Abstract
Schizophrenia (Scz) is a brain disorder that has a typical onset in early adulthood but otherwise maintains unknown disease origins. Unfortunately, little progress has been made in understanding the molecular mechanisms underlying neurodevelopment of Scz due to ethical and technical limitations in accessing developing human brain tissue. To overcome this challenge, we have previously utilized patient-derived Induced Pluripotent Stem Cells (iPSCs) to generate self-developing, self-maturating, and self-organizing 3D brain-like tissue known as cerebral organoids. As a continuation of this prior work, here we provide an architectural map of the developing Scz organoid proteome. Utilizing iPSCs from n = 25 human donors (n = 8 healthy Ctrl donors, and n = 17 Scz patients), we generated 3D cerebral organoids, employed 16-plex isobaric sample-barcoding chemistry, and simultaneously subjected samples to comprehensive high-throughput liquid-chromatography/mass-spectrometry (LC/MS) quantitative proteomics. Of 3,705 proteins identified by high-throughput proteomic profiling, we identified that just ~2.62% of the organoid global proteomic landscape was differentially regulated in Scz organoids. In sum, just 43 proteins were up-regulated and 54 were down-regulated in Scz patient-derived organoids. Notably, a range of neuronal factors were depleted in Scz organoids (e.g., MAP2, TUBB3, SV2A, GAP43, CRABP1, NCAM1 etc.). Based on global enrichment analysis, alterations in key pathways that regulate nervous system development (e.g., axonogenesis, axon development, axon guidance, morphogenesis pathways regulating neuronal differentiation, as well as substantia nigra development) were perturbed in Scz patient-derived organoids. We also identified prominent alterations in two novel GWAS factors, Pleiotrophin (PTN) and Podocalyxin (PODXL), in Scz organoids. In sum, this work serves as both a report and a resource that researchers can leverage to compare, contrast, or orthogonally validate Scz factors and pathways identified in observational clinical studies and other model systems.
Subject terms: Neuroscience, Schizophrenia
Introduction
Schizophrenia (Scz) is a debilitating brain disorder that occurs in approximately ~1% of the population [1]. While Scz onset typically occurs in early adulthood, subtle brain changes and symptoms often begin emerging years prior to onset during the so-called “prodromal period” [2, 3]. In spite of this, it has remained unclear when Scz neuropathology actually begins to unfold in the brain [1]. For instance, does Scz neuropathology begin a couple of years prior to onset in adolescence when prodromal features progressively emerge? Or does Scz neuropathology begin much earlier in neurodevelopment at a scale that is not yet resolvable? Following decades of investigation, there is now strong epidemiological evidence that indicates risk of Scz may begin to accumulate during in utero brain development [4–7]. This includes data from numerous, independent, large-scale population studies [4–7]. Critically, it remains unclear if in utero risk factors for later Scz onset, such as maternal immune activation, famine, or hormonal/steroid factors, elicit risk by inducing neurodevelopmental alterations or promoting rates of de novo mutation [8]. While the latter can’t be ruled out as a potential etiological contributor, the former hypothesis holds strong merit given the highly-regulated nature of cortical development in utero and the fact that innumerous Scz risk factors exhibit known roles in central nervous system development. Indeed, some novel biological intermediaries are starting to be discovered which link in utero environmental risk factors to potential genetic factors, alterations, and/or vulnerabilities [9]. However, resolving these neurodevelopmental hypotheses of Scz has been difficult. Critically, ethical and technical constraints in accessing human primary brain tissue have arrested progress in delineating the neurodevelopmental trajectory of Scz. These ethical and technical limitations are further compounded by our inability to identify prospective cases of Scz, which has further sequestered our understanding of neurodevelopmental mechanisms of psychosis and has caused a rift between the known epidemiology and the presumed neurobiology of Scz. For instance, in the largest GWAS conducted to date a total of 108 loci of risk were identified – yet, many of these loci (e.g. PTN or PODXL) had unknown disease relevance as well as ambiguously defined neurobiology. Without a means to dissect these factors in human-derived tissue, it is possible that identifying the molecular mediators underlying the ontogeny of disease onset in Scz may continue to be protracted.
Recently, we attempted to overcome these technical and ethical limitations by generating human-derived tissue using stem cells. Namely, we modeled the neurodevelopmental pathology of Scz by harnessing human induced pluripotent stem cells (iPSCs) from healthy adults (Ctrls) and idiopathic Scz patients to generate 3D brain-like tissue known as “cerebral organoids” [1]. Cerebral organoids allow human-specific mechanisms of neural development to be studied while capturing the entirety of the molecular-genetic background of patients. This is a particularly useful model system with respect to “black box” diseases such as Scz, whose neurodevelopmental origins have remained unclear, as it allows self-organizing and self-maturing human neural tissue to be spontaneously generated. Thus, 3D stem cell derived methodologies provide access to a limitless supply of human-derived tissue which can be used to dissect complex diseases defined by “daunting polygenicity” [10] under controlled laboratory conditions [11]. Cerebral organoids mimic trimester 1 of early brain development and putatively recapitulate the epigenetic [12], transcriptomic [13, 14], and proteomic [1, 11] architecture that is expected of the developing mammalian brain. This also includes the recapitulation of cortical cell-type diversity and cellular events such as migration [15] and evolutionary mechanisms that support neocortical neurogenesis [16]. Because of this, cerebral organoids have already been used to model prenatal drug/narcotic effects [11], microcephaly [17], macrocephaly [18], Zika virus effects [19, 20], features of autism [21–23], microdeletion syndromes [24] including 22q11 deletion syndrome [25], hypoxic injury [26], and novel neuropathology of Scz [1, 27–31]. In the case of the latter, Scz-related organoid models have revealed a range of novel phenotypes that may be associated with early neurodevelopmental alterations. This includes diminished responses to electrophysiological stimulation and depolarization [27], alterations in growth factor pathways (e.g., FGFR1 [28] including neurotrophic growth factors and their receptors in Scz progenitors and neurons [1]), immune-related alterations (e.g., TNFα [29] and IFITM3 as well as IL6ST in Scz neurons [1]), potential developmental effects in excitation and inhibition [30], and DISC1 effects on neurodevelopment [31, 32]. Recently, we added to this developing literature by being the first to discover that Scz neuropathology is encoded on a cell-by-cell basis and is defined by multiple novel mechanisms in Scz patient-derived organoids [1]. However, we have also predicted that further mechanisms related to neurodevelopment of Scz remain to be discovered [1], thus requiring deeper analysis in larger samples and populations.
Here we sought to expand our existing knowledge of Scz by providing a deep, unbiased, analysis of molecular factors regulating central nervous system development in human-derived 3D tissue. To do this, we generated cerebral organoids from a relatively large pool of human donors (n = 25; n = 8 Ctrl donors and n = 17 Scz donors) and adapted cutting-edge isobaric barcoding chemistry so that samples could be condensed and analytically deconstructed simultaneously via liquid-chromatography/mass-spectrometry (LC/MS). This yielded a large dataset that we have made freely available for other human, mouse, and cellular researchers to analyze. Notably, here we emphasize large-scale changes identified in this dataset, which included a broad reduction in neuronal molecules important for neural cell-type identity and development as well as metabolic and novel GWAS factors. This work and dataset may thus provide insight for other researchers and labs that have an interest in biological data from human-derived 3D stem cell systems but otherwise employ or use other model systems.
Results
To study the molecular architecture of developing human brain-like tissue, we generated 3D cerebral organoids from human iPSC donors banked by the NIMH. In sum, biologics from n = 25 human donors were sampled comprising n = 8 healthy Ctrls and n = 17 Scz patients. Briefly, iPSCs from human donors were grown in 2D culture atop vitronectin-coated plates before being dissociated with Accutase to yield single-cell iPSC suspensions. Stem cell suspensions were correspondingly cultured into 3D aggregates, known as embryoid bodies, before being subjected to a chemically minimalist neural induction media for up to 7 days in vitro (DIV). After exhibiting evidence of neuroepithelial expansions and/or other morphological evidence of neural induction, tissue was impregnated into a matrigel droplet as a scaffold for further tissue expansion. Developing organoids were then maturated under constant agitation atop an orbital shaker. Following this, at approximately 35-40 DIV, organoids from all 25 human donors were sampled for TMT quantitative proteomics. Briefly, this involved dissociating organoids, preparing peptide suspensions (digestion, reduction, and alkylation), barcoding samples with isobaric TMTpro 16-plex chemistry, and then multiplexing samples for simultaneous detection and analysis via nano high-sensitivity proteome profiling (for a simplified schematic of our experimental pipeline, see Fig. 1).
Fig. 1. Schematic of cerebral organoid and TMT-LC/MS analytical pipeline.
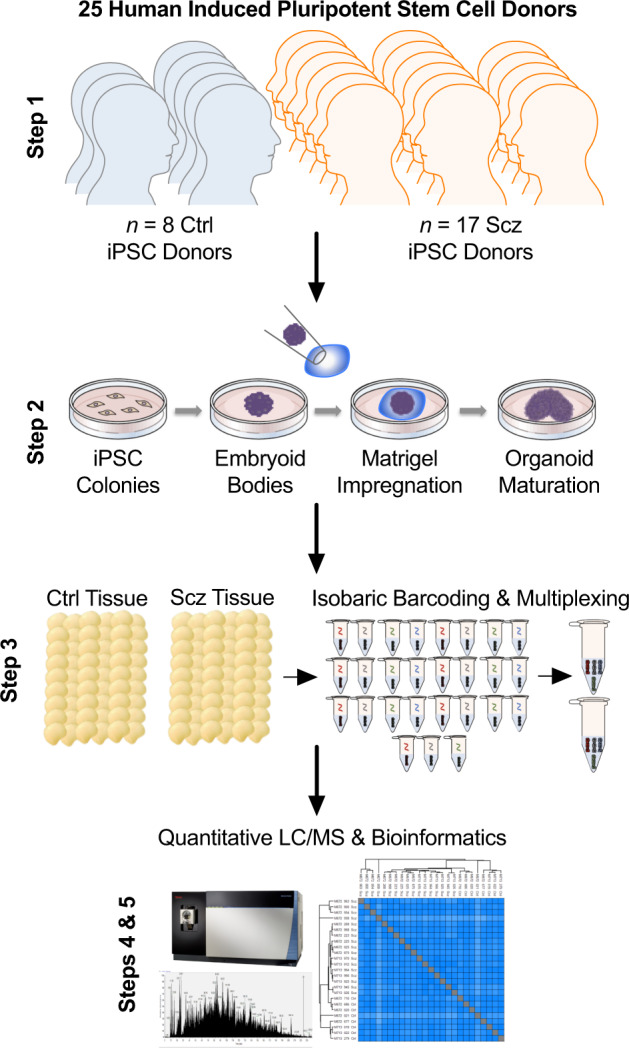
Briefly, 25 distinct human iPSCs were obtained from both healthy Control (Ctrl) donors and Schizophrenia (Scz) patients. Each line represented a biologically unique sample from a specific individual, and lines were predominantly obtained from NIH repositories. Following this, iPSCs were expanded and utilized to generate patient-derived cerebral organoids that mimic the 1st trimester of brain assembly (see Methods, [17, 87] for protocol information, and [1] for our previous application of 3D Scz patient-derived organoids). This process involved dissociating iPSC colonies to generate 3D embryoid body aggregates that could be pushed towards a neural fate via chemically minimalist media cocktails [17, 87]. Following neural induction, organoids were implanted into a matrigel droplet as a scaffold to support tissue expansion and (broadly consistent with our prior study [54]) maturated to a primary endpoint of 35-40 DIV. Following this, samples were individually subjected to protein lysis and tryptic-based enzymatic digestion. For proteomic analysis of cerebral organoids, peptides were isobarically barcoded using TMTpro 16-Plex chemistry that allowed samples to be multiplexed for simultaneous detection of different samples via liquid chomatography–mass spectrometry (LC/MS). This allowed up to 15 samples (+1 pool) to be condensed into a single tube for simultaneous detection via LC/MS analysis, resulting in a total of 27 samples (n = 25 human donor organoids, + n = 2 internal reference pools). Bioinformatics were subsequently conducted in accordance with the parameters described in our Methods as well as two prior manuscripts that have incorporated LC/MS proteomic analysis of human-derived organoid samples [1, 11].
Analysis of organoid proteomes revealed sufficient peptide coverage for high-confidence quantitative analysis of 3705 proteins (peptide >1; intensity >0) across all 25 human donor samples. Based on Log2 transformed protein intensities, the Coefficient of Variation (CV) of Scz and Ctrl proteome groups was highly stringent; Median CV for Ctrls was 1.07% and for Scz 1.23%. This provided confidence in both the degree of neural induction achieved between samples, and that organoids were overall of a very similar and thus comparable composition between iPSC donors and within groups.
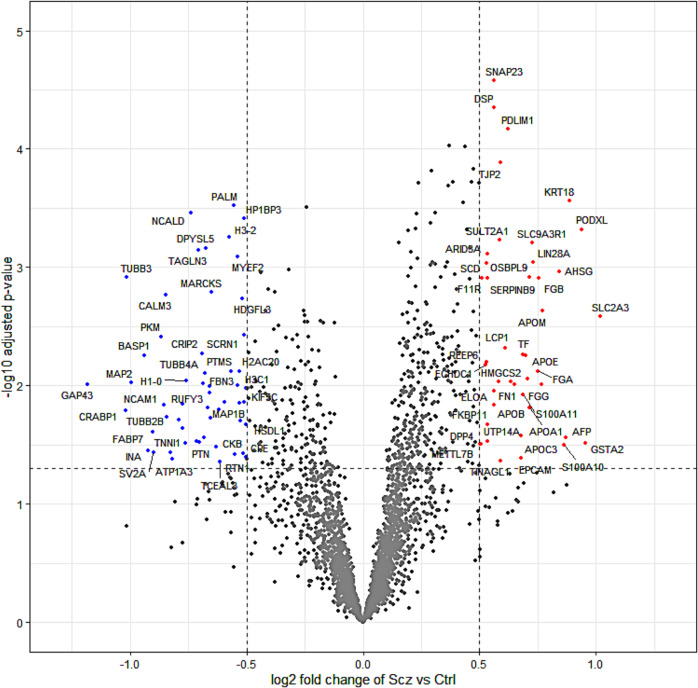
To gain insight into differences between Scz and Ctrl organoids, we next sought to determine which proteins (based on their expression) differed between these groups. Further analysis revealed the significant differential expression of peptide fragments belonging to 97 proteins in Scz organoids, of which 43 were up-regulated (p value < 0.05, Log2FC > 0.05) and 54 were down-regulated (p value < 0.05, Log2FC < −0.05). Thus, in sum, ~2.62% of the total organoid proteome was differentially expressed in Scz organoids, with equivalent (~1.16% vs. ~1.46%) proportions of differentially expressed proteins being up- and down-regulated, respectively.
Deeper examination of significantly down-regulated proteins in Scz organoids, sorted by Log2FC values (see Table 1), revealed several important changes. Notably, we detected a depletion of factors that support neuronal development, differentiation, identity and/or function. Down-regulated neuronal development factors in Scz organoids comprised Neuromodulin (GAP43; Log2FC = −1.183, p = 0.010), Cellular Retinoic Acid-Binding Protein 1 (CRABP1; Log2FC = −1.018, p = 0.016), Neural Cell Adhesion Molecule (NCAM1; Log2FC = −0.854, p < 0.014), and expression of the myelin-modulating factor Myelin Expression Factor 2 (MYEF2; Log2FC = −0.537, p < 0.001). Likewise, down-regulated expression of several other neuronal factors – involved in both neuronal identity and prototypic function – included Microtubule-Associated Protein 2 (MAP2), Tubulin Beta-3 Chain (TUBB3, or β3), Synaptic Vesicle Glycoprotein 2 A (SV2A), and other neuron-specific markers (see Fig. 2). In addition to these changes, we also screened our dataset against novel, yet statistically prominent, Scz GWAS factors identified in the largest population genetic dataset reported to date [33]. One important Scz GWAS factor to emerge from our analysis of down-regulated proteins in Scz organoids was Pleiotrophin (PTN). In our prior work [1], we also detected the differential expression of PTN at both the protein and RNA level in Scz organoids, including in both Scz progenitors and neurons. This better powered analysis therefore replicates this previous finding, and further establishes PTN as a potentially important Scz risk factor during early brain assembly.
Table 1.
54 Down-regulated proteins in Scz organoids (<−0.5 Log2FC, p < 0.05).
| Gene Name | Protein Name | Uniprot ID | Log2FC | P Value |
|---|---|---|---|---|
| GAP43 | Neuromodulin | P17677 | −1.183 | 0.010 |
| CRABP1 | Cellular retinoic acid-binding protein 1 | P29762 | −1.018 | 0.016 |
| TUBB3 | Tubulin beta-3 chain | Q13509 | −1.015 | 0.001 |
| MAP2 | Microtubule-associated protein 2 | P11137-3 | −0.996 | 0.009 |
| BASP1 | Brain acid soluble protein 1 | P80723 | −0.939 | 0.006 |
| INA | Alpha-internexin | Q16352 | −0.921 | 0.035 |
| FABP7 | Fatty acid-binding protein, brain | O15540 | −0.903 | 0.025 |
| SV2A | Synaptic vesicle glycoprotein 2A | Q7L0J3-2 | −0.899 | 0.037 |
| PKM | Pyruvate kinase PKM | P14618-2 | −0.866 | 0.004 |
| NCAM1 | Neural Cell Adhesion Molecule | A0A087WTF6 | −0.854 | 0.014 |
| CALM3 | Calmodulin-3 | P0DP25 | −0.847 | 0.002 |
| TUBB2B | Tubulin beta-2B chain | Q9BVA1 | −0.840 | 0.019 |
| TNNI1 | Troponin I 1 | G3V489 | −0.827 | 0.037 |
| ATP1A3 | Sodium/potassium-transporting ATPase subunit alpha-3 | P13637 | −0.817 | 0.042 |
| CRMP1 | Dihydropyrimidinase-related protein 1 | Q14194 | −0.789 | 0.019 |
| RUFY3 | Protein RUFY3 | Q7L099 | −0.774 | 0.014 |
| ATAT1 | Alpha-tubulin N-acetyltransferase 1 | Q5SQI0-7 | −0.773 | 0.023 |
| PEA15 | Astrocytic phosphoprotein PEA-15 | Q15121 | −0.764 | 0.030 |
| H1-0 | Histone H1.0 | P07305-2 | −0.760 | 0.009 |
| NCALD | Neurocalcin-delta | P61601 | −0.738 | 0.000 |
| PPM1B | Protein phosphatase 1B | O75688 | −0.714 | 0.030 |
| TAGLN3 | Transgelin-3 | Q9UI15 | −0.705 | 0.001 |
| PTN | Pleiotrophin | P21246 | −0.700 | 0.030 |
| CRIP2 | Cysteine-rich protein 2 | P52943 | −0.690 | 0.005 |
| RAB6B | Ras-related protein Rab-6B | Q9NRW1 | −0.684 | 0.010 |
| ENO2 | Gamma-enolase | P09104-2 | −0.682 | 0.027 |
| TUBB4A | Tubulin beta-4A chain | P04350 | −0.676 | 0.008 |
| DPYSL5 | Dihydropyrimidinase-related protein 5 | Q9BPU6 | −0.673 | 0.001 |
| SEPTIN3 | Neuronal-specific septin-3 | Q9UH03-2 | −0.667 | 0.015 |
| GDI1 | Rab GDP dissociation inhibitor alpha | P31150 | −0.659 | 0.011 |
| FHL1 | Four and a half LIM domains protein 1 | Q13642-1 | −0.658 | 0.010 |
| TUBA1A | Tubulin alpha-1A chain | Q71U36-2 | −0.653 | 0.019 |
| MARCKS | Myristoylated alanine-rich C-kinase substrate | P29966 | −0.650 | 0.002 |
| UCHL1 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | P09936 | −0.629 | 0.033 |
| LAMA4 | Laminin subunit alpha-4 | Q16363-2 | −0.619 | 0.016 |
| TCEAL3 | Transcription elongation factor A protein-like 3 | Q969E4 | −0.614 | 0.044 |
| TUBB4B | Tubulin beta-4B chain | P68371 | −0.595 | 0.014 |
| H3-2 | Histone HIST2H3PS2 | Q5TEC6 | −0.574 | 0.001 |
| PTMS | Parathymosin | P20962 | −0.565 | 0.008 |
| PALM | Paralemmin-1 | O75781-2 | −0.552 | 0.000 |
| RTN1 | Reticulon-1 | Q16799-3 | −0.551 | 0.038 |
| FBN3 | Fibrillin-3 | Q75N90 | −0.538 | 0.010 |
| MYEF2 | Myelin Expression Factor 2 | A0A087WUT0 | −0.537 | 0.001 |
| H2AC20 | Histone H2A type 2-C | Q16777 | −0.531 | 0.008 |
| DPYSL2 | Dihydropyrimidinase-related protein 2 | Q16555 | −0.529 | 0.014 |
| MAP1B | Microtubule-associated protein 1B | P46821 | −0.527 | 0.020 |
| HDGFL3 | Hepatoma-derived growth factor-related protein 3 | Q9Y3E1 | −0.517 | 0.002 |
| CKB | Creatine kinase B-type | P12277 | −0.513 | 0.037 |
| KIF5C | Kinesin heavy chain isoform 5C | O60282 | −0.512 | 0.014 |
| SCRN1 | Secernin-1 | Q12765 | −0.510 | 0.004 |
| HP1BP3 | Heterochromatin protein 1-binding protein 3 | Q5SSJ5 | −0.509 | 0.000 |
| H3C1 | Histone H3.1 | P68431 | −0.502 | 0.010 |
| CPE | Carboxypeptidase E | D6RF88 | −0.501 | 0.040 |
| HSDL1 | Inactive hydroxysteroid dehydrogenase-like protein 1 | Q3SXM5-2 | −0.501 | 0.021 |
Fig. 2. Differential expression in the Scz cerebral organoid proteome.
Principal component analysis of the cerebral organoid proteome indicated data grouping based on phenotype, and protein expression distributions indicated data correlation across all samples. This statistical baseline allowed us to consider the differentially expressed proteins present in Scz patient-derived cerebral organoids, which are shown here as a volcano plot split by log2 fold change and –log10 adjusted p values. In sum, ~2.62% of 3705 proteins (peptide >1; intensity >0) identified exhibited differential expression. Significantly up-regulated proteins that surpassed log2 fold change thresholding are depicted to the right in red (p value <0.05, Log2FC > 0.05), whereas down-regulated proteins (p value < 0.05, Log2FC < -0.05) are presented to the left of the plot in blue. Notable Scz GWAS factors (see 108 loci identified in [33]) included the up-regulation of PODXL and down-regulation of PTN, which replicated our previous findings in a smaller cohort [1]. Note also the down-regulation of the neural stem cell proliferation factor CRABP1 [93] as well as canonical neuronal development markers (e.g. NCAM1 [94], NCALD [95], and CPE [78]), neuronal markers (e.g. MAP2, TUBB3, MAP1B), synaptic markers (e.g., SV2A). Conversely, a range of apolipoproteins (APOE, APOA1, APOB, APOC3) were found to be up-regulated in Scz patient-derived cerebral organoids.
Similar to our review of down-regulated proteins, we also identified a number of biologically interesting observations in our up-regulated Scz protein set list (see Table 2). This included up-regulation of numerous fibrinogens (FGG, FGB, FGA; Log2FC = 0.749-0.768, p = 0.008–0.010) and apolipoproteins (APOM, APOA1, APOE, APOC3, APOB; Log2FC = 0.562–0.771, p = 0.001–0.015). However, one of the most notable up-regulated protein was another Scz GWAS factor [33] that (like PTN) we had also identified in our prior Scz patient-derived organoid work [1]; namely, Podocalyxin (PODXL; Log2FC = 0.939, p < 0.001). Therefore, similar to our replication of down-regulated PTN expression in Scz organoids, this analysis in a larger pool of patients confirms that PODXL is another high-confidence candidate that may play a role in modulating Scz risk during early brain development.
Table 2.
43 Up-regulated proteins in Scz organoids (>0.5 Log2FC, p < 0.05).
| Gene Name | Protein Name | Uniprot ID | Log2FC | P-value |
|---|---|---|---|---|
| SLC2A3 | Solute carrier family 2, facilitated glucose transporter member 3 | P11169 | 1.019 | 0.003 |
| GSTA2 | Glutathione S-transferase A2 | P09210 | 0.954 | 0.030 |
| PODXL | Podocalyxin | O00592-2 | 0.939 | 0.000 |
| KRT18 | Keratin, type I cytoskeletal 18 | P05783 | 0.884 | 0.000 |
| AFP | Alpha-fetoprotein | P02771 | 0.868 | 0.027 |
| S100A10 | Protein S100-A10 | P60903 | 0.861 | 0.032 |
| AHSG | Alpha-2-HS-glycoprotein | P02765 | 0.843 | 0.001 |
| APOM | Apolipoprotein M | O95445-2 | 0.771 | 0.002 |
| FGG | Fibrinogen gamma chain | P02679-2 | 0.768 | 0.010 |
| FGB | Fibrinogen beta chain | P02675 | 0.753 | 0.001 |
| FGA | Fibrinogen alpha chain | P02671-2 | 0.749 | 0.008 |
| LIN28A | Protein lin-28 homolog A | Q9H9Z2 | 0.731 | 0.001 |
| SLC9A3R1 | Na(+)/H( + ) exchange regulatory cofactor NHE-RF1 | O14745 | 0.726 | 0.001 |
| APOA1 | Apolipoprotein A-I | P02647 | 0.715 | 0.015 |
| SERPINB9 | Serpin B9 | P50453 | 0.712 | 0.001 |
| SERPINA1 | Alpha-1-antitrypsin | P01009 | 0.705 | 0.009 |
| APOE | Apolipoprotein E | P02649 | 0.698 | 0.006 |
| TF | Serotransferrin | P02787 | 0.687 | 0.005 |
| S100A11 | Protein S100-A11 | P31949 | 0.685 | 0.012 |
| APOC3 | Apolipoprotein C-III | P02656 | 0.678 | 0.027 |
| EPCAM | Epithelial cell adhesion molecule | P16422 | 0.677 | 0.041 |
| FN1 | Fibronectin | P02751-5 | 0.650 | 0.010 |
| APOA4 | Apolipoprotein A-IV | P06727 | 0.634 | 0.009 |
| PDLIM1 | PDZ and LIM domain protein 1 | O00151 | 0.624 | 0.000 |
| LCP1 | Plastin-2 | P13796 | 0.611 | 0.005 |
| TINAGL1 | Tubulointerstitial nephritis antigen-like | Q9GZM7-3 | 0.591 | 0.043 |
| TJP2 | Tight junction protein ZO-2 | Q9UDY2-5 | 0.591 | 0.000 |
| SULT2A1 | Sulfotransferase 2A1 | Q06520 | 0.588 | 0.001 |
| HMGCS2 | Hydroxymethylglutaryl-CoA synthase, mitochondrial | P54868-2 | 0.580 | 0.009 |
| SNAP23 | Synaptosomal-associated protein 23 | O00161 | 0.563 | 0.000 |
| DSP | Desmoplakin | P15924 | 0.562 | 0.000 |
| APOB | Apolipoprotein B-100 | P04114 | 0.562 | 0.015 |
| ELOA | Elongin-A | Q14241 | 0.560 | 0.011 |
| UTP14A | U3 small nucleolar RNA-associated protein 14 homolog A | Q9BVJ6-3 | 0.536 | 0.029 |
| FKBP11 | Peptidyl-prolyl cis-trans isomerase FKBP11 | Q9NYL4-2 | 0.534 | 0.021 |
| F11R | Junctional adhesion molecule A | Q9Y624 | 0.534 | 0.001 |
| ARID3A | AT-rich interactive domain-containing protein 3A | Q99856 | 0.532 | 0.001 |
| OSBPL9 | Oxysterol-binding protein-related protein 9 | Q96SU4-7 | 0.531 | 0.001 |
| REEP6 | Receptor expression-enhancing protein 6 | Q96HR9-2 | 0.530 | 0.006 |
| ECHDC1 | Ethylmalonyl-CoA decarboxylase | Q9NTX5-2 | 0.524 | 0.007 |
| SCD | Acyl-CoA desaturase | O00767 | 0.509 | 0.001 |
| METTL7B | Methyltransferase-like protein 7B | Q6UX53 | 0.505 | 0.031 |
| DPP4 | Dipeptidyl peptidase 4 | P27487 | 0.500 | 0.031 |
We next sought to understand the potential functionality of our differentially expressed protein targets by parsing these factors into pathways, which may also unveil broader changes in regulatory networks underscoring disease-related phenotypes. We principally examined Gene Ontology (GO) pathways, parsed by annotations belonging to biological (Tables 3–4) and molecular (Tables 5–6) function of differentially expressed proteins. We first considered down-regulated GO biological pathways. Down-regulated GO biological pathways essential for normative brain assembly, development, and maturation overwhelmingly defined Scz patient-derived organoids. This included down-regulated expression of factors that map to axonogenesis, axon development, axon guidance, morphogenesis pathways regulating neuronal differentiation, and, broadly speaking, central nervous system development (due the sheer number of pathways involved here, please refer to Table 3 for statistical values). Another interesting down-regulated GO biological process pathway in Scz organoids was specific enrichment for factors regulating substantia nigra development (GO:0021762, adjusted p = 0.0182, Neg Log10 = 1.74), which is of interest given that this midbrain region belongs to the basal ganglia which holds broad relevance to Scz neuropathology and its treatment (e.g., dopamine and monoamine hypotheses of Scz development and symptoms). Contrary to down-regulated GO biological pathways, up-regulated pathways in Scz organoids broadly reflected pathways involved in cellular metabolism, chylomicron assembly and remodeling, sterol and steroid pathways, as well as lipoprotein remodeling and metabolism-related pathways (refer to Table 4 for statistical values).
Table 3.
Down-regulated GO biological processes in Scz organoids (p < 0.05).
| Biological process | GO:BP Term_ID | Adjusted p-value | Neg Log10 adjusted p |
|---|---|---|---|
| Axon Development | GO:0061564 | 1.88E−07 | 6.725 |
| Nervous System Development | GO:0007399 | 2.98E−07 | 6.525 |
| Plasma Membrane Bounded Cell Projection Organization | GO:0120036 | 7.32E−07 | 6.136 |
| Axonogenesis | GO:0007409 | 8.26E−07 | 6.083 |
| Cell Projection Organization | GO:0030030 | 1.16E−06 | 5.937 |
| Cell Morphogenesis Involved in Neuron Differentiation | GO:0048667 | 1.29E−06 | 5.889 |
| Neuron Projection Morphogenesis | GO:0048812 | 4.63E−06 | 5.335 |
| Plasma Membrane Bounded Cell Projection Morphogenesis | GO:0120039 | 6.03E−06 | 5.219 |
| Cell Projection Morphogenesis | GO:0048858 | 6.50E−06 | 5.187 |
| Cell Part Morphogenesis | GO:0032990 | 8.89E−06 | 5.051 |
| Cell Morphogenesis Involved in Differentiation | GO:0000904 | 2.09E−05 | 4.680 |
| Neuron Differentiation | GO:0030182 | 4.18E−05 | 4.379 |
| Cellular Component Morphogenesis | GO:0032989 | 4.21E−05 | 4.375 |
| Neuron Development | GO:0048666 | 9.53E−05 | 4.021 |
| Neuron Projection Development | GO:0031175 | 0.000125503 | 3.901 |
| Cell Morphogenesis | GO:0000902 | 0.000170319 | 3.769 |
| Generation of Neurons | GO:0048699 | 0.000192896 | 3.715 |
| System Development | GO:0048731 | 0.000289587 | 3.538 |
| Neurogenesis | GO:0022008 | 0.00059592 | 3.225 |
| Multicellular Organism Development | GO:0007275 | 0.00099934 | 3.000 |
| Anatomical Structure Development | GO:0048856 | 0.002063881 | 2.685 |
| Axon Guidance | GO:0007411 | 0.002117016 | 2.674 |
| Neuron Projection Guidance | GO:0097485 | 0.002173862 | 2.663 |
| Negative Regulation of Microtubule Polymerization or Depolymerization | GO:0031111 | 0.011544881 | 1.938 |
| Microtubule-Based Process | GO:0007017 | 0.01300057 | 1.886 |
| Cytoskeleton Organization | GO:0007010 | 0.01330451 | 1.876 |
| Anatomical Structure Morphogenesis | GO:0009653 | 0.014459529 | 1.840 |
| Regulation of Axon Extension | GO:0030516 | 0.015068023 | 1.822 |
| Developmental Process | GO:0032502 | 0.01628204 | 1.788 |
| Substantia Nigra Development | GO:0021762 | 0.018211464 | 1.740 |
| Microtubule Cytoskeleton Organization | GO:0000226 | 0.023833988 | 1.623 |
| Regulation of Extent of Cell Growth | GO:0061387 | 0.027856692 | 1.555 |
| Axon Extension | GO:0048675 | 0.047873583 | 1.320 |
Table 4.
Up-regulated GO biological processes in Scz organoids (p < 0.05).
| Biological process | GO:BP Term_ID | Adjusted p value | Neg Log10 adjusted p |
|---|---|---|---|
| Chylomicron Remodeling | GO:0034371 | 1.03E−08 | 7.988 |
| Chylomicron Assembly | GO:0034378 | 3.76E−08 | 7.425 |
| Plasma Lipoprotein Particle Assembly | GO:0034377 | 1.15E−07 | 6.938 |
| Triglyceride-Rich Lipoprotein Particle Remodeling | GO:0034370 | 1.62E−07 | 6.790 |
| Plasma Lipoprotein Particle Remodeling | GO:0034369 | 1.73E−07 | 6.762 |
| Protein-Lipid Complex Remodeling | GO:0034368 | 1.73E−07 | 6.762 |
| Protein-Containing Complex Remodeling | GO:0034367 | 2.53E−07 | 6.598 |
| Protein-Lipid Complex Assembly | GO:0065005 | 2.53E−07 | 6.598 |
| High-Density Lipoprotein Particle Remodeling | GO:0034375 | 6.89E−07 | 6.162 |
| Reverse Cholesterol Transport | GO:0043691 | 2.10E−06 | 5.677 |
| Plasma Lipoprotein Particle Organization | GO:0071827 | 3.08E−06 | 5.511 |
| Protein-Lipid Complex Subunit Organization | GO:0071825 | 4.88E−06 | 5.312 |
| Cholesterol Efflux | GO:0033344 | 1.47E−05 | 4.832 |
| Terpenoid Metabolic Process | GO:0006721 | 1.85E−05 | 4.733 |
| Very-Low-Density Lipoprotein Particle Remodeling | GO:0034372 | 1.97E−05 | 4.706 |
| Platelet Degranulation | GO:0002576 | 2.31E−05 | 4.636 |
| Sterol Transport | GO:0015918 | 2.44E−05 | 4.612 |
| Phospholipid Efflux | GO:0033700 | 2.84E−05 | 4.547 |
| Isoprenoid Metabolic Process | GO:0006720 | 5.53E−05 | 4.257 |
| Positive Regulation of Substrate Adhesion-Dependent Cell Spreading | GO:1900026 | 6.58E−05 | 4.182 |
| High-Density Lipoprotein Particle Assembly | GO:0034380 | 7.18E−05 | 4.144 |
| Cell-Cell Adhesion | GO:0098609 | 8.60E−05 | 4.066 |
| High-Density Lipoprotein Particle Clearance | GO:0034384 | 0.000120368 | 3.919 |
| Cholesterol Homeostasis | GO:0042632 | 0.000156831 | 3.805 |
| Post-Translational Protein Modification | GO:0043687 | 0.000163138 | 3.787 |
| Sterol Homeostasis | GO:0055092 | 0.000166565 | 3.778 |
| Retinoid Metabolic Process | GO:0001523 | 0.00024961 | 3.603 |
| Regulation of Plasma Lipoprotein Particle Levels | GO:0097006 | 0.000263855 | 3.579 |
| Regulation of Substrate Adhesion-Dependent Cell Spreading | GO:1900024 | 0.000344919 | 3.462 |
| Diterpenoid Metabolic Process | GO:0016101 | 0.000345502 | 3.462 |
| Cholesterol Transport | GO:0030301 | 0.000383496 | 3.416 |
| Heterotypic Cell-Cell Adhesion | GO:0034113 | 0.000409755 | 3.387 |
| Cholesterol Biosynthetic Process | GO:0006695 | 0.000568257 | 3.245 |
| Secondary Alcohol Biosynthetic Process | GO:1902653 | 0.000568257 | 3.245 |
| Regulation of Heterotypic Cell-Cell Adhesion | GO:0034114 | 0.000580431 | 3.236 |
| Regulation of Cdc42 Protein Signal Transduction | GO:0032489 | 0.000667476 | 3.176 |
| Sterol Biosynthetic Process | GO:0016126 | 0.000893117 | 3.049 |
| Plasma Lipoprotein Particle Clearance | GO:0034381 | 0.000959187 | 3.018 |
| Lipoprotein Metabolic Process | GO:0042157 | 0.001034387 | 2.985 |
| Chylomicron Remnant Clearance | GO:0034382 | 0.001066283 | 2.972 |
| Triglyceride-Rich Lipoprotein Particle Clearance | GO:0071830 | 0.001066283 | 2.972 |
| Steroid Metabolic Process | GO:0008202 | 0.00122924 | 2.910 |
| Cholesterol Metabolic Process | GO:0008203 | 0.001569372 | 2.804 |
| Positive Regulation of Cholesterol Esterification | GO:0010873 | 0.001596909 | 2.797 |
| Regulated Exocytosis | GO:0045055 | 0.00171245 | 2.766 |
| Positive Regulation of Cell Morphogenesis Involved in Differentiation | GO:0010770 | 0.00174483 | 2.758 |
| Very-Low-Density Lipoprotein Particle Clearance | GO:0034447 | 0.002277712 | 2.643 |
| Secondary Alcohol Metabolic Process | GO:1902652 | 0.002312476 | 2.636 |
| Homotypic Cell-Cell Adhesion | GO:0034109 | 0.002650901 | 2.577 |
| Triglyceride Catabolic Process | GO:0019433 | 0.002810211 | 2.551 |
| Sterol Metabolic Process | GO:0016125 | 0.002987054 | 2.525 |
| Acylglycerol Homeostasis | GO:0055090 | 0.003467959 | 2.460 |
| Triglyceride Homeostasis | GO:0070328 | 0.003467959 | 2.460 |
| Lipid Homeostasis | GO:0055088 | 0.003686054 | 2.433 |
| Vesicle-Mediated Transport | GO:0016192 | 0.003686287 | 2.433 |
| Regulation of Triglyceride Metabolic Process | GO:0090207 | 0.004233532 | 2.373 |
| Regulation of Cell Morphogenesis Involved in Differentiation | GO:0010769 | 0.004547582 | 2.342 |
| Secretion | GO:0046903 | 0.004648136 | 2.333 |
| Cell Adhesion | GO:0007155 | 0.00503037 | 2.298 |
| Biological Adhesion | GO:0022610 | 0.005314483 | 2.275 |
| Organic Hydroxy Compound Transport | GO:0015850 | 0.005357541 | 2.271 |
| Intermembrane Lipid Transfer | GO:0120009 | 0.006131955 | 2.212 |
| Exocytosis | GO:0006887 | 0.006499986 | 2.187 |
| Steroid Biosynthetic Process | GO:0006694 | 0.006623642 | 2.179 |
| Cdc42 Protein Signal Transduction | GO:0032488 | 0.006865718 | 2.163 |
| Regulation of Cholesterol Ssterification | GO:0010872 | 0.006865718 | 2.163 |
| Regulation of Triglyceride Catabolic Process | GO:0010896 | 0.006865718 | 2.163 |
| Acylglycerol Catabolic Process | GO:0046464 | 0.007287829 | 2.137 |
| Neutral Lipid Catabolic Process | GO:0046461 | 0.007287829 | 2.137 |
| Substrate Adhesion-Dependent Cell Spreading | GO:0034446 | 0.008081591 | 2.093 |
| Negative Regulation of Plasma Lipoprotein Oxidation | GO:0034445 | 0.008917017 | 2.050 |
| Regulation of Plasma Lipoprotein Oxidation | GO:0034444 | 0.008917017 | 2.050 |
| Secretion by Cell | GO:0032940 | 0.009039373 | 2.044 |
| Triglyceride Metabolic Process | GO:0006641 | 0.00964557 | 2.016 |
| Positive Regulation of Cell Adhesion | GO:0045785 | 0.009890818 | 2.005 |
| Regulation of Cell Morphogenesis | GO:0022604 | 0.01018014 | 1.992 |
| Positive Regulation of Heterotypic Cell-Cell Adhesion | GO:0034116 | 0.010529465 | 1.978 |
| Regulation of Cell-Cell Adhesion | GO:0022407 | 0.011281044 | 1.948 |
| Negative Regulation of Blood Coagulation | GO:0030195 | 0.01262872 | 1.899 |
| Negative Regulation of Hemostasis | GO:1900047 | 0.013577968 | 1.867 |
| Export from Cell | GO:0140352 | 0.014773845 | 1.831 |
| Cholesterol Esterification | GO:0034435 | 0.015294749 | 1.815 |
| Steroid Esterification | GO:0034433 | 0.015294749 | 1.815 |
| Sterol Esterification | GO:0034434 | 0.015294749 | 1.815 |
| Positive Regulation of Cell-Substrate Adhesion | GO:0010811 | 0.01579924 | 1.801 |
| Negative Regulation of Coagulation | GO:0050819 | 0.019135052 | 1.718 |
| Lipid Catabolic Process | GO:0016042 | 0.020745205 | 1.683 |
| Platelet Aggregation | GO:0070527 | 0.023183729 | 1.635 |
| Plasma Lipoprotein Particle Oxidation | GO:0034441 | 0.026712669 | 1.573 |
| Acylglycerol Metabolic Process | GO:0006639 | 0.028359777 | 1.547 |
| Neutral Lipid Metabolic Process | GO:0006638 | 0.029346144 | 1.532 |
| Supramolecular Fiber Organization | GO:0097435 | 0.029625 | 1.528 |
| Cell Activation | GO:0001775 | 0.029657104 | 1.528 |
| Macromolecule Localization | GO:0033036 | 0.029741556 | 1.527 |
| Transport | GO:0006810 | 0.030983148 | 1.509 |
| Organic Hydroxy Compound Biosynthetic Process | GO:1901617 | 0.031015934 | 1.508 |
| Regulation of Blood Coagulation | GO:0030193 | 0.033129732 | 1.480 |
| Alcohol Biosynthetic Process | GO:0046165 | 0.035839906 | 1.446 |
| Regulation of Hemostasis | GO:1900046 | 0.037051615 | 1.431 |
| Plasminogen Activation | GO:0031639 | 0.037580747 | 1.425 |
| Regulation of Lipoprotein Lipase Activity | GO:0051004 | 0.037580747 | 1.425 |
| Regulation of Localization | GO:0032879 | 0.04005842 | 1.397 |
| Glycerolipid Catabolic Process | GO:0046503 | 0.041304441 | 1.384 |
| Vascular Process in Circulatory System | GO:0003018 | 0.041522396 | 1.382 |
| Regulation of Vesicle-Mediated Transport | GO:0060627 | 0.044575903 | 1.351 |
| Regulation of Cholesterol Transport | GO:0032374 | 0.045905316 | 1.338 |
| Regulation of Sterol Transport | GO:0032371 | 0.045905316 | 1.338 |
| Fibrinolysis | GO:0042730 | 0.048124122 | 1.318 |
| Regulation of Coagulation | GO:0050818 | 0.048341707 | 1.316 |
Table 5.
Down-regulated GO molecular functions in Scz organoids (p < 0.05).
| Molecular function | GO:MF Term_ID | Adjusted p-value | Neg Log10 adjusted p |
|---|---|---|---|
| Structural Constituent of Cytoskeleton | GO:0005200 | 0.000173652 | 3.760 |
| Cytoskeletal Protein Binding | GO:0008092 | 0.005488124 | 2.261 |
| GTPase Activity | GO:0003924 | 0.007451524 | 2.128 |
| Nucleoside-Triphosphatase Activity | GO:0017111 | 0.008195516 | 2.086 |
| Pyrophosphatase Activity | GO:0016462 | 0.020438022 | 1.690 |
| Hydrolase Activity, Acting on Acid Anhydrides, in Phosphorus-Containing Anhydrides | GO:0016818 | 0.023962071 | 1.620 |
| Hydrolase Activity, Acting on Acid Anhydrides | GO:0016817 | 0.024306171 | 1.614 |
| Tubulin Binding | GO:0015631 | 0.034081605 | 1.467 |
| GTP Binding | GO:0005525 | 0.034081605 | 1.467 |
| Microtubule Binding | GO:0008017 | 0.043893561 | 1.358 |
| Structural Molecule Activity | GO:0005198 | 0.047624863 | 1.322 |
| Guanyl Ribonucleotide Binding | GO:0032561 | 0.047641356 | 1.322 |
| Guanyl Nucleotide Binding | GO:0019001 | 0.047641356 | 1.322 |
Table 6.
Up-regulated GO molecular functions in Scz organoids (p < 0.05).
| Molecular function | GO:MF Term_ID | Adjusted p-value | Neg Log10 adjusted p |
|---|---|---|---|
| Sterol Transporter Activity | GO:0015248 | 4.01E−06 | 5.397 |
| Cadherin Binding Involved in Cell-Cell Adhesion | GO:0098641 | 2.28E−05 | 4.642 |
| Cell-Cell Adhesion Mediator Activity | GO:0098632 | 2.68E−05 | 4.571 |
| Cholesterol Transfer Activity | GO:0120020 | 5.42E−05 | 4.266 |
| Cell Adhesion Mediator Activity | GO:0098631 | 6.36E−05 | 4.197 |
| Sterol Transfer Activity | GO:0120015 | 6.55E−05 | 4.184 |
| Phosphatidylcholine-Sterol O-Acyltransferase Activator Activity | GO:0060228 | 7.46E−05 | 4.127 |
| Cell Adhesion Molecule Binding | GO:0050839 | 7.67E−05 | 4.115 |
| Lipoprotein Particle Receptor Binding | GO:0070325 | 0.000150248 | 3.823 |
| Lipid Transporter Activity | GO:0005319 | 0.000343266 | 3.464 |
| Lipid Transfer Activity | GO:0120013 | 0.001163791 | 2.934 |
| Sterol Binding | GO:0032934 | 0.003401382 | 2.468 |
| High-Density Lipoprotein Particle Receptor Binding | GO:0070653 | 0.005374955 | 2.270 |
| Steroid Binding | GO:0005496 | 0.025672825 | 1.591 |
| Signaling Receptor Binding | GO:0005102 | 0.031882878 | 1.496 |
Broadly speaking, these changes were also reflected in our analysis of GO pathways annotated for molecular functionality. Specifically, down-regulated GO molecular functions in Scz organoids comprised cytoskeletal structural, binding, and activity, as well as metabolic pathways relevant to neurodevelopment such GTP binding and GTPase activity (see Table 5; also identified in our prior prenatal drug modeling organoid work [11]). Similarly, up-regulated GO molecular function pathways in Scz organoids were typically related to sterol activity, cell adhesion, and lipoprotein binding/transfer/activity (see Table 6). In sum, these data provide additional veracity to the idea that there are metabolic functions underscoring the depletion of neuronal development factors in Scz organoids.
Lastly, we also considered whether Reactome pathways might unveil other novel biology in Scz organoids. Overall, an analysis of down-regulated (Table 7) and up-regulated (Table 8) Reactome pathways in Scz organoids revealed broadly similar pathway enrichment to those identified via GO analysis, with some notable exceptions. First, in our down-regulated Reactome pathway analysis, we noted that there were numerous significant pathways involved in NMDA receptor activation and assembly, ER to Golgi transport, as well as synaptic transmission (see Table 7 for a comprehensive list and statistical values). Contrary to this, and in addition to a convergent detection of lipoprotein-related metabolism pathways, unique Reactome pathways that were up-regulated in Scz organoids comprised post-translational protein phosphorylation, pathways related to MAPK signaling, and IGF-related pathways. Overall, these data suggest that ying-and-yang alterations in Scz organoids exist, whereby the disruption of neuronal-development factors and pathways yields enrichment for pathways presumably involved in either compensation or other disease-related neuropathology including phenotypes that have possibly not yet been articulated in human-derived tissue (e.g. specific metabolic changes).
Table 7.
Down-regulated reactome pathways in Scz organoids (p < 0.05).
| Reactome pathway | Reactome Term_ID | Adjusted p-value | Neg Log10 adjusted p |
|---|---|---|---|
| L1CAM Interactions | REAC:R-HSA-373760 | 4.04E−07 | 6.393 |
| Microtubule-Dependent Trafficking of Connexons from Golgi to the Plasma Membrane | REAC:R-HSA-190840 | 7.37E−07 | 6.133 |
| Transport of Connexons to the Plasma Membrane | REAC:R-HSA-190872 | 9.65E−07 | 6.015 |
| Recycling Pathway of L1 | REAC:R-HSA-437239 | 1.35E−06 | 5.869 |
| Post-Chaperonin Tubulin Folding Pathway | REAC:R-HSA-389977 | 2.00E−06 | 5.698 |
| COPI-Independent Golgi-to-ER Retrograde Traffic | REAC:R-HSA-6811436 | 2.51E−06 | 5.601 |
| Formation of Tubulin Folding Intermediates by CCT/TriC | REAC:R-HSA-389960 | 3.09E−06 | 5.510 |
| Activation of AMPK Downstream of NMDARs | REAC:R-HSA-9619483 | 4.59E−06 | 5.338 |
| Prefoldin Mediated Transfer of Substrate to CCT/TriC | REAC:R-HSA-389957 | 4.59E−06 | 5.338 |
| Sealing of the Nuclear Rnvelope (NE) by ESCRT-III | REAC:R-HSA-9668328 | 9.34E−06 | 5.030 |
| RHO GTPases Activate IQGAPs | REAC:R-HSA-5626467 | 9.34E−06 | 5.030 |
| Cooperation of Prefoldin and TriC/CCT in Actin and Tubulin Folding | REAC:R-HSA-389958 | 1.10E−05 | 4.959 |
| Gap Junction Assembly | REAC:R-HSA-190861 | 2.30E−05 | 4.638 |
| HCMV Early Events | REAC:R-HSA-9609690 | 3.03E−05 | 4.518 |
| Assembly and Cell Surface Presentation of NMDA Receptors | REAC:R-HSA-9609736 | 4.36E−05 | 4.360 |
| Aggrephagy | REAC:R-HSA-9646399 | 4.91E−05 | 4.309 |
| Carboxyterminal Post-Translational Modifications of Tubulin | REAC:R-HSA-8955332 | 6.18E−05 | 4.209 |
| Gap Junction Trafficking | REAC:R-HSA-190828 | 8.54E−05 | 4.069 |
| HCMV Infection | REAC:R-HSA-9609646 | 9.31E−05 | 4.031 |
| Gap Junction Trafficking and Regulation | REAC:R-HSA-157858 | 9.47E−05 | 4.024 |
| Intraflagellar Transport | REAC:R-HSA-5620924 | 0.000140206 | 3.853 |
| HSP90 Chaperone Cycle for Steroid Hormone Receptors (SHR) | REAC:R-HSA-3371497 | 0.000184495 | 3.734 |
| Kinesins | REAC:R-HSA-983189 | 0.000259999 | 3.585 |
| Nuclear Envelope (NE) Reassembly | REAC:R-HSA-2995410 | 0.000519297 | 3.285 |
| Translocation of SLC2A4 (GLUT4) to the Plasma Membrane | REAC:R-HSA-1445148 | 0.000597876 | 3.223 |
| Golgi-to-ER Retrograde Transport | REAC:R-HSA-8856688 | 0.000664661 | 3.177 |
| Axon Guidance | REAC:R-HSA-422475 | 0.00067359 | 3.172 |
| Post NMDA Receptor Activation Events | REAC:R-HSA-438064 | 0.000783019 | 3.106 |
| The Role of GTSE1 in G2/M Progression after G2 Checkpoint | REAC:R-HSA-8852276 | 0.000949362 | 3.023 |
| Nervous System Development | REAC:R-HSA-9675108 | 0.001007002 | 2.997 |
| Selective Autophagy | REAC:R-HSA-9663891 | 0.001010583 | 2.995 |
| Activation of NMDA Receptors and Postsynaptic Events | REAC:R-HSA-442755 | 0.00171304 | 2.766 |
| Recruitment of NuMA to Mitotic Centrosomes | REAC:R-HSA-380320 | 0.002242316 | 2.649 |
| Chaperonin-Mediated Protein Folding | REAC:R-HSA-390466 | 0.002362049 | 2.627 |
| Factors Involved in Megakaryocyte Development and Platelet Production | REAC:R-HSA-983231 | 0.002639065 | 2.579 |
| COPI-Dependent Golgi-to-ER Retrograde Traffic | REAC:R-HSA-6811434 | 0.003037982 | 2.517 |
| COPI-Mediated Anterograde Transport | REAC:R-HSA-6807878 | 0.003347233 | 2.475 |
| Protein Folding | REAC:R-HSA-391251 | 0.003347233 | 2.475 |
| CRMPs in Sema3A Signaling | REAC:R-HSA-399956 | 0.003988868 | 2.399 |
| Hedgehog ‘off’ State | REAC:R-HSA-5610787 | 0.005515212 | 2.258 |
| Neurotransmitter Receptors and Postsynaptic Signal Transmission | REAC:R-HSA-112314 | 0.005949918 | 2.225 |
| EML4 and NUDC in Mitotic Spindle Formation | REAC:R-HSA-9648025 | 0.006005753 | 2.221 |
| Cilium Assembly | REAC:R-HSA-5617833 | 0.006863941 | 2.163 |
| Intra-Golgi and Retrograde Golgi-to-ER traffic | REAC:R-HSA-6811442 | 0.007259898 | 2.139 |
| Resolution of Sister Chromatid Cohesion | REAC:R-HSA-2500257 | 0.008317829 | 2.080 |
| MHC Class II Antigen Presentation | REAC:R-HSA-2132295 | 0.008317829 | 2.080 |
| RHO GTPase Effectors | REAC:R-HSA-195258 | 0.0094632 | 2.024 |
| Developmental Biology | REAC:R-HSA-1266738 | 0.011666055 | 1.933 |
| Macroautophagy | REAC:R-HSA-1632852 | 0.012565891 | 1.901 |
| RHO GTPases Activate Formins | REAC:R-HSA-5663220 | 0.013491876 | 1.870 |
| Signaling by Hedgehog | REAC:R-HSA-5358351 | 0.02087328 | 1.680 |
| Autophagy | REAC:R-HSA-9612973 | 0.02087328 | 1.680 |
| ER to Golgi Anterograde Transport | REAC:R-HSA-199977 | 0.025184333 | 1.599 |
| Transmission across Chemical Synapses | REAC:R-HSA-112315 | 0.028395357 | 1.547 |
| M Phase | REAC:R-HSA-68886 | 0.044247896 | 1.354 |
Table 8.
Up-regulated reactome pathways in Scz organoids (p < 0.05).
| Reactome pathway | Reactome Term_ID | Adjusted p-value | Neg Log10 adjusted p |
|---|---|---|---|
| Post-Translational Protein Phosphorylation | REAC:R-HSA-8957275 | 8.31E−09 | 8.080 |
| Chylomicron Assembly | REAC:R-HSA-8963888 | 1.82E−08 | 7.739 |
| Chylomicron Remodeling | REAC:R-HSA-8963901 | 1.82E−08 | 7.739 |
| Regulation of Insulin-like Growth Factor (IGF) Transport and Uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | REAC:R-HSA-381426 | 3.18E−08 | 7.498 |
| Plasma Lipoprotein Assembly | REAC:R-HSA-8963898 | 6.10E−07 | 6.215 |
| Retinoid Metabolism and Transport | REAC:R-HSA-975634 | 1.25E−06 | 5.902 |
| Metabolism of Fat-Soluble Vitamins | REAC:R-HSA-6806667 | 2.16E−06 | 5.665 |
| Plasma Lipoprotein Remodeling | REAC:R-HSA-8963899 | 9.87E−06 | 5.006 |
| Platelet Degranulation | REAC:R-HSA-114608 | 3.04E−05 | 4.517 |
| Response to Elevated Platelet Cytosolic Ca2+ | REAC:R-HSA-76005 | 3.98E−05 | 4.400 |
| Regulation of TLR by Endogenous Ligand | REAC:R-HSA-5686938 | 0.000100533 | 3.998 |
| Visual Phototransduction | REAC:R-HSA-2187338 | 0.000156361 | 3.806 |
| Metabolism of Vitamins and Cofactors | REAC:R-HSA-196854 | 0.000455116 | 3.342 |
| Plasma Lipoprotein Assembly, Remodeling, and Clearance | REAC:R-HSA-174824 | 0.000616778 | 3.210 |
| HDL remodeling | REAC:R-HSA-8964058 | 0.000786847 | 3.104 |
| Hemostasis | REAC:R-HSA-109582 | 0.001130796 | 2.947 |
| GRB2:SOS Provides Linkage to MAPK Signaling for Integrins | REAC:R-HSA-354194 | 0.003373717 | 2.472 |
| Platelet Activation, Signaling and Aggregation | REAC:R-HSA-76002 | 0.003972412 | 2.401 |
| p130Cas Linkage to MAPK Signaling for Integrins | REAC:R-HSA-372708 | 0.004208218 | 2.376 |
| Scavenging by Class A Receptors | REAC:R-HSA-3000480 | 0.008886469 | 2.051 |
| Common Pathway of Fibrin Clot Formation | REAC:R-HSA-140875 | 0.014033493 | 1.853 |
| Integrin Signaling | REAC:R-HSA-354192 | 0.023493017 | 1.629 |
| Chylomicron Clearance | REAC:R-HSA-8964026 | 0.032311883 | 1.491 |
| Scavenging by Class B Receptors | REAC:R-HSA-3000471 | 0.032311883 | 1.491 |
| Integrin Cell Surface Interactions | REAC:R-HSA-216083 | 0.043417684 | 1.362 |
| Plasma Lipoprotein Clearance | REAC:R-HSA-8964043 | 0.044251662 | 1.354 |
Discussion
The aim of the current study was to further our knowledge of Scz by providing a deep, unbiased, analysis of molecular factors regulating central nervous system development in human-derived 3D tissue. To circumvent ethical and technical limitations in being able to access developing neural tissue from Scz patients [11], we generated 3D iPSC-derived cerebral organoids from n = 25 human donors (n = 8 Ctrl donors and n = 17 Scz donors). This approach allowed us to generate a theoretically limitless supply of self-regulating 3D neural tissue that recapitulated hallmark features of early brain assembly and corticogenesis [34, 35]. Samples were correspondingly subjected to cutting-edge isobaric barcoding chemistry that allowed up to 15 human donor samples (+ 1 pool for normalization) to be condensed into a single tube that could then be deconstructed via high-sensitivity, online, nano liquid-chromatography/mass-spectrometry proteomics. This allowed us to generate a posttranslational molecular map of factors in Scz patient-derived tissue/organoid samples. Consequently, we were able to identify that Scz organoids principally differed from healthy Ctrls due to differences in the total quantity of molecular factors (rather than their diversity), the altered expression of an ensemble of neuronal factors, and the differential regulation of specific GWAS-implicated [33] disease candidates (namely, PTN and PODXL).
Convergence upon depletion of neuronal factors in Scz organoids
The first phenotype to arise in our molecular mapping of Scz organoids was the extent to which canonical neuron identity and development factors were depleted in Scz patient-derived organoids. For several decades, numerous theories have emerged which link neuronal and synaptic function with Scz [36–38], particularly as it relates to cortical dysfunction [39–41] and the cognitive symptoms [42, 43] observed in clinical cases [44]. Recently, progress has been made in understandingly early-arising changes within the developing brain that may influence novel neurodevelopmental factors with putative links to Scz [45]. This has led to numerous investigations of early-arising biological phenomenon in various model systems. Human-derived models, usually leveraging the power of gene edited or patient-derived iPSCs, have consequently revealed alterations in neuronal differentiation [46], mitochondrial metabolic function [47, 48], catecholamine levels [49], neuron-glia interactions [50], synaptogenesis [51], and synaptic function [52]. Thus, patient-derived iPSCs have proven to be a powerful tool in tracing early neurodevelopmental features of Scz [53]. However, iPSCs can be further exploited if used to generate human-derived organoids, a model system of human brain development which recapitulates endogenous self-regulatory mechanisms associated with cortical patterning [11]. Building upon prior Scz organoid work [1, 27–29], here we report lower levels of an ensemble of neuron-related development factors comprising GAP43, CRABP1, NCAM1, and MYEF2 as well as identity factors comprising MAP2, TUBB3, and SV2A. Broadly speaking, these molecular findings are consistent with our prior work which reported disrupted neurogenesis and lower total neuron numbers within Scz cerebral organoids [1, 54, 55] – a phenotype which has also been independently reported by other groups [28]. Thus, fewer neurons will result in less MAP2, TUBB3, and SV2A expression, which is consistent with the molecular outcomes of this independent investigation. Our detection of lower NCAM1 protein levels in Scz organoids is also consistent with a prior report that found decreased NCAM1 expression in Scz neural progenitor cells [56]. Alterations in the growth-associated factor GAP43 have also been observed across multiple brain regions and independent studies that have evaluated postmortem Scz patient tissue [57–61]. When combined, these data support the idea [1, 28] that a loss of factors which support neuronal development yields an upstream depletion of neurons within Scz patient-derived organoids [1, 28].
Regulation of novel GWAS factors (PTN & PODXL) in Scz organoids
The other major phenotype identified in our molecular mapping of Scz cerebral organoids was the differential expression of two novel GWAS factors, namely PTN and PODXL. This analysis comprised us cross-referencing the highest-confident GWAS factors identified in unbiased clinical samples (see [33]) with our complete list of differentially expressed proteins. In our prior report utilizing a smaller TMT-LC/MS cohort design [1], we identified the differential expression of four GWAS candidates in Scz cerebral organoids at the protein level (PTN, COMT, PLCL1, and PODXL). Of these candidates, we were able to detect and replicate the differential expression of two of these factors in our much larger sample of n = 25 reported here. This specifically comprised alterations in PTN (down-regulated) and PODXL (up-regulated). These factors represent high-confidence GWAS factors associated with Scz, but otherwise have relatively unknown disease relevance. PTN has also been reported to be depleted in neural progenitors and shown to regulate both neurogenesis and survival phenotypes in Scz cerebral organoids [1], providing the first functional molecular data related to this candidate within the Scz literature. Other groups have also recently identified that PTN secreted from neural stem cells supports the maturation of new-born neurons [62], and can function as a neurotrophic growth factor in vivo to modulate neuronal loss [63] and long-term potentiation induction [64]. PTN has also since been implicated in a novel amphetamine-model of relevance to Scz [65], a recent computational protein-network analysis underlying Scz [66], as well as at least one nascent Scz gene-association study (n = 1,823 humans) [67]. On the other hand, little work has been completed on the role of PODXL in Scz, probably because PODXL is a renal-enriched factor most often associated with kidney podocytes and mesothelial cells [68]. Of note, PODXL has recently been shown to play a role in neurite outgrowth, branching, axonal fasciculation, and synapse number [69], supporting a potential role for this factor in synaptic plasticity. Additionally, PODXL was recently shown to be an apical determinant that may alter lumen size of neural progenitor cell rosettes during morphogenesis [70]. Thus, PODXL may be a fruitful target for future investigations seeking to deconvolute the role of novel Scz GWAS factors within the developing brain.
Other novel differentially expressed candidates in Scz organoids
Lastly, it is worth emphasizing several other differentially expressed molecular candidates observed in Scz cerebral organoids hold biological interest. First and foremost, we identified that Carboxypeptidase E (CPE) was downregulated in Scz cerebral organoids. CPE is a prohormone-processing enzyme [71] and regulated secretory pathway receptor [72], possibly best known for regulating the sorting and activity-dependent secretion of BDNF [73, 74] as well as TrkB surface insertion [75] in neurons. However, CPE was recently suggested to also function as a growth factor independently of its enzymatic and sorting activities [76]. Indeed, amongst other reports suggesting a role in neuroprotection [77], it has recently been shown that CPE regulates cortical neuron migration and dendritic morphology [78]. However, the degree to which these effects is dependent upon its cargo, which includes other growth factors (e.g. BDNF), remains unclear. Lastly, the other notable differentially expressed candidates worthy of discussion comprised alterations within the apolipoprotein family, specifically APOM, APOA1, APOE, APOC3, and APOB. Apolipoproteins have been previously investigated as potential metabolic-related biomarkers [79] in peripherally accessible biological fluids (e.g. CSF [80] or plasma [81]). This specifically includes alterations in APOE and APOA1 in Scz patients [82]. These findings are broadly related to cholesterol [83], fatty acid [84], phospholipid metabolism [85], as well as other membrane-related [86] hypotheses of Scz (which are all somewhat related and/or derived from similar evidence pools). Nonetheless, it is interesting that evidence related to these hypotheses was detectable and reproducible across our sample of patients, and may indicate that further work on potential metabolic factors may also be a further avenue of fruitful research.
Conclusion
In closing, we identified a broad reduction in molecules important for neuronal identity and development as well as specific alterations in novel GWAS and other disease-relevant molecules previously implicated in Scz. This work collectively supports the idea that Scz is a complex disease underscored by multifaceted changes that likely yield cell-specific as well as multiple mechanisms [54]. In closing, the authors hope that the current dataset may provide insight for other researchers and labs that have an interest in biological data from human-derived 3D stem cell systems but otherwise employ other model systems.
Methods
Induced pluripotent stem cells
Briefly, human stem cells were principally acquired from NIH deposits at the Rutgers University Cell and DNA Repository. The benefit of utilizing NIH deposited lines is that all biologics have been characterized for identity, pluripotency, exogenous reprogramming factor expression, genetic stability, and viability. In sum, we sampled a total of 25 different iPSC lines comprising both healthy Ctrls and idiopathic Scz patients. Cerebral organoids were generated from all donors in this study, and each iPSC line was biologically independent (representing a unique human donor). Ctrl iPSC lines utilized for cellular experiments included MH0159019, MH0159020, MH0159021, MH0159022, MH0167170, MH0174677, and MH0174686. One Ctrl line (GM23279) was sourced from the Coriell Institute for Medical Research. Scz iPSC lines included MH0159025, MH0159026, MH0185223, MH0185225, MH0200865, MH0217268, MH0185900, MH0185954, MH0185958, MH0185963, MH0185970, MH0185912, MH0185945, MH0185964, MH0185966, MH0185925, and MH0185928. Clinical information for Scz patients is available in Table S1 of our prior publication [1]. All Scz samples were derived from idiopathic cases, which we define here as schizophrenia cases that maintained unknown disease origins and do not meet a genetic/syndrome-based diagnosis (as listed in NIH/NIMH notes). Ctrl iPSC lines were screened for both personal, and family history, of major mental illnesses. All iPSC lines were maintained on Vitronectin-coated plates and fed with Essential 8 (E8) + E8 supplement media (ThermoFisher, CAT#: A1517001).
3D cerebral organoid tissue generation
We adapted the same undirected-differentiation organoid system that we used in our previous, more extensive, analysis of Scz neurodevelopmental mechanisms [1], which had been previously published by Lancaster et al. in Nature [17] and Nature Protocols [87]. Briefly, 2D iPSC colonies were dissociated and cultured into 3D embryoid bodies in ultra-low attachment plates (Corning; CAT#: 3474). Rock inhibitor (1:1000; Stem Cell Tech, CAT#: 72304) and basic fibroblast growth factor (Pepro Tech, CAT#: 100-18B) are included in media for the first 2-4 days of embryoid body culturing to promote stem cell aggregation and survival. Following this, healthy embryoid bodies are isolated and transferred to Nunclon Sphera 24 well plates (Thermo Scientific, CAT#: 174930) for neural fate specification, using neural induction media. Successful early ‘organoids’ were embedded in a 30 µl Matrigel (Corning, CAT#: 354234) spheroid-droplet and polymerized at 37 °C for 20-30 min which provided a matrix for subsequent neural expansion. Organoids suspended in matrigel droplets were next cultured in terminal organoid media for 4-6 days without agitation, and then cultured with agitation at 60-70RPM until harvested for experiments. For further organoid protocol detail, including QC steps, please refer to our previous publication [1]. Likewise, for further insight into organoid handling for proteomic analysis, please refer to our other organoid manuscript [11].
Proteomics sample preparation, TMT labeling and LC/MS
Isobaric stable isotope labeling was achieved viaTandem Mass Tag pro (TMTpro) chemistry and Liquid-Chromatography/Mass-Spectrometry (LC/MS) proteomics as previously described [1, 11, 65]. Briefly, intact organoids were reduced with dithiotreitol and underwent alkylation with iodoacetamide before tryptic digestion at 37 °C overnight. For barcoding chemistry, we employed TMTpro 16-plex labeling according to the manufacturer’s instructions (Thermo Fisher Scientific, CAT# A44521). Each multi-plex experiment contained relevant organoid samples with an additional pooled isobaric reference label made up of the same peptide digest from the pooled mix of organoids (for data normalization between runs; TMT Tag 134 N for both TMT-LC/MS runs). A list of sample labeling strategies and replicates is available in the PRIDE proteomics exchange repository. TMT-labeled peptides were desalted using C18’ stage-tips prior to LC-MS analysis. An EASY-nLC 1200, which was coupled to a Fusion Lumos mass spectrometer, (Thermo Fisher Scientific) was utilized in positive, data-dependent acquisition mode, with samples analyzed in technical duplicate. Buffer A (0.1% FA in water) and buffer B (0.1% FA in 80% ACN) were used as mobile phases for gradient separation. TMT-labeled peptides were analyzed on a 75 μm I.D. column (ReproSil-Pur C18-AQ, 3μm, Dr. Maisch GmbH, German) was packed in-house. A separation gradient of 5–10% buffer B over 1 min, 10%-35% buffer B over 229 min, and 35%-100% B over 5 min at a flow rate of 300 nL/min was adapted. An Orbitrap mass analyzer acquired Full MS scans over a range of 350-1500 m/z with resolution 120,000 at m/z 200. The top 20 most-abundant precursors were selected with an isolation window of 0.7 Thomsons and fragmented by high-energy collisional dissociation with normalized collision energy of 40. The Orbitrap mass analyzer was also used to acquire MS/MS scans. The automatic gain control target value was 1e6 for full scans and 5e4 for MS/MS scans respectively, and the maximum ion injection time was 54 ms for both.
Data processing and bioinformatics pipeline for quantitative analysis
Mass spectra were pre-processed as described [1, 11, 65] and processed using MaxQuant [88] (1.5.5.1). Spectra were searched against the full set of human protein sequences annotated in UniProt (sequence database Sep-2017) using Andromeda. Data was searched as described [1, 11] as a separate and single (combined) batches, with fixed modification, cysteine carbamidomethylation and variable modifications, N-acetylation and methionine oxidation. Searches were performed using a 20 ppm precursor ion tolerance for total protein level analysis. Further modifications included TMT tags on peptide N termini/lysine residues (+229.16293 Da) set as static modifications. Data was processed using trypsin/P as the proteolytic enzyme with up to 2 missed cleavage sites allowed. Peptides less than seven amino acids were not considered for further analysis because of lack of uniqueness, and a 1% False-Discovery Rate (FDR) was used to filter at peptide and protein levels. Protein identification required at least two unique or razor peptides per protein group. Contaminants, and reverse identification were excluded from further data analysis. Quantification was performed with the reporter ion quantification normalization in MaxQuant. Protein intensities were log2 transformed using Perseus [89] (1.x.10). The violin plots of log2 transformed protein intensity distribution and the boxplot of coefficient of variations per sample group were visualized using R package ggplot2. Proteins quantified in at least 70% of samples in at least one sample group were subjected to downstream visualization (principal component analysis, volcano plot) and statistical analysis using Perseus. For principal component analysis, missing values were imputed from normal distribution (downshift 1.8, width 0.3) using Perseus. For differential expression analysis proteins were subjected to Welch’s t-test; p-value < 0.05 and |log2FC | >0.5 visualized in volcano plot and subjected to downstream functional enrichment analysis using g:Profiler, including Gene Ontology, KEGG and Reactome databases (as described, [90, 91]).
Acknowledgements
MN was the recipient of a NHMRC CJ Martin Fellowship that supported mRNA degradation and stem cell training completed at Weill Cornell Medical College of Cornell University.
Author contributions
MN and DC conceived the project and designed experiments. MN generated all 3D tissue from human stem cells, supervised biological interrogation of datasets, and wrote the manuscript. Our technician, AL, provided important logistical support by assisting with the generation and processing of 3D human-derived tissue. Lastly, HF and DG completed all computational analysis, with DG serving as the senior author overseeing bioinformatics analyses.
Data availability
The MS proteomics raw data and MaxQuant search parameters have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org/) via the PRIDE partner repository [92] with the data set identifier PXD027812.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
David Greening, Email: david.greening@baker.edu.
Dilek Colak, Email: dic2009@med.cornell.edu.
References
- 1.Notaras, M, et al., Schizophrenia is defined by cell-specific neuropathology and multiple neurodevelopmental mechanisms in patient-derived cerebral organoids. Mol Psychiatry, 2021. [DOI] [PMC free article] [PubMed]
- 2.Klosterkötter J, Hellmich M, Steinmeyer EM, Schultze-Lutter F. Diagnosing schizophrenia in the initial prodromal phase. Arch Gen Psychiatry. 2001;58:158–164. doi: 10.1001/archpsyc.58.2.158. [DOI] [PubMed] [Google Scholar]
- 3.Cornblatt BA, Lencz T, Smith CW, Correll CU, Auther AM, Nakayama E. The schizophrenia prodrome revisited: a neurodevelopmental perspective. Schizophr Bull. 2003;29:633–651. doi: 10.1093/oxfordjournals.schbul.a007036. [DOI] [PubMed] [Google Scholar]
- 4.Brown AS, Susser ES. In utero infection and adult schizophrenia. Ment Retard Dev Disabil Res Rev. 2002;8:51–57. doi: 10.1002/mrdd.10004. [DOI] [PubMed] [Google Scholar]
- 5.Kunugi H, et al. Schizophrenia following in utero exposure to the 1957 influenza epidemics in Japan. Am J Psychiatry. 1995;3:450–452. doi: 10.1176/ajp.152.3.450. [DOI] [PubMed] [Google Scholar]
- 6.Takei N, Mortensen PB, Klaening U, Murray RM, Sham PC, O'Callaghan E, et al. Relationship between in utero exposure to influenza epidemics and risk of schizophrenia in Denmark. Biol Psychiatry. 1996;40:817–824. doi: 10.1016/0006-3223(95)00592-7. [DOI] [PubMed] [Google Scholar]
- 7.Procopio M, Davies RJ, Marriott P. The hormonal environment in utero as a potential aetiological agent for schizophrenia. Eur Arch Psychiatr Clin Neurosci. 2006;256:77–81. doi: 10.1007/s00406-005-0604-8. [DOI] [PubMed] [Google Scholar]
- 8.McClellan JM, Susser E, King M-C. Maternal famine, de novo mutations, and schizophrenia. JAMA. 2006;296:582–584. doi: 10.1001/jama.296.5.582. [DOI] [PubMed] [Google Scholar]
- 9.Boks MP, Houtepen LC, Xu Z, He Y, Ursini G, Maihofer AX, et al. Genetic vulnerability to DUSP22 promoter hypermethylation is involved in the relation between in utero famine exposure and schizophrenia. NPJ Schizophr. 2018;4:1–8. doi: 10.1038/s41537-018-0058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyman SE. The daunting polygenicity of mental illness: making a new map. Philos Trans R Soc B: Biol Sci. 2018;373:20170031. doi: 10.1098/rstb.2017.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notaras, M, et al., Neurodevelopmental signatures of narcotic and neuropsychiatric risk factors in 3D human-derived forebrain organoids. Mol Psychiatry, 2021, 1–24. [DOI] [PMC free article] [PubMed]
- 12.Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR. Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep. 2016;17:3369–3384. doi: 10.1016/j.celrep.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jourdon A, Scuderi S, Capauto D, Abyzov A, Vaccarino FM. PsychENCODE and beyond: transcriptomics and epigenomics of brain development and organoids. Neuropsychopharmacol. 2021;46:70–85. doi: 10.1038/s41386-020-0763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci USA. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klaus J, Kanton S, Kyrousi C, Ayo-Martin AC, Di Giaimo R, Riesenberg S, et al. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med. 2019;25:561–568. doi: 10.1038/s41591-019-0371-0. [DOI] [PubMed] [Google Scholar]
- 16.Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell. 2019;176:743–756.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Ma L, Yang M, Shao Q, Xu J, Lu Z, et al. Cerebral organoid and mouse models reveal a RAB39b–PI3K–mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev. 2020;34:580–597. doi: 10.1101/gad.332494.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19:258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 21.Ilieva M, Fex Svenningsen Å, Thorsen M, Michel TM. Psychiatry in a dish: stem cells and brain organoids modeling autism spectrum disorders. Biol Psychiatry. 2018;83:558–568. doi: 10.1016/j.biopsych.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsen, B, et al., Human brain organoids reveal accelerated development of cortical neuron classes as a shared feature of autism risk genes. Preprint at https://www.biorxiv.org/content/10.1101/2020.11.10.376509v1.
- 24.Wegscheid ML, Anastasaki C, Hartigan KA, Cobb OM, Papke JB, Traber JN, et al. Patient-derived iPSC-cerebral organoid modeling of the 17q11. 2 microdeletion syndrome establishes CRLF3 as a critical regulator of neurogenesis. Cell Rep. 2021;36:109315. doi: 10.1016/j.celrep.2021.109315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan TA, Revah O, Gordon A, Yoon SJ, Krawisz AK, Goold C, et al. Neuronal defects in a human cellular model of 22q11. 2 deletion syndrome. Nat Med. 2020;26:1888–1898. doi: 10.1038/s41591-020-1043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daviaud N, Chevalier C, Friedel RH, Zou H. Distinct vulnerability and resilience of human neuroprogenitor subtypes in cerebral organoid model of prenatal hypoxic injury. Front Cell Neurosci. 2019;13:336. doi: 10.3389/fncel.2019.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kathuria A, Lopez-Lengowski K, Jagtap SS, McPhie D, Perlis RH, Cohen BM, et al. Transcriptomic landscape and functional characterization of induced pluripotent stem cell–derived cerebral organoids in schizophrenia. JAMA Psychiatry. 2020;77:745–754. doi: 10.1001/jamapsychiatry.2020.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stachowiak EK, Benson CA, Narla ST, Dimitri A, Chuye LEB, Dhiman S, et al. Cerebral organoids reveal early cortical maldevelopment in schizophrenia—computational anatomy and genomics, role of FGFR1. Transl Psychiatry. 2017;7:1–24.. doi: 10.1038/s41398-017-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson CA, Powell HR, Liput M, Dinham S, Freedman DA, Ignatowski TA, et al. Immune factor, TNFα, disrupts human brain organoid development similar to schizophrenia—schizophrenia increases developmental vulnerability to TNFα. Front Cell Neurosci. 2020;14:1–18.. doi: 10.3389/fncel.2020.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawada T, Chater TE, Sasagawa Y, Yoshimura M, Fujimori-Tonou N, Tanaka K, et al. Developmental excitation-inhibition imbalance underlying psychoses revealed by single-cell analyses of discordant twins-derived cerebral organoids. Mol Psychiatry. 2020;25:2695–2711. doi: 10.1038/s41380-020-0844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srikanth P, Lagomarsino VN, Muratore CR, Ryu SC, He A, Taylor WM, et al. Shared effects of DISC1 disruption and elevated WNT signaling in human cerebral organoids. Transl Psychiatry. 2018;8:1–14.. doi: 10.1038/s41398-017-0025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, et al. DISC1 regulates neurogenesis via modulating kinetochore attachment of Ndel1/Nde1 during mitosis. Neuron. 2017;96:1041–1054.e5. doi: 10.1016/j.neuron.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ripke S, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason JO, Price DJ. Building brains in a dish: prospects for growing cerebral organoids from stem cells. Neuroscience. 2016;334:105–118. doi: 10.1016/j.neuroscience.2016.07.048. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, Wu Q, Wang X. Modeling brain development and diseases with human cerebral organoids. Curr Op Neurobiol. 2021;66:103–115. doi: 10.1016/j.conb.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Mehta UM, Thirthalli J, Aneelraj D, Jadhav P, Gangadhar BN, Keshavan MS. Mirror neuron dysfunction in schizophrenia and its functional implications: a systematic review. Schizophr Res. 2014;160:9–19. doi: 10.1016/j.schres.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freedman R, Waldo M, Bickford-Wimer P, Nagamoto H. Elementary neuronal dysfunctions in schizophrenia. Schizophr Res. 1991;4:233–243. doi: 10.1016/0920-9964(91)90035-P. [DOI] [PubMed] [Google Scholar]
- 38.Olney JW, Farber NB. Glutamate receptor dysfunction and schizophrenia. Arch Gen Psychiatry. 1995;52:998–1007. doi: 10.1001/archpsyc.1995.03950240016004. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez-Burgos G, Lewis DA. GABA neurons and the mechanisms of network oscillations: implications for understanding cortical dysfunction in schizophrenia. Schizophr Bull. 2008;34:944–961. doi: 10.1093/schbul/sbn070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis DA. GABAergic local circuit neurons and prefrontal cortical dysfunction in schizophrenia. Brain Res Revs. 2000;31:270–276. doi: 10.1016/S0165-0173(99)00042-9. [DOI] [PubMed] [Google Scholar]
- 41.Curley AA, Lewis DA. Cortical basket cell dysfunction in schizophrenia. J Physiol. 2012;590:715–724. doi: 10.1113/jphysiol.2011.224659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis DA. Inhibitory neurons in human cortical circuits: substrate for cognitive dysfunction in schizophrenia. Curr Op Neurobiol. 2014;26:22–26. doi: 10.1016/j.conb.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee A, Carvalho F, Eliez S, Caroni P. Long-lasting rescue of network and cognitive dysfunction in a genetic schizophrenia model. Cell. 2019;178:1387–1402.e14. doi: 10.1016/j.cell.2019.07.023. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Burgos G, Fish KN, Lewis DA. GABA neuron alterations, cortical circuit dysfunction and cognitive deficits in schizophrenia. Neural Plasticity. 2011;2011:2011–24. doi: 10.1155/2011/723184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura JP, Schroeder A, Hudson M, Jones N, Gillespie B, Du X, et al. The maternal immune activation model uncovers a role for the Arx gene in GABAergic dysfunction in schizophrenia. Brain Behav Immun. 2019;81:161–171. doi: 10.1016/j.bbi.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Robicsek O, Karry R, Petit I, Salman-Kesner N, Müller FJ, Klein E, et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry. 2013;18:1067–1076. doi: 10.1038/mp.2013.67. [DOI] [PubMed] [Google Scholar]
- 47.Ni P, Noh H, Park GH, Shao Z, Guan Y, Park JM, et al. iPSC-derived homogeneous populations of developing schizophrenia cortical interneurons have compromised mitochondrial function. Mol Psychiatry. 2020;25:2873–2888. doi: 10.1038/s41380-019-0423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Ryan SK, Deboer E, Cook K, Fitzgerald S, Lachman HM, et al. Mitochondrial deficits in human iPSC-derived neurons from patients with 22q11. 2 deletion syndrome and schizophrenia. Transl Psychiatry. 2019;9:1–10.. doi: 10.1038/s41398-018-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hook V, Brennand KJ, Kim Y, Toneff T, Funkelstein L, Lee KC, et al. Human iPSC neurons display activity-dependent neurotransmitter secretion: aberrant catecholamine levels in schizophrenia neurons. Cell Rep. 2014;3:531–538. doi: 10.1016/j.stemcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Windrem MS, Osipovitch M, Liu Z, Bates J, Chandler-Militello D, Zou L, et al. Human iPSC glial mouse chimeras reveal glial contributions to schizophrenia. Cell Stem Cell. 2017;21:195–208.e6. doi: 10.1016/j.stem.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Habela CW, Song H. and G.-l. Ming, Modeling synaptogenesis in schizophrenia and autism using human iPSC derived neurons. Mol Cell Neurosci. 2016;73:52–62. doi: 10.1016/j.mcn.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kathuria A, Lopez-Lengowski K, Watmuff B, McPhie D, Cohen BM, Karmacharya R. Synaptic deficits in iPSC-derived cortical interneurons in schizophrenia are mediated by NLGN2 and rescued by N-acetylcysteine. Transl Psychiatry. 2019;9:1–13.. doi: 10.1038/s41398-019-0660-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahmad R, Sportelli V, Ziller M, Spengler D, Hoffmann A. Tracing early neurodevelopment in schizophrenia with induced pluripotent stem cells. Cells. 2018;7:2–25.. doi: 10.3390/cells7090140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Notaras M, Lodhi A, Dundar F, Collier P, Sayles N, Tilgner H, et al. Multiple neurodevelopmental mechanisms of schizophrenia in patient-derived cerebral organoids. Biol Psychiatry. 2021;89:S100. doi: 10.1016/j.biopsych.2021.02.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Notaras, M, et al. BRN2 and PTN unveil multiple neurodevelopmental mechanisms in Schizophrenia patient-derived cerebral organoids. Preprint at https://www.biorxiv.org/content/10.1101/2021.06.10.447949v1. [DOI] [PMC free article] [PubMed]
- 56.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eastwood SL, Harrison PJ. Synaptic pathology in the anterior cingulate cortex in schizophrenia and mood disorders. A review and a Western blot study of synaptophysin, GAP-43 and the complexins. Brain Res Bull. 2001;55:569–578. doi: 10.1016/S0361-9230(01)00530-5. [DOI] [PubMed] [Google Scholar]
- 58.Perrone-Bizzozero NI, Sower AC, Bird ED, Benowitz LI, Ivins KJ, Neve RL. Levels of the growth-associated protein GAP-43 are selectively increased in association cortices in schizophrenia. Proc Natl Acad Sci USA. 1996;93:14182–14187. doi: 10.1073/pnas.93.24.14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sower AC, Bird ED, Perrone-Bizzozero NI. Increased levels of GAP-43 protein in schizophrenic brain tissues demonstrated by a novel immunodetection method. Mol Cell Neuropathol. 1995;24:1–11. doi: 10.1007/BF03160108. [DOI] [PubMed] [Google Scholar]
- 60.Chambers JS, Thomas D, Saland L, Neve RL, Perrone-Bizzozero NI. Growth-associated protein 43 (GAP-43) and synaptophysin alterations in the dentate gyrus of patients with schizophrenia. Prog Neuropsycho Pharm. 2005;29:283–290. doi: 10.1016/j.pnpbp.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 61.Weickert CS, Webster MJ, Hyde TM, Herman MM, Bachus SE, Bali G, et al. Reduced GAP-43 mRNA in dorsolateral prefrontal cortex of patients with schizophrenia. Cereb Cortex. 2001;11:136–147. doi: 10.1093/cercor/11.2.136. [DOI] [PubMed] [Google Scholar]
- 62.Tang C, Wang M, Wang P, Wang L, Wu Q, Guo W. Neural stem cells behave as a functional niche for the maturation of newborn neurons through the secretion of PTN. Neuron. 2019;101:32–44.e6. doi: 10.1016/j.neuron.2018.10.051. [DOI] [PubMed] [Google Scholar]
- 63.Nikolakopoulou AM, Montagne A, Kisler K, Dai Z, Wang Y, Huuskonen MT, et al. Pericyte loss leads to circulatory failure and pleiotrophin depletion causing neuron loss. Nat Neurosci. 2019;22:1089–1098. doi: 10.1038/s41593-019-0434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pavlov I, Võikar V, Kaksonen M, Lauri SE, Hienola A, Taira T, et al. Role of heparin-binding growth-associated molecule (HB-GAM) in hippocampal LTP and spatial learning revealed by studies on overexpressing and knockout mice. Mol Cell Neurosci. 2002;20:330–342. doi: 10.1006/mcne.2002.1104. [DOI] [PubMed] [Google Scholar]
- 65.Greening, DW, Notaras M, Chen M, Xu R, Smith JD, Cheng L, et al., Chronic methamphetamine interacts with BDNF Val66Met to remodel psychosis pathways in the mesocorticolimbic proteome. Mol Psychiatry. 2019: 1–17. [DOI] [PubMed]
- 66.Chang X, Lima LA, Liu Y, Li J, Li Q, Sleiman P, et al. Common and rare genetic risk factors converge in protein interaction networks underlying schizophrenia. Front Genet. 2018;9:434. doi: 10.3389/fgene.2018.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lv Y, Sun Y, Dai D, Luan ZL, Lu HY, Li CJ, et al. Positive association between PTN polymorphisms and schizophrenia in Northeast Chinese Han population. Psychiatr Genet. 2020;30:141–149. doi: 10.1097/YPG.0000000000000262. [DOI] [PubMed] [Google Scholar]
- 68.Kang HG, Lee M, Lee KB, Hughes M, Kwon BS, Lee S, et al. Loss of podocalyxin causes a novel syndromic type of congenital nephrotic syndrome. Exp Mol Med. 2017;49:e414. doi: 10.1038/emm.2017.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vitureira N, Andrés R, Pérez-Martínez E, Martínez A, Bribián A, Blasi J, et al. Podocalyxin is a novel polysialylated neural adhesion protein with multiple roles in neural development and synapse formation. PloS ONE. 2010;5:e12003. doi: 10.1371/journal.pone.0012003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Townshend RF, Shao Y, Wang S, Cortez CL, Esfahani SN, Spence JR, et al. Effect of cell spreading on rosette formation by human pluripotent stem cell-derived neural progenitor cells. Front Cell Dev Biol. 2020;8:1–15.. doi: 10.3389/fcell.2020.588941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hook VY, Eiden LE, Brownstein MJ. A carboxypeptidase processing enzyme for enkephalin precursors. Nature. 1982;295:341–342. doi: 10.1038/295341a0. [DOI] [PubMed] [Google Scholar]
- 72.Cool DR, Normant E, Shen F, Chen HC, Pannell L, Zhang Y, et al. Carboxypeptidase E is a regulated secretory pathway sorting receptor: genetic obliteration leads to endocrine disorders in Cpefat mice. Cell. 1997;88:73–83. doi: 10.1016/S0092-8674(00)81860-7. [DOI] [PubMed] [Google Scholar]
- 73.Lou H, Kim SK, Zaitsev E, Snell CR, Lu B, Loh YP. Sorting and activity-dependent secretion of BDNF require interaction of a specific motif with the sorting receptor carboxypeptidase e. Neuron. 2005;45:245–255. doi: 10.1016/j.neuron.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 74.Notaras M, van den Buuse M. Brain-derived neurotrophic factor (BDNF): novel insights into regulation and genetic variation. Neuroscientist. 2019;25:434–454. doi: 10.1177/1073858418810142. [DOI] [PubMed] [Google Scholar]
- 75.Li N, Teng SW, Zhao L, Li JR, Xu JL, Li N, et al. Carboxypeptidase E regulates activity-dependent TrkB neuronal surface insertion and hippocampal memory. J Neurosci. 2021;41:6987–7002. doi: 10.1523/JNEUROSCI.0236-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng Y, Cawley NX, Loh YP. Carboxypeptidase E (NF-α1): a new trophic factor in neuroprotection. Neurosci Bull. 2014;30:692–696. doi: 10.1007/s12264-013-1430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cong L, Cheng Y, Cawley NX, Murthy SR, Loh YP. A novel single nucleotide T980C polymorphism in the human carboxypeptidase E gene results in loss of neuroprotective function. PLoS One. 2017;12:e0170169. doi: 10.1371/journal.pone.0170169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang C, Carrel D, Omelchenko A, Kim H, Patel A, Fanget I, et al. Cortical neuron migration and dendrite morphology are regulated by carboxypeptidase E. Cereb Cortex. 2019;29:2890–2903. doi: 10.1093/cercor/bhy155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xuan J, Pan G, Qiu Y, Yang L, Su M, Liu Y, et al. Metabolomic profiling to identify potential serum biomarkers for schizophrenia and risperidone action. J Proteome Res. 2011;10:5433–5443. doi: 10.1021/pr2006796. [DOI] [PubMed] [Google Scholar]
- 80.Huang JT, Wang L, Prabakaran S, Wengenroth M, Lockstone HE, Koethe D, et al. Independent protein-profiling studies show a decrease in apolipoprotein A1 levels in schizophrenia CSF, brain and peripheral tissues. Mol Psychiatry. 2008;13:1118–1128. doi: 10.1038/sj.mp.4002108. [DOI] [PubMed] [Google Scholar]
- 81.Dean B, Digney A, Sundram S, Thomas E, Scarr E. Plasma apolipoprotein E is decreased in schizophrenia spectrum and bipolar disorder. Psychiatr Res. 2008;158:75–78. doi: 10.1016/j.psychres.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 82.Martins-De-Souza D, Wobrock T, Zerr I, Schmitt A, Gawinecka J, Schneider-Axmann T, et al. Different apolipoprotein E, apolipoprotein A1 and prostaglandin-H2 D-isomerase levels in cerebrospinal fluid of schizophrenia patients and healthy controls. World J Biol Psychiatry. 2010;11:719–728. doi: 10.3109/15622971003758748. [DOI] [PubMed] [Google Scholar]
- 83.Woods AG, Sokolowska I, Taurines R, Gerlach M, Dudley E, Thome J, et al. Potential biomarkers in psychiatry: focus on the cholesterol system. J Cell Mol Med. 2012;16:1184–1195. doi: 10.1111/j.1582-4934.2012.01543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fenton WS, Hibbeln J, Knable M. Essential fatty acids, lipid membrane abnormalities, and the diagnosis and treatment of schizophrenia. Biol Psychiatry. 2000;47:8–21. doi: 10.1016/S0006-3223(99)00092-X. [DOI] [PubMed] [Google Scholar]
- 85.Opler MG, Opler LA. Abnormal phospholipid metabolism in schizophrenia: evidence from epidemiological findings, clinical observations, and preliminary clinical trials. Front Biosci. 2001;6:e61–65. doi: 10.2741/opler. [DOI] [PubMed] [Google Scholar]
- 86.Horrobin DF, Glen AIM, Vaddadi K. The membrane hypothesis of schizophrenia. Schizophr Res. 1994;13:195–207. doi: 10.1016/0920-9964(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 87.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 89.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 90.Carli A, Afshar-Sterle S, Rai A, Fang H, O'Keefe R, Tse J, et al. Cancer stem cell marker DCLK1 reprograms small extracellular vesicles toward migratory phenotype in gastric cancer cells. Proteomics. 2021;21:2000098. doi: 10.1002/pmic.202000098. [DOI] [PubMed] [Google Scholar]
- 91.Kompa AR, Greening DW, Kong AM, McMillan PJ, Fang H, Saxena R, et al. Sustained subcutaneous delivery of secretome of human cardiac stem cells promotes cardiac repair following myocardial infarction. Cardiovasc Res. 2021;117:918–929. doi: 10.1093/cvr/cvaa088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nuc Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin Y-L, Persaud SD, Nhieu J, Wei LN. Cellular retinoic acid–binding protein 1 modulates stem cell proliferation to affect learning and memory in male mice. Endocrinol. 2017;158:3004–3014. doi: 10.1210/en.2017-00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frese CK, Mikhaylova M, Stucchi R, Gautier V, Liu Q, Mohammed S, et al. Quantitative map of proteome dynamics during neuronal differentiation. Cell Rep. 2017;18:1527–1542. doi: 10.1016/j.celrep.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Upadhyay A, Hosseinibarkooie S, Schneider S, Kaczmarek A, Torres-Benito L, Mendoza-Ferreira N, et al. Neurocalcin delta knockout impairs adult neurogenesis whereas half reduction is not pathological. Front Mol Neurosci. 2019;12:1–15.. doi: 10.3389/fnmol.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MS proteomics raw data and MaxQuant search parameters have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org/) via the PRIDE partner repository [92] with the data set identifier PXD027812.