Abstract
PRKACB (Protein Kinase CAMP-Activated Catalytic Subunit Beta) is predominantly expressed in the brain, and regulation of this gene links to neuroprotective effects against tau and Aβ-induced toxicity. Here we studied a (GCC)-repeat spanning the core promoter and 5′ UTR of this gene in 300 human subjects, consisting of late-onset neurocognitive disorder (NCD) (N = 150) and controls (N = 150). We also implemented several models to study the impact of this repeat on the three-dimensional (3D) structure of DNA. While the PRKACB (GCC)-repeat was strictly monomorphic at 7-repeats, we detected two 7/8 genotypes only in the NCD group. In all examined models, the (GCC)7 and its periodicals had the least range of divergence variation on the 3D structure of DNA in comparison to the 8-repeat periodicals and several hypothetical repeat lengths. A similar inert effect on the 3D structure was not detected in other classes of short tandem repeats (STRs) such as GA and CA repeats. In conclusion, we report monomorphism of a long (GCC)-repeat in the PRKACB gene in human, its inert effect on DNA structure, and enriched divergence in late-onset NCD. This is the first indication of natural selection for a monomorphic (GCC)-repeat, which probably evolved to function as an “epigenetic knob”, without changing the regional DNA structure.
Subject terms: DNA, Structural biology, Evolution, Genetics, Molecular biology, Neuroscience
Introduction
Short tandem repeats (STRs) are the most polymorphic genetic elements in the vertebrate genomes. Because of their polymorphic nature and plasticity, these elements provide an efficient source of variation at the inter- and intraspecies levels1–6. Accumulating evidence indicates that certain STRs may be associated with selective advantage in human and other species7–10. Among the long STRs spanning the core promoter and 5′ untranslated region (UTR) of human protein-coding genes, the protein kinase cAMP-activated catalytic subunit beta (PRKACB) gene contains a (GCC)-repeat of 7-repeats11. Across human tissues, PRKACB has the highest level of expression in the brain12 (https://www.proteinatlas.org/ENSG00000142875-PRKACB/tissue). Furthermore, in comparison with fourteen non-human primates, the brain expression of this gene reaches highest quantity in human (https://www.ncbi.nlm.nih.gov/ieb/research/acembly/av.cgi?db=human&term=PRKACB&submit=Go). The kinase encoded by PRKACB is involved in tau phosphorylation at Alzheimer’s disease (AD)-related sites, and regulation of this gene has been linked to neuroprotective effects against tau and Aβ-induced toxicity13,14.
The length of the PRKACB (GCC)-repeat in human is among the rare long lengths for this type of STR7. We hypothesized that this length may be of selective advantage to human. In view of the pathways in which PRKACB is involved in, this (GCC)-repeat may have a role in the higher order brain functions, such as cognition. Here, we studied the (GCC)-repeat in a sample of human subjects, consisting of late-onset neurocognitive disorder (NCD) and controls, and implemented several models to analyze its impact on DNA three-dimensional (3D) structure.
Materials and methods
Subjects
Three hundred unrelated Iranian subjects, consisting of late-onset NCD patients (N = 150) and controls (N = 150) were recruited from the provinces of Tehran, Qazvin, and Rasht. In each NCD case, the Persian version of the Abbreviated Mental Test Score (AMTS)15,16 was implemented (AMTS of < 7 was an inclusion criterion for NCD), medical records were reviewed in all participants, and CT-scans were taken where possible. The AMTS is currently one of the most accurate primary screening instruments to increase the probability of NCD17, and the Persian version of the AMTS is a valid cognitive assessment tool for older Iranian adults, which can be used for NCD screening in Iran15. The control group was selected based on cognitive AMTS of > 7, lack of major medical history, and CT-scan where possible. The cases and controls were matched based on age, gender, and residential district. The subjects' informed consent was obtained (from their guardians where necessary) and their identities remained confidential throughout the study. The research was approved by the Ethics Committee of the University of Social Welfare and Rehabilitation Sciences, Tehran, Iran, and was consistent with the principles outlined in an internationally recognized standard for the ethical conduct of human research. All methods were performed in accordance with the relevant guidelines and regulations.
Allele and genotype analysis of the PRKACB (GCC)-repeat
Genomic DNA was obtained from peripheral blood using a standard salting out method. PCR reactions for the amplification of the PRKACB (GCC)-repeat were set up with the following primers Forward: CGCCTGCGAAGATACAGTC, Reverse: CAACTCACCGCTCTCCAC.
PCR reactions were carried out using High GC buffer (30% of the final volume per PCR reaction) in a thermocycler (PEQSTAR, model: PEQLAB) under the following conditions: 95 °C for 4 min, 35 cycles of denaturation at 94 °C for 30 s, annealing for 30 s at 58 °C and extension at 72 °C for 40 s, and a final extension of 72 °C for 5 min. All the samples included in this study were sequenced by the forward primer, using an ABI PRISM 3130 DNA sequencer. The divergent genotypes were sequenced by the reverse primer as well (Supplementary 1).
Analysis of the PRKACB (GCC)7 across vertebrates
The interval between + 1 and + 100 of the transcription start site (TSS) of the PRKACB was searched across several orders of vertebrates based on the Ensembl Release 102 (https://asia.ensembl.org/index.html). Alignment was performed using CodonCode Aligner 9.0.1.
In silico DNA reconstruction of various (GCC)-repeat lengths in human
The DNA 3D structure across the human PRKACB (GCC)-repeat was studied using several models, including AA-Wedge, DNase, Bolshoy, Cacchione, and Calladine (Supplementary 2)18–23. For each reconstruction, the 100 nucleotide flanking sequences to the repeats were also included. As an instance, based on the twist, roll and tilt, the AA-Wedge model predicts experimental A-tract curvature as measured by gel retardation and cyclization kinetics18. In comparison to a number of existing DNA predicting models, such as the Crothers, Dickerson, Jernigan, Tung-Harvey, and Zhurkin, the AA-Wedge model has been reported as the most consistent and accurate19. Following obtaining the coordinates of the nucleotides in a 3D space, the DNA structure was visualized using plot3D package in R software24.
Furthermore, three additional STRs, SMAD9 (GCC)-STR, RASGEF1 (GGC)-STR, and GPM6B (GA)-STR, were reconstructed according to the AA-Wedge model18,19. The additional genes were studied as sample, for comparison between GCC and non-GCC STR effects.
Divergence calculation across DNA constructs of various periodical (GCC)-repeat lengths
Let for be coordinates of points for each repeat, in the first step we scaled these coordinates as follows
Parametric equations of a segment line that passes through ’s were
where if and if . To measure similarity between two scaled lines, we defined integrated Euclidean distance (IED). Let and for are two lines then IED is as follows
)i.j)-th elements revealed the divergence scores obtained from the above method between the i-th and j-th diagrams.
The accuracy of data was validated by two-by-two comparison of constructs of identical lengths.
Gene network reconstruction
The STRING database (https://string-db.org) was used to find the interactions of PRKACB with other genes. STRING is a biological database of known molecular interactions containing information from experimental, computationally predicted, and public text collections25. The minimum required interaction score was set at 0.7, and the maximum number of interactions was set at 20. To ensure the most important and reliable interactions, we selected only data from experimental studies as the active interaction sources. Subsequently, the interactive network was reconstructed, using Cytoscape version 3.8.2 and according to the interactions found26.
Results
Predominant monomorphism of the PRKACB (GCC)-repeat in human, at 7-repeats, and divergence from monomorphism in two patients afflicted with late-onset NCD
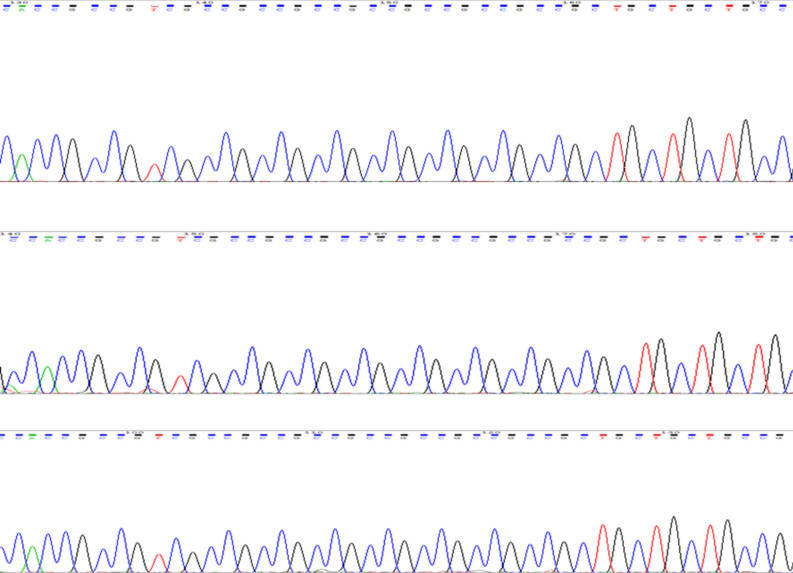
We sequenced the PRKACB (GCC)-repeat in 300 human subjects, and found strict monomorphism of this STR at 7-repeats in this human sample (Fig. 1). Exceptions included two 7/8 genotypes in two patients with late-onset NCD (Fig. 2). The two patients harboring the 7/8 genotypes were females of 78 (Patient 1) and 83 (Patient 2) years of age and AMTS of 4 and 3, respectively.
Figure 1.
Strict monomorphism of the PRKACB (GCC)-repeat, at 7-repeats, in human. Only four samples are represented as examples.
Figure 2.
Two cases of late-onset NCD with 7/8 genotypes.
In patient 1 the progression of neurocognitive symptoms was gradual, strengthening the possible diagnosis of AD. In patient 2, symptoms of neurocognitive impairment including aphonia and dementia occurred abruptly and subsequent to a cerebrovascular accident two years before the interview, strengthening the possible diagnosis of vascular dementia. Other causes of late-onset neurological disorders were ruled out in both patients by neurologists.
Status of the PRKACB (GCC)-repeat across vertebrates
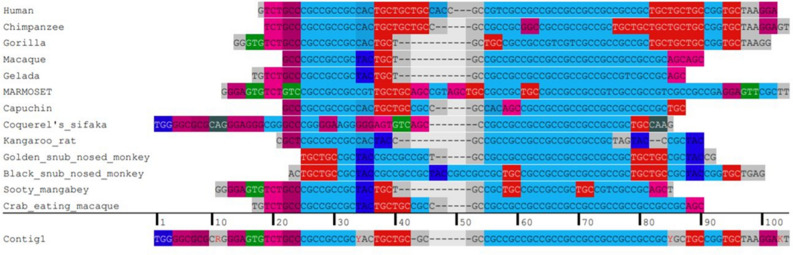
The PRKACB (GCC)-repeat was highly conserved in numerous orders of vertebrates (Table 1). This repeat probably emerged in Birds and Reptiles, as we did not detect it in Amphibians, Fish, and other eukaryotes. In human, the interval encompassing the + 1 to + 100 to the TSS contains two (GCC)-repeats of (GCC)3 and (GCC)7 formula (Table 1). We detected a long ancestral trace of (GCC)-repeats, identifiable by triplet nucleotides of (GCC) or non-(GCC) in various species (Fig. 3). This long trace of (GCC) resulted in complex and unique (GCC) blocks in each species.
Table 1.
Status of the PRKACB (GCC)-repeat across vertebrates.
| Species | Transcript ID | STR formula | Species | Transcript ID | STR formula |
|---|---|---|---|---|---|
| Primates | Leopard | ENSPPRT00000024708.1 | (GCC)3 | ||
| Human | ENST00000370689.6 | (GCC)3 (GCC)7 | Dingo | – | – |
| Bonobo | – | – | Shrew | – | – |
| Chimpanzee | ENSPTRT00000109165.1 | (GCC)3 (GCC)4 | |||
| Gorilla | ENSGGOT00000045976.1 | (GCC)3 (GCC)3 | Lamprey | – | – |
| Orangutan | – | – | Coelacanth | – | – |
| Gibbon | ENSNLET00000012651.2 | (GCC)2 | Elephant shark | ENSCMIT00000019194.1 | (GCC)2 |
| Crab-eating macaque | ENSMFAT00000033375.1 | (GCC)2 (GCC)13 | Hagfish | – | – |
| Macaque | ENSMMUT00000076404.2 | (GCC)2 (GCC)10 | Afrotheria | ||
| Golden snub-nosed monkey | ENSRROT00000025377.1 | (GCC)2 (GCC)8 | Elephent | – | – |
| Gelada | ENSTGET00000029662.1 | (GCC)2 (GCC)8 | Hyrax | – | – |
| Mouse lemur | ENSMICT00000059697.1 | (GCC)5 | Lesser hedgehog tenrec | – | – |
| Vervet-AGM | – | – | Xenatrthra | ||
| Marmoset | ENSCJAT00000115300.1 | (GCC)3 (GCC)4 | Armadilo | – | – |
| Sooty mangabey | ENSCATT00000063374.1 | (GCC)2 (GCC)3 | Sloth | – | – |
| Drill | ENSMLET00000056274.1 | (GCC)7 (GCC)1 | Other mammals | ||
| Pig-tailed macaque | ENSMNET00000056095.1 | (GCC)2 (GCC)6 | Common wombat | – | – |
| Angola colobus | – | – | Koala | – | – |
| Ugandan red Colobus | ENSPTET00000046092.1 | (GCC)11 | Opossum | ENSMODT00000083179.1 | (GCC)2 |
| Black snub-nosed monkey | ENSRBIT00000058758.1 | (GCC)2 (GCC)6 | Platypus | ENSOANT00000057701.1 | (GCC)2 |
| Ma's night monkey | ENSANAT00000031038.1 | (GCC)3 (GCC)2 | Birds and reptiles | ||
| Capuchin | ENSCCAT00000025557.1 | (GCC)3 (GCC)8 | Bengalese finch | ENSLSDT00000018202.1 | (GCC)2 (GCC)3 |
| Bolivian squirrel monkey | ENSSBOT00000050020.1 | (GCC)2 | Blue tit | – | – |
| Tarsier | – | – | Burrowing owl | – | – |
| Greater bamboo lemur | ENSPSMT00000037282.1 | (GCC)5 | Chicken | ENSGALT00000062352.2 | (GCC)3 |
| Coquerel's sifaka | ENSPCOT00000017095.1 | (GCC)7 | Common canary | – | – |
| Bushbaby | – | – | Golden eagle | ENSACCT00020005753.1 | (GCC)4 |
| Rodent | Kakapo | ENSSHBT00005016149.1 | (GCC)4 | ||
| Mouse | ENSMUST00000102515.9 | (GCC)3 (GCC)4 | Anole lizard | ENSACAT00000030613.1 | (GCC)4 |
| Long-tailed chinchilla | ENSCLAT00000008409.1 | (GCC)6 | Eastern brown snake | ENSPTXT00000009104.1 | (GCC)2 |
| Kangaroo rat | ENSDORT00000033480.1 | (GCC)3 (GCC)7 | Amphibians | ||
| Naked mole-rat female | ENSHGLT00000016208.1 | (GCC)5 | Tropical clawed frog | – | – |
| Rabbit | ENSOCUT00000051257.1 | (GCC)5 (GCC)10 | Fish | ||
| Laurasiatheria | Asian bonytongue | – | – | ||
| Cat | ENSFCAT00000050575.2 | (GCC)3 | Zebrafish | – | – |
| Cow | ENSBTAT00000074614.1 | (GCC)4 (GCC)5 | Common crap | – | – |
| Goat | – | – | Zig-zag eel | – | – |
| Blue whale | ENSBMST00010016111.1 | (GCC)3 | Other eukaryotes | ||
| Horse | ENSECAT00000063662.1 | (GCC)8 | Saccharomyces cerevisiae | – | – |
| Siberian musk deer | ENSMMST00000013945.1 | (GCC)6 | Drosophila melanogaster | – | – |
| Red Fox | ENSVVUT00000055345.1 | (GCC)2 | Caenorhabditis elegans | – | – |
| PIG | – | – | Other cordates | ||
| Dog | – | – | C.intestinalis | – | – |
| Panda | – | – | C.savignyi | – | – |
Figure 3.
Sequence alignment of the PRKACB region encompassing the (GCC)-repeat in various primates. Each species revealed complex and unique stretches of (GCC)-repeats. Contig1 indicates the possible ancestral sequence across the selected species. Triplet nucleotides which may be remnants of GCC sequences are detectable in the ancestral sequence.
In silico reconstruction of the human PRKACB 5′ UTR sequence encompassing the (GCC)-repeat
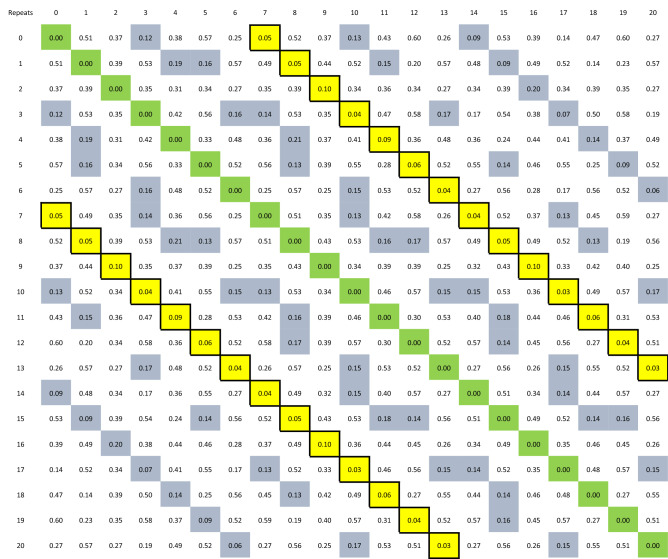
We detected only two alleles in the human population studied (7 and 8-repeats). We reconstructed DNA for those repeat lengths and their periodicals in order to analyze their effect on the 3D structure of the region. Across all studied models, the (GCC)7 periodicals e.g. (0, 7, 14), (1, 8, 15), (2, 9, 16), had a significantly less effect on the regional DNA structure in comparison to the 8-repeat periodicals e.g. (0, 8, 16), (1, 9, 17), (2, 10, 18) (Fig. 4, Table 2, Supplementary 2). While the 7-repeat periodicals were strictly consistent in respect of having the least amount of divergence variation, all other repeat length periodicals, including the 8-repeat, were noncontinuous and interrupted by high divergence scores.
Figure 4.
3D construction of human DNA encompassing the PRKACB (GCC)-repeat based on the AA-Wedge model. Striking similarity was detected in repeats where subtraction or addition of 7-repeats was included.
Table 2.
Divergence scores of existing (7 and 8-repeat) and hypothetical PRKACB repeat periodicals in human.
The highlighted areas represent the 30 percentile of the entire scores (the lowest 30% of the entire scores). The 7-repeat periodical (solid yellow cells) revealed continuous and strictly narrow divergence score variation in comparison to the 8-repeat and all other periodicals. Only the 7 and 8 repeats were detected in the human population studied.
To examine the specificity of the PRKACB (GCC)-repeat inert effect, we performed DNA reconstruction of additional GCC/GGC-repeats in the 5′ UTR of the SMAD9 and RASGEF1C genes, and also a non-GCC STR, such as a GA-repeat in the 5′ UTR of human GPM6B (Supplementary 3). Strikingly, while the reconstructed 3D structures were significantly divergent at various lengths in the instance of the GPM6B 5′ UTR GA-repeat, the SMAD9 and RASGEF1C 3D structures were almost identical for all repeat lengths studied. We also studied the interactive effect of divergent non-repeat flanking sequences with the (GCC)-repeats by studying a different species than human, such as capuchin. The inert effect of the (GCC)-block was consistent, regardless of the differential flanking non-repeat sequences in capuchin versus human.
Gene network reconstruction
Based on the available experimental evidence, the reconstructed interactive network consisted of 16 nodes and 45 edges (Fig. 5). Within this network, PRKACB interacts with genes of other subunits of the cAMP-dependent protein kinase complex, as well as other genes such as MAPT, FOXO1, and PDE5A, which are central to maintaining cell structure and function.
Figure 5.

The interactive network reconstructed based on experimental evidence. The network consists of 16 nodes, including genes of the cAMP-dependent protein kinase subunits, as well as other structurally and functionally important genes such as MAPT, FOXO1, and PDE5A. The 16 genes interact with each other through 45 edges.
Discussion
Because of containing internal CpG dinucleotides, susceptible to methylation and subsequent base substitutions, (GCC)-repeats are potential mutation hotspots27–29, and this may be the reason why this class of repeats do not expand beyond certain lengths unless where selected, such as in the gene regulatory regions30. Large expansions of (GCC)-repeats in the 5′ UTR of a number of genes are associated with hypermethylation and intellectual disability31. Based on the Ensembl database, the PRKACB gene is in the 10th percentile of genes in respect of containing long (GCC)-repeats14. Here we show that this (GCC)-repeat is predominantly monomorphic in human at 7-repeats, and there may be enrichment of divergence from this monomorphism in late-onset NCD. We propose that there was selective advantage in human at this particular STR to reach to, and stabilize at that length. The above proposition is supported by the observations that mutations that have negative fitness consequences tend to be eliminated from the population32. This STR was found to be highly conserved across various mammalian species, indicating its important role in growth and development. In general, STRs near the TSSs of genes are often highly conserved, and distance from a STR to the nearest TSS is a good predictor of the STR conservation score33.
We also found that (GCC)7 periodicals had the least effect on the 3D structure in contrast to the more significant effects that various other repeats inserted, which probably confers an inert effect to (GCC)7 without changing the structure of DNA or RNA. The above was deduced from the exceptionally continuous trend of those periodicals (uninterrupted by high divergence scores) in contrast to all other repeats. Interestingly, the divergence score between the 7 and 8 repeat (the latter was an allele of the two divergent genotypes) was among the highest in all examined models. While we also detected an inert effect of GCC/GGC repeats in two additional genes, SMAD9 and RASGEF1C, this effect was not observed in the GA class of STRs, where various lengths of GA-repeats resulted in significant variation in the 3D structure of DNA in the present study and in a previously reported research in the instance of human ZMYM311. Significant divergence has also been observed with various CA-repeat lengths in the human NHLH2 gene9. The above findings indicate that long GCC/GGC repeats might have evolved to exert epigenetic effects, as epigenetic knobs, without changing the structure of the regional DNA.
GCC motifs of STRs significantly overlap with G-quadraplex (G4) non-B structures34. Recent research indicates that organisms may have evolutionarily developed G4 into a novel reversible and elaborate transcriptional regulatory mechanism benefiting multiple physiological activities of higher organisms35,36.
PRKACB is involved in a number of physiological and pathological pathways, the outcomes of which directly affect cognition in human12,13,37–39. Future studies are warranted to sequence the PRKACB (GCC)-repeat in a large number of patients with neurological disorders. Considering the pivotal role of PRKACB in the brain, CRISPR/Cas9 methods may also be ideal to edit this STR at the genomic level, and investigate differentiation of human stem cells into neural cells.
Conclusion
In conclusion, we report the first instance of predominant monomorphism of a long (GCC)-repeat in human and enriched divergent genotypes from this monomorphism in human disease. We also propose that (GCC)-repeats might have evolved in the genome as regulatory elements (such as epigenetic regulation) without dramatically changing the 3D structure of the regional DNA. Further experimental research is warranted to examine the proposed model.
Supplementary Information
Abbreviations
- 3D
Three-dimensional
- NCD
Neurocognitive disorder
- PRKACB
Protein Kinase CAMP-Activated Catalytic Subunit Beta
- STR
Short tandem repeat
- TSS
Transcription start site
- UTR
Untranslated region
Author contributions
S.K. did the molecular experiments. Z.J. performed part of the DNA modeling analyses. A.B. conceived and performed the DNA modeling and mathematics analyses, and wrote part of the manuscript. M.T. performed part of the mathematics analyses. H.A. collected the human samples and their clinical information. M.K. and R.M. provided critical comments and contributed to data collection. M.O. conceived and supervised the project, and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Safoura Khamse and Zahra Jafarian.
Contributor Information
Ali Bozorgmehr, Email: bozorgmehr.a@iums.ac.ir.
Mina Ohadi, Email: mi.ohadi@uswr.ac.ir.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-99932-3.
References
- 1.Jakubosky D, D’Antonio M, Bonder MJ, Smail C, Donovan MKR, Greenwald WWY, Matsui H, D’Antonio-Chronowska A, Stegle O, Smith EN. Properties of structural variants and short tandem repeats associated with gene expression and complex traits. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-020-16482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fotsing SF, Margoliash J, Wang C, Saini S, Yanicky R, Shleizer-Burko S, Goren A, Gymrek M. The impact of short tandem repeat variation on gene expression. Nat. Genet. 2019;51:1652–1659. doi: 10.1038/s41588-019-0521-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Press MO, Hall AN, Morton EA, Queitsch C. Substitutions are boring: Some arguments about parallel mutations and high mutation rates. Trends Genet. 2019;35:253–264. doi: 10.1016/j.tig.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valipour E, Kowsari A, Bayat H, Banan M, Kazeminasab S, Mohammadparast S, Ohadi M. Polymorphic core promoter GA-repeats alter gene expression of the early embryonic developmental genes. Gene. 2013;531:175–179. doi: 10.1016/j.gene.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 5.Ohadi M, Valipour E, Ghadimi-Haddadan S, Namdar-Aligoodarzi P, Bagheri A, Kowsari A, Rezazadeh M, Darvish H, Kazeminasab S. Core promoter short tandem repeats as evolutionary switch codes for primate speciation. Am. J. Primatol. 2015;77:34–43. doi: 10.1002/ajp.22308. [DOI] [PubMed] [Google Scholar]
- 6.Mohammadparast S, Bayat H, Biglarian A, Ohadi M. Exceptional expansion and conservation of a CT-repeat complex in the core promoter of PAXBP1 in primates. Am. J. Primatol. 2014;76:747–756. doi: 10.1002/ajp.22266. [DOI] [PubMed] [Google Scholar]
- 7.Namdar-Aligoodarzi P, Mohammadparast S, Zaker-Kandjani B, Talebi Kakroodi S, Jafari Vesiehsari M, Ohadi M. Exceptionally long 5' UTR short tandem repeats specifically linked to primates. Gene. 2015;569(1):88–94. doi: 10.1016/j.gene.2015.05.053. [DOI] [PubMed] [Google Scholar]
- 8.Watts PC, Kallio ER, Koskela E, Lonn E, Mappes T, Mokkonen M. Stabilizing selection on microsatellite allele length at arginine vasopressin 1a receptor and oxytocin receptor loci. Proc. R. Soc. B Biol. Sci. 2017;284:20171896. doi: 10.1098/rspb.2017.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afshar H, Khamse S, Alizadeh F, Delbari A, Najafipour R, Bozorgmehr A, Khazaei M, Adelirad F, Alizadeh A, Kowsari A. Evolving evidence on a link between the ZMYM3 exceptionally long GA-STR and human cognition. Sci. Rep. 2020;10:1–7. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afshar H, Adelirad F, Kowsari A, Kalhor N, Delbari A, Najafipour R, Foroughan M, Bozorgmehr A, Khamse S, Nazaripanah N. Natural selection at the NHLH2 core promoter exceptionally long CA-repeat in human and disease-only genotypes in late-onset neurocognitive disorder. Gerontology. 2020;66:514–522. doi: 10.1159/000509471. [DOI] [PubMed] [Google Scholar]
- 11.Alizadeh F, Moharrami T, Mousavi N, Yazarlou F, Bozorgmehr A, Shahsavand E, Delbari A, Ohadi M. Disease-only alleles at the extreme ends of the human ZMYM3 exceptionally long 5′ UTR short tandem repeat in bipolar disorder: A pilot study. J. Affect. Disord. 2019;251:86–90. doi: 10.1016/j.jad.2019.03.056. [DOI] [PubMed] [Google Scholar]
- 12.London E, Noguchi A, Springer D, Faidas M, Gavrilova O, Eisenhofer G, Stratakis CA. The catalytic subunit β of PKA affects energy balance and catecholaminergic activity. J. Endocr. Soc. 2019;3:1062–1078. doi: 10.1210/js.2019-00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Liu J, Wang Q, Jiang H, Zeng L, Li Z, Liu R. MicroRNA-200a-3p mediates neuroprotection in Alzheimer-related deficits and attenuates amyloid-beta overproduction and tau hyperphosphorylation via coregulating BACE1 and PRKACB. Front. Pharmacol. 2019;10:806. doi: 10.3389/fphar.2019.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vázquez-Higuera JL, Mateo I, Sánchez-Juan P, Rodríguez-Rodríguez E, Pozueta A, Calero M, Dobato JL, Frank-García A, Valdivieso F, Berciano J. Genetic variation in the tau kinases pathway may modify the risk and age at onset of Alzheimer's disease. J. Alzheimers Dis. 2011;27:291–297. doi: 10.3233/JAD-2011-110794. [DOI] [PubMed] [Google Scholar]
- 15.Foroughan M, Wahlund LO, Jafari Z, Rahgozar M, Farahani IG, Rashedi V. Validity and reliability of Abbreviated Mental Test Score (AMTS) among older Iranian. Psychogeriatr. Off. J Jpn. Psychogeriatr. Soc. 2017;17:460–465. doi: 10.1111/psyg.12276. [DOI] [PubMed] [Google Scholar]
- 16.Hodkinson H. Evaluation of a mental test score for assessment of mental impairment in the elderly. Age Ageing. 1972;1:233–238. doi: 10.1093/ageing/1.4.233. [DOI] [PubMed] [Google Scholar]
- 17.Carpenter CR, Banerjee J, Keyes D, Eagles D, Schnitker L, Barbic D, Fowler S, LaMantia MA. Accuracy of dementia screening instruments in emergency medicine: A diagnostic meta-analysis. Acad. Emerg. Med. 2019;26:226–245. doi: 10.1111/acem.13573. [DOI] [PubMed] [Google Scholar]
- 18.Tan RKZ, Harvey SC. A comparison of six DNA bending models. J. Biomol. Struct. Dyn. 1987;5:497–512. doi: 10.1080/07391102.1987.10506410. [DOI] [PubMed] [Google Scholar]
- 19.Goodsell DS, Dickerson RE. Bending and curvature calculations in B-DNA. Nucleic Acids Res. 1994;22:5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarovici A, Zhou T, Shafer A, Dantas Machado AC, Riley TR, Sandstrom R, Sabo PJ, Lu Y, Rohs R, Stamatoyannopoulos JA, Bussemaker HJ. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. U. S. A. 2013;110(16):6376–6381. doi: 10.1073/pnas.1216822110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolshoy A, McNamara P, Harrington R, Trifonov E. Curved DNA without A-A: Experimental estimation of all 16 DNA wedge angles. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cacchione S, De Santis P, Foti D, Palleschi A, Savino M. Periodical polydeoxynucleotides and DNA curvature. Biochemistry. 1989;28(22):8706–8713. doi: 10.1021/bi00448a006. [DOI] [PubMed] [Google Scholar]
- 23.Calladine CR, Drew HR, McCall MJ. The intrinsic curvature of DNA in solution. J. Mol. Biol. 1988;201(1):127–137. doi: 10.1016/0022-2836(88)90444-5. [DOI] [PubMed] [Google Scholar]
- 24.Soetaert, K. plot3Drgl: Tools for plotting 3-D and 2-D data in openGL.
- 25.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang HS, Shin WJ, Lee JE, Do JT. CpG and Non-CpG methylation in epigenetic gene regulation and brain function. Genes (Basel) 2017;8(6):148. doi: 10.3390/genes8060148.PMID:28545252;PMCID:PMC5485512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li QS, Sun Y, Wang T. Epigenome-wide association study of Alzheimer’s disease replicates 22 differentially methylated positions and 30 differentially methylated regions. Clin. Epigenet. 2020;12:1–14. doi: 10.1186/s13148-019-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fryxell KJ, Moon W-J. CpG mutation rates in the human genome are highly dependent on local GC content. Mol. Biol. Evol. 2005;22(3):650–658. doi: 10.1093/molbev/msi043. [DOI] [PubMed] [Google Scholar]
- 30.Sawaya S, Bagshaw A, Buschiazzo E, Kumar P, Chowdhury S, Black MA, Gemmell N. Microsatellite tandem repeats are abundant in human promoters and are associated with regulatory elements. PLoS ONE. 2013;8:e54710. doi: 10.1371/journal.pone.0054710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesca G, Biancalana V, Brunel M-J, Quack B, Calender A, Lespinasse J. Clinical, cytogenetic, and molecular description of a FRAXE French family. Psychiatr. Genet. 2003;13:43–46. doi: 10.1097/00041444-200303000-00007. [DOI] [PubMed] [Google Scholar]
- 32.Gymrek M, Willems T, Reich D, Erlich Y. Interpreting short tandem rpeat variations in humans using mutational constraint. Nat. Genet. 2017;49:1495. doi: 10.1038/ng.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawaya SM, Lennon D, Buschiazzo E, Gemmell N, Minin VN. Measuring microsatellite conservation in mammalian evolution with a phylogenetic birth–death model. Genome Biol. Evol. 2012;4:636–647. doi: 10.1093/gbe/evs050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiegel J, Adhikari S, Balasubramanian S. The structure and function of DNA G-quadruplexes. Trends Chem. 2020;2:123–136. doi: 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niu K, Xiang L, Jin Y, Peng Y, Wu F, Tang W, Zhang X, Deng H, Xiang H, Li S. Identification of LARK as a novel and conserved G-quadruplex binding protein in invertebrates and vertebrates. Nucleic Acids Res. 2019;47:7306–7320. doi: 10.1093/nar/gky958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu F, Niu K, Cui Y, Li C, Lyu M, Ren Y, Chen Y, Deng H, Huang L, Zheng S. Genome-wide analysis of DNA G-quadruplex motifs across 37 species provides insights into G4 evolution. Commun. Biol. 2021;4:1–11. doi: 10.1038/s42003-020-01566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu YQ, Liu Q, Wang HB, Chen C, Huang H, Sun YM, Ma LH, Wan J, Sun YY, Miao HH. Microarray analysis identifies key differentially expressed circular RNAs in aged mice with postoperative cognitive dysfunction. Front Aging Neurosci. 2021;16(13):716383. doi: 10.3389/fnagi.2021.716383.PMID:34483886;PMCID:PMC8415796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Shen Q, Wu X, Zhang D, Xing D. Activation of PKA/SIRT1 signaling pathway by photobiomodulation therapy reduces Aβ levels in Alzheimer’s disease models. Aging Cell. 2020;19:e13054. doi: 10.1111/acel.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dagda RK, Das BT. Role of protein kinase a in regulating mitochondrial function and neuronal development: Implications to neurodegenerative diseases. Rev. Neurosci. 2015;26:359–370. doi: 10.1515/revneuro-2014-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.