case 1 confusion matrix for VGG16(a) and proposed model CovXmlc (b).
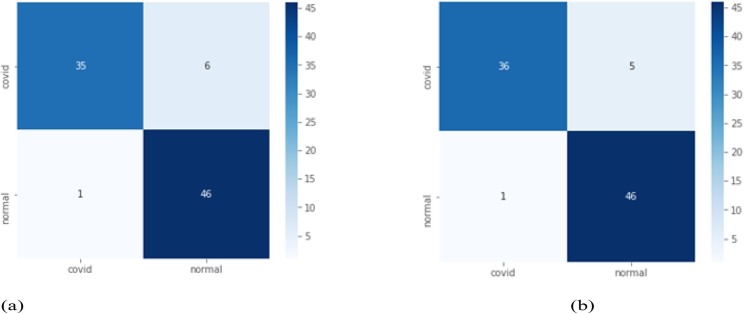
Case 2: In this, we have performed the experiment using a total of 75 epochs in CNN. From performance Table 4 , it can be noted that CovXmlc having reasonable improvement in the case of accuracy with all the other models. In case while comparing with VGG16 for example precision and F1-Score is 0.03 better in the proposed model. Specificity is marginally higher by 0.02. Fig. 5 shows better results of the proposed model in all four quadrants.
Table 4.
Case 2 with 75 epochs.
| Model | Accuracy | Recall | Specificity | F1-Score | Precision |
|---|---|---|---|---|---|
| VGG16 | 0.89 | 0.87 | 0.91 | 0.90 | 0.90 |
| CovXmlc | 0.93 | 0.93 | 0.93 | 0.93 | 0.93 |
| VGG19 | 0.85 | 0.85 | 0.99 | 0.85 | 0.85 |
| RestNet50 | 0.82 | 0.83 | 0.81 | 0.83 | 0.83 |
| InceptionV3 | 0.90 | 0.91 | 0.85 | 0.91 | 0.91 |
| DenseNet121 | 0.92 | 0.92 | 0.96 | 0.93 | 0.93 |
Fig. 5.
Case 2 confusion matrix for VGG16(a) and proposed model CovXmlc (b).
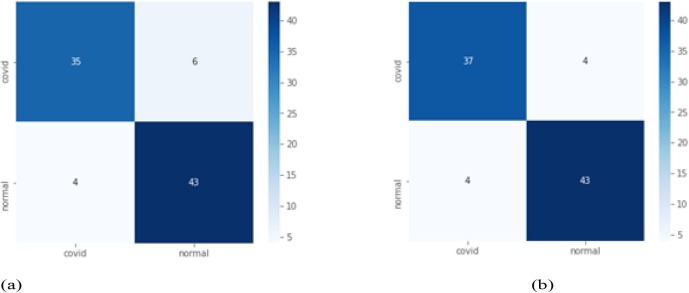
Case 3: 100 epochs are considered here for experimental evaluation. For instance while comparing with VGG16 (Table 5 ) shows 0.08, 0.01 and 0.03 improvement in recall, accuracy and F1-score respectively. While precision and Specificity remain same. In addition to this Fig. 6 shows 1 unit of increase in True Positive while 1 unit of decrease in False Positive.
Table 5.
Case 3 with 100 epochs.
| Model | Accuracy | Recall | Specificity | F1-Score | Precision |
|---|---|---|---|---|---|
| VGG16 | 0.92 | 0.85 | 0.97 | 0.90 | 0.97 |
| CovXmlc | 0.93 | 0.93 | 0.97 | 0.93 | 0.97 |
| VGG19 | 0.9 | 0.91 | 0.95 | 0.91 | 0.91 |
| RestNet50 | 0.83 | 0.83 | 0.80 | 0.83 | 0.80 |
| InceptionV3 | 0.91 | 0.91 | 0.87 | 0.91 | 0.91 |
| DenseNet121 | 0.92 | 0.92 | 0.95 | 0.92 | 0.92 |
Fig. 6.
Case 3 confusion matrix for VGG16(a) and proposed model CovXmlc (b).
Case 4: This case considers 125 epochs for the experimental setup. The evaluated Table 6 shows significant improvement in accuracy by 0.03 units while a vast improvement in case of recall by 0.8 units while others evaluation is marginally higher than the existing one, in case of comparing with VGG16.
Table 6.
Case 4 with 125 epochs.
| Model | Accuracy | Recall | Specificity | F1-Score | Precision |
|---|---|---|---|---|---|
| VGG16 | 0.88 | 0.85 | 0.91 | 0.89 | 0.89 |
| CovXmlc | 0.91 | 0.91 | 0.92 | 0.91 | 0.93 |
| VGG19 | 0.90 | 0.91 | 0.95 | 0.91 | 0.91 |
| RestNet50 | 0.79 | 0.79 | 0.85 | 0.79 | 0.80 |
| InceptionV3 | 0.91 | 0.91 | 0.85 | 0.91 | 0.91 |
| DenseNet121 | 0.91 | 0.91 | 0.95 | 0.91 | 0.93 |
Fig. 7 shows 2 units of increase in True Positive and 2 units of decrease in False Positive. While False Negative and False Positive remains unchanged.
Fig. 7.
Case 4 confusion matrix for VGG16(a) and proposed model CovXmlc (b).
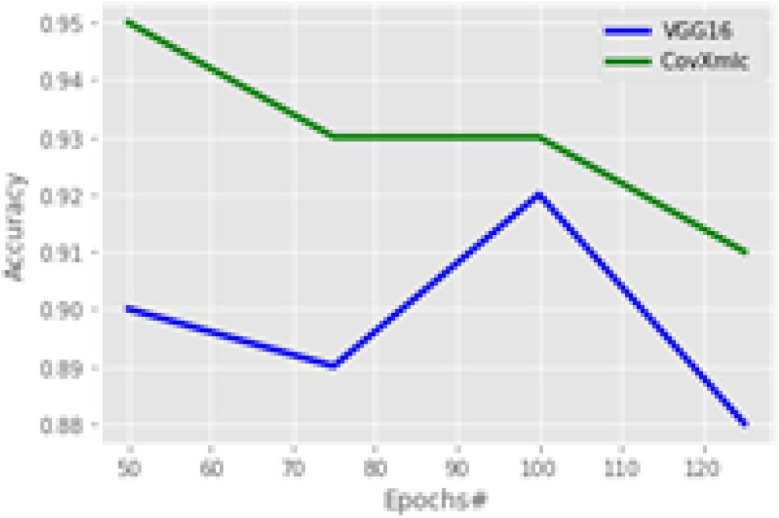
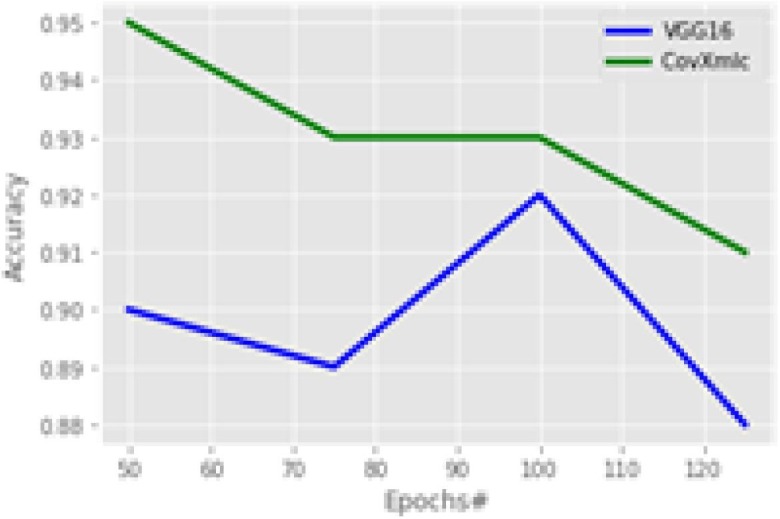
Fig. 8 shows the accuracy of the models considering a variable number of epochs. It is the primary measure of performance for any classification problems. It can be seen from the figure that the proposed model is having higher accuracy with all the epoch sizes. However, the gap in the accuracy is decreased from 100 epochs, but still, it is significant. The proposed model outperforms with an average increase in accuracy by 3.5 % while minimum with 1 % at 100 epochs and maximum is 5.5 % at 50 epochs. The precision plot is shown in Fig. 9 ,
Fig. 8.
Accuracy Comparison.
Fig. 9.
Precision Comparison.
it mentions the fraction of COVID-19 instances among the retrieved instances. It can be seen from the plot the proposed model provides better precision in all the epochs but with 100 epochs there is no improvement. The maximum increase in precision is 5.4 % with 50 epochs, the minimum is 0 % at 100 epochb and the average is 2.7 % overall.
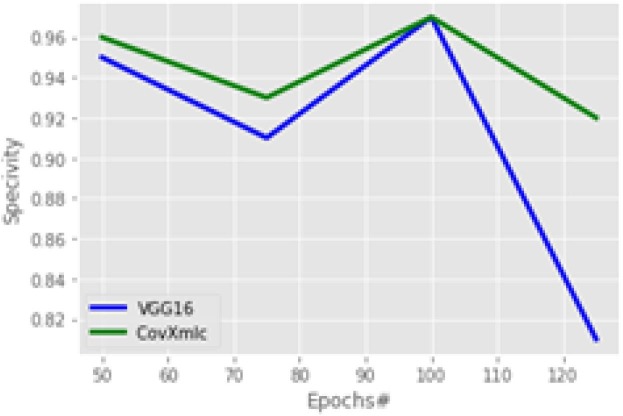
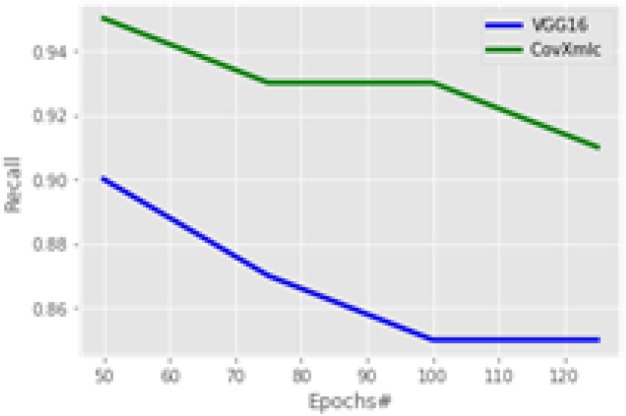
In the experiment specificity plot at Fig. 10 is almost contrary to that of accuracy as here with a smaller number of epochs the increase in the proposed model is minimum. Overall, the average increase in specificity with the proposed model is 3.3 % while a minimum increase is 0 % with 100 epochs and the maximum increase is 13 % with 125 epochs. To evaluate the proportion of COVID-19 positive cases identified correctly recall is calculated on various number of echoes.
Fig. 10.
Specificity Comparison.
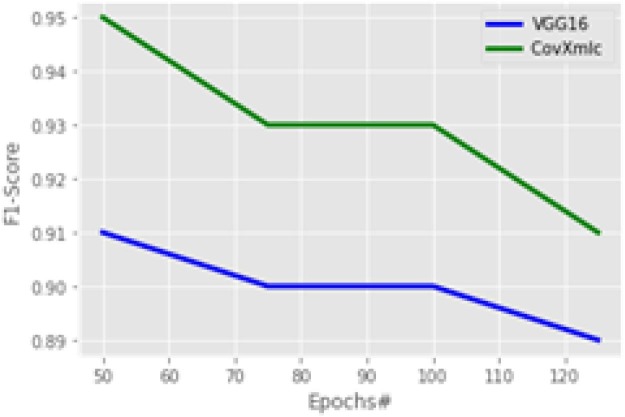
Recall comparison is shown in Fig. 11 for the proposed model with a competitive one. The proposed model significantly much higher in terms of recall, it reveals that it is better by 7.2 % on average while having a maximum improvement of 9.4 % at 100 epochs and 5.5 % is at the minimum with 50 epochs. A weighted harmonic mean known as F1-Score is plotted in Fig. 12 . It is most suitable in case of uneven class distributions. The plot reveals that the proposed model outperforms by 3.3 % on an average case. The maximum increase is 4.4 % with 50 epochs while the minimum increase is 2.2 % at 125 epochs.
Fig. 11.
Recall Comparison.
Fig. 12.
F1-Score Comparison.
4. Conclusion
In this study, a deep neural network architecture CovXmlc is proposed to detect and classify COVID-19 cases. For this, X-ray images are used to train the model. The strength of the proposed model is to add SVM on top of the VGG16 model, it significantly improved accuracy to 95 % from 90 %. Our model shows promising results even by utilizing a limited public dataset. Our result shows with the limited number of epochs all the performance parameters were significantly higher than the existing ones, but with a higher number of epochs, our results are marginally better. This is due to the overfitting in the learning phase. So, in the experiment, we can conclude that CovXmlc is performing significantly excellent with lower epochs even using a small dataset. In the current scenario, different variants of COVID-19 are existing. Our research does not take this under consideration, in the future we will try to incorporate the classification using these variants. SVM will also be incorporated and analyze with transfer learning models other than VGG16. Also, our model will be tested with other diseases as well in which radiology images are available as a public dataset.
CRediT authorship contribution statement
Sourabh Singh Verma: Conceptualization, Methodology, Software. Ajay Prasad: Software, Validation, Data curation, Writing – original draft. Anil Kumar: Supervision, Visualization, Investigation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.World Health Organization, Novel Coronavirus(2019-nCoV) Situation Report-30. https://www.who.int/docs/default-source/coronaviruse/situationreports/20200219-sitrep-30-covid-19.pdf?sfvrsn=6e50645\_2. Accessed 24 March 2020.
- 2.Zhang Y (2020) The epidemiological characteristics of an outbreak of 2019 novel coronavirus (COVID-19)-China CCDC. Zhonghua liu xing bing xue za zhi=Zhonghuauxingbingxue zazhi 41(2):145. [DOI] [PubMed]
- 3.Xie Z. Pay attention to SARS-CoV-2 infection in children. Pediatr Invest. 2020;4(1):1–4. doi: 10.1002/ped4.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., Hu B.o., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan. China. Jama. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chih-Cheng Lai, Tzu-Ping Shih, Wen-Chien Ko, Hung-Jen Tang, Po-Ren Hsueh, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges, International Journal of Antimicrobial Agents, Volume 55, Issue 3, 2020, 105924, ISSN 0924-8579, https://doi.org/10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed]
- 6.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem 2020;66:549–55. https://doi.org/10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed]
- 7.Peacock W.F., Dzieciatkowski T.J., Chirico F., Szarpak L. Self-testing with antigen tests as a method for reduction SARS-CoV-2. Am J Emerg Med. 2021 doi: 10.1016/j.ajem.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chirico F., Nucera G., Magnavita N. Hospital infection and COVID-19: Do not put all your eggs on the “swab” tests. Infect. Control Hosp. Epidemiol. 2021;42(3):372–373. doi: 10.1017/ice:2020.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Y. Zu, M.D. Jiang, P.P. Xu, W. Chen, Q.Q. Ni, G.M. Lu, L.J. Zhang, Coronavirus disease 2019 (COVID-19): a perspective from China, Radiology (2020), https://doi. org/10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed]
- 10.Chan J.F.W., Yuan S., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection, Radiology: Cardiothoracic. Imaging. 2020;2(1):e200028. doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee E.Y., Ng M.Y., Khong P.L. COVID-19 pneumonia: what has CT taught us? Lancet Infect. Dis. 2020;20(4):384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H., Han X., et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;24(4):425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao W., Zhong Z., Xie X., Yu Q., Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. Am. J. Roentgenol. 2020;214(5):1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Xia L., Disease C. (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2019;2020:1–7. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 16.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 17.Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks, in. Advances in Neural Information Processing Systems. 2012:1097–1105. [Google Scholar]
- 18.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talo M., Yildirim O., Baloglu U.B., Aydin G., Acharya U.R. Convolutional neural networks for multi-class brain disease detection using MRI images. Comput. Med. Imag. Graph. 2019;78 doi: 10.1016/j.compmedimag.2019.101673. [DOI] [PubMed] [Google Scholar]
- 20.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A.W.M., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Cheng J.-Z., Ni D., Chou Y.-H., Qin J., Tiu C.-M., Chang Y.-C., Huang C.-S., Shen D., Chen C.-M. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in us images and pulmonary nodules in ct scans. Sci Rep. 2016;6(1):1–13. doi: 10.1038/srep24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S.K. Lakshmanaprabu, Mohanty Sachi Nandan, K. Shankar, N. Arunkumar, Ramirez Gustavo. Optimal deep learning model for classification of lung cancer on ct images. Future Generat Comput Syst. 2019;92:374–382. [Google Scholar]
- 23.Wang L, Wong A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest Radiography Images. ArXiv 2200309871 2020. [DOI] [PMC free article] [PubMed]
- 24.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19). MedRxiv 2020:2020.02.14.20023028. doi:10.1101/2020.02.14.20023028. [DOI] [PMC free article] [PubMed]
- 25.L. Wang, A. Wong, COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest Radiography Images, 2020 arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- 26.Verma S.S., Vishwakarma S.K., Sharma A.K. A Review on COVID-19 DiAgnosis Using ImAging And ArtificiAl Intelligence. Springer; Cham: 2021. [DOI] [Google Scholar]
- 27.Rahimzadeh Mohammad, Attar Abolfazl. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inf. Med. Unlocked. 2020;19:100360. doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammed K. Automated detection of COVID-19 coronavirus cases using deep neural networks with X-ray images. Al-Azhar Univ. J. Virus Res. Stud. 2020 [Google Scholar]
- 29.Ucar Ferhat, Korkmaz Deniz. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahmud, T., Rahman, M. A., & Fattah, S. A. (2020). CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Computers in Biology and Medicine, 122, 103869. (2020). [DOI] [PMC free article] [PubMed]
- 31.Karen Simonyan, Andrew Zisserman, Very Deep Convolutional Networks for Large-Scale Image Recognition, arXiv:1409.1556,2015.
- 32.Moeskops Pim, Viergever Max A., Mendrik Adrienne M., de Vries Linda S., Benders Manon J.N.L., Isgum Ivana. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans MedImaging. 2016;35(5):1252–1261. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
- 33.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2015.
- 34.Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. 2016.
- 35.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. Boston, MA, 2015,: IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015. p. 1–9.
- 36.Xception Chollet F. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Deep Learning With Depthwise Separable Convolutions; pp. 1251–1258. [Google Scholar]
- 37.Joseph Paul Cohen and Paul Morrison and Lan Dao COVID-19 image data collection, arXiv:2003.11597, 2020 https://github.com/ieee8023/covid-chestxray-dataset.
- 38.Burges C. A tutorial on support vector machines for pattern recognition. Data Mining Knowledge Discovery. 1998;2(2):121–167. [Google Scholar]
- 39.Vapnik V. John Wiley; New York: 1998. Statistical Learn Theory. [Google Scholar]