Abstract
In the present study, we report for the first time on the detection of bovine herpesvirus type 1 (BHV-1) in whole-blood samples derived from naturally infected cattle. Sensitive PCR assays specific for glycoprotein B (gB), gC, and gE of BHV-1 allow the detection of one BHV-1 DNA copy in 105 to 107 peripheral blood leukocytes (PBLs). The incidence of BHV-1-positive PBLs in naturally infected cattle appears to be quite high (92.2% positive PBLs among all samples tested), although in most cases only between 10−5 and 10−7 positive leukocytes were present. The results demonstrate that the viral DNA is detectable not only in the peripheral blood of acutely infected animals but, more importantly, also in the peripheral blood of subclinically infected cattle. The gE-specific PCR described in the report allows discrimination between wild-type (WT) virus-infected and vaccinated animals, which is of importance for control programs that use the recently introduced vaccination strategy with a gE-negative virus. The results further show that doubtful serological results can be verified or falsified and that individual animals can be monitored for the presence or absence of WT BHV-1 or gE-negative virus in cattle herds. The PCR protocols allow the detection of BHV-1 prior to seroconversion or in BHV-1-seronegative cattle. Finally, the results indicate the simultaneous presence of WT and gE-negative vaccine virus in the PBLs of several cattle. Therefore, investigations of viremia in naturally and experimentally infected cattle and on the identification of infected cell types of bovine PBLs can be now performed.
Bovine herpesvirus type 1 (BHV-1), a member of the family Alphaherpesvirinae, is an important pathogen for ruminants and is classified into three subtypes according to the major clinical manifestations (52). BHV-1.1 (subtype 1) is predominantly associated with infections of the respiratory tract (infectious bovine rhinotracheitis [IBR]), whereas BHV-1.2 infects the genital tract (infectious pustular vulvovaginitis and infectious balanoposthitis) and the respiratory tract and BHV-1.3, which has been reclassified as BHV-5 (31), causes encephalitis. BHV-1 infection is a major threat to the health of cattle herds and leads to economically significant losses. Therefore, vaccination is frequently practiced to protect cattle against the disease and to reduce the level of virus spread. One important improvement in vaccine development was the introduction of so-called marker vaccines that allow the serological differentiation between vaccinated and field virus- or wild-type (WT) virus-infected animals, as practiced in the control of Aujeszky’s disease virus (pseudorabies virus [PRV]) infection of pigs (44). By following the same concept used for PRV, a live vaccine against BHV-1 was recently introduced. In that vaccine the glycoprotein E (gE) gene is deleted (38, 45). BHV-1 without gE is reported to be of reduced virulence, although it displays the same organ and tissue tropism as WT BHV-1, including the ability to establish latency (41, 42). The gE-negative vaccine protects cattle against the disease (17, 38), but occasional outbreaks cannot be prevented (3).
The availability of reliable companion diagnostic tests is important for a successful control program based on the use of live and inactivated marker vaccines. As the method of choice for routine diagnosis, enzyme-linked immunosorbent assays (ELISAs) that allow serological discrimination between gE antibody-positive (WT BHV-1-infected) and gE antibody-negative (vaccinated) animals have been developed. BHV-1-infected animals can be recognized serologically approximately 7 days postinfection by an ELISA for the detection of antibodies specific for glycoprotein gB (21), whereas gE-specific serum antibodies are first detectable between 11 and 17 days postinfection (45, 46) and persist for at least 2 to 3 years (18, 46). Differences in the sensitivities of commercially available BHV-1 ELISAs can lead to various results in different laboratories, at least with sera containing low titers of specific antibodies (22). This can become a major concern and might be due to a generally weaker or delayed anti-gE response or to a lesser sensitivity of the gE ELISA that is used (17, 46).
The PCR represents an excellent tool for the fast and very sensitive detection of viral genomes in biological and clinical specimens. Various PCR assays for the recognition of BHV-1 have been described. Primers were selected to amplify parts of the gB gene (25, 36, 47), the gC gene (13, 42, 43), the gD gene (8, 50), and the thymidine kinase gene of BHV-1 (20) with various sensitivities. The possibility of detection of BHV-1 by PCR in nasal swabs and mucosa (8, 36, 43), in the lungs, tracheae, lymph nodes, and tonsils (36, 43), and in the sacral and trigeminal ganglia (16a) of experimentally infected cattle has also been described. Until now, however, information on the applicability of these PCR assays with field samples has been scarce (8) because only nasal swabs or blood samples are usually available for routine diagnosis. Detection of BHV-1 in nasal swabs requires virus excretion, which is limited to 1 to 2 weeks during productive virus infection.
The presence of BHV-1 in peripheral blood of infected animals had already been suggested by Nyaga and McKercher (28) and was supported by the demonstration of viremia before antibodies were detectable (4, 29), by the isolation of virus from leukocytes of infected calves (5), and by the detection of BHV-1 DNA in peripheral blood leukocytes of infected animals (26). Therefore, a role for infected blood monocytes is suggested in the systemic spread of BHV-1 (12, 39). Infection of peripheral blood mononuclear cells by alphaherpesviruses appears to be not uncommon. Viral replication in peripheral blood mononuclear cells predominantly in monocytes has also been demonstrated for PRV (11, 33, 51) and for equid herpesvirus type 1 (6).
Here we report for the first time on the detection of BHV-1 in whole-blood samples derived from naturally infected cattle. To this end, sensitive PCR assays that amplify different parts of the viral genome (gB, gC, and gE) were established. These assays allow the specific detection of a single copy of BHV-1 DNA in 105 to 107 leukocytes. In addition, this is the first report to introduce a highly sensitive gE-specific PCR which is suitable for discrimination of WT BHV-1-infected and vaccinated animals. The results presented here demonstrate that the PCR assays represent excellent alternative or additional tools for the detection of BHV-1 since questionable or doubtful serological results can be ascertained. Individual animals in cattle herds displaying clinical signs suspicious for BHV-1 infection can be monitored for the presence or absence of WT BHV-1 or gE-negative vaccine virus.
MATERIALS AND METHODS
Cells and virus strains.
BHV-1.1 (IBR-like) LA, SP (Spiel), and VD7 and BHV-1.2 (IPV-like) SCH (Schönböken) and K22 were originally obtained from O. C. Straub (Federal Research Centre for Virus Diseases of Animals, Tübingen, Germany). The gE-negative BHV-1 vaccine strains Difivac (Bayer AG, Wuppertal, Germany) and Rhinobovin Marker (Hoechst Veterinär GmbH, Unter-Schleißheim, Germany) were used. BHV-5 N569, BHV-4, and caprine herpesvirus type 1 (CapHV-1) McK/US were kindly provided by M. Engels, Institute for Virology, University of Zürich, Zürich, Switzerland. BHV-3 (ovine herpesvirus 2) was obtained from H. Reid, Moredun Research Institute, Edinburgh, United Kingdom. Virulent PRV (Suid herpesvirus 1) Phylaxia has been described previously (34). All virus strains were propagated in Madin-Darby bovine kidney cells, and viral DNA was isolated essentially as described previously (34).
Restriction enzyme analysis, gel electrophoresis, and Southern blot hybridization.
Viral DNA was digested with HindIII according to the recommendations of the manufacturer (New England BioLabs, Schwalbach, Germany), and the fragments were separated in horizontal 0.8% agarose gel (Gibco BRL Life Technologies, Karlsruhe, Germany). PCR products were separated in 1% agarose containing 0.3 μg of ethidium bromide per ml. Alkaline transfer onto a positively charged nylon membrane (Amersham Life Sciences, Freiburg, Germany) was achieved by the method of Reed and Mann (30). The membrane was hybridized with 32P-labelled probes (2 × 104 to 4 × 106 cpm per ml) in 1.5× SSPE (1× SSPE is 180 mM NaCl, 10 mM sodium phosphate, and 1 mM disodium EDTA [pH 7.4]), 0.5% (wt/vol) nonfat milk powder, 1.0% (wt/vol) sodium dodecyl sulfate (SDS), 0.5 mg of denatured calf thymus DNA per ml, and 50% (vol/vol) deionized formamide at 52°C (53), followed by exposure to X-ray film (Kodak AR) at −70°C with an intensifying screen (Curix; Agfa-Gevaert, Munich, Germany). For rehybridization, the blot was stripped off by boiling in 0.5% (wt/vol) SDS.
Hybridization probes were prepared by random priming with [α-32P]dCTP (>3,000 Ci/mM; ICN Biochemicals, Eschwege, Germany) and the Rediprime labelling kit (Amersham Life Sciences) to obtain a specific activity of approximately 109 cpm per μg of DNA. As internal probes the nested PCR products gBN1/2 and gEN1/2 (see below) were used after isolation from low-melting-temperature agarose gel (LMP; Gibco BRL Life Technologies). The gC-specific internal probe was prepared after gel electrophoretic isolation of a 301-bp DNA fragment obtained after BssHII cleavage of the cloned gC1/2 PCR product (pMUH-2; see below).
Blood specimens.
Heparin- or EDTA-treated blood was obtained from animals of herds (i) that displayed a confirmed BHV-1 infection, (ii) that were suspected of having the disease because of clinical signs, or (iii) that had serological results indicative of a previous virus infection, as detailed in Results. In addition, blood from BHV-1-negative animals was kindly provided by D. Müller-Doblies (Institute for Virology, University of Zürich, Zürich, Switzerland).
Serum samples were tested for BHV-1 antibodies with commercially available ELISA kits (Screening [IDEXX GmbH, Woerstadt, Germany]; Checkit Trachitest-screeningtest [Bommeli AG, Hoechst Roussel Vet., Unter-Schleißheim, Germany) and for the presence of gE-specific serum antibodies (HerdCheck; IDEXX GmbH). BHV-1 serum neutralizing antibodies were determined in the absence of complement with a 24-h serum-virus incubation period.
DNA extraction from leukocytes.
DNA was isolated from 1 to 10 ml of heparinized or EDTA-treated blood by a method modified from that of Xu et al. (54). Erythrocytes were lysed by incubation in 0.15 M NH4Cl–0.01 M KHCO3–0.001 M disodium EDTA for 15 min on ice. After centrifugation of the lysate at 1,600 to 2,000 × g for 5 to 10 min, the supernatant was decanted and the cell pellet was washed twice with phosphate-buffered saline. The cells were lysed with 2 ml of 6 M guanidinium-HCl solution (containing 0.1 M sodium acetate), and the lysate was carefully layered under 10 ml of absolute ethanol. The DNA was recovered by rolling the tube horizontally and then gently inverting the tube several times. After one more extraction with ethanol, the DNA was solved in 2 ml of lysis buffer (10 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 2% SDS, 1 mg of RNase per ml, 1 mg of proteinase K per ml) and incubated at 37°C for at least 30 min (or overnight). Thereafter, the DNA was extracted by adding 10 ml of ethanol, completely solved in 0.1 ml of TE (10 mM Tris, 0.1 mM EDTA [pH 8.0]) at 65°C for approximately 30 min, and chilled on ice. The DNA concentration was determined spectroscopically.
Alternatively, the PUREGENE DNA isolation kit (Biozyme, Oldendorf, Germany) was used as a more practical procedure for the preparation of template DNA. We found that leukocytes prepared from only 0.3 ml of EDTA-treated blood (after lysis of erythrocytes as described above) routinely resulted in a sufficient amount of DNA for at least 10 PCR assays of equal quality.
Primer selection.
PCR primers were selected according to the DNA sequence published for glycoproteins gB (BHV-1.1 Cooper; accession no. M21474), gC (BHV-1.1 Cooper; accession no. M27491), and gE (BHV-1.1 FM; accession no. U06934). To enable detection of various BHV-1 subtypes, those gene regions which were found to be mostly conserved among BHV-1 strains according to the alignment of BHV sequences available in GenBank were selected. The oligonucleotides were synthesized on a DNA synthesizer (Biosearch 8700; New Brunswick, Nürtingen, Germany) and were subsequently purified by denaturing gel electrophoresis and reversed-phase chromatography on SepPack columns (Millipore Waters, Eschborn, Germany). The sequences and nucleotide positions of the primers and the size of each PCR product are given in Table 1. The gE-specific primers are located in the N-terminal part of the gE-encoding region, which is deleted from the vaccine strain (17). To exclude false-negative PCR results caused by failure of amplification, primer pair NF1 and NF2 (37) was routinely used to amplify the cellular nuclear factor gene (accession no. X12764), which allows one to survey the success of the individual PCR.
TABLE 1.
Sequences of synthetic oligonucleotide primers and their locations in the respective glycoprotein genes
| Oligonucleotide | Sequence | Primer location (nucleotide position) | PCR product | PCR product length (bp) |
|---|---|---|---|---|
| gB-1a | 5′-TACGACTCGTTCGCGCTCTC-3′ | 883–902 | ||
| gB-2 | 5′-GGTACGTCTCCAAGCTGCCC-3′ | 1341–1360 | gB1/2 | 478 |
| gBN-1 | 5′-TCTCGACCGGGGACATTATC-3′ | 899–918 | ||
| gBN-2 | 5′-GCCTCTTCGATCACGCAGTC-3′ | 1264–1283 | gBN1/2 | 385 |
| gC-1b | 5′-AGGAGCGCAAGTGGATGCTCTG-3′ | 563–581 | ||
| gC-2 | 5′-GTAGCCGTTGCGGAACCAGTGC-3′ | 1068–1087 | gC1/2 | 527 |
| gE-1c | 5′-GCTTCGGTCGACACGGTCTT-3′ | 501–520 | ||
| gE-2 | 5′-CTTTGTCGCCCGTTGAGTCG-3′ | 746–765 | gE1/2 | 265 |
| gEN-1 | 5′-CGCGCCCGTCTTTCTCCCA-3′ | 536–554 | ||
| gEN-2 | 5′-CACGTCGGTGAAGCACTCG-3′ | 655–674 | gEN1/2 | 139 |
Conditions of PCR.
PCR was performed in a total volume of 50 μl containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl (Gibco BRL Life Technologies), 0.02 mM deoxynucleoside triphosphates (Pharmacia Biotech, Freiburg, Germany), 45 to 90 nM each primer, and 1.5 mM MgCl2. After the addition of template DNA (0.4 to 3.0 μg of DNA from blood) and boiling for 3 min, 2.5 U of Taq polymerase (Gibco BRL Life Technologies) was added at 80°C in order to avoid premature amplification. The following cycling program was performed in a thermocycler (Biometra Trio, Göttingen, Germany): 10 cycles of 60 s at 96°C, 45 s at 65°C, and 45 s at 72°C, followed by 15 cycles of 60 s at 96°C, 45 s at 54°C, and 45 s at 72°C and 10 cycles of 60 s at 96°C, 45 s at 60°C, and 45 s at 72°C and a final extension of 10 min at 72°C. In preliminary tests we found that the addition of 5% (vol/vol) dimethyl sulfoxide improved specific amplifications in the presence of primer pairs gE-1–gE-2 and gEN-1–gEN-2.
The optimal cycling conditions of the NF1-NF2 PCR comprised one cycle of 120 s at 98°C, 45 s at 55°C, and 45 s at 72°C, followed by 35 cycles of 60 s at 96°C, 45 s at 55°C, and 45 s at 72°C and a final extension of 10 min at 72°C. Therefore, the NF1-NF2 PCR was usually performed in a separate reaction. Preliminary results demonstrated that the NF1-NF2 PCR can be also performed in a single reaction together with the gB-, gC-, or gE-specific primers by using the BHV-1-specific PCR parameters; however, this leads to at least a 10-fold decrease in sensitivity (data not shown).
Cloning and sequencing of PCR products.
The PCR products obtained with strain LA DNA as the template were cloned into the plasmid vector pCRII according to the instructions of the manufacturer (TA cloning kit; Invitrogen, Groningen, The Netherlands). Plasmid DNA was purified as described previously (16) and was used for DNA sequencing either by the dideoxy chain termination method (35) with T7 polymerase (Pharmacia Biotech) or automatic sequencing (model 377; Applied Biosystems, Weiterstadt, Germany) by using a dye terminator cycle sequencing kit (Applied Biosystems, Perkin-Elmer Corp., Weiterstadt, Germany). Sequence data were analyzed with the Wisconsin Package (version 9.1; Genetics Computer Group, Madison, Wis.) and the Basic Alignment Search Tool (BLAST) provided by the National Center for Biotechnology Information.
Nucleotide sequence accession number.
The DNA sequence of the amplified region of gC of BHV-1 LA has been deposited in GenBank under accession no. AF135441.
RESULTS
BHV-1 specificity.
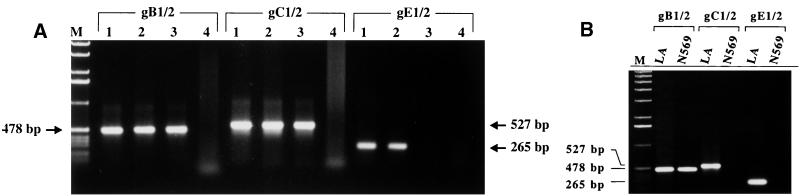
The different PCR assays were first established with genomic DNA of prototype strain BHV-1.1 LA. gB, gC, and gE amplifications were optimized by numerous test experiments with different primers and primer combinations. According to specificity and efficacy, the use of the following primer pairs showed the best results: gB-1–gB-2 (478 bp), gC-1–gG-2 (527 bp), and gE-1–gE-2 (265 bp) (Table 1; Fig. 1). By using template DNAs of laboratory strains BHV-1.1 LA and SP and BHV-1.2 K22 and SCH, identical fragments of the expected size were amplified by each PCR assay, as represented in Fig. 2A. Amplification of DNA isolated from the commercially available gE-negative vaccine strains Rhinobovin Marker (data not shown) and Difivac with primers gB-1–gB-2 and gC-1–gG-2 also resulted in the expected DNA fragments, whereas PCR with primers gE-1 and gE-2 remained negative (Fig. 2A, lanes 3). No amplification was obtained in each PCR with DNA of the closely related virus PRV (both genomic PRV DNA and DNA isolated from organ tissues acutely infected with PRV) (Fig. 2A, lanes 4). In addition, 35 different virus isolates obtained from field-virus-infected animals (isolated from 1994 to 1996), which have been typed by restriction analysis as BHV-1.1 or BHV-1.2 (16a), were all correctly detected by PCR with primers gB-1–gB-2, gC-1–gC-2, and gE-1–gE-2 (data not shown).
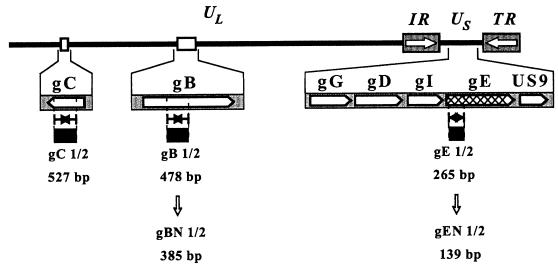
FIG. 1.
Map of the BHV-1 genome showing the unique long (UL), unique short (US), and inverted repeat (IR and TR) regions. The locations of the indicated glycoprotein genes (not drawn to scale) are depicted on the map. The amplified regions and the respective sizes of the different PCR products are indicated, as are the sizes of the nested PCR products gBN1/2 and gEN1/2.
FIG. 2.
Specificity of the indicated PCRs. (A) DNAs (100 ng each) of BHV-1.1 LA (lanes 1), BHV-1.2 K22 (lanes 2), gE-negative vaccine virus strain Difivac (lanes 3), and PRV (lanes 4) were used as templates. (B) Results of the indicated PCRs with DNA from BHV-5 N569 compared to the results obtained with DNA from BHV-1.1 LA. BHV-5 is not detected by the gC- and gE-specific PCR. The PCR products (10 μl) were separated in a 1.0% agarose gel. The sizes of the amplified products are indicated (lane M, 1-kb ladder as a size marker).
To test the specificity of each PCR for BHV-1, DNAs of the related herpesviruses BHV-3, BHV-4, BHV-5 N569, and CapHV-1 (BHV-6) were analyzed. No amplification by any of the three PCR tests was obtained with BHV-3 or BHV-4 DNA (data not shown). With primer pair gB-1–gB-2 and BHV-5 DNA as the template, a fragment of the same size as that obtained with BHV-1 DNA was amplified, whereas PCRs with primers gG-1–gC-2 and gE-1–gE-2 were negative (Fig. 2B). CapHV-1 DNA could be amplified only by the PCR with gC-1–gC-2, resulting in a DNA fragment of the same size as that from BHV-1 DNA (data not shown). Taken together, these results show that BHV-1 can be detected specifically by using primer pair gE-1–gE-2 and, with the exception of BHV-5, primer pair gB-1–gB-2. WT BHV-1 can be reliably discriminated from the gE-negative vaccine strains.
Blot hybridization and DNA sequence analysis.
To prove the specificity and authenticity of the PCR products, the PCR fragments obtained with strain LA DNA and primer pairs gB-1–gB-2, gC-1–gC-2, and gE-1–gE-2 were cloned. The resulting plasmids, pMUH-1, pMUH-2, and pMUH-3, respectively, were subsequently used as 32P-labelled probes for Southern blot hybridization experiments (data not shown). After HindIII cleavage of the genomic DNA, the radioactively labelled gB1/2 fragment (pMUH-1) hybridized strongly with fragment G of BHV-1.1 and BHV-1.2 and with fragment B of BHV-5. The gC-specific product (pMUH-2) detected the expected fragments (fragment 1 of the BHV-1 strains and fragment B of BHV-5 strains). The radioactively labelled gE1/2 PCR product (pMUH-3), which represents the N-terminal part of the gE gene, hybridized with fragment K of LA, fragment L of K22, and fragment A of N569 obtained by cleavage with HindIII. Due to the gE deletion, no signal was obtained with cleaved DNA of vaccine strain Difivac. These results were all in agreement with the published genomic maps (10, 23).
Plasmids pMUH-1, pMUH-2, and pMUH-3 were used for DNA sequencing, and the sequences were subsequently compared with the corresponding sequences of other BHV strains available in the gene data bank. The amplified parts of the gB, gC, and gE genes were found to be 100% identical to the published sequences of BHV-1.1 Cooper (accession no. M21474). Compared to BHV-1.2 P8-2 (accession no. M23257) the sequence of the amplified gB region of strain LA DNA differed at one nucleotide (99.8% identity), which also led to one different amino acid (the amino acid at position 114; S to T). The gE sequence available for BHV-1.2 ST (accession no. Z23068) demonstrated two silent base exchanges compared to the sequence of pMUH-3 (99.2% nucleotide sequence identity).
The comparison of gC sequences between the amplified regions of strain LA and BHV-1.1, BHV-1.2, BHV-5, and CapHV-1 E/CH again demonstrated 100% homology between the two BHV-1.1 strains, strains LA (pMUH-1) and Cooper (accession no. M27491). Compared to the gC sequence of BHV-1.2 K22 (accession no. Z49223), 10 base differences (leading to four different amino acids) accounted for 98.1% homology. The gC DNA sequences of two different BHV-5 strains, strains N569 (accession no. Z49224) and TX-89 (accession no. U35883), displayed homologies with strain LA (pMUH-2) of 90.1 and 90.3%, respectively. Despite this relatively high degree of homology, no amplification was obtained with primer pair gC-1–gC-2 and N569 DNA, which might be due to 4 base mismatches in primer gC-1. The available CapHV-1 gC sequence (strain E/CH; accession no. Z49225) was found to be 76.8% homologous to the sequence of the LA PCR product, reflecting a more distant relationship of CapHV-1 to BHV-1.
Determination of sensitivity.
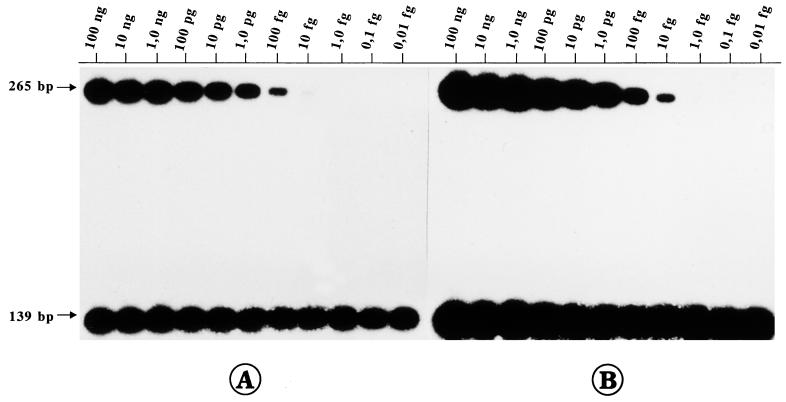
For potential diagnostic use of the PCR, it is mandatory to know the limit of detection of viral genomes. Therefore, in a series of reconstruction experiments, 10-fold dilutions of strain LA DNA (100 ng to 10 ag) were prepared with 1 μg of calf thymus (CT) DNA per μl and were used as templates in each PCR. Furthermore, to increase the sensitivity and to improve specific amplification, we established a nested PCR for gB1/2 and gE1/2, respectively. Amplification with primer pair gBN-1–gBN-2 resulted in a DNA fragment with a size of 385 bp, and amplification with primer pair gEN-1–gEN-2 generated a 139-bp DNA fragment (Table 1). Separation of the PCR products in an ethidium bromide-stained gel allowed the detection of 1.0 to 0.1 pg of viral DNA/μg of CT DNA by gB-1–gB-2 PCR and 1.0 to 0.1 fg of BHV-1 DNA after nested PCR with gBN-1–gBN-2 (data not shown). By the gE-specific PCR the sensitivity could be increased approximately 100-fold. The gE-1–gE-2 PCR detected 10 to 1 fg of LA DNA/μg of CT DNA, and less than 10 ag of viral DNA was amplified specifically after the nested PCR with gEN-1–gEN-2 (data not shown). The gC-1–gC-2 PCR exhibited the lowest limit of detection (10 pg of BHV-1 DNA/μg of CT DNA; data not shown). Finally, 32P-labelled internal probes were used for Southern blot hybridization of the PCR products, which resulted in an additional 10-fold increase in sensitivity (Fig. 3). Thus, the detection of 1.0 fg of BHV-1 DNA by gE-1–gE-2 PCR and less than 10 ag of BHV-1 DNA per μg of CT DNA by the nested gEN-1–gEN-2 PCR (Fig. 3B) was possible. According to the reported size of BHV-1 DNA (135 kbp; accession no. AJ004801), 1 fg of BHV-1 DNA corresponds to approximately 7 genomic molecules, and consequently, it can be calculated that one copy of BHV-1 DNA was detectable in approximately 5 × 104 cells (gBN-1–gBN-2) and up to at least 5 × 106 cells by gEN-1–gEN-2 PCR.
FIG. 3.
Reconstruction experiment to determine sensitivities of PCRs. Strain LA DNA was mixed together with CT DNA to obtain the indicated concentration of BHV-1 DNA per microgram of CT DNA. Southern blot hybridization of the gE1/2 and gEN1/2 PCR products with the internal probe (derived from the gEN1/2 PCR product) radioactively labelled with 32P was performed. The sizes of the specific amplification products are indicated. (A) Exposure of X-ray film for 30 min. A total of 10 fg of strain LA DNA per μg of CT DNA is detectable after gE-1–gE-2 PCR (upper part), and at least 10 ag of viral DNA per μg of CT DNA is detectable after nested gEN-1–gEN-2 PCR (lower part). (B) An increase in the time of exposure to X-ray film to 2 h demonstrates a 10-fold increase in the limit of detection.
In order to mimic the analysis of blood samples, similar reconstruction experiments were performed with blood derived from an uninfected cow. To this end, decreasing amounts of BHV-1 were mixed together with 1.0 ml of blood prior to template preparation. Results (data not shown) similar to those obtained in the reconstruction experiments with 1 μg of CT DNA (equaling approximately 105 cells) were obtained. On the average, 1.0 ml of blood from cattle contains 4 × 106 to 10 × 106 leukocytes (1). Thus, the amount of template DNA routinely used for PCR (5 μl of 100 μl of total DNA prepared from blood) corresponds to 3.5 × 105 leukocytes. This demonstrates the comparable sensitivities of the PCR assays with DNA from blood and no significant loss of BHV-1 DNA during the extraction of DNA from blood cells.
Detection of BHV-1 DNA in leukocyte samples from naturally infected cattle.
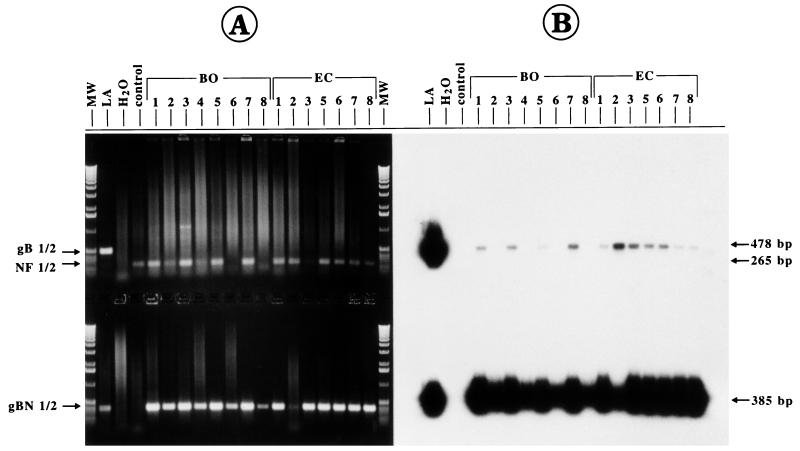
The PCR assays established for the present study were used to test blood samples derived from animals in cattle stocks considered to be infected with BHV-1. In the first case, blood was taken from animals that either showed clinical signs of IBR (animals EC-1 to EC-8; Table 2) or that were positive for serum neutralizing antibodies (titers between 1:32 and 1:128) but that were negative for virus reisolation from nasal swabs. After gB-1–gB-2 PCR and separation of the PCR products in ethidium bromide-stained gels, no gB-specific amplification could be detected (Fig. 4A, upper part). The failure to amplify a visible, specific PCR product, however, was not due to insufficient PCR of the template DNA extracted from the blood samples, as demonstrated by successful amplification of the cellular nuclear factor gene (Fig. 4A, upper part). The presence of small amounts of the gB1/2 PCR product could be revealed by Southern blot hybridization with the 32P-labelled internal gBN-1–gBN-2 probe (Fig. 4B). The diluted material of the gB-1–gB-2 PCR was then used for nested PCR with gBN-1–gBN-2, resulting in the specific DNA fragment (385 bp) in the ethidium bromide-stained gel (Fig. 4A, lower part). In addition to the water control (PCR mixture without template), DNA extracted in parallel from the blood of an uninfected animal was included as a negative control in all PCR tests (Fig. 4, lanes control). To estimate the amount of BHV-1 DNA present in the positive blood samples, the BHV-1 DNA copy number per cell (viral genomic equivalents) was calculated. This was achieved by comparing densitometrically the hybridization signal strengths of the gB1/2 fragments with those from the reconstruction experiments (as shown in Fig. 3) by using 32P-labelled probes with the same specific activities and identical film exposures. As summarized in Table 2, approximately one BHV-1 DNA copy was present in 102 to 104 leukocytes of samples from animals EC-1 to EC-8. During a productive BHV-1 infection, several hundred to a thousand viral genome copies are present in a single cell (42), and even cells latently infected with herpesvirus can harbor 10 to 100 herpesviral genome copies (34). Taking this into account, it is reasonable to assume that only one infected cell is present in 104 to 107 leukocytes or 1 to 50 infected leukocytes per ml of these blood samples. No correlation was found between the calculated virus load and the neutralizing antibody titers of the respective sera (Tables 2 and 3).
TABLE 2.
PCR revealed different copy numbers of gB and gE in PBLs
| Animal | No. of cells with one BHV-1 DNA copy by PCR witha:
|
ELISA resultb | SNT dilutionc | |
|---|---|---|---|---|
| gB-1–gB-2 | gE1–gE2 | |||
| EC-1 | 1 × 104 | 1 × 106 | NTd | 1:64 |
| EC-2 | 1 × 102 | 1 × 102 | NT | 1:128 |
| EC-3 | 5 × 102 | 5 × 104 | NT | 1:32 |
| EC-5 | 1 × 103 | 1 × 103 | NT | 1:128 |
| EC-6 | 1 × 103 | 1 × 105 | NT | 1:128 |
| EC-7 | 1 × 104 | 1 × 104 | NT | 1:32 |
| EC-8 | 1 × 104 | 1 × 104 | NT | 1:64 |
| BO-1 | 1 × 103 | 1 × 105 | Pos ? | NT |
| BO-2 | 1 × 105 | Neg | Neg | NT |
| BO-3 | 1 × 103 | 1 × 105 | Pos | NT |
| BO-4 | 1 × 105 | 1 × 106 | Neg | NT |
| BO-5 | 1 × 104 | 1 × 105 | Pos | NT |
| BO-6 | 1 × 105 | Neg | Neg | NT |
| BO-7 | 5 × 102 | 5 × 104 | Pos | NT |
| BO-8 | 1 × 105 | 1 × 106 | Neg | NT |
The calculation was performed after Southern blot hybridization, as described in Materials and Methods. Neg, negative result.
BHV-1-specific ELISA (Screening [IDEXX GmbH] and Checkit Trachitest-screeningtest [Bommeli AG, Hoechst Roussel Vet.], performed according to the instructions of the manufacturer. Pos, positive result; Pos ?, doubtful positive result; neg, negative result.
SNT, serum neutralization test without complement; with the indicated serum dilution, 100% neutralization of BHV-1.1 was obtained.
NT, not tested.
FIG. 4.
Detection of BHV-1 DNA in blood samples from field-virus-infected cattle (animals BO-1 to BO-8 and EC-1 to EC-8) after gB-1–gB-2 PCR (upper part) and gBN-1–gBN-2 PCR (lower part). As a positive control, 10 ng of strain LA DNA was used as a template (lane LA), and as negative controls, amplifications were performed with DNA derived from noninfected cattle (lane control) and in the absence of template (lane H2O). Lane MW, separation of molecular size markers (1-kbp ladder). The control NF1-NF2 PCR (265 bp) was performed separately, but the PCR products were separated together with the gB-1–gB-2 PCR products in the same gel slots. (A) The PCR products were analyzed by electrophoresis in an ethidium bromide-stained agarose gel (1.0%). No visible gB1/2 product (478 bp) was amplified by the first PCR, although in most cases a successful PCR could be demonstrated by a positive NF1-NF2 amplification (upper panel). Nested PCR (gBN-1–gBN-2; 385 bp) showed the presence of BHV-1 DNA in blood samples from all animals in herds BO and EC (lower part). (B) Southern blot hybridization of the gel shown in panel A with the radioactively labelled internal probe already revealed specific amplification in some of the blood specimens after the first PCR (upper part) and corroborated the positive results of the nested PCR (lower part).
TABLE 3.
Comparison of PCR and serological results
| Animal | gEN-1–gE-2 PCR result | ELISA result fora
|
SNT dilutionb | No. of leukocytes with one BHV-1 DNA copyc | |
|---|---|---|---|---|---|
| BHV-1 | gE | ||||
| NI-3 | Pos | Pos | Pos | 1:256 | 1 × 106 |
| NI-4 | Pos | Pos | Pos | 1:32 | 1 × 106 |
| NI-5 | Pos | Pos | Pos | 1:64 | 5 × 104 |
| FO-2 | Pos | Pos | Neg | 1:2 | 5 × 105 |
| FO-5 | Pos | Pos | Neg | Neg | 5 × 104 |
| SP-2 | Pos | Pos | Neg | 1:8 | 1 × 106 |
| SP-3 | Pos | Pos | Neg | Neg | 1 × 106 |
| FO-3 | Pos | Neg | Neg | Neg | 1 × 105 |
| FO-6 | Pos | Neg | Neg | Neg | 1 × 107 |
| FO-7 | Pos | Neg | Neg | Neg | 5 × 105 |
| FO-9 | Pos | Neg | Neg | Neg | 5 × 105 |
| FO-10 | Pos | Neg | Neg | 1:16 | 1 × 107 |
| NI-1 | Pos | Neg | Neg | 1:3 | 1 × 106 |
| NI-2 | Pos | Neg | Neg | Neg | 1 × 106 |
| SP-1 | Pos | Neg | Neg | Neg | 1 × 106 |
| SP-4 | Pos | Neg | Neg | 1:2 | 1 × 106 |
| FO-8 | Neg | Pos | Neg | Neg | |
| FO-1 | Neg | Neg | Neg | Neg | |
| FO-4 | Neg | Neg | Neg | Neg | |
A BHV-1-specific ELISA (Screening [IDEXX GmbH] and Checkit Trachitest-screeningtest [Bommeli AG, Hoechst Roussel Vet.]) and a gE-specific ELISA (HerdCheck; IDEXX GmbH) were performed. Pos, positive result; Neg, negative result.
See footnote c of Table 2. Neg, negative result.
See footnote a of Table 2.
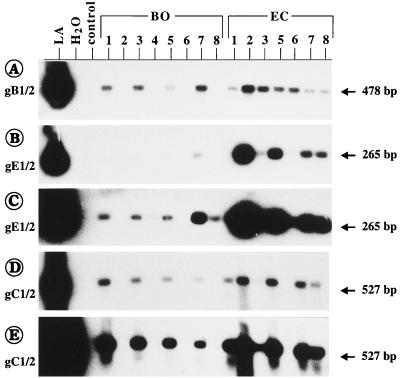
Since vaccination was performed with gE-negative live vaccine 4 to 5 months prior to this investigation, it was important to differentiate between the presence of WT BHV-1 or gE-negative vaccine virus. As shown in Fig. 4B and 5C, the samples from animals EC-1 to EC-8 were all positive after gE-1–gE-2 PCR and nested PCR with gEN-1–gEN-2 (data not shown), indicating the presence of virulent WT BHV-1.1. On the basis of the hybridization results of the gE-1–gE-2 PCR, the BHV-1 DNA copy number per cell was also estimated. Interestingly, in animals EC-1, EC-3, and EC-6 the amount of gE sequences was approximately 100-fold lower than the estimate for gB (Fig. 5; Table 2). This might indicate the simultaneous presence of WT BHV-1 and gE-negative vaccine virus in the blood of these animals.
FIG. 5.
Different copy numbers of gB and gE were found in individual blood samples (from animals BO-1 to BO-8 and EC-1 to EC-8). The amplification products obtained by the indicated PCRs (A to E; the size of each PCR fragment is indicated to the right) were blot hybridized with the respective internal probes. The X-ray films were exposed for 2 h (A), 1 h (B), 20 h (C), 7 h (D), and 68 h (D). It can be seen that the positive samples from animals in herd BO as well as the samples from animals EC-1, EC-3, and EC-6 contained 10- to 100-fold larger amounts of gB than gE.
Blood samples were investigated from animals of another herd (herd BO), which was suspected to contain BHV-1-infected animals according to positive or doubtful positive ELISA results (animals BO-1, BO-3, BO-5, and BO-7; Table 2). Blood derived from four ELISA-negative animals in this herd (animals BO-2, BO-4, BO-6, and BO-8) was also included in the analysis. None of the animals had clinical signs of IBR or infectious pustular vulvovaginitis, no previous vaccination was known, and no animal serologically positive for BHV-1 was reported in the previous year. Again, gB-1–gB-2 PCR with ethidium bromide-stained gels remained negative, but the nested PCR unequivocally demonstrated the presence of BHV-1 in all samples (Fig. 4A, lower part, lanes BO-1 to BO-8). Interestingly, Southern blot hybridization of the gB-1–gB-2 PCR products revealed positive signals only with the blood samples derived from the ELISA-positive or doubtfully positive animals (Fig. 4B, upper part, lanes BO-1, BO-3, BO-5, and BO-7), showing the presence of higher concentrations of the BHV-1 genome than the concentrations in the ELISA-negative blood samples. This is also reflected by the stronger hybridization signals of these samples after nested PCR (Fig. 4B, lower part). After blot hybridization it could be calculated from the gB-specific PCR tests that indeed only approximately one DNA copy was present in 1 × 104 to 5 × 104 leukocytes of ELISA-negative animals BO-2, BO-4, BO-6, and BO-8, whereas 5- to 100-fold higher loads of viral DNA were found in the other samples (Table 2). After gE-1–gE-2 and repeated gEN-1–gEN-2 PCR, only two animals, animals BO-2 and BO-6, respectively, remained negative (data not shown). All other animals in herd BO were positive for gE. Again, the blood samples from animals in herd BO displayed 10- to 100-fold larger amounts of gB-specific products than gE-specific products (Fig. 5; Table 2). All gB-1–gB-2 PCR-positive blood samples were also positive by gC-1–gC-2 PCR (Fig. 5D and E; Table 2).
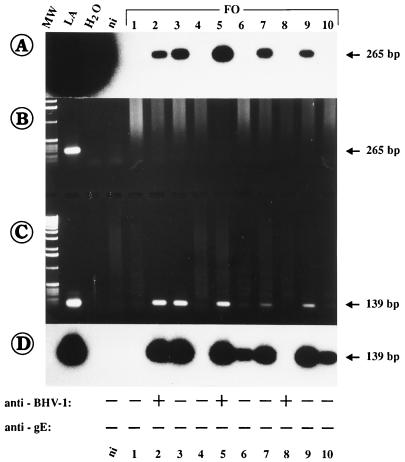
Finally, 19 different blood samples from other animals derived from three different herds (cattle stocks FO, NI, and SP) which had been vaccinated routinely were investigated. Some animals were serologically (ELISA) positive for BHV-1, but most of them were negative for BHV-1 gE-specific antibodies (Table 3), as expected after vaccination with the gE-negative live virus vaccine. Similar to the results described above, no gE1/2 amplification products were visible in ethidium bromide-stained gels (Fig. 6B). However, after gEN-1–gEN-2 PCR, amplification products with different staining intensities were obtained from most samples (Fig. 6C), and the products were clearly detectable after Southern blot hybridization (Fig. 6A and D). In addition, several of these samples were also positive after gBN-1–gBN-2 and gC-1–gC-2 PCRs (data not shown). By comparison of the ELISA results for the corresponding serum samples derived from these animals, it was found that all blood samples from animals in herd FO were positive for gE by nested PCR but were negative by the gE-specific ELISA (Fig. 6, bottom). Only three of these animals (Fig. 6, animals FO-2, FO-5, and FO-8) were serologically positive by the anti-BHV-1 ELISA. As summarized in Table 3, the PCR results demonstrated the presence of WT BHV-1 in the blood of 16 of the 19 (84.2%) animals tested.
FIG. 6.
Detection of BHV-1 DNA in blood samples (herd FO) by gE-1–gE-2 (A and B) and nested gEN-1–gEN-2 PCR (C and D). Lane ni, the result obtained with blood from noninfected cattle. The sizes of the PCR products are indicated to the right. The ELISA results for the detection of serum antibodies against BHV-1 and against gE of BHV-1 are given at the bottom for each animal. As can be seen, samples 2, 3, 5, 6, 7, 9, and 10 were clearly positive after nested gEN-1–gEN-2 PCR, but the corresponding serum samples were all negative by gE-specific ELISA.
The estimated BHV-1 DNA copy number per cell ranged between one molecule in 1 × 105 to 5 × 105 leukocytes and, in animals FO-6, FO-10, NI-1 to NI-3, and SP-1 to SP-3, which were found to be positive only after nested gEN-1–gEN-2 PCR, only one molecule in less than 1 × 106 leukocytes (Table 3). In a comparison of the PCR and serological analyses, for seven PCR-positive animals, anti-BHV-1 antibodies were also found by ELISA, but only three of them (15.8%) were also positive for gE-specific antibodies (Table 3). The remaining nine gE PCR-positive animals were found to be free of BHV-1 by both ELISAs. In total, at the time of investigation 11 of the 27 (40.7%) PCR-positive animals were recognized serologically by ELISA to be infected with BHV-1 (Table 2, animals BO-1 to BO-8, and Table 3). Notably, the blood of only one serologically positive animal was found to be reproducibly negative by PCR (Table 3, animal FO-8). For this animal the possibility that BHV-1 was present in amounts below the detection limit of the gEN-1–gEN-2 PCR (i.e., less than 10−7 infected leukocytes) cannot be excluded.
DISCUSSION
In the present study we demonstrate for the first time the successful detection of BHV-1 DNA in peripheral blood of naturally infected cattle. To achieve this goal, at first we focused on the establishment of a very sensitive PCR by testing various pairs of primers specific for different BHV-1 genes. These experiments revealed that the gE-specific PCR was the most sensitive, followed by the approximately 100 times less sensitive gB PCR and the again 10 times less sensitive gC PCR. The limits of detection were thoroughly evaluated by performing reconstruction experiments in the presence of cellular DNA and also by spiking noninfected whole blood with decreasing amounts of BHV-1 prior to DNA template preparation. The results confirmed that during DNA template preparation no significant loss of the BHV-1 genome occurs and that the methods described here can be reliably used for analysis of peripheral blood without affecting the specificity and the sensitivity of the PCR. In summary, the nested PCR protocols presented here detect a single BHV-1 DNA copy in at least 5 × 104 leukocytes (gBN-1–gBN-2 PCR) to 5 × 106 to 10 × 106 leukocytes (gEN-1–gEN-2 PCR). To our knowledge, this sensitivity is superior to those of other BHV-1-specific PCR tests reported so far. By a different gC-specific PCR, the limit of detection, as determined in the presence of DNA from noninfected cells, ranged between one molecule in approximately 102 to 103 cells (13), or three copies in 105 cells (42).
Southern blot hybridization of the gB1/2 and gE1/2 PCR products with the internal probes allowed a semiquantitative determination of the BHV-1 DNA copy number present in the different blood samples. Although less accurate than the use of an internal standard PCR product (42) or the addition of a competitor DNA to the PCR mixtures (11a), comparison of the intensities of the hybridization signals described here allows an easy and quick evaluation of the amount of infected peripheral blood leukocytes (PBLs) in each blood sample. This assessment of all 35 blood samples investigated (Tables 2 and 3) demonstrates that about 53% (18 samples) contained only between 10−5 and 10−7 infected PBLs. Even in the blood derived from animals in a herd showing acute BHV-1 infection (animals EC-1 to EC-8; Table 2) relatively small amounts of infected PBLs (10−4 to 10−2) were detectable. All samples containing more than 10−4 positive PBLs were also found to be positive by the gC-1–gC-2 PCR. Thus, the detection of genes located at the 5′ end, in the middle, and at the 3′ end of the BHV-1 genome strongly indicates the presence of the complete viral genome and, therefore, of potentially infectious virus. Our findings can now explain (i) that only rare cases of successful reisolation of BHV-1 from PBLs of infected cattle are reported (4, 5) and (ii) the recently reported failure of detection of BHV-1 DNA in a few blood samples from experimentally infected cattle by a gC-specific PCR with a detection limit lower than that of the PCR described here (42). Infection of PBLs appears to be quite common, since 92.2% of the PBLs analyzed in this study were found to harbor viral DNA. Because we analyzed exclusively naturally infected (field-virus-infected) animals, it seems unlikely that infection of PBLs depends a great deal on the virulence and the infectious dose of a given BHV-1 strain or on the route of infection (42).
The results presented here prove for the first time and substantiate earlier suggestions that only a very small number of leukocytes from infected cattle contain BHV-1 (28, 29, 39). Furthermore, the data strongly support the idea that BHV-1 can be transported via the bloodstream to target organs and confirm the tendency of the virus to accumulate in various lymphoid tissues of acutely infected animals. This is corroborated by the regular detection of viral DNA in the lungs, tonsils, tracheae, and lymph nodes of both experimentally infected (36, 43) and naturally infected (48) cattle. Monocytes are the population of PBLs that are suspected to become productively infected (28, 39); however, this remains to be proven in vivo. During BHV-1 infection lymphocytes and monocytes of the peripheral blood commonly infiltrate the submucosa of the respiratory tract, and by this route mononuclear and lymphoid cells might become infected with and disseminate the virus. By the highly sensitive PCR tests described here, these predictions can be now investigated in more detail.
The semiquantitative determination of the BHV-1 DNA copy number revealed the presence of approximately 10 to 100 times more copies of the gB gene relative to the number of copies of the gE gene in several PBLs (Fig. 5 and Table 2). These reproducible results were not due to the different amplification efficiencies of the gB and gE PCRs for these individual samples, as controlled by the NF1-NF2 PCR. Since the gE PCR is about 100 times more sensitive than the gB PCR, these data indicate the simultaneous presence of gE-positive WT and gE-negative BHV-1 strains in those PBLs. In addition, even the gC-1–gC-2 PCR, which displayed the lowest sensitivity, resulted in larger amounts of specific amplification products compared to the amounts obtained by the gE PCR for some PBL samples (Fig. 5). This is in agreement with the history of animals EC-1 to EC-8, which belonged to a cattle herd vaccinated with live gE-negative vaccine 4 to 5 months before analysis of their PBLs. One can conclude that despite vaccination, the animals became infected with BHV-1 and even suffered from IBR. It cannot be determined, however, whether BHV-1 was introduced into this herd by primary infection or reactivation of latent BHV-1. An explanation for the IBR outbreak in this herd, despite vaccination, could be that the animals were exposed to virus in the field just before or shortly after vaccination. If this assumption is correct, vaccination of already infected cattle might have an adverse effect, since both WT and gE-negative BHV-1 (live as well as inactivated virus) are known to immunosuppress the animals (2) and, consequently, might contribute to severe IBR outbreaks. With experimentally infected cattle it has been documented that live vaccination per se does not prevent WT virus infection, establishment of latency, or reactivation of latent virus (14, 21, 27, 39). Except for animals EC-1 to EC-8, all other cattle investigated in the present study did not suffer from marked, typical IBR symptoms. Therefore, it can be assumed that a subclinical BHV-1 infection had occurred in these herds. A subclinical infection that lasted for at least 6 months has been reported previously (39). Consequently, the spread of undetected virus can lead to infection of the complete herd within 7 weeks (14), which can result in BHV-1 outbreaks even in vaccinated cattle stocks (3).
The simultaneous presence of WT and gE-negative vaccine viruses has not yet been observed. This can be explained since cocultivation or explant culture techniques are used to reisolate BHV-1 from organ tissues. In the presence of both virus types, multiplication of WT BHV-1 would be favored, since the level of replication of gE-negative virus is known to be reduced significantly compared to that of WT BHV-1 (41, 45). Consequently, in the case of the presence of a mixture of WT and gE-negative BHV-1, a selective growth advantage of WT virus can be expected, and the detection of gE-negative vaccine virus might fail. Because of the lack of a gE-specific PCR, the detection of small amounts of the gE-negative vaccine virus in organs or cells is not yet possible. The incidence of two different BHV-1 strains in the same tissues of latently infected cattle has been demonstrated previously (49). According to the data presented here, the gE-negative vaccine virus was present in the PBLs of animals EC-1 to EC-8 even 5 months postvaccination, and it could also be reisolated from nasal swabs from another animal in the same herd (data not shown). The continuous presence of the gE-negative vaccine virus over such a long period of time is surprising, since vaccine virus could no longer be isolated from various organ tissues beyond 2 to 4 weeks after vaccination (42, 45). The viral genome, however, was detectable in various organs of cattle until at least 2 months after vaccination with the gE-negative live vaccine (43). The reasons for a prolonged presence of the gE-negative vaccine virus under field conditions cannot be determined from the present data.
Animals EC-1 to EC-8 exhibited clinical symptoms of progressive IBR and displayed virus neutralizing serum antibodies; therefore, their sera were not tested by ELISA. Surprisingly, 13 of 24 animals whose PBL samples were positive for BHV-1 by PCR were serologically negative for BHV-1 by ELISA (complete BHV-1 antigen) at the time of investigation (Tables 2 and 3). One explanation for this discrepancy is that WT BHV-1 infection had occurred during the first 3 weeks before the analysis of the respective PBLs, at a time when no specific antibodies or only very low antibody titers had developed, and the antibodies were not detectable by both anti-BHV-1- and gE-specific ELISAs (21, 46). Indeed, the sera of animals BO-2, BO-4, BO-6, and BO-8 (Table 2) were positive by the BHV-1 ELISA approximately 2 weeks later. Numerous animals which were identified as vaccinated cattle according to the BHV-1-positive but gE-negative ELISA results were found. However, the PCR results demonstrated the presence of gE-positive WT virus in their PBLs (Table 3). The titers of anti-gE serum antibodies were probably still too low to be detected by ELISA, as reported previously (9, 22). Whether these animals had seroconverted is not known, because later serum samples from most of the animals tested were not yet available; samples were available from two animals. For these animals (animals NI-1 and NI-2) later ELISAs remained negative. Therefore, it remains to be shown that under field conditions seronegative animals are also BHV-1 infected and that those carrier animals can represent an important source for the unnoticed spread of BHV-1. Recently, reactivation of latent BHV-1 was shown to occur in BHV-1-seronegative animals (15). It is possible that either adult cattle can become seronegative (40) or infected calves containing maternal antibodies until up to 9 months of age become virtually seronegative (24).
Taken together, the PCR protocols described here enable the identification of BHV-1-infected cattle before detectable seroconversion has occurred. Further studies are now needed to define more precisely the time window after infection during which BHV-1 DNA can be found in PBLs, before specific antibodies are generated, and how long after infection BHV-1 DNA can be detected in PBLs. Finally, the results presented here encourage further investigations on the duration and importance of viremia in naturally and experimentally infected cattle and should allow the identification of distinct infected populations of bovine PBLs.
ACKNOWLEDGMENTS
This study was initiated and supported by the Bundesministerium für Ernährung, Landwirtschaft und Forsten, of the Federal Republic of Germany.
Primers gE-1 and gE-2 were kindly provided by Tobias Schlapp, Bayer AG. We thank Mathias Büttner and Lothar Stitz (Federal Research Centre for Virus Diseases of Animals, Tübingen, Germany) for valuable discussions, and Rebecca Sparks-Thissen (Princeton University, Princeton, N.J.) for critically commenting on the manuscript.
REFERENCES
- 1.Ayoub I A, Yang T J. Age-dependent changes in peripheral blood lymphocyte subpopulations in cattle: a longitudinal study. Dev Comp Immunol. 1996;20:353–363. doi: 10.1016/s0145-305x(96)00024-9. [DOI] [PubMed] [Google Scholar]
- 2.Babiuk L A, van Drunen Little-van den Hurk S, Tikoo S K. Immunology of bovine herpesvirus 1 infection. Vet Microbiol. 1996;53:31–42. doi: 10.1016/s0378-1135(96)01232-1. [DOI] [PubMed] [Google Scholar]
- 3.Bosch J C, DeJong M C M, Franken P, Frankena K, Hage J J, Kaashoek M J, Maris-Veldhuis M A, Noordhuizen J P T M, Van der Poel W H M, Verhoeff J, Weerdmeester K, Zimmer G M, Van Oirschot J T. An inactivated gE-negative marker vaccine and an experimental gD-subunit vaccine reduce the incidence of bovine herpesvirus 1 infection in the field. Vaccine. 1998;16:265–271. doi: 10.1016/s0264-410x(97)00166-7. [DOI] [PubMed] [Google Scholar]
- 4.Brenner J, Ungar-Waron H, Elad D, Abraham A. Clinical and serological follow up of young calves experimentally infected with virulent bovine herpesvirus (BHV-1) Isr J Vet Med. 1989;45:21–25. [Google Scholar]
- 5.Castrucci G, Ferrari M, Traldi V, Tartaglione E. Effects in calves of mixed infections with bovine viral diarrhea virus and several other bovine viruses. Comp Immunol Microbiol Infect Dis. 1992;15:261–270. doi: 10.1016/0147-9571(92)90005-c. [DOI] [PubMed] [Google Scholar]
- 6.Chester P M, Allsop R, Purewal A, Edington N. Detection of latency-associated transcripts of equid herpesvirus 1 in equine leukocytes but not in trigeminal ganglia. J Virol. 1997;71:3437–3443. doi: 10.1128/jvi.71.5.3437-3443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinsakchai S, Molitor T W. Immunobiology of pseudorabies virus infection in swine. Vet Immunol Immunpathol. 1994;43:107–116. doi: 10.1016/0165-2427(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 8.De Gee A L W, Wagter L H A, Hage J J. The use of polymerase chain reaction assay for the detection of bovine herpesvirus 1 in semen during a natural outbreak of infectious bovine rhinotracheitis. Vet Microbiol. 1996;53:163–168. doi: 10.1016/s0378-1135(96)01244-8. [DOI] [PubMed] [Google Scholar]
- 9.De Wit J J, Hage J J, Brinkhof J, Westenbrink F. A comparative study of serological tests for use in the bovine herpesvirus 1 eradication programme in The Netherlands. Vet Microbiol. 1998;61:153–163. doi: 10.1016/s0378-1135(98)00166-7. [DOI] [PubMed] [Google Scholar]
- 10.Engels M, Loepfe E, Wild P, Schraner E, Wyler R. The genome of caprine herpesvirus 1: genome structure and relatedness to bovine herpesvirus 1. J Gen Virol. 1987;68:2019–1023. doi: 10.1099/0022-1317-68-7-2019. [DOI] [PubMed] [Google Scholar]
- 11.Enquist L W. Infection of the mammalian nervous system by pseudorabies virus (PRV) Semin Virol. 1994;5:221–231. [Google Scholar]
- 11a.Fischer, T., and H.-J. Rziha. Unpublished data.
- 12.Forman A J, Babiuk L A. Effect of infectious bovine rhinotracheitis virus infection on bovine alveolar macrophage function. Infect Immun. 1982;35:1041–1047. doi: 10.1128/iai.35.3.1041-1047.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galeota J A, Flores E F, Kit S, Kit M, Osorio F A. A quantitative study of the efficacy of a deletion mutant bovine herpesvirus-1 differential vaccine in reducing the establishment of latency by wildtype virus. Vaccine. 1997;15:123–128. doi: 10.1016/s0264-410x(96)00165-x. [DOI] [PubMed] [Google Scholar]
- 14.Hage J J, Schukken Y H, Barkeman H W, Benedictus G, Rijsewijk F A M, Wentink G H. Population dynamics of bovine herpesvirus 1 infection in a dairy herd. Vet Microbiol. 1996;53:169–180. doi: 10.1016/s0378-1135(96)01245-x. [DOI] [PubMed] [Google Scholar]
- 15.Hage J J, Glas R D, Westra H H, Maris-Veldhuis M A, van Oirschot J T, Rijsewijk F A M. Reactivation of latent bovine herpesvirus 1 in cattle seronegative to glycoproteins gB and gE. Vet Microbiol. 1998;60:87–98. doi: 10.1016/s0378-1135(97)00152-1. [DOI] [PubMed] [Google Scholar]
- 16.Hattori M, Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986;152:232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- 16a.Hübert, P. Unpublished data.
- 17.Kaashoek M J, Moerman A, Madic J, Rijsewijk F A M, Quak J, Gielkens A L J, Van Oirschot J T. A conventionelly attenuated glycoprotein E-negative strain of bovine herpesvirus type 1 is an efficacious and safe vaccine. Vaccine. 1994;12:439–444. doi: 10.1016/0264-410x(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 18.Kaashoek M J, Rijsewijk F A M, Van Oirschot J T. Persistence of antibodies against bovine herpesvirus 1 and virus reactivation two or three years after infection. Vet Microbiol. 1996;53:103–110. doi: 10.1016/s0378-1135(96)01238-2. [DOI] [PubMed] [Google Scholar]
- 19.Kaashoek M J, Straver P J, Van Rooji E M A, Quak J, Van Oirschot J T. Virulence, immunogenicity and reactivation of seven bovine herpesvirus 1.1 strains: clinical and virological aspects. Vet Rec. 1996;139:416–421. doi: 10.1136/vr.139.17.416. [DOI] [PubMed] [Google Scholar]
- 20.Kibenge F S B, Harris L M, McKenna P K, Wadowska D, Yason C V. Amplification of strains of bovine herpesvirus 1 by use of polymerase chain reaction with primers in the thymidine kinase region. Am J Vet Res. 1994;55:1206–1212. [PubMed] [Google Scholar]
- 21.Kramps J A, Magdalena J, Quak J, Weerdmeester K, Kaashoek M J, Maris-Veldhuis M A, Rijsewijk F A M, Keil G, van Oirschot J T. A simple, specific, and highly sensitive blocking enzyme-linked immunosorbent assay for detection of antibodies to bovine herpesvirus 1. J Clin Microbiol. 1994;32:2175–2181. doi: 10.1128/jcm.32.9.2175-2181.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramps J A, Perrin B, Edwards S, Van Oirschot J T. A European interlaboratory trial to evaluate the reliability of serological diagnosis of bovine herpesvirus 1 infections. Vet Microbiol. 1996;53:153–161. doi: 10.1016/s0378-1135(96)01243-6. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence W C, D’Urso R C, Kunkel C A, Whitbeck J C, Bello L J. Map location of the gene for a 130,000-dalton glycoprotein of bovine herpesvirus 1. J Virol. 1986;60:405–414. doi: 10.1128/jvi.60.2.405-414.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaire M, Meyer G, Ernst E, Vanherrwege V, Limbourg B, Pastoret P P, Thiry E. Latent bovine herpesvirus 1 infection in calves protected by colostral immunity. Vet Rec. 1995;137:70–71. doi: 10.1136/vr.137.3.70. [DOI] [PubMed] [Google Scholar]
- 25.Lyaku J R S, Vilcek S, Nettleton P F, Marsden H S. The distinction of serologically related ruminant alphaherpesviruses by the polymerase chain reaction (PCR) and restriction endonuclease analysis. Vet Microbiol. 1996;48:135–142. doi: 10.1016/0378-1135(95)00136-0. [DOI] [PubMed] [Google Scholar]
- 26.Mweene A S, Okazaki K, Kida H. Detection of viral genome in non-neural tissue of cattle experimentally infected with bovine herpesvirus 1. Jpn J Vet Res. 1996;44:165–174. [PubMed] [Google Scholar]
- 27.Nettleton P F, Sharp J M, Herring A J. Infectious bovine rhinotracheitis virus excretion after vaccination, challenge and immunosuppression. In: Wittmann G, Gaskell R M, Rziha H-J, editors. Latent herpesvirus infections in veterinary medicine. Boston, Mass: Martinus Nijhoff; 1984. pp. 191–209. [Google Scholar]
- 28.Nyaga P N, McKercher D G. Pathogenesis of bovine herpesvirus-1 (BHV-1) infections: interactions of the virus with peripheral bovine blood cellular components. Comp Immunol Microbiol Infect Dis. 1980;2:587–602. doi: 10.1016/0147-9571(79)90100-0. [DOI] [PubMed] [Google Scholar]
- 29.Pastoret P-P, Thiry E, Brochier B, Derboven G, Vindevogel H. The role of latency in the epizootiology of infectious bovine rhinotracheitis. In: Wittmann G, Gaskell R, Rziha H-J, editors. Latent herpesvirus infections in veterinary medicine. Boston, Mass: Martinus Nijhoff; 1984. pp. 211–228. [Google Scholar]
- 30.Reed K C, Mann K I D A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985;13:7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roizman B. Family Herpesviridae. Classification and nomenclature of viruses. 5th Report of the International Committee on Taxonomy of Viruses. Arch Virol. 1991;2(Suppl.):103–110. [Google Scholar]
- 32.Rossi C R, Kiesel G K. Susceptibility of bovine macrophages and tracheal ring cultures to bovine viruses. Am J Vet Res. 1977;38:1705–1708. [PubMed] [Google Scholar]
- 33.Rziha H-J, Mettenleiter T C, Wittmann G. Occurrence of pseudorabies virus DNA in latently infected pigs. Curr Top Vet Med Anim Sci. 1984;27:429–444. [Google Scholar]
- 34.Rziha H-J, Mettenleiter T C, Ohlinger V, Wittmann G. Herpesvirus (pseudorabies virus) latency in swine: occurrence and physical state of viral DNA in neural tissues. Virology. 1986;155:600–613. doi: 10.1016/0042-6822(86)90220-5. [DOI] [PubMed] [Google Scholar]
- 35.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain- terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santurde G, Da Silva N, Villares R, Tabares E, Solana A, Bautista J M, Castro J M. Rapid and high sensitivity test for direct detection of bovine herpesvirus-1 genome in clinical samples. Vet Microbiol. 1996;49:81–92. doi: 10.1016/0378-1135(95)00169-7. [DOI] [PubMed] [Google Scholar]
- 37.Schang L M, Kutish G F, Osorio F A. Correlation between precolonization of trigeminal ganglia by attenuated strains of pseudorabies virus and resistance to wild-type virus latency. J Virol. 1994;68:8470–8476. doi: 10.1128/jvi.68.12.8470-8476.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strube W, Auer S, Block W, Heinen E, Kretzdorn D, Rodenbach C, Schmeer N. A gE deleted infectious bovine rhinotracheitis marker vaccine for use in improved bovine herpesvirus 1 control programs. Vet Microbiol. 1996;53:181–189. doi: 10.1016/s0378-1135(96)01246-1. [DOI] [PubMed] [Google Scholar]
- 39.Tikoo S K, Campos M, Babiuk L A. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis and control. Adv Virus Res. 1995;45:191–223. doi: 10.1016/s0065-3527(08)60061-5. [DOI] [PubMed] [Google Scholar]
- 40.Van der Poel W H M, Kramps J A, Quak J, Brand A, van Oirschot J T. Persistence of bovine herpesvirus 1-specific antibodies in cattle after intranasal vaccination with a live virus vaccine. Vet Rec. 1995;137:347–348. doi: 10.1136/vr.137.14.347. [DOI] [PubMed] [Google Scholar]
- 41.Van Engelenburg F A C, Kaashoek M J, Rijsewijk F A M, Van den Burg L, Moerman A, Gielkens A L J, Van Oirschot J T. A glycoprotein E deletion mutant of bovine herpesvirus 1 is avirulent in calves. J Gen Virol. 1994;75:2311–2318. doi: 10.1099/0022-1317-75-9-2311. [DOI] [PubMed] [Google Scholar]
- 42.Van Engelenburg F A C, Kaashoek M J, Van Oirschot J T, Rijsewijk F A M. A glycoprotein E deletion mutant of bovine herpesvirus 1 infects the same limited number of tissues in calves as wild type virus, but for a shorter period. J Gen Virol. 1995;76:2387–2392. doi: 10.1099/0022-1317-76-9-2387. [DOI] [PubMed] [Google Scholar]
- 43.Van Engelenburg F A C, Van Schie F W, Rijsewijk F A M, Van Oirschot J T. Excretion of bovine herpesvirus 1 in semen is detected much longer by PCR than by virus isolation. J Clin Microbiol. 1995;33:308–312. doi: 10.1128/jcm.33.2.308-312.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Oirschot J T, Rziha H-J, Moonen P J L, Pol J M A, van Zaane D. Differentiation of serum antibodies from pigs vaccinated or infected with Aujeszky’s disease virus by a competitive enzyme immunoassay. J Gen Virol. 1986;67:1179–1182. doi: 10.1099/0022-1317-67-6-1179. [DOI] [PubMed] [Google Scholar]
- 45.Van Oirschot J T, Kaashoek M J, Rijsewijk F A M, Stegeman J A. The use of marker vaccines in eradication of herpesviruses. J Biotechnol. 1996;44:75–81. doi: 10.1016/0168-1656(95)00129-8. [DOI] [PubMed] [Google Scholar]
- 46.Van Oirschot J T, Kaashoek M J, Maris-Veldhuis M A, Weerdmester K, Rijsewijk F A M. An enzyme-linked immunosorbent assay to detect antibodies against glycoprotein gE of bovine herpesvirus 1 allows differentiation between infected and vaccinated cattle. J Virol Methods. 1997;67:23–34. doi: 10.1016/s0166-0934(97)00073-6. [DOI] [PubMed] [Google Scholar]
- 47.Vilcek S. Detection of bovine herpesvirus-1 (BHV-1) genome by PCR. J Virol Methods. 1993;41:245–248. doi: 10.1016/0166-0934(93)90132-b. [DOI] [PubMed] [Google Scholar]
- 48.Vilcek S, Nettleton P F, Herring A J. Detection of bovine herpesvirus 1 in clinical samples by the polymerase chain reaction. Dtsch Tierärztl Wochenschr. 1995;102:219–258. [PubMed] [Google Scholar]
- 49.Whetstone C, Miller J. Two different strains of an alphaherpesvirus can establish latency in the same tissue of the host animal: evidence from bovine herpesvirus 1. Arch Virol. 1989;107:27–34. doi: 10.1007/BF01313875. [DOI] [PubMed] [Google Scholar]
- 50.Wiedmann M, Brandon R, Wagner P, Dubovi E J, Bat C A. Detection of bovine herpesvirus 1 in bovine semen by a nested PCR assay. J Virol Methods. 1993;44:129–140. doi: 10.1016/0166-0934(93)90015-j. [DOI] [PubMed] [Google Scholar]
- 51.Wittmann G, Rziha H-J. Aujeszky’s disease (Pseudorabies) in pigs. In: Wittmann G, editor. Herpesvirus diseases of cattle, horses, and pigs. Boston, Mass: Kluwer Academic Publishers; 1989. pp. 230–325. [Google Scholar]
- 52.Wyler R, Engels M, Schwyzer M. Infectious bovine rhinotracheitis/vulvovaginitis (BHV 1) In: Wittmann G, editor. Herpesvirus diseases of cattle, horses, and pigs. Boston, Mass: Kluwer Academic Publishers; 1989. pp. 1–72. [Google Scholar]
- 53.Xia J Q, Lofstedt R M, Yason C V, Kibenge F S B. Detection of bovine herpesvirus-1 in the semen of experimentally infected bulls by dot-blot hybridisation, polymerase chain reaction and virus isolation. Res Vet Sci. 1995;59:183–185. doi: 10.1016/0034-5288(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 54.Xu A M, Jevnikar V E, Rubin-Kelley E. A simple method for the preparation of chromosomal DNA from cell culture. Nucleic Acids Res. 1990;18:4943. doi: 10.1093/nar/18.16.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]