Abstract
Objective
Increased fatty acid and triglyceride synthesis in liver, majorly modulated by Sterol Regulator Elementing Binding Protein 1c (SREBP1c), is one of the main features of non-alcoholic fatty liver disease (NAFLD). In the present study, we aimed to identify the relation between SREBP1c and autophagy mediated lipid droplet (LD) catabolism in oleic acid (OA) induced lipid accumulation.
Methods
Increased LD formation and SREBP1c induction were identified in hepatocytes (AML12 cells) following the OA administration. SREBP1c level was reduced through siRNA against SREBP1c. The amount and the size of LDs were determined by BODIPY, while protein and mRNA expressions were identified by immunoblotting and qRT-PCR, respectively. LD-lysosome colocalization was determined with immunofluorescence.
Results
Increased LD formation and SREBP1c levels were determined at 0.06 mM OA concentration. SREBP1c silencing reduced the number of LDs, while increasing mRNA levels of PPARα. On the other hand, SREBP1c silencing in non-OA and OA treated cells enhanced autophagy mediated LD catabolism.
Conclusion
Our results implicate the effect of SREBP1c deficiency in modulating PPARα signaling and autophagy mediated LD catabolism against OA induced lipid accumulation.
Keywords: Lipid accumulation, SREBP1c, Autophagy, Non-alcoholic fatty liver disease
Abbreviations: FASN, Fatty acid synthase; LAMP1, Lysosomal-associated membrane protein 1; LC3, Microtubule-Associated Protein Light Chain 3; LD, Lipid droplet; NAFLD, Nonalcoholic fatty liver disease; OA, Oleic acid; PA, Palmitic acid; PPARα, Peroxisome proliferator activated receptor alpha; SCD-1, Stereoyl-CoA desaturase-1; SREBP, Sterol regulatory element binding protein
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is a metabolic disorder in which lipid accumulation occurs in the liver independent of viral infection or alcohol consumption [1]. The increasing prevelence of disease, especially in western countries, is related to liver-dependent disorders as well as affecting extrahepatic organs and cellular regulatory mechanism [2]. Simple steatosis, which is an early phase of NAFLD development, refers the triglyceride accumulation in hepatocytes that drives the transition to nonalcoholic steatohepatitis (NASH), fibrosis and cirrhosis [3]. Hence, the underlying mechanisms that enhance hepatic lipid accumulation have a crucial role in steatosis development. Oleic acid (OA) and palmitic acid (PA), which are found at the highest levels in the diet, are the fatty acids found at the highest levels in individuals with NAFLD and contribute to the pathogenesis of the disease. OA is a fatty acid that stimulates steatosis and production of reactive oxygen species that causes the progression of NAFLD. Studies have determined that a number of hepatocyte phenotypes exposed to OA application exhibit steatotic morphologies [4].
The sterol regulatory element binding proteins (SREBPs) encoded by the SREBF1 and SREBF2 genes are transcription factors that control lipid metabolism. SREBPs have three isoforms, each with unique effects on lipid metabolism: SREBP1a, SREBP1c, and SREBP2 [5]. SREBP1c regulates the transcription of genes that modulate the fatty acid and triglyceride synthesis, including FASN and SCD-1. SREBP2 is specifically responsible for cholesterol synthesis, while SREBP1a targets the genes of both pathways [6]. Both animal and cell culture studies have shown that deficiency of SREBP1c attenuates the risk of metabolic diseases such as atherosclerosis, obesity and NAFLD [7].
Autophagy is an intracellular degradation system that enables the removal of macromolecules and organelles by transporting them into lysosomes. In addition to being an intracellular degradation system, autophagy is a system that has roles on metabolic pathways such as lipid catabolism [8]. Autophagy-mediated degradation of cellular lipids is defined as lipophagy. It is known that autophagic activity increases in the liver with steatosis in order to reduce the excess lipid concentration [9]. On the contrary, abnormalities in autophagic system might lead the steatosis development in the liver. In addition, liver biopsy samples obtained from individuals with NAFLD have demonstrated an impairment in lipophagic activity [8].
Despite the importance of understanding the mechanisms modulating steatosis in NAFLD pathogenesis, current literature is not fully enough [10]. Although SREBP1c is a well-identified transcription factor involving in NAFLD progression by induction of fatty acid and triglyceride synthesis, its interaction with lipid droplet (LD) catabolism in OA induced steatosis has not been determined yet. In this study we aimed to contribute to literature by clarifying the association between autophagy and SREBP1c by using OA induced steatosis model.
2. Materials and methods
2.1. Cell culture and treatments
AML12 (mouse hepatocyte) cells were maintained in Dulbeco's Modified Eagle's Medium (DMEM) supplemented with 10% FBS (Gibco), 100 U/mL penicillin and 100 mg/mL streptomycin (Gibco) at 37 °C with 5% CO2. Intracellular lipid accumulation was stimulated by treating the cells with 0.06 mM OA:BSA complex (Sigma Aldrich) for 24 h.
2.2. SREBP1c siRNA transfection
AML12 were seeded and transfected with siRNA specific for SREBP1c (Thermo Fisher Scientific, siRNA ID: 151861) by using Lipofectamine RNAiMax Reagent (Thermo Fisher Scientific, catalog No. 13778075) according to manufacturer's instructions. Briefly, 80 pmol SREBP1c siRNA in 1:3 ratio of siRNA: Lipofectamine RNAiMax Reagent was prepared in serum-free culture medium (OptiMEM) and incubated for 5 min at room temperature. The mixture was gently added dropwise to the cells in OptiMEM and then incubated at 37 °C with 5% CO2. qRT-PCR and immunoblot experiments were applied to confirm the siRNA efficacy.
Following the siRNA transfection, cells were treated with OA:BSA and divided into four groups totally; i) Control, ii) OA, iii) SREBP1c siRNA, iv) SREBP1c siRNA + OA.
2.3. BODIPY staining
Following the siRNA and/or OA administrations, cells were fixed with 4% paraformaldehyde, washed three times with PBS, and stained with BODIPY 493/503 (0.3 μg/ml, Invitrogen) for 20 min. Following the wash with PBS, cells were stained with DAPI, and photographed using a Zeiss LSM700 confocal microscope (Amsterdam, Netherlands). LD diameter was measured using ImageJ software. LDs with a diameter of less than 3 μm were called “small LDs”, while the droplets larger than 3 μm were called “large LDs”. Quantification was applied in at least thirty cells for each group.
2.4. Gene expression analysis
RNA was isolated using PureLink RNA Mini Kit (Thermo Fisher) and reverse transcribed with High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher) according to manufacturer's instruction. cDNA samples were then applied to quantitative reverse transcriptase PCR with Power UP SYBR Green Master Mix (Thermo Fisher) and Rotor Gene Q-RT PCR system (Qiagen). The threshold cycle (CT) was determined, and the relative gene expression subsequently was calculated as follows: fold change = 2−Δ(ΔCT), where ΔCT = CT-CT target housekeeping (β-actin) and Δ(ΔCT) = ΔCT-CT treated control. Primer sequences are provided in Supplementary Table 1.
2.5. Immunoblot analysis
AML12 was lysed in RIPA buffer (Cell Signalling) and protein concentration was determined using BCA assay (Thermo Scientific) according to manufacturer's protocol. Total 20 μg of protein was separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. Following the blocking with 5% BSA in TBST, membrane was incubated with primary antibodies against SREBP1c (Novus Biologicals, catalog No. NB100-2215), beclin-1 (Cell Signaling; catalog No. 3495), LC3 (Cell Signaling; catalog No. 3868) and β-actin (Cell Signaling, catalog No. 4967) overnight. After washing of unbound primary antibodies with TBST and incubation with HRP-conjugated secondary antibody, blots were visualized with chemiluminescence kit (Cell Signaling). The density of bands was quantified and normalized with β-actin using Image J software.
2.6. Determination of lipid droplet fusion with lysosome by LAMP1/BODIPY co-staining
After the OA incubation with or without SREBP1c silencing, cells were fixed in 4% paraformaldehyde, blocked in 10% goat serum, and incubated with LAMP1 (Novus Biologicals, catalog No. NBP2-25154) primary antibody for overnight at 4 °C. Following use of Alexa Fluor 594 secondary antibody and DAPI, BODIPY 493/503 was applied as indicated above. Cells were photographed using a Zeiss LSM700 confocal microscope (Amsterdam, Netherlands), while analysis was established using ImageJ software. LDs co-localized with LAMP1 were determined in at least thirty cells for group.
2.7. Statistical analysis
Statistical analysis was performed using Prism 4 (Graph-Pad) software. For determination of statistical significances of differences, one-way ANOVA was performed followed by multiple comparisons using the Student-Newman-Keuls test. P-value less than 0.05 has been accepted to be statistically significant.
3. Results
3.1. Oleic acid induced lipid accumulation and evaluation of SREBP1c
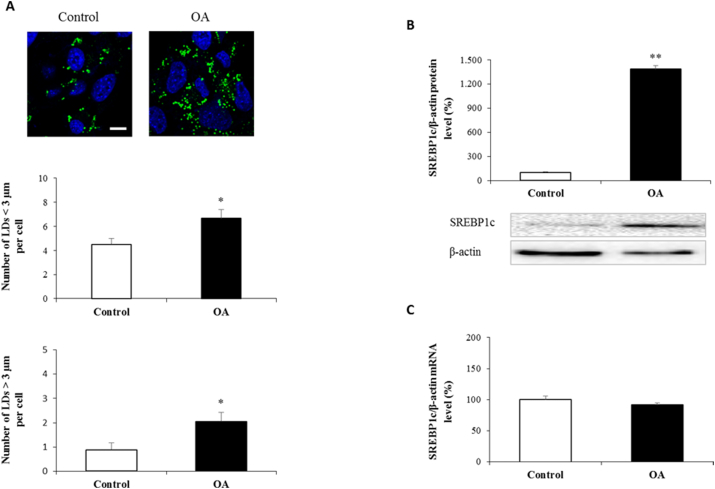
Increased LD formation in hepatocytes is a typical finding of hepatic steatosis. The water-soluble OA:BSA complex is shown to efficiently stimulate lipid accumulation and used in the literature [11,12]. Here in our study, AML12 was treated with 0.06 mM OA to stimulate LD formation. We measured the number and diameter of all BODIPY-labeled structures by confocal microscopy and revealed an increase in the abundance and diameter of LDs when AML12 was administrated with OA (Fig. 1A). SREBP1c is a transcription factor involve in the regulation of fatty acid and triglyceride synthesis. As shown in Fig. 1B, OA administrated cells exhibited a robust induction in SREBP1c protein expression. However, mRNA levels of SREBP1c did not affected by OA (Fig. 1C).
Fig. 1.
Oleic acid induced lipid accumulation and evaluation of SREBP1c
AML12 cells were treated with OA:BSA complex at 0.06 mM concentration for 24 h. Represantative confocal microscopy images show lipid droplet formation following BODIPY staining. Quantification of the numbers of lipid droplet formation per cell (A). SREBP1c protein and mRNA expressions were analyzed by western blotting (B) and qRT-PCR (C) following the normalization to β-actin. Scale bar = 10 μm
Data are expressed as mean ± S.D.
**p < 0.01, and *p < 0.05 vs. control,
##p < 0.01 vs. OA, (n = 3).
3.2. SREBP1c inhibition and its effect on lipid accumulation
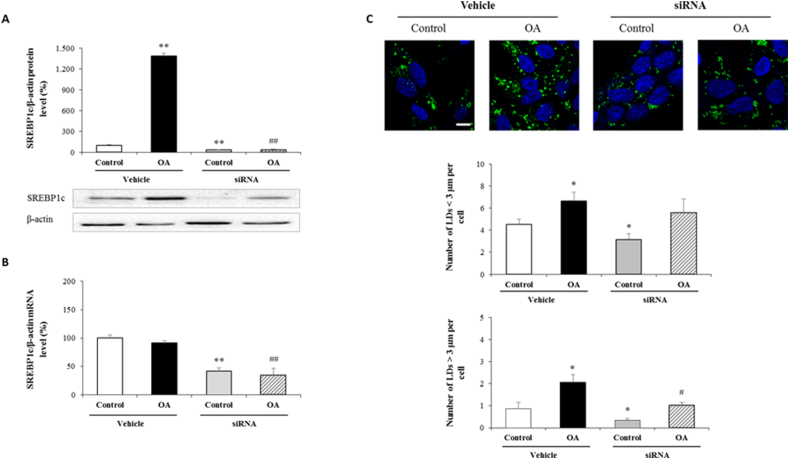
We next asked whether SREBP1c suppression in OA-treated AML12 was associated with a change in LD expansion. To assess the role of SREBP1c in our study, we transfected AML12 with siRNA specific for SREBP1c. As shown in Fig. 2A and B, siRNA transfection significantly blocked SREBP1c expression in both transcriptional and translational manner (Fig. 2A and B). Additionally, SREBP1c inhibition without OA administration reduced lipid droplet formation in all diameters. Differently from non-OA treated ones, siRNA transfection followed by OA administration only decreased the number of LDs that were >3 μm in diameter compared to its vehicle (Fig. 2C). Our findings so far reveal the possible effect of SREB1c silencing on LD expansion under normal or OA induced steatosis.
Fig. 2.
SREBP1c inhibition and its effect on lipid accumulation
Cells were transfected with siRNA specific for SREBP1c and then treated with OA:BSA complex (0.5 mM) for 24 h to divide into following groups; i) control, ii) OA, iii) SREBP1c siRNA, iv) SREBP1c siRNA + OA. SREBP1c protein and mRNA expressions were analyzed by western blotting (A) and qRT-PCR (B) following the normalization to β-actin. Represantative confocal microscopy images show lipid droplet formation following BODIPY staining. Quantification of the numbers of lipid droplet formation per cell (C). Scale bar = 10 μm
Data are expressed as mean ± S.D.
**p < 0.01, and *p < 0.05 vs. control,
##p < 0.01, and #p < 0.05 vs. OA, (n = 3).
3.3. Evaluation of lipid metabolism following SREBP1c silencing and OA administration
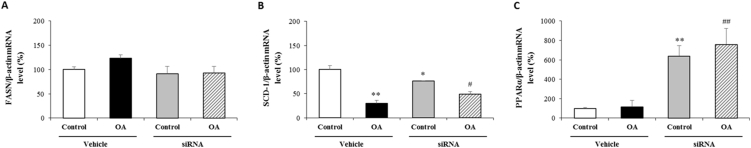
Based on the alterations in LD formation following SREBP1c suppression, we tested the mechanisms that regulate lipid metabolism. Activation of SREBP1c during steatosis is shown to enhance the transcription of fatty-acid synthesis pathway enzymes, including FASN and SCD-1 [7]. In our study, neither OA application nor siRNA tranfection had any effect on FASN (Fig. 3A), while SCD-1 expression was significantly reduced in cells loaded with OA. However, OA administration in SREBP1c silenced cells was significantly induced SCD-1 expression compared to its vehicle (Fig. 3B). PPARα is a nuclear receptor protein that stimulates the expression of genes involved in fatty acid catabolism in the liver [13]. As shown in Fig. 3C, cells transfected with siRNA exhihibed a robust induction of PPARα expression compared to their vehicle, suggesting that the reduction of LD formation following SREBP1c inhibition might be dependent on PPARα-driven lypolysis.
Fig. 3.
Evaluation of lipid metabolism following SREBP1c silencing and OA administration
Cells were transfected with siRNA specific for SREBP1c and then treated with OA:BSA complex (0.5 mM) for 24 h to divide into following groups; i) control, ii) OA, iii) SREBP1c siRNA, iv) SREBP1c siRNA + OA. mRNA expressions of each group were measured by RT-PCR and normalized to β-actin. Relative mRNA expressions of FASN (A), SCD-1 (B) and PPARα (C).
Data are expressed as mean ± S.D.
**p < 0.01, and *p < 0.05 vs. control,
##p < 0.01, and #p < 0.05 vs. OA, (n = 3).
3.4. Evaluation of autophagic activity following SREBP1c silencing and OA administration
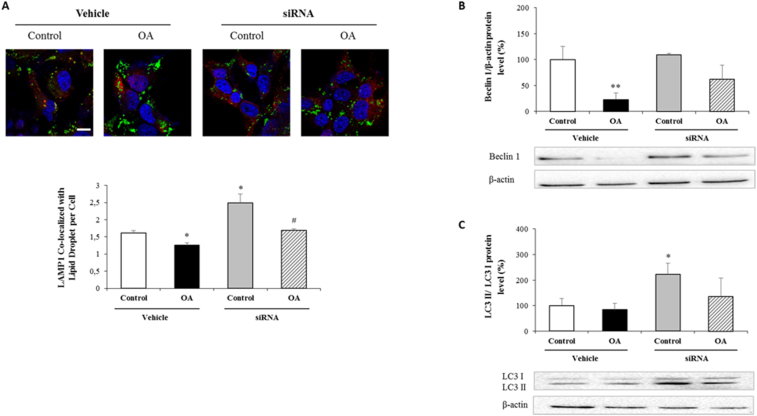
Selective degradation of LDs by autophagy (lipophagy) is a critical mechanism against steatosis in preventing lipid accumulation as well as providing fatty acids during nutrient stress [14]. In the present study, siRNA transfected and OA administrated cells were labeled with LAMP1, a glycoprotein mainly resides in lysosomal membrane, and BODIPY which led us to determine lipophagy at late stage when lipid droplets are fused with lysosomes. Quantification of LD colocalizing with LAMP1 revealed a significant decrease in OA administrated control cells. In addition, SREBP1c silencing in both OA treated and non-OA treated groups revealed its effect by increasing LD/LAMP1 double positive structures compared to their vehicles (Fig. 4A). Beclin-1 and LC3-II/I ratio are the most-studied autophagosome indicators used in monitoring autophagy. In the present study, Beclin-1 was only affected in OA administrated control cells (Fig. 4B). However, SREBP1c silencing under normal conditions significantly increased LC3-II/I ratio (Fig. 4C). Unlike non-OA treated cells, siRNA transfection followed by OA administration had no significant effect on neither Beclin-1 (Fig. 4B) nor LC3-II/I ratio (Fig. 4C), which might reflect that the increase in lipophagy occurs through other autophagy parameters.
Fig. 4.
Evaluation of autophagic activity following SREBP1c silencing and OA administration
Cells were transfected with siRNA specific for SREBP1c and then treated with OA:BSA complex (0.5 mM) for 24 h to divide into following groups; i) control, ii) OA, iii) SREBP1c siRNA, iv) SREBP1c siRNA + OA. Representative BODIPY/LAMP1 co-stained confocal microscopic images from each group. Cells were fixed, and then stained for BODIPY (green), LAMP1 (red) and nuclei by DAPI (blue). The graph shows quantification of the numbers of lipid droplet co-localized with LAMP1 per cell as explained in Methods section (A). Protein expressions of each group were measured by western blotting following the normalization to β-actin against Beclin 1 (B) and LC3 antibodies (C). Scale bar = 10 μm
Data are expressed as mean ± S.D.
**p < 0.01, and *p < 0.05 vs. control,
#p < 0.05 vs. OA, (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
NAFLD, identified by accumulation of lipids in more than 5% of liver weight, is one of the most common metabolic diseases. Due to the increased prevalence of type 2 diabetes mellitus and obesity all around the world, NAFLD patients are rising continuously and if untreated, they progress from steatosis to NASH and liver cirrhosis [15,16]. Neutral lipids, including cholesterol esters and triglycerides, are mainly stored in the form of LDs that can undergo dynamic changes reflecting the status of metabolism [17,18]. Increased accumulation of LDs in hepatocytes is observed in patients with NAFLD and characterized as a typical finding of steatosis [19,20]. Therefore, understanding the mechanisms that modulate the expansion and catabolism of LDs is a subject of intense study and great interest. PA and OA are two most abundant long-chain free fatty acids in the fatty liver specimens that is shown to enhance the steatosis in a time- and dose-dependent manner [21]. In accordance, OA administration has been shown to enhance steatosis development in human hepatocellular carcinoma (HepG2) cell line [22]. Similarly, OA administration induced the abundance of LDs more than PA in both HepG2 cells and primary mouse hepatocytes [23]. Another study in rat primary hepatocytes determined the induction of lipid accumulation and cytotoxicity following PA and OA administration in combination [24]. In the present study, due to their stable phenotype and replicative ability, we used AML12 and established a steatosis model by administrating 0.06 mM OA for 24 h. We also determined increased expression of SREBP1c in OA administrated cells, consistent with the finding of lipid accumulation. Thus, 0.06 mM OA administration for 24 h is likely to lead SREBP1c induction and LD expansion in AML12.
SREBP1c is a highly expressed transcription factor that is located in a number of tissues, including liver, and is involved in a variety conditions, including, cell growth, inflammation and nutrition. Capacity of SREBP1c in inducing fatty acid and triacylglycerol synthesis through the transcription of genes makes SREBP1c an important candidate to study steatosis [7]. In this scope, both in vitro and in vivo studies determined a strong link between SREBP1c overexpression and insulin resistance, diabetes and NAFLD [[25], [26], [27]]. In our study, SREBP1c silencing (without OA administration) in AML12 resulted in a decrease in both small and large LD formations. Although, SREBP1c suppression only decreased the number of LDs with a diameter higher than 3 μm which stated its effect against OA induced steatosis.
In the current knowledge, two major mechanisms modulate the catabolism of LDs; lipolysis and autophagy [28]. In adipocytes, lipolysis activation resulted in the accumulation of smaller LDs formed by free fatty acid re-esterification [29]. Mechanistically, SREBP1c inhibition by sauchinone, an anti-inflammatory and antioxidant lignan found in Saururus chinensis, inhibited steatosis development and reduced FASN and SCD-1 mRNA levels [30]. PPARα agonists, crucial stimulators of fatty acid catabolism, are indicated to be used as promising therapeutic molecules in NAFLD patients [13]. In vitro studies using hepatocytes demonstrated the inhibitory effect of PPARα agonists on SREBP1c expression [31,32]. This inverse correlation between PPARα and SREBP1c is also determined in liver samples of obese individuals with NAFLD and the SREBP-1c/PPARα ratio of the patients was determined to be higher than the control group [33]. Abdelmegeed and colleagues have been shown that PPARα activation in mice inhibits the steatosis development by increasing the expression of genes responsible for fatty acid oxidation in the liver [34]. In another study using HepG2 cells, supplementation of PA but not OA was associated with an induction in PPARα compared to control [35]. According to the literature and the negative correlation between LD content and PPARα levels in the present study, we hypothesized that the lesser steatosis extent observed in siRNA transfected cells is due to PPARα induction which might lead to increased fatty acid catabolism. Such interaction might protect against OA induced lipotoxicity when LD content are enhanced.
Activation of lipophagy is another process that modulates the breakdown of LDs, representing an alternative to lipolysis mediated LD catabolism [36,37]. Studies using mouse models has shown the crucial role of lipophagy in regulating lipid metabolism by breaking down triglycerides and reducing fatty liver development [14]. Additional studies have identified the reduced autophagy activity in obese mice fed a high-fat diet [38,39]. Accordingly, a number of treatments identified to activate autophagy are currently being studied as therapeutic approaches against NAFLD development [40]. Nguyen and colleagues [41] investigated the interaction between SREBP1c and autophagy, and reported that high fat mediated SREBP1c impairs autophagic lipid catabolism to promote hepatic steatosis. However, we determined a significant increase in LAMP1-BODIPY double positive structures when cells were transfected with SREBP1c and treated with OA. Our finding lead us to propose that SREBP1c suppression causes the catabolism of larger-sized LDs by increasing their fusion with lysosomes. Based on these findings, we tested whether components of autophagosome formation are associated with lipophagy induction. LC3-II/I ratio and Beclin-1 are two of the best studied components of autophagic activity involve in autophagosome formation. In the present study, OA administration alone reduced Beclin-1 levels, whereas SREBP1c silencing in non-OA treated cells increased LC3-II/I ratio. Collectively, further experiments are needed to determine the proteins involve in SREBP1c mediated lipophagy induction.
Taken together, our findings suggest a role of SREBP1c suppression in restoring the accumulation of LDs >3 μm in diameter in OA-treated AML12 cells. Increase in PPARα expression in SREBP1c siRNA + OA group suggested a possible role of SREBP1c silencing in activating lipolysis. Enhanced lipid droplet/LAMP1 co-localization in SREBP1c siRNA + OA group is also highlighted that lipophagy might be involve in reduction of LD expansion. Therefore, further investigations are needed to clarify the interaction between SREBP1c and autophagy. Our data collected so far in OA induced steatosis model gain insight for therapeutic potential of SREBP1c to reverse fatty liver development by means of lipolysis and lipophagy.
Financial Support
This study was funded by Marmara University Research Fund SAG-A-241018-0564.
CRediT authorship contribution statement
Erdi Sozen: Conceptualization, Methodology, Visualization, Writing – original draft, preparation. Tugce Demirel-Yalciner: Methodology, Visualization, Investigation, Writing – review & editing. Dyana Sari: Visualization, Investigation. Ceren Avcilar: Visualization, Investigation. Tuna Felix Samanci: Visualization, Investigation. Nesrin Kartal Ozer: Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare no conflict of interest to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.metop.2021.100138.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cohen J.C., Horton J.D., Hobbs H.H. Human fatty liver disease: old questions and new insights. Science. 2011;332(6037):1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne C.D., Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62(1 Suppl):S47–S64. doi: 10.1016/j.jhep.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Buzzetti E., Pinzani M., Tsochatzis E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65(8):1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Rafiei H., Omidian K., Bandy B. Dietary polyphenols protect against oleic acid-induced steatosis in an in vitro model of NAFLD by modulating lipid metabolism and improving mitochondrial function. Nutrients. 2019;11(3) doi: 10.3390/nu11030541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y. Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell Biol. 2015;16(11):678–689. doi: 10.1038/nrm4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neuschwander-Tetri B.A. Non-alcoholic fatty liver disease. BMC Med. 2017;15(1):45. doi: 10.1186/s12916-017-0806-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moslehi A., Hamidi-Zad Z. Role of SREBPs in liver diseases: a mini-review. J Clin Transl Hepatol. 2018;6(3):332–338. doi: 10.14218/JCTH.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carotti S. Lipophagy impairment is associated with disease progression in NAFLD. Front Physiol. 2020;11:850. doi: 10.3389/fphys.2020.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao Y. Autophagy: a new target for nonalcoholic fatty liver disease therapy. Hepat Med. 2016;8:27–37. doi: 10.2147/HMER.S98120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ipsen D.H., Lykkesfeldt J., Tveden-Nyborg P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell Mol Life Sci. 2018;75(18):3313–3327. doi: 10.1007/s00018-018-2860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L. Programming and regulation of metabolic homeostasis by HDAC11. EBioMedicine. 2018;33:157–168. doi: 10.1016/j.ebiom.2018.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thayer T.E. The role of bone morphogenetic protein signaling in non-alcoholic fatty liver disease. Sci Rep. 2020;10(1):9831. doi: 10.1038/s41598-020-66770-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pawlak M., Lefebvre P., Staels B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol. 2015;62(3):720–733. doi: 10.1016/j.jhep.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 14.Singh R. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams L.A. NAFLD as a risk factor for the development of diabetes and the metabolic syndrome: an eleven-year follow-up study. Am J Gastroenterol. 2009;104(4):861–867. doi: 10.1038/ajg.2009.67. [DOI] [PubMed] [Google Scholar]
- 16.Williams C.D. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140(1):124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 17.Ducharme N.A., Bickel P.E. Lipid droplets in lipogenesis and lipolysis. Endocrinology. 2008;149(3):942–949. doi: 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- 18.Goodman J.M. The gregarious lipid droplet. J Biol Chem. 2008;283(42):28005–28009. doi: 10.1074/jbc.R800042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross D.A., Silver D.L. Cytosolic lipid droplets: from mechanisms of fat storage to disease. Crit Rev Biochem Mol Biol. 2014;49(4):304–326. doi: 10.3109/10409238.2014.931337. [DOI] [PubMed] [Google Scholar]
- 20.Polyzos S.A. Current and emerging pharmacological options for the treatment of nonalcoholic steatohepatitis. Metabolism. 2020;111S:154203. doi: 10.1016/j.metabol.2020.154203. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Lechon M.J. A human hepatocellular in vitro model to investigate steatosis. Chem Biol Interact. 2007;165(2):106–116. doi: 10.1016/j.cbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Cui W., Chen S.L., Hu K.Q. Quantification and mechanisms of oleic acid-induced steatosis in HepG2 cells. Am J Transl Res. 2010;2(1):95–104. [PMC free article] [PubMed] [Google Scholar]
- 23.Mei S. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Therapeut. 2011;339(2):487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moravcova A. The effect of oleic and palmitic acid on induction of steatosis and cytotoxicity on rat hepatocytes in primary culture. Physiol Res. 2015;64(Suppl 5):S627–S636. doi: 10.33549/physiolres.933224. [DOI] [PubMed] [Google Scholar]
- 25.Vitto M.F. Reversion of steatosis by SREBP-1c antisense oligonucleotide did not improve hepatic insulin action in diet-induced obesity mice. Horm Metab Res. 2012;44(12):885–890. doi: 10.1055/s-0032-1321819. [DOI] [PubMed] [Google Scholar]
- 26.Muraoka T. Ezetimibe decreases SREBP-1c expression in liver and reverses hepatic insulin resistance in mice fed a high-fat diet. Metabolism. 2011;60(5):617–628. doi: 10.1016/j.metabol.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Karasawa T. Sterol regulatory element-binding protein-1 determines plasma remnant lipoproteins and accelerates atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2011;31(8):1788–1795. doi: 10.1161/ATVBAHA.110.219659. [DOI] [PubMed] [Google Scholar]
- 28.Schott M.B. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J Cell Biol. 2019;218(10):3320–3335. doi: 10.1083/jcb.201803153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paar M. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J Biol Chem. 2012;287(14):11164–11173. doi: 10.1074/jbc.M111.316794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y.W. Inhibition of SREBP-1c-mediated hepatic steatosis and oxidative stress by sauchinone, an AMPK-activating lignan in Saururus chinensis. Free Radic Biol Med. 2010;48(4):567–578. doi: 10.1016/j.freeradbiomed.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Konig B. Activation of PPARalpha and PPARgamma reduces triacylglycerol synthesis in rat hepatoma cells by reduction of nuclear SREBP-1. Eur J Pharmacol. 2009;605(1–3):23–30. doi: 10.1016/j.ejphar.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Yoshikawa T. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARs suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. Mol Endocrinol. 2003;17(7):1240–1254. doi: 10.1210/me.2002-0190. [DOI] [PubMed] [Google Scholar]
- 33.Pettinelli P. Enhancement in liver SREBP-1c/PPAR-alpha ratio and steatosis in obese patients: correlations with insulin resistance and n-3 long-chain polyunsaturated fatty acid depletion. Biochim Biophys Acta. 2009;1792(11):1080–1086. doi: 10.1016/j.bbadis.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Abdelmegeed M.A. PPARalpha expression protects male mice from high fat-induced nonalcoholic fatty liver. J Nutr. 2011;141(4):603–610. doi: 10.3945/jn.110.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricchi M. Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J Gastroenterol Hepatol. 2009;24(5):830–840. doi: 10.1111/j.1440-1746.2008.05733.x. [DOI] [PubMed] [Google Scholar]
- 36.Singh R., Cuervo A.M. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu K., Czaja M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20(1):3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metabol. 2010;11(6):467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cursio R. The role of autophagy in liver diseases: mechanisms and potential therapeutic targets. BioMed Res Int. 2015;2015:480508. doi: 10.1155/2015/480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni H.M. Targeting autophagy for the treatment of liver diseases. Pharmacol Res. 2012;66(6):463–474. doi: 10.1016/j.phrs.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen T.T.P. SREBP-1c impairs ULK1 sulfhydration-mediated autophagic flux to promote hepatic steatosis in high-fat-diet-fed mice. Mol Cell. 2021 doi: 10.1016/j.molcel.2021.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.