Abstract
Rat-1 cells are used in many studies on transformation, cell cycle, and apoptosis. Whereas UV treatment of Rat-1 cells results in apoptosis, X-ray treatment does not induce either apoptosis or a cell cycle block. X-ray treatment of Rat-1 cells results in both an increase of p53 protein and expression of the p53-inducible gene MDM2 but not the protein or mRNA of the p53-inducible p21WAF1/CIP1 gene, which in other cells plays an important role in p53-mediated cell cycle block. The lack of p21WAF1/CIP1 expression appears to be the result of hypermethylation of the p21WAF1/CIP1 promoter region, as p21WAF1/CIP1 protein expression could be induced by growth of Rat-1 cells in the presence of 5-aza-2-deoxycytidine. Furthermore, sequence analysis of bisulfite-treated DNA demonstrated extensive methylation of cytosine residues in CpG dinucleotides in a CpG-rich island in the promoter region of the p21WAF1/CIP1 gene. Stable X-ray-induced p53-dependent p21WAF1/CIP1 expression and cell cycle block were restored to a Rat-1 clone after transfection with a P1 artificial chromosome (PAC) DNA clone containing a rat genomic copy of the p21WAF1/CIP1 gene. The absence of expression of the p21WAF1/CIP1 gene may contribute to the suitability of Rat-1 cells for transformation, cell cycle, and apoptosis studies.
A number of tissue culture cell lines can be transformed by single oncogenes, and many of these cell lines are used to study aspects of cell cycle regulation and apoptosis. These cell lines usually have genetic alterations which make them amenable for these types of studies. A number of these alterations have been found to affect the p53 pathway. Rat-1 cells are widely used for assessing transformation and for cell cycle and apoptosis studies (13, 16, 18, 20, 22, 32). Despite being wild type for p53 (22), Rat-1 cells can be transformed by single oncogenes, suggesting that some stage in the p53 pathway may be abrogated in these cells.
The p53-inducible p21WAF1/CIP1 gene encodes a protein which binds to and inhibits a broad range of cyclin–cyclin-dependent kinase complexes which function to promote cell cycle progression (19, 44). Thus, the general consequence of p21WAF1/CIP1 activity is growth arrest, which is particularly evident following exposure of cells to DNA-damaging agents such as γ radiation or adriamycin (9, 42). p21WAF1/CIP1 null mice are deficient in this response (4, 7), and in human cells devoid of p21WAF1/CIP1 expression this response is absent altogether (42). It is well established that DNA damage brings about p21WAF1/CIP1-induced growth arrest via transcriptional upregulation of p21WAF1/CIP1 by the p53 tumor suppressor gene (10, 24). Indeed, p53 null cells exposed to γ radiation fail to exhibit either induction of p21WAF1/CIP1 expression or G1 arrest (24). Furthermore, the p21WAF1/CIP1 promoter region has been shown to contain two conserved p53-binding sites through which p53 can regulate p21WAF1/CIP1 transcription (10, 11).
In addition to its role in cell cycle regulation, p21WAF1/CIP1 is also believed to inhibit DNA replication through its ability to bind proliferating cell nuclear antigen (PCNA), which is required for both replicative DNA synthesis and DNA repair. However, p21WAF1/CIP1 has no inhibitory effect on the DNA repair function of PCNA (21, 41). Thus, p21WAF1/CIP1 may play a central role in preventing the replication of mutations incurred after exposure of cells to DNA damage.
As a consequence of its importance in cell cycle control and its possible role in maintaining genome fidelity, the p21WAF1/CIP1 gene might be predicted to be a frequent target for mutation in the neoplastic process. However, p21WAF1/CIP1 mutations are extremely rare (2, 25, 34). Furthermore, p21WAF1/CIP1 null mice develop normally (4, 7) and fail to exhibit any increase in tumor incidence (4). Recently many studies have focused on the potential role of p21WAF1/CIP1 in apoptosis, and induction of p21WAF1/CIP1 expression has been associated with apoptosis in some instances (9, 28, 40). However, p21WAF1/CIP1 appears to be dispensable for apoptosis since p21WAF1/CIP1-deficient cells exhibit a full apoptotic response (4, 7) and most recent studies report a protective role for p21WAF1/CIP1 against apoptosis (3, 5, 17, 29, 30).
We have shown previously that U2OS, a human osteosarcoma cell line which is wild type for p53, responds differentially to two distinct forms of radiation, X ray and UVC, and that the response may correlate with level of p21WAF1/CIP1 expression (1). To investigate further the p53-induced p21WAF1/CIP1 response to irradiation, we analyzed Rat-1 cells which are also wild type for p53. We report here that X irradiation failed to induce either apoptosis or G1 growth arrest in Rat-1 cells. In addition, after X irradiation there was no up-regulation of p21WAF1/CIP1 expression at either the protein or RNA level, despite induction of p53 transactivation activity. Rescue of p21WAF1/CIP1 protein expression by 5-aza-2-deoxycytidine (5-AzaC) treatment and sequence analysis of bisulfite-treated DNA (6, 14) of the p21WAF1/CIP1 5′ untranslated region (5′UTR) indicated that the failure of Rat-1 cells to express p21WAF1/CIP1 was due to methylation of the p21WAF1/CIP1 promoter region. We were able to restore stable p53-dependent X-ray induction of p21WAF1/CIP1 expression and subsequent G1 arrest after transfecting Rat-1 cells with a 120-kb P1 artificial chromosome (PAC) containing the rat p21WAF1/CIP1 gene and extensive surrounding genomic DNA sequence. These results strongly indicate that the lack of endogenous p21WAF1/CIP1 expression in Rat-1 cells is solely responsible for the absence of a G1 arrest after X irradiation in these cells. To our knowledge, this is the first report of inactivation of p21WAF1/CIP1 by promoter methylation and has implications for transformation, apoptosis, and cell cycle studies which utilize the Rat-1 cell line.
MATERIALS AND METHODS
Cell culture, radiation treatment, plasmids, and transfections.
Rat-1 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum (DMEM–10% FCS), penicillin (100 μg/ml), and streptomycin (100 μg/ml). Radiation treatment was carried out as described previously (1). The DN10 and P13.5-DN.A1 clones were established by transfection of Rat-1 and P13.5 (see below) cells, respectively, with the p53 dominant negative construct p53.302-90, which contained a C-terminal fragment of p53 encoding amino acids 302 to 390 and a puromycin selectable marker (gift from T. Littlewood). Expression of the transfected construct was confirmed by Western blotting. The P13.5 clone was established by cotransfection of Rat-1 with a rat PAC DNA clone, 277G17 (Human Genome Mapping Project [HGMP] Resource Centre, Hinxton, United Kingdom), containing a genomic copy of rat p21WAF1/CIP1 at a ratio of 10:1 with pBABEPuro (gift from T. Littlewood). Clones DN10, P13.5, and P13.5-DN.A1 were selected in puromycin (2.5 μg/ml). Transfections were carried out in 5-cm-diameter dishes using 2 μg of CsCl-purified plasmid DNA or Qiagen maxiprep-purified PAC DNA and 15 μl of Superfect in DMEM–10% FCS as instructed by the manufacturer (Qiagen).
Analysis of cell cycle distribution and apoptosis.
Cell cycle distribution was analyzed by flow cytometry. At times indicated after irradiation, cells were incubated for 30 min with bromodeoxyuridine (BrdU; 10 μM), harvested, washed twice with phosphate-buffered saline, and fixed in 70% ethanol. Subsequently, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated anti-BrdU antibody and with propidium iodide (PI). DNA synthesis (FITC) and DNA content (PI) were determined with a fluorescence-activated cell sorting (FACS) analyzer (FACSCalibur; Becton Dickinson). For analysis of apoptosis, cells were harvested, treated with PI, and assayed by flow cytometry as described above. The percentage of cells with a sub-G1 DNA content was used as a measure of apoptosis.
Western analysis of protein expression.
Western analysis was carried out as described previously (1). Protein concentrations were determined using the DC protein assay (Bio-Rad); 50 μg of total protein was loaded per lane, and loading was also assessed by Ponceau S staining of nitrocellulose filters. Blots were probed for p53 (R-19, 0.5 μg/ml; Santa Cruz), p21WAF1/CIP1 (sx-118, 1:4, tissue culture supernatant; gift from X. Lu) and MDM2 (2A10, 1:50, tissue culture supernatant; gift from A. Levine). The appropriate secondary horseradish peroxidase-conjugated antibodies were obtained from Santa Cruz (anti-goat for R-19) or Amersham (anti-mouse for sx-118 and 2A10).
Northern analysis of RNA.
Total RNA was prepared using 1 ml of Trizol reagent/2 × 106 cells as instructed by the manufacturer (Gibco BRL). Northern analysis was carried out as described previously (1). The blot was probed with a PstI fragment (approximately 300 bp) of rat p21WAF1/CIP1 cDNA (gift from P. Jat).
Sequencing and reverse transcription (RT)-PCR.
For sequencing of the two p53 response elements (REs) in the p21WAF1/CIP1 promoter, the two p53 REs were amplified by PCR separately, using Rat-1 genomic DNA as a template. For p53 RE-1, the primers were Rp21A (5′-CTCAGCCTCAGAGGGTACCTGC) and Rp21C (5′-CCTTCACCTGGTACATATCAC). The primers used for p53 RE-2 were Rp21D (5′-GACTGGATGGTTCAGGAGCTGG) and Rp21B (5′-CTGGCCTAGGTTACAGGAGACCC). Three independent PCR products were cloned into the T-Easy vector (Promega) and sequenced using a BigDye terminator cycle sequencing kit (Perkin-Elmer) and the vector-specific primers T7 and SP6.
For RT-PCR and sequencing of p21WAF1/CIP1 cDNA, RNA was prepared from Rat-1 cells 24 h after X irradiation with 12 Gy as described above for Northern analysis. RT was carried out using 5 μg of total RNA as template, 10 pmol of the 3′ gene-specific primer Rp21T (5′ GAATTGCACGAGGGGAGG), and 200 U of Superscript II in a total volume of 20 μl as instructed by the manufacturer (Gibco BRL). One microliter of the RT reaction was used as the template for PCR with primers Rp21E (5′ AATTGGAGGCAGGCGCCGATCC) and p21-G (5′ GGCAGAAGATGGGGAAGAGGCC). Purified PCR products (QIAquick gel extraction kit; Qiagen) were then used as templates for direct sequencing using the same primers (Rp21E and p21-G). Two independent RT reactions were used in three independent PCRs for sequencing.
Calculation of CpG frequency.
The MacVector program (Oxford Molecular Group) was used to calculate CpG frequencies for consecutive 800-bp intervals of a contig comprising the rat p21WAF1/CIP1 promoter, exon 1, and the 5′ end of intron 1. This software was also used to compare observed/expected CpG frequencies for this region.
Sequencing of bisulfite-treated DNA.
A modified version of the technique described by Clark et al. (6) was used. Ten micrograms of genomic DNA was digested with SacI and purified by extraction with phenol (twice), chloroform-isoamylalcohol (24:1), and ethanol precipitation. For bisulfite treatment of DNA, 5 μg of SacI-digested DNA was diluted to 20 μl with distilled water; 2 μl of 3 M NaOH (fresh) was added, and the reaction mixture was incubated at 75°C for 15 min to denature the DNA prior to snap cooling on ice. To the reaction mixture were added (i) 250 μl of sodium bisulfite (freshly prepared 4.8 M Na2S2O5 [sodium metabisulfite; pH 5.0] was used as a source of sodium bisulfite) and (ii) 14 μl of 10 mM hydroquinone (freshly prepared). The reaction mixture was overlaid with mineral oil and incubated at 55°C for 4 h in the dark. Bisulfite was removed from the DNA with a Wizard DNA cleanup kit (Promega). The reaction mixture was desulfonated by addition of NaOH to a final concentration of 0.3 M and incubation at 37°C for 15 min and neutralized by the addition of ammonium acetate (pH 7.0) to a final concentration of 3 M, and the DNA was precipitated with 3 volumes of ethanol overnight at −20°C. The DNA pellet was then dissolved in distilled water to give approximately 100 ng/μl. Bisulfite-treated DNA was used as template for subsequent PCR and sequencing using primer sequences based on the rat p21WAF1/CIP1 intron 1 sequence (L. Allan and T. Duhig, unpublished data) and specific for fully modified DNA. Primers for the sense strand were BS.A1 (5′-GTTGGGTTTTAGATTTTTGTGGATTAGGTG) and BS.A4 (5′-ATACTACCTCTCTACAATACAAACTCCTCC); primers for the antisense strand were BS.B1 (5′-ATACCTCTAAATCCCCTACCCTTATAAACC) and BS.B4 (5′-GAGGGGTTTTTAGTTTAGGGGAGTTTGATG). PCR was carried out using a hot start and then the following cycling parameters: two cycles of 94°C (30 s), 58°C (30 s), and 72°C (30 s), followed by repetitions of this cycle but decreasing the annealing temperature by 1°C every second cycle until it reached 51°C. This was followed by 25 cycles of 94°C (30 s), 51°C (30 s), and 72°C (30 s), followed by 72°C (5 min) and 8°C to cool. Sequencing was carried out directly on the purified PCR products as described for RT-PCR sequencing (see above), using the same primers as used for the PCRs. Two independent PCR products were sequenced in both directions and on both strands to confirm the sequence.
Isolation of p21WAF1/CIP1 rat PAC clones.
A rat genomic DNA PAC library, RPC131, was supplied as a series of seven gridded filters by the HGMP Resource Centre. These were probed by standard procedures (8) with a 700-bp fragment corresponding to the 5′ end of the rat p21WAF1/CIP1 intron 1. Nineteen positive clones were obtained as Escherichia coli stocks from the HGMP Resource Centre and screened by PCR for the presence of the 5′ end of the promoter region and for exon 2 of the rat p21WAF1/CIP1 gene. One clone, 277G17, which was positive for both reactions was used in subsequent experiments.
RESULTS
Effect of γ radiation on the cell cycle in Rat-1 cells.
Rat-1, a cell line known to be wild type for p53 (22), undergoes apoptosis readily in response to ectopic expression of the c-myc oncogene (13). To evaluate the response of this cell line to radiation-induced DNA damage, the effect of X radiation on the viability and cell cycle profile of Rat-1 cells was assessed. X-ray (12 Gy) treatment failed to induce apoptosis in these cells as determined by a lack of sub-G1 peak (Fig. 1A), and cells remained intact and attached to the culture dish. In contrast, treatment with a 20-J/m2 dose of UVC caused rapid apoptosis (>30% by 24 h postirradiation) (Fig. 1A). Furthermore, BrdU analysis showed that exposure to X rays also had little effect on the cell cycle in Rat-1 cells, with only a minor proportion appearing to accumulate in G2. Notably, cells did not arrest in G1 and continued to enter S phase (Fig. 1B). Similar treatment of another wild-type p53 rat fibroblast cell line, REF52, has been shown previously to cause a G1 arrest (27).
FIG. 1.
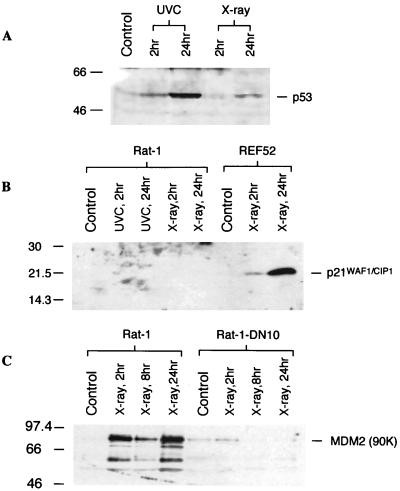
Effect of X rays on viability and cell cycle profile in Rat-1 cells. (A) X-ray (12 Gy) treatment does not cause apoptosis in Rat-1 cells, shown by the absence of cells with a sub-G1 content (M1). In contrast, UVC treatment (20 J/m2) results in apoptosis, with more than 30% of the cells exhibiting a sub-G1 DNA content. (B) X-ray treatment has little effect on the cell cycle distribution in Rat-1 cells at either 7 or 24 h after irradiation. Although there appears to be a minor accumulation in G2, there is no G1 arrest and cells continue to cycle into S phase.
Rat-1 cells fail to induce p21WAF1/CIP1 following γ irradiation.
We assessed the effect of X irradiation on p53 protein levels and transactivation activity. UVC treatment is known to stabilize p53 considerably (23) and was included as a control. Figure 2A shows that X-ray treatment stabilized p53 protein levels, suggesting that the radiation-induced DNA damage signaling pathway to p53 is intact in Rat-1 cells.
FIG. 2.
p53 expression and transcriptional activity after X irradiation. (A) X-ray (12 Gy) treatment stabilized p53 in Rat-1 cells 24 h after irradiation, although to a lesser extent than UVC. (B) Rat-1 cells fail to express p21WAF1/CIP1 protein after X irradiation or UVC treatment, whereas REF52 cells, which arrest in G1 after X irradiation, exhibit increasing induction of p21WAF1/CIP1 at 2 and 24 h postirradiation, respectively. (C) X-ray (12 Gy) treatment induces MDM2 in Rat-1 cells in a p53-dependent manner, as shown by the abrogation of this response by expression of a dominant negative p53 in Rat-1-DN10 cells. Sizes are indicated in kilodaltons.
In an attempt to determine why Rat-1 cells fail to undergo growth arrest, we studied the expression profile of p21WAF1/CIP1, a p53-inducible gene whose induction is believed to be primarily responsible for p53-mediated G1 arrest. As shown in Fig. 2B, p21WAF1/CIP1 was undetectable in Rat-1 cells, even after treatment with X rays. This is in marked contrast to the induction of p21WAF1/CIP1 in REF52 cells after X irradiation. To confirm that the p53 protein in Rat-1 cells is functionally wild type, we analyzed its ability to induce expression of another p53-inducible gene, MDM2. Following exposure to X rays, MDM2 protein was induced in Rat-1 cells in a p53-dependent manner since this induction was abrogated by expression of dominant negative p53 (Rat-1-DN10 cells) (Fig. 2C). Thus, Rat-1 p53 protein is competent for transcriptional activation following X irradiation.
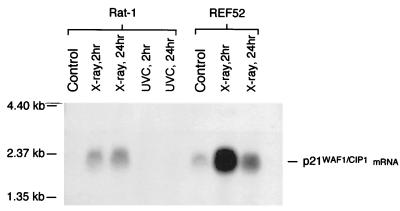
Next we assessed the effect of X irradiation on p21WAF1/CIP1 RNA levels in Rat-1 cells. X-ray treatment induced p21WAF1/CIP1 mRNA in Rat-1 cells, but only to a level marginally higher than that observed in untreated REF52 cells (Fig. 3), consistent with the lack of detectable rat p21WAF1/CIP1 protein (Fig. 2B). In contrast REF52 cells showed a substantial induction of rat p21WAF1/CIP1 mRNA following X irradiation (Fig. 3), in agreement with the increase in p21WAF1/CIP1 protein levels (Fig. 2B). This suggests that the lack of p21WAF1/CIP1 induction in Rat-1 after X irradiation either occurs at the transcriptional level or may be due to RNA instability.
FIG. 3.
Northern blot analysis of MDM2 mRNA after X-ray treatment. X-ray (12 Gy) treatment induces only very low levels of p21WAF1/CIP1 mRNA in Rat-1 cells at either 2 or 24 h postirradiation. This induced level reflects the background level of expression observed in REF52 cells, which show highly elevated levels of p21WAF1/CIP1 mRNA after X irradiation.
Sequencing Rat-1 p21WAF1/CIP1 regulatory elements and cDNA.
The rat p21WAF1/CIP1 promoter contains two p53 REs through which p53 is believed to regulate the expression of p21WAF1/CIP1 in response to radiation-induced DNA damage (11). Mutation of either site could prevent p53 binding and abrogate p53-dependent induction of p21WAF1/CIP1 transcription. Therefore, we sequenced each p53 RE and approximately 100 bp of Rat-1 genomic DNA on either side of each site. Results (not shown) confirmed that the sequences of the two p53 RE sites in Rat-1 are identical to the published sequences (11), indicating that the lack of p21WAF1/CIP1 expression in Rat-1 is not due to mutation of the promoter at the p53 binding sites.
Many studies have shown that p21WAF1/CIP1 gene mutations are rare (2, 25, 34) in tumor cells. However, it is possible that Rat-1 cells harbor a mutation which, for example, may reduce p21WAF1/CIP1 RNA stability, leading to the low level of p21WAF1/CIP1 RNA observed in these cells. To address this possibility, we analyzed Rat-1 p21WAF1/CIP1 cDNA by RT-PCR, using the very low level of p21WAF1/CIP1 RNA after X irradiation (see above), followed by direct sequencing of three independent PCR products. Results confirmed that Rat-1 p21WAF1/CIP1 cDNA sequence is identical to the published wild-type coding sequence (GenBank accession no. U24174) (11). Thus, mutation in this region is unlikely to account for the lack of p21WAF1/CIP1 expression in Rat-1 cells.
The p21WAF1/CIP1 5′UTR contains a putative CpG island which is methylated in Rat-1 cells.
An increasing amount of evidence suggests that tumor suppressor gene inactivation by promoter methylation may be a frequent occurrence in tumor cells (12, 31, 33, 35, 36). Therefore, we wanted to determine whether p21WAF1/CIP1 expression may be abrogated by this mechanism in Rat-1 cells. In general the CpG dinucleotide is underrepresented in mammalian DNA with a typical observed/expected CpG ratio of 0.3 or less as a result of deamination of 5-methylcytosine to thymidine. CpG-rich islands characteristically exhibit an observed/expected CpG ratio of greater than 0.6. The 5′UTR of the human p21WAF1/CIP1 gene (GenBank accession no. Z85996) is believed to contain a CpG island spanning approximately 2 kb which encompasses the 3′ end of the promoter, the first exon, and the 5′ end (approximately one-third) of the first intron. Analysis of published sequences for the rat p21WAF1/CIP1 promoter and coding region and our further analysis of approximately 2.4 kb of the 5′ end of intron 1 support the existence of a CpG island in a similar position in the rat p21WAF1/CIP1 gene (Fig. 4). The frequency of CpG dinucleotides (observed/expected) was calculated over 800-bp intervals for a contig comprising the promoter, exon 1, and the 5′ end of intron 1. The highest frequency of CpG, 0.81, was observed for the interval containing the last 20 bp of the noncoding exon 1 and the first 780 bp of intron 1 (Fig. 4, interval 4800-5600), compared with only 0.31 for the 5′ end of the promoter (Fig. 4, interval 1-800).
FIG. 4.
Quantitation of CpG frequency (observed/expected [obs/exp]) in a contig encompassing the promoter, exon 1, and the 5′ end of intron 1 of the rat p21WAF1/CIP1 gene. CpG frequency was computed over 800-bp intervals and shows a putative CpG island spanning bp 4800 to 7201 comprising the last 20 bp of exon 1 (located in the interval from bp 4800 to 5600) and the 5′ end of intron 1.
These observations suggest that p21WAF1/CIP1 expression could be inhibited by methylation of its promoter region. To investigate this possibility we made use of 5-AzaC, a cytosine analogue which is refractory to methylation and which can be used to relieve the inhibitory effects of methylation on gene expression. Rat-1 cells were cultured in the presence of 5-AzaC (5 μM), with fresh drug added every 24 h. Expression of p21WAF1/CIP1 protein was analyzed 4 days after initial addition of 5-AzaC. In the absence of 5-AzaC, cells failed to express p21WAF1/CIP1 protein even after X-ray treatment (Fig. 2B and 5). However, following 4 days of 5-AzaC treatment, p21WAF1/CIP1 expression was detected in Rat-1 cells (Fig. 5). X-ray treatment failed to elicit a further increase in p21WAF1/CIP1 expression, suggesting that 5-AzaC treatment alone was sufficient to cause a DNA damage response (37) and corresponding induction of elevated levels of p21WAF1/CIP1 expression. These results suggest that the lack of p21WAF1/CIP1 expression in Rat-1 cells is due to a block on transcription by methylation which can be relieved by 5-AzaC.
FIG. 5.
Effect of 5-AzaC treatment on p21WAF1/CIP1 expression in Rat-1 cells. Incubation of Rat-1 cells with 5-AzaC for 4 days resulted in high levels of p21WAF1/CIP1. X irradiation of 5-AzaC-treated Rat-1 cells showed little increase in p21WAF1/CIP1 expression.
To investigate this further, part of the Rat-1 p21WAF1/CIP1 intron 1 within the region of highest CpG frequency (3′ end of interval 4800-5600 and 5′ end of interval 5600-6400) (Fig. 4) was analyzed for methylation by sequencing of bisulfite-treated Rat-1 cellular DNA. Bisulfite treatment of single-stranded DNA converts unmethylated cytosine residues to uracil, while methylated cytosines remain nonreactive. Subsequent PCR amplifies uracil as thymine with only methylated cytosines being amplified as cytosine, and these changes can be identified by direct sequencing. Figure 6 shows the sequence generated for a 120-bp region containing 15 CpG dinucleotides. The total region analyzed comprised 330 bp encompassing 37 CpG dinucleotides. Bisulfite sequencing results for Rat-1 DNA were compared with those for REF52 DNA since REF52 cells express p21WAF1/CIP1 and, therefore, would not be expected to exhibit methylation of the p21WAF1/CIP1 promoter region. Both sequences were also compared with untreated DNA (Fig. 6, upper sequence). As expected, bisulfite-treated REF52 DNA (Fig. 6, REF52.bs, lower sequence) showed no methylation in the region analyzed since 100% of the cytosine residues were converted to uracil compared with the untreated sequence. In contrast, although many cytosine residues were also converted in bisulfite-treated Rat-1 DNA, all of the treated Rat-1 cytosines which were part of a CpG dinucleotide in 330 bp analyzed (total of 37 CpG) remained unreacted and, therefore, are methylated (Fig. 6, Rat-1.bs, middle sequence). Thus, the 5′UTR of the p21WAF1/CIP1 gene in Rat-1 cells appears to be extensively methylated exclusively at cytosine residues which form part of CpG dinucleotides. Together with induction of p21WAF1/CIP1 expression in Rat-1 by 5-AzaC, these results suggest that the lack of p21WAF1/CIP1 expression in Rat-1 cells, and hence the absence of a G1 arrest after X irradiation, is the result of transcriptional inhibition by extensive promoter methylation.
FIG. 6.
Bisulfite sequencing of part of the putative CpG island in the p21WAF1/CIP1 gene in Rat-1 cells. A 120-bp sequence containing 15 CpGs out of a total of 330 bp and 37 CpGs analyzed shown in a comparison of bisulfite-treated Rat-1 DNA (Rat-1.bs, middle sequence) with bisulfite-treated REF52 DNA (REF52.bs, lower sequence) and untreated DNA (upper sequence). Rat-1 cells contain methylated cytosine (detected as cytosine) at all the CpG dinucleotides analyzed, while all other cytosines were unmethylated (detected as thymine [boxed]). Bisulfite-treated REF52 exhibited only unmethylated cytosines (detected as thymine [boxed]) even at the CpG dinucleotides in this region.
Restoration of stable p53-dependent p21WAF1/CIP1 expression and G1 growth arrest to Rat-1 cells by a rat genomic clone containing the p21WAF1/CIP1 gene.
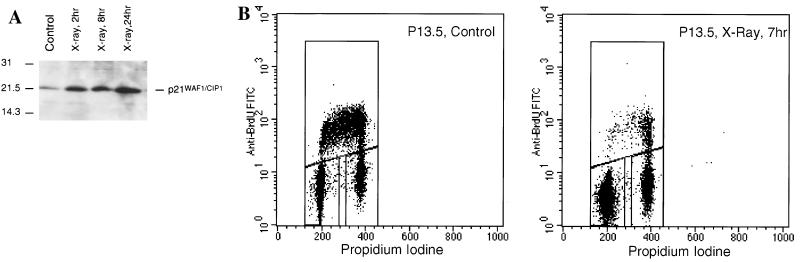
A PAC (277G17) containing the p21WAF1/CIP1 gene was isolated from the rat genomic DNA PAC library, RPC131 (obtained from the HGMP Resource Centre), after screening with a 700-bp probe corresponding to the 5′ end of intron 1 of the rat p21WAF1/CIP1 gene (see Materials and Methods). The 120-kb 277G17 PAC was cotransfected with pBABEpuro into Rat-1 cells, and stable transformants were selected by resistance to puromycin. Out of 16 puromycin-resistant Rat-1 clones isolated, one (P13.5) showed restoration of p21WAF1/CIP1 gene activity after X-ray treatment. Whereas p21WAF1/CIP1 protein was undetectable in parental Rat-1 cells (Fig. 2B) and was expressed at low levels in untreated P13.5 cells, its expression was induced as early as 2 h after X-ray treatment in P13.5 cells and continued to increase at 8 and 24 h after irradiation (Fig. 7A). Cell cycle analysis showed that untreated P13.5 cells exhibited a cell cycle profile similar to that of parental Rat-1 cells (compare Fig. 7B with Fig. 1B). However, after X irradiation, the majority of P13.5 cells were arrested in G1 (Fig. 7B), a finding not observed in parental Rat-1 cells (Fig. 1B). Furthermore, the induction of p21WAF1/CIP1 activity after X-ray treatment was p53 dependent, as both p21WAF1/CIP1 expression and the cell cycle block were lost after X-ray treatment of P13.5 cells expressing a dominant negative C-terminal fragment of p53 comprising amino acids 302 to 390 (clone P13.5-DN.A1) (Fig. 8). Similar results were obtained for another clone expressing a different dominant negative p53 construct containing an Arg→His mutation at position 175 (data not shown). Clearly, Rat-1 cells are competent for the induction of p53-dependent G1 growth arrest downstream of p21WAF1/CIP1 in response to radiation-induced DNA damage. These results clearly implicate the inhibition of p21WAF1/CIP1 expression by promoter methylation as the direct cause of the lack of growth arrest response in Rat-1 cells.
FIG. 7.
p21WAF1/CIP1 expression and a G1 arrest are restored after X-ray treatment of P13.5 cells, a clone of Rat-1 cells stably transfected with a 120-kb PAC containing a genomic copy of the rat p21WAF1/CIP1 gene. (A) A low level of background p21WAF1/CIP1 expression was observed in P13.5 cells which is significantly increased after X irradiation (12 Gy). Sizes are indicated in kilodaltons. (B) P13.5 cells exhibit a G1 arrest 7 h after X irradiation (12 Gy).
FIG. 8.
Expression and activity of p21WAF1/CIP1 in P13.5 cells are p53 dependent. (A) Induction of p21WAF1/CIP1 expression by X irradiation (12 Gy) in P13.5 cells is abrogated by expression of dominant negative p53 (P13.5-DN.A1 cells). Sizes are indicated in kilodaltons. (B) The cell cycle arrest exhibited by parental P13.5 cells at 7 h after X irradiation (12 Gy) is absent in P13.5-DN.A1 cells.
DISCUSSION
Rat-1 is a cell line which contains wild-type p53 (22) and is used frequently to assess transformation and for apoptosis studies (13, 16, 18, 20, 22, 32). We report here that the p53-inducible p21WAF1/CIP1 gene is not expressed in Rat-1 cells. The lack of a p53-induced cell cycle block via p21WAF1/CIP1 expression may contribute to the suitability of Rat-1 cells for such studies.
The effects of two forms of radiation, X ray and UVC, on Rat-1 cells were analyzed. Initially, the response of Rat-1 cells to X-ray and UVC at the cellular level was assessed. Whereas treatment with UVC caused rapid apoptosis, X irradiation had no effect on cell viability and failed to induce a G1 arrest, causing only a small proportion of cells to accumulate in G2. This is in contrast to our previous findings in U2OS, where X ray caused a p53-dependent growth arrest (1). However, X irradiation of Rat-1 cells was shown to stabilize the p53 protein, suggesting that the pathway responsible for signaling the presence of X-ray-induced DNA damage to p53 is intact in Rat-1 cells. X-ray treatment also induced expression of MDM2 protein in a p53-dependent manner, indicating that the p53 protein in Rat-1 cells is competent for transactivation of its target genes. However, X-ray treatment failed to induce expression of p21WAF1/CIP1 at either the protein or RNA level in Rat-1 cells. Previous reports have also shown a lack of p21WAF1/CIP1 induction in Rat-1 cells after X radiation (22) or during Myc-induced apoptosis (20). The lack of p21WAF1/CIP1 protein expression offers a potential explanation for the lack of radiation-induced growth arrest observed in this cell line. Similar results have been observed in CHO cells, commonly used in toxicology and transformation studies. Radiation treatment failed to induce p21WAF1/CIP1 expression or cause a growth arrest despite the presence of functionally wild-type p53 protein (39), raising the possibility that the p21WAF1/CIP1 promoter may also be hypermethylated in the CHO cell line.
We considered the possibility that the Rat-1 p21WAF1/CIP1 gene may contain a mutation in either of the p53 REs which could preclude p53 binding and prevent induction of p21WAF1/CIP1 expression by p53. However, sequence analysis confirmed that both p53 REs were identical to the published wild-type sequence (11). Furthermore, the possibility of a mutation in the p21WAF1/CIP1 coding region, which could potentially reduce the stability of the RNA, was also excluded by the demonstration of a wild-type Rat-1 p21WAF1/CIP1 sequence.
This led us to evaluate whether the lack of p21WAF1/CIP1 expression might be the result of hypermethylation of its promoter region. Analysis of the rat p21WAF1/CIP1 5′UTR sequence showed that a region encompassing the 3′ end of exon 1 and the 5′ end of intron 1 exhibited a frequency of CpG dinucleotide consistent with that of a CpG island (15). Although we have not formally identified the boundaries of the putative CpG island in rat cells, our findings are supported by comparison with the human p21WAF1/CIP1 gene in which a putative CpG island is believed to span essentially the same region of the gene (GenBank accession no. Z85996). To investigate whether the 5′UTR of the p21WAF1/CIP1 gene is methylated in Rat-1 cells, we assessed the effect on p21WAF1/CIP1 expression in these cells after treatment with 5-AzaC, a cytosine analogue which is refractory to methylation. Following treatment with 5-AzaC, Rat-1 cells expressed high levels of p21WAF1/CIP1, directly implicating hypermethylation as the cause of the p21WAF1/CIP1 transcriptional repression.
Part of the putative CpG island within the region of highest CpG frequency and containing 37 CpG dinucleotides was analyzed by sequencing bisulfite-treated DNA, which can distinguish between methylcytosine and unmethylated cytosines. Comparison of bisulfite-treated Rat-1 DNA with untreated DNA identified methylcytosine residues at all 37 CpG dinucleotides analyzed, whereas all other cytosine residues were unmethylated. Furthermore, REF52, a cell line which expresses p21WAF1/CIP1, was shown to contain unmethylated cytosines at these 37 CpG dinucleotides. These results support those obtained with 5-AzaC (above) that p21WAF1/CIP1 transcriptional expression is functionally inactivated in Rat-1 cells by extensive methylation of the putative CpG island in the 5′UTR. Similarly, promoter hypermethylation has also been reported to cause the transcriptional silencing of many genes whose functional inactivation may contribute to the neoplastic process (12, 31, 33, 35, 36). To our knowledge, however, this is the first report of functional inactivation of the p21WAF1/CIP1 gene by this mechanism.
We attempted to restore p53-dependent p21WAF1/CIP1 activity to Rat-1 cells. To this end we generated the P13.5 cell line by the transfection of Rat-1 cells with a 120-kb rat PAC containing the p21WAF1/CIP1 gene and extensive surrounding DNA sequence. Following treatment of P13.5 cells with X rays, we observed a rapid p53-dependent induction of p21WAF1/CIP1 protein and subsequent cell cycle arrest. Clearly, this indicates that the X-radiation-induced growth arrest pathway is intact in Rat-1 cells downstream of p21WAF1/CIP1. When considered together with our results indicating that the signaling pathway to p53 is functional and that p53 is competent for transactivation following X-radiation, these results specifically define the defect in the Rat-1 growth arrest pathway as the inhibition of p21WAF1/CIP1 expression by promoter methylation.
To date, almost no coding region p21WAF1/CIP1 mutations have been found in tumor cells, despite extensive screening of hundreds of various tumors (2, 25, 34, 38, 43). Hypermethylation of the p21WAF1/CIP1 promoter region may represent an alternative mechanism by which the p21WAF1/CIP1 gene can be inactivated. Rat-1 cells are used extensively for transformation, cell cycle, and apoptosis analyses. Exactly what role the absence of p21WAF1/CIP1 activity plays in these studies with Rat-1 cells is not clear. However, in light of our previous results with the U2OS cell line, in which sensitivity to UVC-induced apoptosis appeared to correlate with the level of p21WAF1/CIP1 expression (1), it is tempting to speculate that the Rat-1 cell line may be particularly amenable to the induction of apoptosis by virtue of its inability to express p21WAF1/CIP1. Interestingly, Ling et al. (22) reported that Rat-1 cells expressing exogenous Myc fail to express p21 and are more susceptible to radiation- induced apoptosis than are rat embryo cells which express p21 following radiation treatment (22). It would be of interest to see if expression of the p21WAF1/CIP1 gene is altered in other continuous cell lines used for apoptosis studies.
It is also interesting that our preliminary studies indicate that the polyomavirus oncoprotein middle T antigen generates smaller transformed colonies with the p21WAF1/CIP1-expressing P13.5 cells than with parental Rat-1 cells in an overgrowth focus assay. Whether this is due to the presence of p21WAF1/CIP1 or other genes present on the transfected PAC or other reasons is not clear at this time. In NIH 3T3 cells ectopic p21WAF1/CIP1 expression has also been observed to inhibit the formation of transformed foci by the ras oncogene (26). With respect to Rat-1 cells, it would be interesting to assess whether the p21WAF1/CIP1-expressing P13.5 cells and the p21WAF1/CIP1-negative parental Rat-1 cells differ in their transforming, cell cycle, and apoptotic responses to other oncogenes and stimuli.
ACKNOWLEDGMENTS
We thank Trevor Littlewood for the p53.302-90 and pBABEPuro plasmids, Xin Lu and Arnold Levine for the p21WAF1/CIP1 and MDM2 antibodies, respectively, and Parmjit Jat for the rat p21WAF1/CIP1 cDNA probe. We also thank Peng Yeong Woon, Pieter De Jong, and the HGMP Resource Centre, Hinxton, United Kingdom, for the rat PAC library, Derek Davies and Aaron Rae for the FACS analysis, and Gordon Peters and Martine Lomax for help and advice in preparation of the manuscript.
REFERENCES
- 1.Allan L A, Fried M. p53-dependent apoptosis or growth arrest induced by different forms of radiation in U2OS cells: p21WAF1/CIP1 repression in UV induced apoptosis. Oncogene. 1999;18:5403–5412. doi: 10.1038/sj.onc.1202931. [DOI] [PubMed] [Google Scholar]
- 2.Balbin M, Hannon G J, Pendas A M, Ferrando A A, Vizoso F, Fueyo A, Lopez-Otin C. Functional analysis of a p21WAF1,CIP1,SDI1 mutant (Arg94→Trp) identified in a human breast carcinoma. Evidence that the mutation impairs the ability of p21 to inhibit cyclin-dependent kinases. J Biol Chem. 1996;271:15782–15786. doi: 10.1074/jbc.271.26.15782. [DOI] [PubMed] [Google Scholar]
- 3.Bissonnette N, Hunting D J. p21-induced cycle arrest in G1 protects cells from apoptosis induced by UV-irradiation or RNA polymerase II blockage. Oncogene. 1998;16:3461–3469. doi: 10.1038/sj.onc.1201899. [DOI] [PubMed] [Google Scholar]
- 4.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 5.Canman C E, Kastan M B. Small contribution of G1 checkpoint control manipulation to modulation of p53-mediated apoptosis. Oncogene. 1998;16:957–966. doi: 10.1038/sj.onc.1201612. [DOI] [PubMed] [Google Scholar]
- 6.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 8.Duhig T, Ruhrberg C, Mor O, Fried M. The human Surfeit locus. Genomics. 1998;52:72–78. doi: 10.1006/geno.1998.5372. [DOI] [PubMed] [Google Scholar]
- 9.El-Deiry W S, Harper J W, O'Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 10.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry W S, Tokino T, Waldman T, Oliner J D, Velculescu V E, Burrell M, Hill D E, Healy E, Rees J L, Hamilton S R, et al. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- 12.Esteller M, Levine R, Baylin S B, Ellenson L H, Herman J G. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 13.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 14.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 16.Godwin A K, Lieberman M W. Early and late responses to induction of rasT24 expression in Rat-1 cells. Oncogene. 1990;5:1231–1241. [PubMed] [Google Scholar]
- 17.Gorospe M, Cirielli C, Wang X, Seth P, Capogrossi M C, Holbrook N J. p21(Waf1/Cip1) protects against p53-mediated apoptosis of human melanoma cells. Oncogene. 1997;14:929–935. doi: 10.1038/sj.onc.1200897. [DOI] [PubMed] [Google Scholar]
- 18.Han J W, Dionne C A, Kedersha N L, Goldmacher V S. p53 status affects the rate of the onset but not the overall extent of doxorubicin-induced cell death in rat-1 fibroblasts constitutively expressing c-Myc. Cancer Res. 1997;57:176–182. [PubMed] [Google Scholar]
- 19.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 20.Lenahan M K, Ozer H L. Induction of c-myc mediated apoptosis in SV40-transformed rat fibroblasts. Oncogene. 1996;12:1847–1854. [PubMed] [Google Scholar]
- 21.Li R, Waga S, Hannon G J, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371:534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- 22.Ling C C, Guo M, Chen C H, Deloherey T. Radiation-induced apoptosis: effects of cell age and dose fractionation. Cancer Res. 1995;55:5207–5212. [PubMed] [Google Scholar]
- 23.Lu X, Lane D P. Differential induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes? Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 24.Macleod K F, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–944. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 25.Malkowicz S B, Tomaszewski J E, Linnenbach A J, Cangiano T A, Maruta Y, McGarvey T W. Novel p21WAF1/CIP1 mutations in superficial and invasive transitional cell carcinomas. Oncogene. 1996;13:1831–1837. [PubMed] [Google Scholar]
- 26.Michieli P, Li W, Lorenzi M V, Miki T, Zakut R, Givol D, Pierce J H. Inhibition of oncogene-mediated transformation by ectopic expression of p21Waf1 in NIH3T3 cells. Oncogene. 1996;12:775–784. [PubMed] [Google Scholar]
- 27.Mor O, Read M, Fried M. p53 in polyoma virus transformed REF52 cells. Oncogene. 1997;15:3113–3119. doi: 10.1038/sj.onc.1201549. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhyay T, Roth J A. Induction of apoptosis in human lung cancer cells after wild-type p53 activation by methoxyestradiol. Oncogene. 1997;14:379–384. doi: 10.1038/sj.onc.1200835. [DOI] [PubMed] [Google Scholar]
- 29.Poluha W, Poluha D K, Chang B, Crosbie N E, Schonhoff C M, Kilpatrick D L, Ross A H. The cyclin-dependent kinase inhibitor p21 (WAF1) is required for survival of differentiating neuroblastoma cells. Mol Cell Biol. 1996;16:1335–1341. doi: 10.1128/mcb.16.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polyak K, Waldman T, He T C, Kinzler K W, Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 31.Qian X, Jin L, Kulig E, Lloyd R V. DNA methylation regulates p27kip1 expression in rodent pituitary cell lines. Am J Pathol. 1998;153:1475–1482. doi: 10.1016/S0002-9440(10)65735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resnitzky D. Ectopic expression of cyclin D1 but not cyclin E induces anchorage-independent cell cycle progression. Mol Cell Biol. 1997;17:5640–5647. doi: 10.1128/mcb.17.9.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson K D, Jones P A. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiohara M, el-Deiry W S, Wada M, Nakamaki T, Takeuchi S, Yang R, Chen D L, Vogelstein B, Koeffler H P. Absence of WAF1 mutations in a variety of human malignancies. Blood. 1994;84:3781–3784. [PubMed] [Google Scholar]
- 35.Stirzaker C, Millar D S, Paul C L, Warnecke P M, Harrison J, Vincent P C, Frommer M, Clark S J. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229–2237. [PubMed] [Google Scholar]
- 36.Takita J, Hayashi Y, Nakajima T, Adachi J, Tanaka T, Yamaguchi N, Ogawa Y, Hanada R, Yamamoto K, Yokota J. The p16 (CDKN2A) gene is involved in the growth of neuroblastoma cells and its expression is associated with prognosis of neuroblastoma patients. Oncogene. 1998;17:3137–3143. doi: 10.1038/sj.onc.1202232. [DOI] [PubMed] [Google Scholar]
- 37.Taylor E M, McFarlane R J, Price C. 5-Azacytidine treatment of the fission yeast leads to cytotoxicity and cell cycle arrest. Mol Gen Genet. 1996;253:128–137. doi: 10.1007/s004380050305. [DOI] [PubMed] [Google Scholar]
- 38.Terry L A, Boyd J, Alcorta D, Lyon T, Solomon G, Hannon G, Berchuck A, Beach D, Barrett J C. Mutational analysis of the p21/WAF1/CIP1/SDI1 coding region in human tumor cell lines. Mol Carcinog. 1996;16:221–228. doi: 10.1002/(SICI)1098-2744(199608)16:4<221::AID-MC6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 39.Tzang B S, Lai Y C, Hsu M, Chang H W, Chang C C, Huang P C, Liu Y C. Function and sequence analyses of tumor suppressor gene p53 of CHO.K1 cells. DNA Cell Biol. 1999;18:315–321. doi: 10.1089/104454999315376. [DOI] [PubMed] [Google Scholar]
- 40.Vater C A, Bartle L M, Dionne C A, Littlewood T D, Goldmacher V S. Induction of apoptosis by tamoxifen-activation of a p53-estrogen receptor fusion protein expressed in E1A and T24 H-ras transformed p53−/− mouse embryo fibroblasts. Oncogene. 1996;13:739–748. [PubMed] [Google Scholar]
- 41.Waga S, Hannon G J, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 42.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 43.Watanabe H, Fukuchi K, Takagi Y, Tomoyasu S, Tsuruoka N, Gomi K. Molecular analysis of the Cip1/Waf1 (p21) gene in diverse types of human tumors. Biochim Biophys Acta. 1995;1263:275–280. doi: 10.1016/0167-4781(95)00110-3. [DOI] [PubMed] [Google Scholar]
- 44.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]