Abstract
Gastrointestinal stromal tumors (GISTs) are primarily characterized by activating mutations of tyrosine kinase or platelet-derived growth factor receptor alpha. Although the revolutionary therapeutic outcomes of imatinib are well known, the long-term benefits of imatinib are still unclear. The effects of BRD9, a recently identified subunit of noncanonical BAF complex (ncBAF) chromatin remodeling complexes, in GISTs are not clear. In the current study, we evaluated the functional role of BRD9 in GIST progression. Our findings demonstrated that the expression of BRD9 was upregulated in GIST tissues. The downregulation or inhibition of BRD9 could significantly reduce cellular proliferation, and facilitates apoptosis in GISTs. BRD9 inhibition could promote PUMA-dependent apoptosis in GISTs and enhance imatinib activity in vitro and in vivo. BRD9 inhibition synergizes with imatinib in GISTs by inducing PUMA upregulation. Mechanism study revealed that BRD9 inhibition promotes PUMA induction via the TUFT1/AKT/GSK-3β/p65 axis. Furthermore, imatinib also upregulates PUMA by targeting AKT/GSK-3β/p65 axis. In conclusion, our results indicated that BRD9 plays a key role in the progression of GISTs. Inhibition of BRD9 is a novel therapeutic strategy in GISTs treated alone or in combination with imatinib.
Subject terms: Chemotherapy, Gastric cancer
Introduction
Statistical analyses suggest that GISTs (gastrointestinal stromal tumors) are one of the most common types of mesenchymal tumors, and GISTs originate from ICCs (interstitial cells of Cajal), further affecting the gastrointestinal tract [1, 2]. Mutations of KIT (tyrosine kinase) are responsible for 75–80% of GISTs, whereas 5–10% of confirmed cases occur due to activating mutations of PGDFRA (platelet-derived growth factor receptor alpha) [3, 4]. These mutations further affect downstream signaling pathways to promote tumorigenic cellular proliferation. Nevertheless, 10% of cases lack any evidence of mutations [5].
Imatinib is a significant tyrosine kinase inhibitor (TKI) that can be used as the first-line treatment for metastatic or unresectable GISTs, and its use leads to the progression-free survival of patients with GISTs [6, 7]. However, clinical analysis has also confirmed that more than 50% of patients develop imatinib resistance within 2 years of treatment [8, 9]. These patients develop secondary KIT and PDGFRA mutations via activation of alternative oncogenic signaling pathways [10]. These important bottlenecks can be easily overcome by offering second- and third-line treatments, such as the targeted TKI sunitinib [11]. Therefore, it is very important to understand the molecular mechanism underlying GIST malignancy to develop novel therapeutic options.
Bromodomains (BRDs) exhibit a wide range of catalytic and scaffolding functions due to their evolutionarily conserved protein-protein interaction modules [12]. One of the well-known functions of BRDs is the regulation of gene expression through selective recognition and binding associated with acetylated Lys residues [13]. Oncogenic mapping has revealed that these BRD-associated proteins are highly dysregulated in cancers [14]. However, many subunits still require further attention; among them, one of the largely unexplored subunits is the SWI/SNF complex of BRD9 (bromodomain-containing protein 9) [15]. The proteins that form a complex with bromodomain identify acetylated lysine residues on histones and are further responsible for promoting epigenetic changes in expression, such as transcription regulation, chromatin remodeling, and histone modification [16]. BRD9 also identifies butyryl lysine; however, its exact role in epigenetic modification still needs to be explored further [17]. By analyzing these epigenetic signatures, researchers further showed that BRD9 might be involved in gene transcription associated with SWI/SNF, DNA repair, and uncontrolled cellular differentiation [18]. Downregulation of BRD9 expression in AML (acute myeloid leukemia) may result in GI arrest [19]. Mutations in BRD9, as well as mutations in five other proteins associated with the SNF/SWI complex, are also correlated with a higher number of mutations and genomic instability, especially in lung cancer [20]. Previous studies have shown that the mammalian SWI/SNF chromatin remodeling complex exists in three components: canonical BAF (cBAF), PBAF, and a noncanonical complex (ncBAF), and BRD9 only in the ncBAF complex [21, 22]. Numerous studies have established that the SWI/SNF complex has altered in many tumor types [23]. Unlike the cBAF and pBAF complexes, the ncBAF complex has not been reported to have repeated mutations in cancer [23, 24]. In addition, a study has shown that BRD9 acts as a cofactor to stabilize the structure of the SS18-SSX fusion and to maintain its oncogenic transcription in synovial sarcoma [25]. Moreover, due to general toxicity issues with pan-BAF suppression strategies, targeting only the ncBAF complex seems to be highly specific for certain cancer types, including synovial sarcoma [25] and certain types of leukemia [26]. Therefore, in this study, we focused on the role of BRD9 in GISTs.
Targeted molecular therapies can be a novel approach in anticancer therapies. As a Bcl-2 family member, p53-upregulated modulator of apoptosis (PUMA) critically functions as a regulator of apoptotic activity [27]. However, apoptosis can also be induced without PUMA activation using a wide variety of nongenotoxic stimuli, such as UCN-01 (a pan-kinase inhibitor) [28], gefitinib and erlotinib (EGFR tyrosine kinase inhibitors; TKIs) [29], and TNF-α (tumor necrosis factor-α) [30]. The p53-independent induction of PUMA expression can be stimulated with the help of novel transcription factors, such as Forkhead Box O3a (FoxO3a) [28], p73 [31], or nuclear factor κB (NF-κB) [30]. Furthermore, PUMA, when induced, facilitates apoptosis by complementing BCL-XL, an antiapoptotic BCL-2 family member [32]. PUMA also activates pro-apoptotic members, such as Bak and Bax, which results in caspase activation and mitochondrial dysfunction [33].
In the current study, we investigated the correlation between the expression of BRD9 and the inhibition of cellular proliferation by promoting apoptosis. We also evaluated the role of kinases in the progression of GIST. The study further confirmed that the downregulation of BRD9 can serve as a novel therapeutic target.
Material and methods
Cell lines and reagents
The cell lines GIST-882 and GIST-T1 were obtained from the Shanghai Institute of Biological Science (Shanghai, China) and were grown in RPMI-1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% l-glutamine (Gibco, USA) in 5% CO2 at 37 °C. According to the previous study [34], PUMA-KO GIST-882 and GIST-T1 cell lines were generated by CRISPER/Cas 9. GSK602 and imatinib mesylate were purchased from Selleckchem (TX, USA). dBRD9-A was obtained from MedKoo Biosciences (NC, USA).
Immunohistochemistry (IHC)
The present study was approved by The Second Hospital of Jilin University ethics committee. After receiving informed consent from each selected donor, GIST specimens were collected from The Second Hospital of Jilin University. All the specimens were first analyzed through IHC, which was performed using anti-BRD9. The required IHC kit was obtained from Boster (Wuhan, China). The protocol was followed according to the manufacturer’s instructions on the kit. The statistical analysis was conducted using values obtained from estimating the intensity of staining and the staining area. The scale used to assess the staining intensity was negative (0), weak (1), moderate (2), and strong (3). The underlying staining area scores were as follows: none (0), 1–25% (1), 26–50% (2), 51–75% (3), and 76–100% (4). Accordingly, final scores were obtained by multiplying staining intensity scores and staining area scores.
RNA interference
The synthesis of specific BRD9 siRNAs (TGGAACACTTCCTCCGCCAG and TCAAGCTGATGTGTGATAAT) and p65 siRNA (GCTGCAGTTTGATGATGAA) was conducted by RiboBio (Guangzhou, China). Transfection of siRNAs was performed in these cells with Lipofectamine 2000 (Thermo, USA) according to the provided instructions. The cells were incubated for 48 h to study the knockdown efficiency of siRNA, and the same procedure was carried out using western blotting analysis.
Cell viability assay
Cell viability was examined using an MTT (methylthiazolyl diphenyl-tetrazolium bromide) assay kit (Beyotime, Shanghai, China). GIST-882 and GISR-T1 cells were seeded at a density of approximately 5 × 103 cells in each well (96-well plate). BRD9 siRNA transfection in 96-well plates was performed for toxicity analysis using 10 μL of 5 mg/mL MTT solution. The cells were incubated with MTT for 1 h with dimethyl sulfoxide (Sigma, USA). The absorbance was determined at 490 nm. The assessment of the IC50 (half inhibitory concentration) of GSK602 along with the combinational index (CI) of imatinib and GSK602 was performed after 48 h. CompuSyn software was used for the calculation of CI based on the equation of Chou-Talalay CI, which indicated the antagonistic effect.
Colony formation
GIST-882 and GIST-T1 cells were seeded in a six-well plate at a seeding density of 1 × 103 cells in each well and cultured for 14 days. Then, the cells were transfected with siRNA against BRD9. For the assessment of colony formation ability, cells were subjected to treatment with GSK602 for 48 h. The medium was replaced with a fresh medium, and the cells were allowed to grow for two full weeks. To assess colony formation, the cells were fixed for 20 min in 4% paraformaldehyde and stained with crystal violet (0.5%). The images of colonies were acquired and counting was performed using Image J software.
Western blot assay
Western blotting was performed as described in previous studies [35, 36]. Briefly, total protein was extracted with RIPA buffer (Sigma, MO, USA), followed by the addition of protease inhibitors from Bimake (TX, USA) and phosphate inhibitors (Service Bio, Wuhan, China). Total protein (50 μg) was separated using SDS-PAGE and transferred to PVDF membranes. Blocking of the membrane was performed using nonfat milk for 1 h. The membrane was then incubated with primary antibodies at 4 °C overnight. After washing with TBST three times, the blots were incubated with secondary antibodies. Detection of the bands was performed using ECL solution. Imaging was performed using a Bio-Rad Imaging system detector. β-actin was used as the control. The primary antibodies are list as follows: BRD9 (ab137245, Abcam, USA), β-actin (A5441, Sigma, USA), Cleaved PARP (5625, Cell Signaling Technology, USA), Cleaved caspase 3 (9661, Cell Signaling Technology), Cleaved caspase 8 (9496, Cell Signaling Technology), Cleaved caspase 9 (9507, Cell Signaling Technology), γH2AX (2595, Cell Signaling Technology), AKT (9272, Cell Signaling Technology), p-AKT (9646, Cell Signaling Technology), GSK-3β (4337, Cell Signaling Technology), p-GSK-3β (9323, Cell Signaling Technology), p65 (6956, Cell Signaling Technology), p-p65 (3033, Cell Signaling Technology), PUMA (4976, Cell Signaling Technology), Bim (2819, Cell Signaling Technology), Bcl-2 (15071, Cell Signaling Technology), Mcl-1 (39224, Cell Signaling Technology), Bax (2774, Cell Signaling Technology), Cox IV (4811, Cell Signaling Technology), cytochrome C (4272, Cell Signaling Technology).
Quantitative PCR (qPCR)
qPCR was performed based on previous studies [37, 38]. Total RNA was extracted with RNAiso Plus from TaKaRa (Dalian, China), and reverse transcription was performed using PrimeScript RT master mix from TaKaRa to form complementary DNA. The expression of genes was assessed through SYBR Premix Ex TaqTM (TaKaRa) on a StepOnePlus™ Real-Time PCR System, and the 2−ΔΔCt method was used for calculations. The expression of GAPDH was used for normalization. The primary primers are list as follows: BRD9, forward: 5′-ATGTTCCATGAAGCCTCCAG-3′, reverse: 5′-AGCTCCTTCTTCACCTTCCC-3′; PUMA, forward: 5′-ACGACCTCAACGCACAGTACG-3′, reverse: 5′-TCCCATGATGAGATTGTACAGGAC-3′; GAPDH forward: 5′-GCACCGTCAAGGCTGAGAAC-3′, reverse: 5′-TGGTGAAGACGCCAGTGGA-3′.
Xenograft
The animal experiments were approved by the local animal ethics committee of The Second Hospital of Jilin University. Female Nu/Nu mice (5–6-week-old) were kept in a sterile animal house with microisolator cages and had free access to sterile water and chow. The mice were injected subcutaneously in both flanks with 4 × 106 WT or PUMA-KO GIST-882 cells. The mice were observed to monitor the growth of the tumor for seven days. After one week, all the mice were randomized into four groups and subjected to GSK602 treatment at 10 mg/kg daily. For combination treatment, mice were randomized into four groups and treated with 2 mg/kg GSK602 and 50 mg/kg imatinib for 10 days. The growth of the tumor was estimated using calipers, and the formula ½ × length × width2 was used to calculate tumor volumes. Once the tumor reached the size of approximately 1 cm3, the mice were euthanized, and tumors were dissected. The separated tumors were fixed in formalin (10%) and paraffin-embedded. Then, 5-μm sections embedded in paraffin were immunostained with TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling) and cleaved caspase 3.
Statistical analysis
Prism VI software was used for statistical analyses. The significant difference between the two groups was assessed using a two-tailed t test, and for multiple groups, one-way ANOVA was used. The data are presented as the mean ± S.D. P < 0.05 indicated statistically significant differences between the groups.
Results
BRD9 is expressed at high levels in GIST tissues
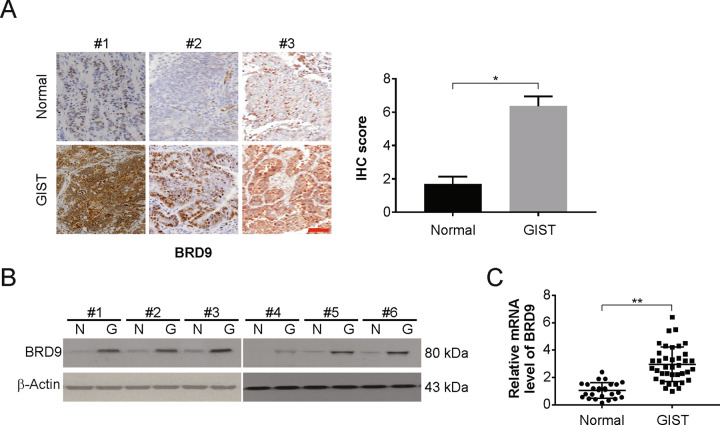
To investigate the functional role of BRD9 in GISTs, we validated the expression of BRD9 in clinically confirmed GIST specimens, which included 38 GIST tissues that were compared with 25 adjacent nontumor tissues. The analysis results of IHC and western blotting demonstrated the increased expression of BRD9 in GIST tissues compared with adjacent healthy, nontumor tissues (Fig. 1A and B). The mRNA levels were also increased in GIST samples (Fig. 1C). Furthermore, the correlation between the BRD9 expression and pathological significance in patients with GISTs was investigated. Positive staining of BRD9 was correlated with the size of tumors and the grade of risk (Table 1). However, the positive staining was not significant for gender, age, or the site of the tumors. These results further confirmed the involvement of BRD9 kinase in GIST progression.
Fig. 1. BRD9 is overexpressed in GISTs.
A BRD9 protein expression in GISTs and adjacent nontumor tissues was examined by IHC. Scale bar: 100 μm. B BRD9 protein expression in GISTs and adjacent nontumor tissues was examined by western blotting. C BRD9 mRNA level in GISTs and adjacent nontumor tissues was examined by real-time PCR. Results were expressed as means ± SD of three independent experiments. *P < 0.05; **P < 0.01.
Table 1.
The correlation between BRD9 level and clinicopathologic features of GIST patients.
| Clinicopathologic features | BRD9 expression | p value | |
|---|---|---|---|
| low (n = 19) | high (n = 19) | ||
| Gender | |||
| Male | 8 | 7 | 0.163 |
| Female | 11 | 12 | |
| Age | |||
| ≥60 | 7 | 9 | 0.278 |
| <60 | 12 | 10 | |
| Tumor site | |||
| Stomach | 13 | 12 | 0.143 |
| Intestinal | 6 | 7 | |
| Tumor size (cm) | |||
| <5 | 11 | 4 | 0.032 |
| ≥5 to <10 | 5 | 13 | |
| ≥10 | 3 | 2 | |
| NIH criteria | |||
| Low risk | 14 | 3 | 0.008 |
| Intermediate risk | 3 | 3 | |
| High risk | 2 | 13 | |
BRD9 inhibition suppressed cell growth and induced DNA damage and apoptosis in GIST cells
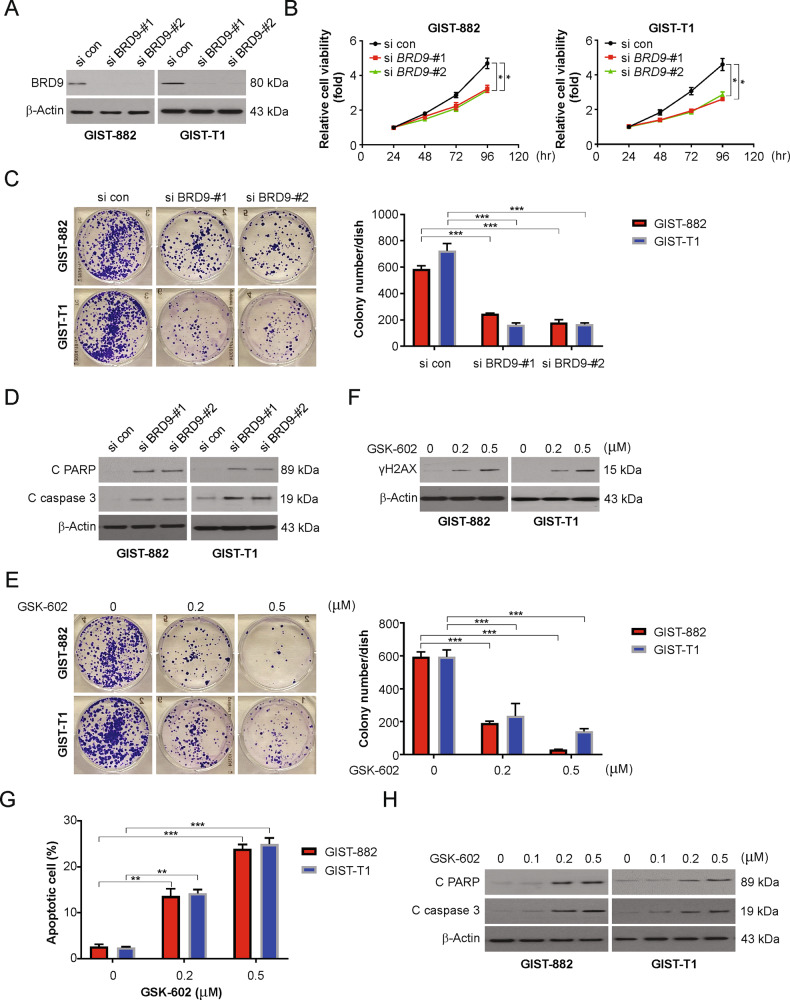
The function of BRD9 in the progression of GISTs was analyzed with two independent siRNAs that were used to silence the BRD9 gene in GIST-882 and GIST-T1 cells. The silencing of BRD9 was efficient in the siRNA-treated groups (Fig. 2A). The MTT assay further confirmed that silencing BRD9 was responsible for the reduced proliferative capacity of GIST-882 and GIST-T1 cells (Fig. 2B). This further confirmed that silencing BRD9 prominently reduced the tumorigenic proliferation of cells (Fig. 2C). Apoptotic activity was confirmed by analyzing the expression of the activity of proteins related to apoptosis and further observations of the increased expression of caspase 3 and cleaved PARP compared to those in the negative control group (Fig. 2D). The results indicate that the inhibition of BRD9 could induce GIST cellular apoptosis.
Fig. 2. BRD9 inhibition reduced GIST cell proliferation.
A Indicated cell lines were transfected with siRNA against BRD9 for 24 h, BRD9 protein level was analyzed by western blotting. B The viability of GIST-882 and GIST-T1 cells transfected with siRNA against BRD9 was determined by the MTT. C The viability of GIST-882 and GIST-T1 cells transfected with siRNA against BRD9 was determined by colony formation assay. D Indicated cell lines were transfected with siRNA against BRD9 for 24 h. Cleaved PARP and caspase 3 level were analyzed by western blotting. E Indicated cell lines were treated increasing concentration of GSK602 for 24 h. Cell viability was analyzed by colony formation assay. F Indicated cell lines were treated increasing concentration of GSK602 for 24 h. γH2AX level was analyzed by western blotting. G Indicated cell lines were treated increasing concentration of GSK602 for 24 h. Apoptosis was analyzed by fragment nuclei assay. H Indicated cell lines were treated increasing concentration of GSK602 for 24 h. Cleaved PARP and caspase 3 level was analyzed by western blotting. Results were expressed as means ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Furthermore, the pharmacological inhibition of BRD9 with the help of a small molecular inhibitor, GSK602, exerts a significant antitumorigenic effect in different types of cancers. The viability analysis showed that GSK602 reduced cellular viability in a dose-dependent manner (Fig. S1A). Furthermore, dBRD9-A, a BRD9 degrader, also inhibited cell viability in GIST cell lines (Fig. S1B). Noncancer cell lines HEK293 and NCM356 cells were highly resistant to BRD9 inhibition (Fig. S1A and B). A previous study showed that the synovial sarcoma cell line SYO1 was sensitive to BRD9 inhibition [25]. Our findings indicated that SYO1 cells were more sensitive to BRD9 inhibition than GIST cell lines (Fig. S1A and B). Colony formation analysis confirmed that GSK602-treated cells exhibited less colony formation than control cells (Fig. 2E). It was further hypothesized that the inhibition of BRD9 may result in cleavage of the DNA double helix during DNA replication [39, 40]. Based on this, the western blotting assay was carried out to analyze the expression of γH2AX, a surrogate marker confirming DNA damage [41]. As expected, the expression of γH2AX was increased in GIST-T1 and GIST-882 cells post-treatment with GSK602 or dBRD9-A, confirming increased damage to the DNA double helix during replication and hence increased cellular apoptosis (Figs. 2F and S1C). The degree of apoptosis was confirmed by treating GIST cells with GSK602 or dBRD9-A, and the proportion of cellular apoptosis was analyzed using nuclear fragmentation. The data suggested that the percentage of apoptosis increased in a dose-dependent manner (Figs. 2G and S1D). As shown in Figs. 2H and S1E, GSK602 or dBRD9-A treatment increased cleaved caspase 3 and PARP in GIST-T1 and GIST-882 cells. Therefore, the above data demonstrate that BRD9 inhibition is responsible for the induction of GIST cellular apoptosis and the reduced rate of proliferation in GIST cells.
PUMA is required for BRD9 inhibition-induced apoptosis in GIST cells
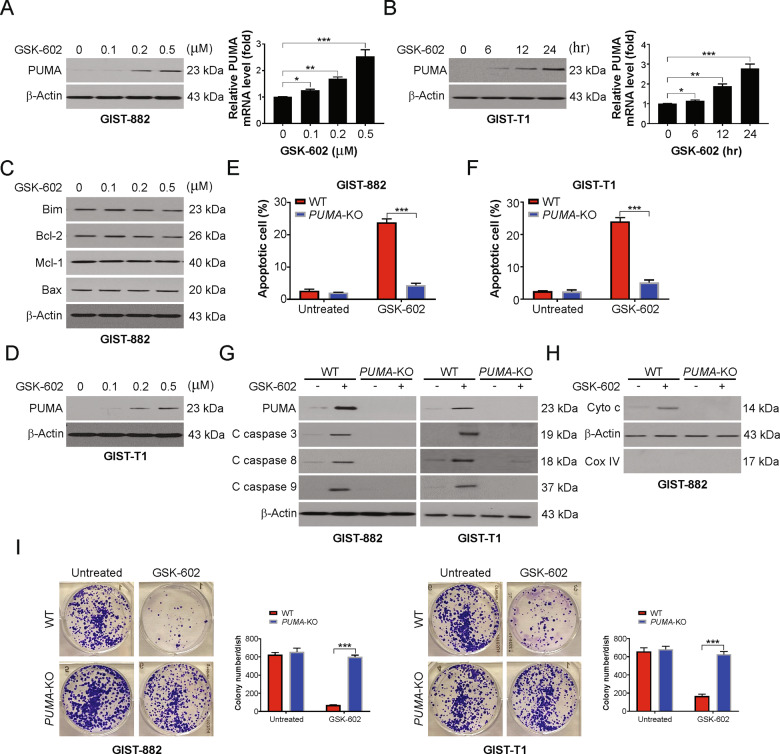
To further explore the mechanism underlying BRD9 inhibition-induced apoptosis, we investigated the protein levels of the Bcl-2 family after treating cells with GSK602. Our findings indicated that GSK602 treatment promotes the upregulation of PUMA (Fig. 3A and B). The highest PUMA protein expression, as well as its mRNA expression, was detected after 24 hr of GIST-882 cells treatment with 0.5 μM GSK602 (Fig. 3A and B). However, GSK602 treatment did not affect the expression of other pro-apoptotic Bcl-2 family members, including but not limited to Bim, Bid, and Bak; however, it certainly caused a decrease in the expression of antiapoptotic proteins, such as Mcl-1 and Bcl-XL (Fig. 3C). Upregulation of PUMA in GIST-T1 cells was also confirmed after treatment with GSK602 (Fig. 3D). Our findings also demonstrated that dBRD9-A induced PUMA in a dose- and time-dependent manner (Fig. S2A and B). Then, the function of PUMA in GSK602-induced apoptosis was investigated with PUMA-KO GIST-882 cell lines. The rate of apoptotic induction using 0.5 μM GSK602 was considerably lower in PUMA-KO cells than in WT GIST-882 cells (Fig. 3E). The GSK602-mediated apoptotic response dependent on PUMA was also observed in GIST-T1 cells (Fig. 3F). The absence of PUMA in GIST-882 after GSK602 induction confirmed the abrogation of mitochondrial events, including activation of caspases 3, 8, and 9 (Fig. 3G) and release of cytochrome c (Fig. 3H). In addition, PUMA depletion attenuated dBRD9-A-induced apoptosis, caspase activation, and cytochrome c release (Fig. S2C–F). Furthermore, the long-term survival of PUMA-KO cells was improved compared to that of WT GIST-882 cells after treatment with GSK602 (Fig. 3I). Thus, PUMA is crucial for the apoptotic induction of BRD9 inhibition in GIST cells.
Fig. 3. PUMA is required for BRD9 inhibition-induced apoptosis in GIST.
A GIST-882 cells were treated with GSK602 at indicated concentrations for 24 h. Left, PUMA expression was analyzed by western blotting. Right, PUMA mRNA level was analyzed by real-time PCR. B GIST-T1 cells were treated with GSK602 at indicated concentrations for 24 h. Left, PUMA expression was analyzed by western blotting. Right, PUMA mRNA level was analyzed by real-time PCR. C GIST-882 cells were treated with GSK602 at indicated concentrations for 24 h. The expression of indicated Bcl-2 family members was analyzed by western blotting. D GIST-T1 cells were treated with GSK602 at indicated concentrations for 24 h. The expression of PUMA was analyzed by western blotting. E WT and PUMA-KO GIST-882 cells were treated with 0.5 μM GSK602 for 24 h. Apoptosis was analyzed by fragment nuclei assay. F WT and PUMA-KO GIST-T1 cells were treated with 0.5 μM GSK602 for 24 h. Apoptosis was analyzed by fragment nuclei assay. G WT and PUMA-KO GIST-882 or GIST-T1 cells were treated with 0.5 μM GSK602 for 24 h. Indicated protein level was analyzed by western blotting. H Cytosolic fractions isolated from WT and PUMA-KO GIST-882 cells treated with 0.5 μM GSK602 for 24 hr were probed for cytochrome c by western blotting. β-actin and cytochrome oxidase subunit IV (Cox IV), which are expressed in cytoplasm and mitochondria, respectively, were analyzed as the control for loading and fractionation. I WT and PUMA-KO GIST-882 or GIST-T1 cell lines were treated 0.5 μM GSK602 for 24 h. Cell viability was analyzed by colony formation assay. Results were expressed as means ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
Next, we investigated how BRD9 inhibition-induced PUMA upregulation. Previous research has shown that BRD9 regulates the TUFT1/AKT signaling pathway in human hepatocellular carcinoma (HCC) [16]. We found the BRD9 inhibition downregulates TUFT1 expression and dephosphorylated AKT (Fig. S2G). Our findings further indicated that BRD9 inhibition suppressed GSK-3β phosphorylation at Ser9 [42], which inhibited GSK-3β kinase activity (Fig. S2G). Previous studies have shown p65 mediates PUMA induction by binding to the PUMA promoter [30]. BRD9 inhibition promoted the phosphorylation of p65 (Fig. S2G and H), and knockdown of p65 attenuated PUMA induction by BRD9 inhibition (Fig. S2I). Therefore, our findings suggest that BRD9 inhibition-induced PUMA upregulation via the TUFT1/AKT/GSK-3β/p65 signaling pathway.
PUMA-mediated antitumor effects of BRD9 inhibition in the xenograft mouse model
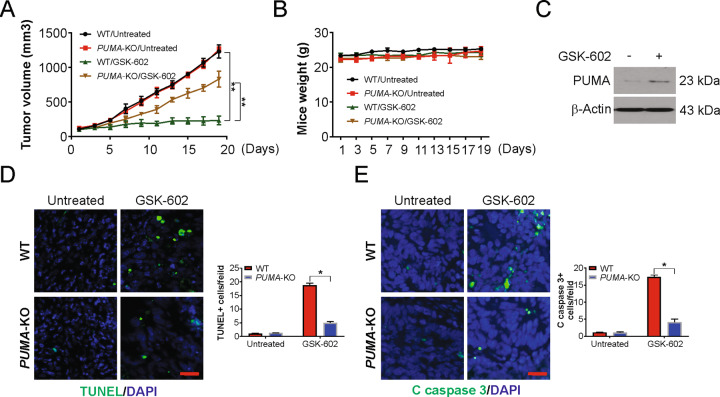
To investigate whether PUMA mediated the antitumor effect of GSK602 in vivo, WT and PUMA-KO cells were injected into nude mice through subcutaneous administration. After treatment with GSK602, we observed that PUMA-KO and WT tumors were not significantly different from those in the control groups (Fig. 4A and B). Our findings further indicated that GSK602 treatment suppressed WT tumor progression by approximately 70–80% (Fig. 4A). In contrast, tumors in the PUMA-KO group were observed to be significantly less sensitive to GSK602 than WT tumors (Fig. 4A and B), indicating that the absence of PUMA inhibited GSK602 antitumor activity. Western blotting results showed PUMA induction upon GSK602 treatment (Fig. 4C). Further analysis using TUNEL staining revealed that the apoptotic rate was significantly higher in WT tumors than in PUMA-KO tumors after treatment with GSK602, but apoptosis was not detected in the control mice (Fig. 4D). Staining of active caspase 3 confirmed PUMA-dependent apoptosis in GSK602-induced tumors (Fig. 4E). Thus, the above data demonstrate that the antiapoptotic, antitumor, and antiangiogenic functions of GSK602 in vivo depend largely on PUMA.
Fig. 4. PUMA is required for the antitumor effects of BRD9 inhibition.
A Nude mice were injected s.c. with 5 × 106 WT or PUMA-KO GIST-882 cells. After 7 days, mice were treated with 10 mg/kg GSK602 or the vehicle control for 10 consecutive days. Tumor volume at indicated time points after treatment was calculated and plotted with p values, n = 6 in each group. B Mice weight after treatment. C Mice with WT GIST-882 xenograft tumors were treated with 10 mg/kg GSK602 or the vehicle as in (A) for 4 consecutive days. The level of PUMA in three randomly selected tumors were analyzed by Western blotting. D Paraffin-embedded sections of WT or PUMA-KO tumor tissues from mice treated as in (C) were analyzed by TUNEL staining. Left, representative TUNEL staining pictures; Right, TUNEL-positive cells were counted and plotted, Scale bar: 25 μm. E Tissue sections from (D) were analyzed by active caspase 3 staining. Left, representative staining pictures; Right, active caspase 3-positive cells were counted and plotted, Scale bar: 25 μm. Results were expressed as means ± SD of three independent experiments. *P < 0.05.
BRD9 inhibition enhances the antitumor activity of imatinib via PUMA induction
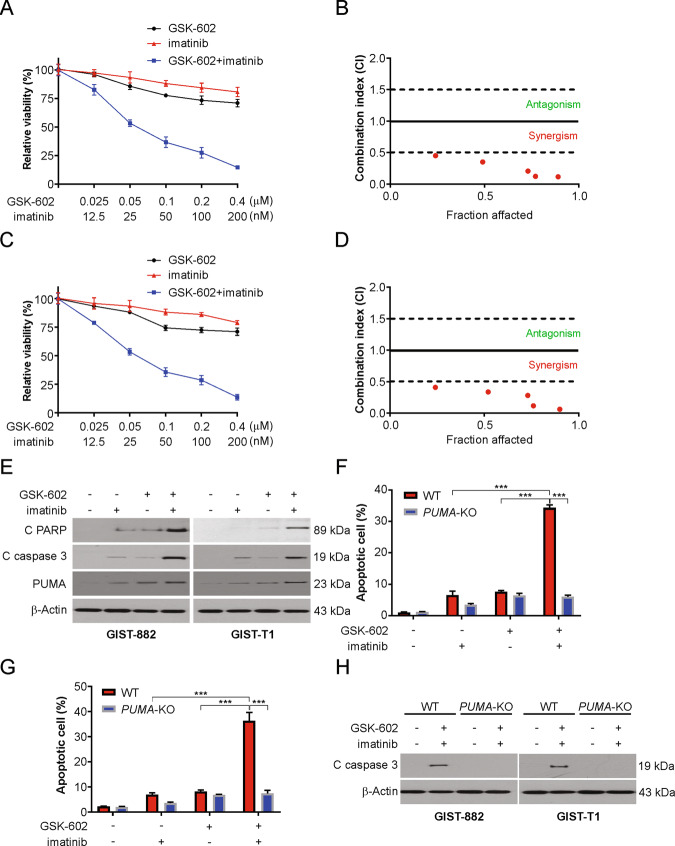
The combinational approach was validated by performing cellular proliferation analysis in both cell types (GIST-T1 and GIST-882 cells). A significant synergism between the BRD9 inhibition and imatinib, as indicated by the combination index (CI), was observed in these cell lines (Fig. 5A–D and S3A–S3D). However, no synergistic effect was found between GSK602 and imatinib in noncancer cells (Fig. S2E and F). Further analysis of the levels of apoptosis-related proteins confirmed the upregulation of cleaved PARP and caspase 3 in cells treated with the combination treatment (Figs. 5E and S5G). The results further confirmed that the combinational approach was more potent with more apoptotic cells (Fig. 5F–H). To assess the molecular mechanism in cells treated with a combinational approach, we evaluated whether induction of PUMA is primarily required before exposure to a combination treatment. Our results confirmed that silencing PUMA reduced apoptotic ability, further confirming that induction of PUMA is important for the combination treatment (Fig. 5F–H). Together, our findings indicate that PUMA is required for the combination of BRD9 inhibition and imatinib.
Fig. 5. BRD9 inhibition enhances the antitumor effect of imatinib in vitro.
A GIST-882 cells were treated with GSK602 combined with imatinib at indicated concentration for 48 h. Cell viability was analyzed by MTT. B Combination index (CI) and fraction affected of GSK602 and imatinib combining at different concentration in GIST-882 cells treated for 48 h were analyzed by the CompuSyn program (ComboSyn). C GIST-T1 cells were treated with GSK602 combined with imatinib at indicated concentration for 48 h. Cell viability was analyzed by MTT. D Combination index (CI) and fraction affected of GSK602 and imatinib combining at different concentration in GIST-T1 cells treated for 48 h were analyzed by the CompuSyn program (ComboSyn). E Indicated cells were treated with 100 nM GSK602 and 50 nM imatinib for 24 h. The indicated proteins were analyzed by western blotting. F WT and PUMA-KO GIST-882 cells were treated with 100 nM GSK602 and 50 nM imatinib for 24 h. Apoptosis was analyzed by fragment nuclei assay. G WT and PUMA-KO GIST-T1 cells were treated with 100 nM GSK602 and 50 nM imatinib for 24 h. Apoptosis was analyzed by fragment nuclei assay. H WT and PUMA-KO GIST-882 or GIST-T1 cells were treated with the combination of 100 nM GSK602 and 50 nM imatinib for 24 h. Caspase 3 level was analyzed by western blotting. Results were expressed as means ± SD of three independent experiments. ***P < 0.001.
Next, we investigated the mechanism by which imatinib promotes PUMA induction. Our results demonstrated that imatinib inhibits the BCR-ABL tyrosine kinase protein downstream signaling pathway [43], AKT, leading to GSK-3β activation and resulting in p65 phosphorylation (Fig. S4A and B). Moreover, cotreatment of GIST cells with BRD9 inhibition and imatinib was much more potent than single treatment (Fig. S4C and D). Therefore, the above results indicate the combined effect of BRD9 inhibition and imatinib in cotargeting the AKT/GSK-3β/p65 axis, and promoting PUMA induction.
PUMA mediates the combined effects of BRD9 inhibition and imatinib in vivo
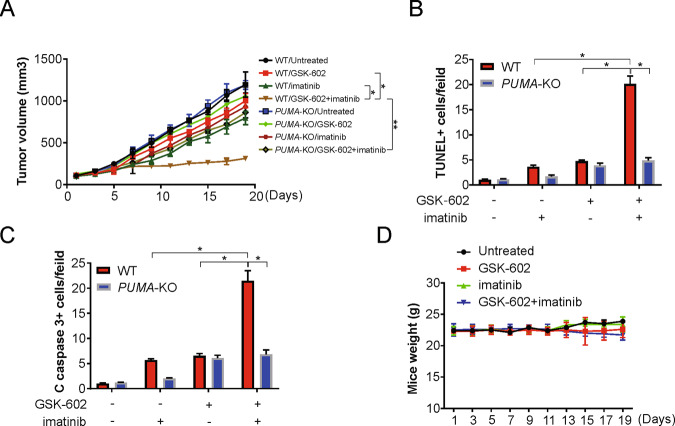
To investigate the induction of PUMA in vivo, we grafted nude mice with WT and PUMA-KO GIST-882 xenografts and found that the combined approach was more effective in reducing the progression of tumors than a single line of treatment. (Fig. 6A). However, it should also be noted that the growth inhibition of the combinational approach was largely abolished in PUMA-KO tumors (Fig. 6A), which was also related to a reduced rate of apoptosis, as detected by TUNEL staining (Fig. 6B) and cleaved caspase 3 staining (Fig. 6C). In conclusion, administration of GSK602 and imatinib alone or in combination did not cause a remarkable change in mouse body weight, indicating that there was no detectable toxicity due to these treatments (Fig. 6D). Thus, these results indicate that the effects of GSK602 in combination with imatinib are mediated by PUMA in vitro and in vivo.
Fig. 6. GSK602 enhances the antitumor effect of imatinib in vivo.
A Nude mice were injected s.c. with 5 × 106 WT or PUMA-KO GIST-882 cells. After 7 days, mice were treated with 2 mg/kg GSK602 daily, 50 mg/kg imatinib, or their combination for 10 consecutive days. Tumor volume at indicated time points after treatment was calculated and plotted with p values for indicated comparisons, n = 6 in each group. B Mice weight after treatment. C Mice with WT or PUMA-KO GIST-882 xenograft tumors were treated with 2 mg/kg GSK602, 50 mg/kg imatinib, or their combination as in (A) for 4 consecutive days. Paraffin-embedded sections were analyzed by TUNEL staining. TUNEL-positive cells were counted and plotted. D Tissue sections from (C) were analyzed by cleaved caspase 3 staining. Cleaved caspase 3-positive cells were counted and plotted. Results were expressed as means ± SD of three independent experiments. *P < 0.05; **P < 0.01.
Discussion
The detection of BRDs was confirmed in almost forty-two different proteins expressed in mammalian cells that are responsible for genetic expression and epigenetic changes through multiple mechanisms [44]. For example, BRD-containing proteins can either function as scaffolds to promote protein assembly, can act as transcription factors, or can be coregulated [45]. Furthermore, proteins harboring BRDs are primarily reported to be dysregulated in many cancers. However, the exact role of BRD9 and the underlying mechanism of action have yet to be fully explored. In the current study, we investigated the role of BRD9 in GISTs. Our results demonstrated that the level of BRD9 was highly increased in GIST specimens, and correlated with the increased size of the tumor. In addition, our results indicated that BRD9 knockdown or inhibition suppresses GIST proliferation and promotes PUMA-dependent apoptosis. Our findings also showed that BRD9 inhibition enhances the antitumor effects of imatinib, and PUMA is required for the combination effects. The chemosensitization effects of BRD9 inhibition and imatinib are mediated by PUMA through cotargeting the AKT/GSK-3β/NF-κB axis.
Bromodomain proteins can further be subdivided into two families, the BET and non-BET families, on the basis of their structural parameters [46]. BET proteins are directly associated with transcription and maintaining the effective progression of the cellular cycle [46]. During the last couple of years, several small molecular inhibitors were detected that primarily target the expression of BET proteins and were developed due to their anticancer effects both in vivo and in vitro [47, 48]. Currently, these inhibitors are being clinically investigated for use in solid and hematopoietic cancers [44]. The inhibition of cellular expression by reduced expression of BRD9 has yet to be fully studied; hence, there are only a few available reports.
Silencing of the expression of BRD9 may result in the reduced progression of tumor development and an increased rate of cellular apoptosis [49]. BRD9 is critical for prostate cancer progression by regulating androgen receptor signaling [50]. In an AML mouse model, BI-9564-treated mice exhibited a significant decrease in tumor growth and improved survival [51]. Inhibition of BRD9 by I-BRD9 in Kasumi-1 cells causes a decrease in the expression of several genes associated with cancer [52, 53]. Mammalian SWI/SNF (mSWI/SNF) complexes are ATP-dependent chromatin remodeling agents that can regulate genomic structure and DNA accessibility, thereby enabling timely and appropriate control of gene expression [24]. They are assembled from the combination of products of 29 total genes into three final forms of complexes: canonical BAF (cBAF), PBAF (polybromo-associated BAF complexes), and noncanonical BAF (ncBAF), with specific subtypes specifying different complexes, such as BRD7, ARID2, and PBRM1 in PBAF complexes, DPF2 and ARID1A/ARID1B in cBAF complexes, and BRD9 and GLTSCR1/GLTSCR1L in ncBAF complexes [21, 22]. Importantly, a noncanonical BRD9-containing BAF chromatin remodeling complex plays a key role in naïve pluripotency regulation in mouse embryonic stem cells [54]. A previous study revealed that BRD9 degradation reverses the expression of oncogenes in synovial sarcoma [25]. Therefore, in this study, we investigated the role of BRD9 in another type of soft tissue sarcoma, GISTs.
Furthermore, we also proposed the role of PUMA induction in facilitating a higher rate of cellular apoptosis, which can be useful in detecting chemosensitivity. We already confirmed that induction of PUMA can increase the differential sensitivity of cancers to EGFR TKIs, especially for cancers of the neck and head, whereas the absence of induction may be associated with resistance to EGFR TKIs [29]. Additionally, enhanced expression of PUMA can aid in better prognosis in patients undergoing chemotherapy [55, 56]. This hypothesis was further confirmed with a recent study, wherein a better response was detected in patients undergoing chemotherapy after PUMA induction, further confirming it to be a better strategy for reducing GIST progression.
Imatinib is a tyrosine kinase (TK) inhibitor that can competitively block the ATP-binding site of the TK receptor, thereby inhibiting a variety of TKs, including PDGFRA and c-Kit, and subsequently suppressing tumor growth and inhibiting signal transduction [57, 58]. Imatinib is now the standard first-line drug for patients with GIST [58]. However, the widespread use of imatinib in clinical cases is related to the emergence of secondary drug resistance, which limits the effectiveness of chemotherapy drugs [58, 59]. These bottlenecks can be resolved with the help of novel targeted therapeutics and alternative medicine approaches [60]. The expression of KITs can be significantly regulated through epigenetic alterations [61]. Previous studies showed that administration of imatinib alone can lead to mild DNA damage [62]. In this study, we investigated the synergistic effect of BRD9 inhibition and imatinib. The results demonstrated that the combination of imatinib and BRD9 inhibition promotes effective cellular apoptosis due to AKT inhibition, leading to GSK-3β/NF-κB activation and resulting in PUMA induction, thus achieving better antitumor activity with increased apoptosis and reduced cellular proliferation. The data obtained from the present study further suggest that the combinational approach can be clinically evaluated for better therapeutic outcomes.
In conclusion, our findings demonstrated the significance of BRD9 expression in GISTs in correlating tumor size and risk grade. Moreover, our study also suggests BRD9 inhibition as a potential mechanism to suppress tumorigenic progression and reduce associated malignancy. BRD9 inhibition sensitized GISTs to imatinib by cotargeting the AKT/GSK-3β/NF-κB axis, leading to PUMA induction. PUMA is required for the antitumor effects of the combination. Thus, targeting BRD9 can be a novel therapeutic approach with better clinical outcomes in the form of reduced progression and increased life expectancy in GIST patients.
Supplementary information
Author contributions
J.M. and P.S. conceived and designed the study. J.M., X.S., Z.Z., H.S., and P.S. carried out the experimental validation. J.M. and P.S. drafted and revised the manuscript. All authors have read and approved the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Ethics statement
This study was approved by the ethical review board of The Second Hospital of Jilin University. All of the patients were given and accepted informed consent form prior to their enrollment.
Footnotes
Edited by Professor Boris Zhivotovsky
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-025-07756-0
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/5/2025
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41419-025-07756-0
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-04186-6.
References
- 1.Parab TM, DeRogatis MJ, Boaz AM, Grasso SA, Issack PS, Duarte DA, et al. Gastrointestinal stromal tumors: a comprehensive review. J Gastrointest Oncol. 2019;10:144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishida T, Yasumasa K. [Target-based therapy againstgastrointestinal stromal tumors–from molecular diagnosis to molecular targettherapy]. Gan To Kagaku Ryoho. 2003;30:1071–8. [PubMed] [Google Scholar]
- 3.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865–78. [DOI] [PubMed] [Google Scholar]
- 4.Mei L, Smith SC, Faber AC, Trent J, Grossman SR, Stratakis CA, et al. Gastrointestinal stromal tumors: the GIST of precision medicine. Trends Cancer. 2018;4:74–91. [DOI] [PubMed] [Google Scholar]
- 5.Oppelt PJ, Hirbe AC, Van Tine BA. Gastrointestinal stromal tumors (GISTs): point mutations matter in management, a review. J Gastrointest Oncol. 2017;8:466–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopes LF, Bacchi CE. Imatinib treatment for gastrointestinal stromal tumour (GIST). J Cell Mol Med. 2010;14:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li GZ, Raut CP. Targeted therapy and personalized medicine in gastrointestinal stromal tumors: drug resistance, mechanisms, and treatment strategies. Onco Targets Ther. 2019;12:5123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H, Liang H, Zhan ZL, Li H, Ren XB, Hao XS. Surgical outcomes of gastrointestinal stromal tumors of the stomach: a single unit experience in the era of targeted drug therapy. Med Oncol. 2012;29:941–7. [DOI] [PubMed] [Google Scholar]
- 9.Eisenberg BL, Trent JC. Adjuvant and neoadjuvant imatinib therapy: current role in the management of gastrointestinal stromal tumors. Int J Cancer. 2011;129:2533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Napolitano A, Vincenzi B. Secondary KIT mutations: the GIST of drug resistance and sensitivity. Br J Cancer. 2019;120:577–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly CM, Gutierrez Sainz L, Chi P. The management of metastatic GIST: current standard and investigational therapeutics. J Hematol Oncol. 2021;14:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer. 2018;17:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein K. Bromodomain protein inhibition: a novel therapeutic strategy in rheumatic diseases. RMD Open. 2018;4:e000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mita MM, Mita AC. Bromodomain inhibitors a decade later: a promise unfulfilled? Br J Cancer. 2020;123:1713–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Liao Y, Tang L. Targeting BRD9 for cancer treatment: a new sStrategy. Onco Targets Ther. 2020;13:13191–13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dou C, Sun L, Wang L, Cheng J, Wu W, Zhang C, et al. Bromodomain-containing protein 9 promotes the growth and metastasis of human hepatocellular carcinoma by activating the TUFT1/AKT pathway. Cell Death Dis. 2020;11:730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flynn EM, Huang OW, Poy F, Oppikofer M, Bellon SF, Tang Y, et al. A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications. Structure. 2015;23:1801–14. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Wang S, Troisi EC, Howard TP, Haswell JR, Wolf BK, et al. BRD9 defines a SWI/SNF sub-complex and constitutes a specific vulnerability in malignant rhabdoid tumors. Nat Commun. 2019;10:1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohmann AF, Martin LJ, Minder JL, Roe JS, Shi J, Steurer S, et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat Chem Biol. 2016;12:672–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H, Wang Y, Li Q, Fei X, Ma H, Hu R. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett. 2019;446:81–89. [DOI] [PubMed] [Google Scholar]
- 21.Mashtalir N, Suzuki H, Farrell DP, Sankar A, Luo J, Filipovski M, et al. A structural model of the endogenous human BAF complex informs disease mechanisms. Cell. 2020;183:802–17. e824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE, et al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018;175:1272–88. e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helming KC, Wang X, Roberts CWM. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell. 2014;26:309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mittal P, Roberts CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol. 2020;17:435–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brien GL, Remillard D, Shi J, Hemming ML, Chabon J, Wynne K, et al. Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. Elife. 2018;7:e41305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel BC, D’Avino AR, Cassel SH, Mashtalir N, McKenzie ZM, McBride MJ, et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol. 2018;20:1410–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27:S71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudgeon C, Wang P, Sun X, Peng R, Sun Q, Yu J, et al. PUMA induction by FoxO3a mediates the anticancer activities of the broad-range kinase inhibitor UCN-01. Mol Cancer Ther. 2010;9:2893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Q, Ming L, Thomas SM, Wang Y, Chen ZG, Ferris RL, et al. PUMA mediates EGFR tyrosine kinase inhibitor-induced apoptosis in head and neck cancer cells. Oncogene. 2009;28:2348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P, Qiu W, Dudgeon C, Liu H, Huang C, Zambetti GP, et al. PUMA is directly activated by NF-kappaB and contributes to TNF-alpha-induced apoptosis. Cell Death Differ. 2009;16:1192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Young A, Zhang J, Chen X. P73 tumor suppressor and its targets, p21 and PUMA, are required for madin-darby canine kidney cell morphogenesis by maintaining an appropriate level of epithelial to mesenchymal transition. Oncotarget. 2015;6:13994–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell KJ, Tait SWG. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018;8:180002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai H, Pang YP, Ramirez-Alvarado M, Kaufmann SH. Evaluation of the BH3-only protein Puma as a direct Bak activator. J Biol Chem. 2014;289:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang C, Shi S, Su Y, Tong JS, Li L. P2X7R promotes angiogenesis and tumour-associated macrophage recruitment by regulating the NF-kappaB signalling pathway in colorectal cancer cells. J Cell Mol Med. 2020;24:10830–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu R, Tong JS. miR-126 reduces trastuzumab resistance by targeting PIK3R2 and regulating AKT/mTOR pathway in breast cancer cells. J Cell Mol Med. 2020;24:7600–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Tan X, Luo J, Yao H, Si Z, Tong JS. The miR-30a-5p/CLCF1 axis regulates sorafenib resistance and aerobic glycolysis in hepatocellular carcinoma. Cell Death Dis. 2020;11:902. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Tan X, Zhang Z, Liu P, Yao H, Shen L, Tong JS. Inhibition of EZH2 enhances the therapeutic effect of 5-FU via PUMA upregulation in colorectal cancer. Cell Death Dis. 2020;11:1061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Zhou Q, Huang J, Zhang C, Zhao F, Kim W, Tu X, et al. The bromodomain containing protein BRD-9 orchestrates RAD51-RAD54 complex formation and regulates homologous recombination-mediated repair. Nat Commun. 2020;11:2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiu LY, Gong F, Miller KM. Bromodomain proteins: repairing DNA damage within chromatin. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–86. [DOI] [PubMed] [Google Scholar]
- 42.Cai G, Wang J, Xin X, Ke Z, Luo J. Phosphorylation of glycogen synthase kinase-3 beta at serine 9 confers cisplatin resistance in ovarian cancer cells. Int J Oncol. 2007;31:657–62. [PubMed] [Google Scholar]
- 43.Atfi A, Abecassis L, Bourgeade MF. Bcr-Abl activates the AKT/Fox O3 signalling pathway to restrict transforming growth factor-beta-mediated cytostatic signals. EMBO Rep. 2005;6:985–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stathis A, Bertoni F. BET proteins as targets for anticancer treatment. Cancer Disco. 2018;8:24–36. [DOI] [PubMed] [Google Scholar]
- 45.Fu LL, Tian M, Li X, Li JJ, Huang J, Ouyang L, et al. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget. 2015;6:5501–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrikopoulou A, Liontos M, Koutsoukos K, Dimopoulos MA, Zagouri F. The emerging role of BET inhibitors in breast cancer. Breast. 2020;53:152–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, Vakoc CR. Targeting cancer cells with BET bromodomain inhibitors. Cold Spring Harb Perspect Med. 2017;7:a026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou S, Zhang S, Wang L, Huang S, Yuan Y, Yang J, et al. BET protein inhibitor JQ1 downregulates chromatin accessibility and suppresses metastasis of gastric cancer via inactivating RUNX2/NID1 signaling. Oncogenesis. 2020;9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bevill SM, Olivares-Quintero JF, Sciaky N, Golitz BT, Singh D, Beltran AS, et al. GSK2801, a BAZ2/BRD9 bromodomain inhibitor, synergizes with BET inhibitors to induce apoptosis in triple-negative breast cancer. Mol Cancer Res. 2019;17:1503–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alpsoy A, Utturkar SM, Carter BC, Dhiman A, Torregrosa-Allen SE, Currie MP, et al. BRD9 Is a critical regulator of androgen receptor signaling and prostate cancer progression. Cancer Res. 2021;81:820–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin LJ, Koegl M, Bader G, Cockcroft XL, Fedorov O, Fiegen D, et al. Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor. J Med Chem. 2016;59:4462–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kataoka TR, Kumanogoh A, Hirata M, Moriyoshi K, Ueshima C, Kawahara M, et al. CD72 regulates the growth of KIT-mutated leukemia cell line Kasumi-1. Sci Rep. 2013;3:2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Del Gaudio N, Di Costanzo A, Liu NQ, Conte L, Migliaccio A, Vermeulen M, et al. BRD9 binds cell type-specific chromatin regions regulating leukemic cell survival via STAT5 inhibition. Cell Death Dis. 2019;10:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gatchalian J, Malik S, Ho J, Lee DS, Kelso TWR, Shokhirev MN, et al. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat Commun. 2018;9:5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knickelbein K, Tong J, Chen D, Wang YJ, Misale S, Bardelli A, et al. Restoring PUMA induction overcomes KRAS-mediated resistance to anti-EGFR antibodies in colorectal cancer. Oncogene. 2018;37:4599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen D, Wei L, Yu J, Zhang L. Regorafenib inhibits colorectal tumor growth through PUMA-mediated apoptosis. Clin Cancer Res. 2014;20:3472–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laurent M, Brahmi M, Dufresne A, Meeus P, Karanian M, Ray-Coquard I, et al. Adjuvant therapy with imatinib in gastrointestinal stromal tumors (GISTs)-review and perspectives. Transl Gastroenterol Hepatol. 2019;4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Essat M, Cooper K. Imatinib as adjuvant therapy for gastrointestinal stromal tumors: a systematic review. Int J Cancer. 2011;128:2202–14. [DOI] [PubMed] [Google Scholar]
- 59.Bauer S, Joensuu H. Emerging Agents for the Treatment of Advanced, Imatinib-Resistant Gastrointestinal Stromal Tumors: Current Status and Future Directions. Drugs. 2015;75:1323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iqbal N, Iqbal N. Imatinib: a breakthrough of targeted therapy in cancer. Chemother Res Pr. 2014;2014:357027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dahl C, Abildgaard C, Riber-Hansen R, Steiniche T, Lade-Keller J, Guldberg P. KIT is a frequent target for epigenetic silencing in cutaneous melanoma. J Invest Dermatol. 2015;135:516–24. [DOI] [PubMed] [Google Scholar]
- 62.Dinis J, Silva V, Gromicho M, Martins C, Laires A, Tavares P, et al. DNA damage response in imatinib resistant chronic myeloid leukemia K562 cells. Leuk Lymphoma. 2012;53:2004–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.