Abstract
The aim of this study is to determine the roles of global histone acetylation (Ac)/methylation (me), their modifying enzymes, and gene-specific histone enrichment in obstructive sleep apnea (OSA). Global histone modifications, and their modifying enzyme expressions were assessed in peripheral blood mononuclear cells from 56 patients with OSA and 16 matched subjects with primary snoring (PS). HIF-1α gene promoter-specific H3K36Ac enrichment was assessed in another cohort (28 OSA, 8 PS). Both global histone H3K23Ac and H3K36Ac expressions were decreased in OSA patients versus PS subjects. H3K23Ac expressions were further decreased in OSA patients with prevalent hypertension. HDAC1 expressions were higher in OSA patients, especially in those with excessive daytime sleepiness, and reduced after more than 6 months of continuous positive airway pressure treatment. H3K79me3 expression was increased in those with high C-reactive protein levels. Decreased KDM6B protein expressions were noted in those with a high hypoxic load, and associated with a higher risk for incident cardiovascular events or hypertension. HIF-1α gene promoter-specific H3K36Ac enrichment was decreased in OSA patients versus PS subjects. In vitro intermittent hypoxia with re-oxygenation stimuli resulted in HDAC1 over-expression and HIF-1α gene promoter-specific H3K36Ac under-expression, while HDAC1 inhibitor, SAHA, reversed oxidative stress through inhibiting NOX1. In conclusions, H3K23/H3K36 hypoacetylation is associated with the development of hypertension and disease severity in sleep-disordered breathing patients, probably through up-regulation of HDAC1, while H3K79 hypermethylation is associated with higher risk of cardiovascular diseases, probably through down-regulation of KDM6B.
Subject terms: Biomarkers, Diseases, Medical research, Molecular medicine
Introduction
Affecting approximately 20–30% of men and 10–15% of women, obstructive sleep apnea (OSA) is characterized by chronic intermittent hypoxia with re-oxygenation (IHR) injury, and associated with the increased risks for cardiovascular disease, metabolic syndrome, and all-cause mortality1–3. Randomized controlled trials show that continuous positive airway pressure (CPAP) treatment can improve hypertension and insulin resistance, but does not reduce risks of cardiovascular outcomes or death for patients with OSA4,5.
In response to external stimuli, such as hypoxia, epigenetic modification can render regulatory elements of genes more or less permissive to interaction with the transcriptional machinery6. In our previous epigenome-wide study, we identified and replicated a specific association between a high hypoxic load and IL1R2 hypomethylation/AR hypermethylation, as well as between excessive daytime sleepiness and NPR2 hypomethylation/SP140 hypermethylation in the DNA samples from OSA patients7. Histone (H) acetylation (Ac) occurs on lysine (K) residues, and promotes relaxation of chromatin structure and transcriptional activation, while histone methylation (me) occurs on lysine (K) and arginine residues on histones H3 and H4, and can be associated with either activation or repression of transcription, depending on the degree (trimethylation (me3); dimethylation (me2); monomethylation (me1)) and location of the modification. Methylation at H3K4, H3K36, H3K79, or H4K20 is associated with transcriptional activation, whereas methylation at H3K9, H3K23, H3K27, and H3K56 is linked with transcriptional repression8–10. Histone modifications are mediated by histone-modifying enzymes, including histone acetyltransferases (HAT), histone deacetylases (HDACs), histone methyltransferases, and histone lysine demethylases (KDMs)11,12.
Based on previous findings of the relationships between histone modifications and persistent hypoxia13, it is hypothesized that OSA patients have aberrant global histone methylation/acetylation patterns and altered expressions of their corresponding enzymes, which may affect disease severity, clinical phenotypes, and outcomes in this chronic IHR-mediated disease. To test this hypothesis, this case–control study checked eleven global histone modification expressions and their corresponding enzymes in the peripheral blood mononuclear cells (PBMCs) from 56 OSA patients and 16 primary snoring (PS) subjects. We correlated these histone markers with the occurrence of prevalent and incident co-morbidities in the following 3 years, and re-checked them in 8 OSA patients who received home CPAP treatment for > 1/2 year (self-reported use for more than 4 h/night)14. Furthermore, hypoxia inducible factor (HIF)-1α and HIF-2α promoter specific H3K36Ac and H3K23Ac enrichment was determined in another cohort of 8 PS subjects and 28 treatment-naïve OSA patients, and an in vitro IHR cell culture model was used to verify these molecular changes.
Results
A total of 16 PS subjects and 56 patients with treatment-naïve OSA were enrolled and analyzed (cohort 1). Demographic, PSG, and blood chemistry data of all the study participants are presented in Table 1. There were no significant differences between two groups in terms of age, gender, BMI, current smoking, alcoholism, co-morbidities, blood cholesterol/triglyceride level, and fasting blood sugar. There were significant differences in PSG parameters and serum hypersensitivity C-reactive protein (hsCRP) levels between two groups.
Table 1.
Demographic, biochemistry, and sleep data of all the 108 study participants.
| Cohort-1 | Cohort-2 | |||||
|---|---|---|---|---|---|---|
| PS subjects (n = 16) | OSA patients(n = 56) | p value | PS subjects (n = 8) | OSA patients (n = 28) | p value | |
| Age, years | 41.1 ± 12.8 | 43.3 ± 8.6 | 0.532 | 44.8 ± 9.3 | 50.2 ± 12.4 | 0.262 |
| Male Sex, n (%) | 14 (87.5) | 50 (89.3) | 0.376 | 6 (75) | 23 (82.1) | 0.653 |
| BMI, kg/m2 | 25.2 ± 2.9 | 25.9 ± 2.8 | 0.383 | 24.6 ± 3 | 26.6 ± 3.1 | 0.114 |
| AHI, events/hour | 3.7 ± 2.5 | 66.9 ± 12 | < 0.001 | 3.2 ± 2.6 | 57.2 ± 2.1 | < 0.001 |
| ODI, events/hour | 2.1 ± 2.2 | 55.9 ± 20.7 | < 0.001 | 1.2 ± 1.2 | 45.7 ± 25.9 | < 0.001 |
| Mean SaO2, % | 96.4 ± 1 | 93.6 ± 2.4 | < 0.001 | 95.8 ± 0.8 | 93.7 ± 2.5 | 0.003 |
| Minimum SaO2, % | 86.1 ± 10.1 | 67.5 ± 14.6 | < 0.001 | 91.2 ± 3.6 | 70.2 ± 10.7 | < 0.001 |
| Snoring index, counts/hour | 107 ± 201.5 | 377.8 ± 197.7 | < 0.001 | 66.4 ± 60.8 | 358.5 ± 174.3 | < 0.001 |
| hs CRP, mg/L | 1.79 ± 1.11 | 3.64 ± 4.73 | 0.013 | NA | NA | |
| ESS | 9.7 ± 4.1 | 10.6 ± 5.5 | 0.553 | 9.6 ± 5.7 | 10.3 ± 4.9 | 0.776 |
| EDS, n (%) | 4 (21.4) | 16 (88.9) | < 0.001 | 2 (33.3) | 10 (41.7) | 0.709 |
| Current smoking, n (%) | 6 (37.5) | 21 (44.7) | 0.616 | 2 (28.6) | 9 (32.1) | 0.856 |
| Alcoholism, n (%) | 0 (0) | 3 (6.4) | 0.3 | 0 (0) | 2 (7.1) | 0.466 |
| Cholesterol, mg/dl | 201.5 ± 42.6 | 196.3 ± 33.5 | 0.616 | 180.4 ± 37.6 | 196.2 ± 42.3 | 0.442 |
| Triglycerides, mg/dl | 166.2 ± 130.7 | 153.5 ± 86.5 | 0.566 | 131.6 ± 137.2 | 137.2 ± 89.3 | 0.9 |
| Fasting sugar, mg/dl | 100.9 ± 18.8 | 100.7 ± 11.8 | 0.944 | 98 ± 6.4 | 100.6 ± 25.5 | 0.823 |
| Hypertension, n (%) | 3 (18.8) | 18 (35.3) | 0.213 | 2 (25) | 9 (32.1) | 0.699 |
| DM, n (%) | 1 (6.3) | 2 (3.9) | 0.694 | 1 (12.5) | 4 (14.3) | 0.898 |
| Heart disease, n (%) | 1 (6.3) | 4 (7.8) | 0.832 | 0 (0) | 0 (0) | 1 |
| Stroke, n (%) | 0 (0) | 4 (7.8) | 0.248 | 1 (12.5) | 0 (0) | 0.058 |
| CKD, n (%) | 0 | 1 (1.8) | 0.59 | 0 (0) | 0 (0) | 1 |
PS primary snoring, OSA obstructive Sleep Apnea, BMI body mass index, AHI apnea hypopnea index, SaO2 arterial oxyhemoglobin saturation, ODI oxygen desaturation index, the number of dips > 4% of basal SaO2%//hour, hsCRP hypersensitivity C-reactive protein, ESS Epworth Sleepiness Scale, EDS excessive daytime sleepiness, DM diabetes mellitus, CKD chronic kidney disease.
Differential global histone methylation/acetylation patterns related to OSA and its clinical phenotypes
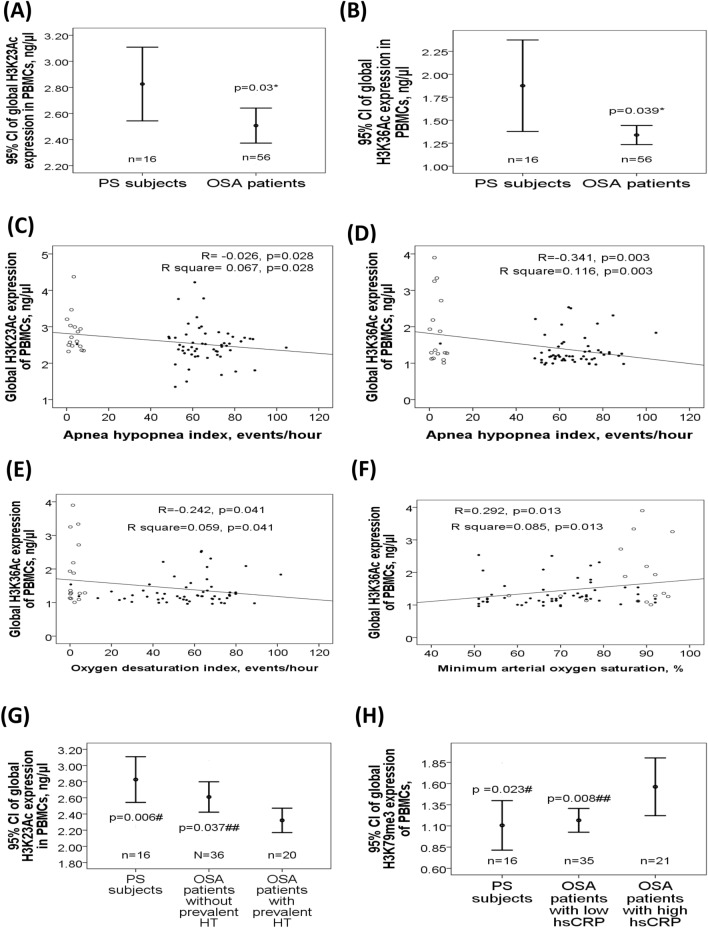
To determine the role of global histone acetylation/methylation in the development of OSA and comorbidities, protein levels of H3K9Ac, H3K14Ac, H3K23Ac, H3K36Ac, H3K56Ac, H4K16Ac, H3K4me3, H3K9me3, H3K27me3, H3K36me3, and H3K79me3 were measured in the PBMC samples from cohort 1. These modification sites were chosen based on their known biological effects on gene expressions and the accessibility of the ELISA kits8–10. Both global histone H3K23Ac (2.5 ± 0.51 versus 2.83 ± 0.53 ng/μl, adjusted p = 0.03, Fig. 1A) and H3K36Ac (1.34 ± 0.39 versus 1.88 ± 0.94 ng/μl, adjusted p = 0.039, Fig. 1B) expressions were significantly decreased in OSA patients as compared with that in PS subjects, while the other global histone modification expressions were not different between OSA and PS groups (Supplementary Fig. S1). H3K23Ac expression was negatively correlated with both AHI (r = − 0.26, p = 0.028, Fig. 1C).The correlation remained statistically significant in multivariate linear regression analysis model 2 (Supplementary Table S1), but did not reach statistical significance in the analysis of separate PS and OSA groups. H3K36Ac expression was negatively correlated with both AHI (r = − 0.341, p = 0.003, Fig. 1D) and ODI (r = − 0.242, p = 0.041, Fig. 1E), and positively correlated with minimum SaO2 (r = 0.292, p = 0.013, Fig. 1F). The three correlations remained statistically significant in multivariate linear regression analyses model 1 or 2 (Supplementary Tables S2, S3, S4), but were reduced in separate PS and OSA groups. Subgroup analysis revealed that global histone H3K23Ac expression was further decreased in OSA patients with prevalent hypertension (n = 20, 2.32 ± 0.32 ng/μl, Fig. 1G) versus those without prevalent hypertension (n = 36, 2.61 ± 0.56 ng/μl, p = 0.042/adjusted p = 0.037) or PS subjects (p = 0.004/adjusted p = 0.006). Furthermore, global histone H3K79me3 expression was increased in OSA patients with high serum hsCRP levels (n = 21, hsCRP > 3; 1.56 ± 0.71 ng/μl; Fig. 1H) versus those with low serum hsCRP levels (n = 35, hsCRP≦3; 1.17 ± 0.4 ng/μl, p = 0.005/adjusted p = 0.008) or PS subjects (1.11 ± 0.51 ng/μl, p = 0.048/adjusted p = 0.023)15.
Figure 1.
Differential global histone methylation/acetylation patterns related to obstructive sleep apnea (OSA) and its clinical phenotypes. Both (A) global histone H3K23Ac and (B) H3K36 Ac expressions were decreased in OSA patients. (C) H3K23Ac expression was negatively correlated with apnea hypopnea index (AHI). H3K36Ac expression was negatively correlated with both (D) AHI and (E) oxygen desaturation index (ODI), and positively with (F) minimum oxygen saturation (SaO2). (G) H3K23Ac expression was further decreased in OSA patients with prevalent hypertension. (H) H3K79me3 expression was increased in OSA patients with high serum hypersensitivity C-reactive protein (hsCRP) levels. *adjust for age, body mass index, gender, smoking history, alcoholism history, and co-morbidities (diabetes mellitus, hypertension, stroke, cardiac disease, chronic kidney disease) by multivariate linear regression analysis. #compared between primary snoring (PS) subjects and obstructive sleep apnea (OSA) patients with a particular phenotype (hypertension or high hsCRP), adjusted by multivariate linear regression analysis. ##compared between OSA patients with a particular phenotype (hypertension or high hsCRP) and those without the phenotype, adjusted by multivariate linear regression analysis. Hollow circle indicates PS subjects; solid star indicates OSA patients. PBMC = peripheral blood mononuclear cell; CI = confidence interval; HT = hypertension.
Gene and protein expression changes of specific histone modifying enzymes responsible for the global histone methylation/acetylation patterns
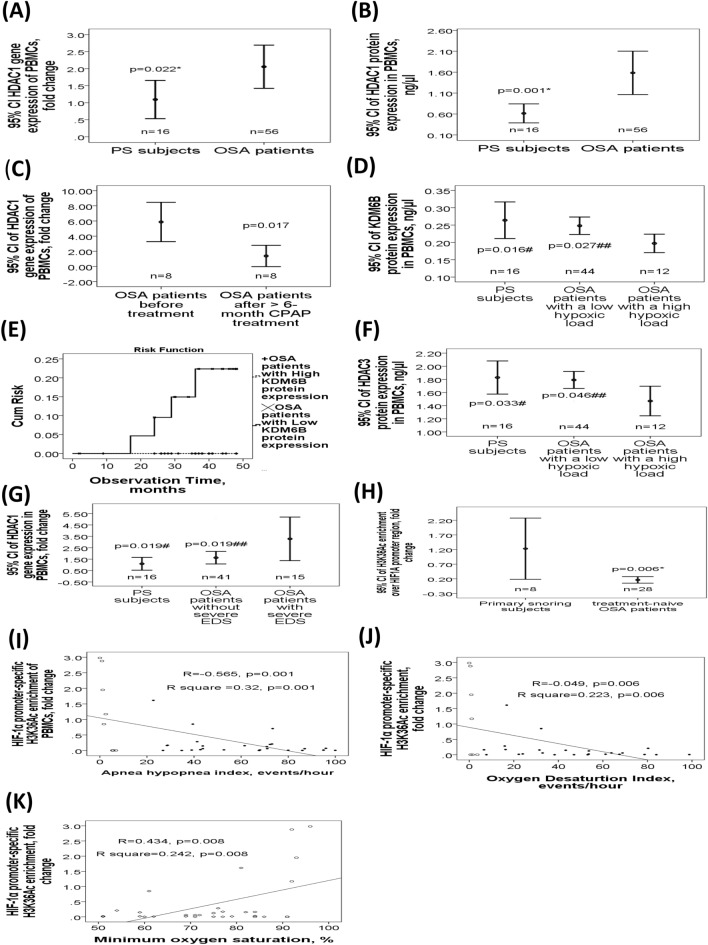
To determine the role of specific histone modifying enzymes in the development of OSA and comorbidities, gene expression levels of the KDM1A, KDM4B, KDM4C, KDM5A-D, KDM6B, DOT1L, HDAC1-7, KAT2A, and KAT6B genes, and protein levels of the KDM1A, KDM4, KDM5, KDM6B, and HDAC1-4 were measured in the PBMC samples. These enzymes were chosen based on their known biological effects on specific histone modification patterns and the accessibility of the ELISA kits. Both HDAC1 gene (2.06 ± 2.33 versus 1.09 ± 0.88 fold change, adjusted p = 0.022; Fig. 2A) and HDAC1 protein (1.58 ± 1.94 versus 0.61 ± 0.43 ng/μl, adjusted p = 0.001; Fig. 2B) expressions were significantly higher in OSA patients than that in PS subjects, while HDAC1 gene expression was significantly reduced after more than 6 months of CPAP treatment in 8 selected OSA patients (1.38 ± 1.52 versus 5.96 ± 3.08 fold change, p = 0.017; Fig. 2C). There was no significant difference of gene or protein expression in the other histone modifying enzymes between two groups (Supplementary Figs. S2, S3 and S4). Subgroup analysis showed that KDM6B protein expression was decreased in OSA patients with a high hypoxic load (n = 12, ODI > 70 events/hour; 0.2 ± 0.04 ng/μl; Fig. 2D) versus those without a high hypoxic load (n = 44; 0.25 ± 0.08 ng/μl, p = 0.017/adjusted p = 0.027) or PS subjects (0.26 ± 0.1 ng/μl, p = 0.036/adjusted p = 0.016). OSA patients with low KDM6B protein expression (< 0.21 ng/μl, n = 23) had higher risk for incident cardiovascular events or hypertension in the following 3 years (p = 0.014 by Log-Rank test, Fig. 2E) than those with high KDM6B protein expression (≧0.21 ng/μl, n = 33). HDAC3 protein expression was decreased in OSA patients with a high hypoxic load (1.5 ± 0.35 ng/μl; Fig. 2F) versus those without a high hypoxic load (1.8 ± 0.43 ng/μl, p = 0.024/adjusted p = 0.046) or PS subjects (1.83 ± 0.47 ng/μl, p = 0.031/adjusted p = 0.033). HDAC1 gene expression was increased in OSA patients with severe excessive daytime sleepiness (EDS) (n = 15, ESS > 14; 3.28 ± 3.31 fold change; Fig. 2G) versus those without severe EDS (n = 41; 1.63 ± 1.72 fold change, p = 0.014/adjusted p = 0.019) or PS subjects (1.09 ± 0.88 fold change, p = 0.01/adjusted p = 0.019).
Figure 2.
Specific gene and protein expression changes of specific histone modifying enzymes related to OSA and its clinical phenotypes. Both (A) HDAC1 gene and (B) HDAC1 protein expressions were higher in OSA patients. (C) HDAC1 gene expression was reduced after more than 6 months of continuous positive airway pressure (CPAP) treatment in 8 selected OSA patients. (D) KDM6B protein expression was significantly decreased in OSA patients with high hypoxic load. (E) OSA patients with low KDM6B protein expression had a higher risk for recent-onset cardiovascular events or hypertension in the following 3 years than those with high KDM6B protein expression. (F) HDAC3 protein expression was decreased in OSA patients with high hypoxic load. (G) HDAC1 gene expression was increased in OSA patients with severe excessive daytime sleepiness (EDS). (H) H3K36Ac enrichment over the HIF-1α gene promoter region was decreased in OSA patients. This histone marker was negatively correlated with (I) apnea hypopnea index (AHI) and (J) oxygen desaturation index (ODI), and positively with (K) minimum oxygen saturation. *adjust for age, body mass index, gender, smoking history, alcoholism history, and co-morbidities (diabetes mellitus, hypertension, stroke, cardiac disease, chronic kidney disease) by multivariate linear regression analysis. #compared between primary snoring (PS) subjects and obstructive sleep apnea (OSA) patients with a particular phenotype (a high hypoxic load or severe EDS), adjusted by multivariate linear regression analysis. ##compared between OSA patients with a particular phenotype (a high hypoxic load or severe EDS) and those without the phenotype, adjusted by multivariate linear regression analysis. Hollow circle indicates PS subjects; solid star indicates OSA patients. PBMC = peripheral blood mononuclear cell; cum = cumulative; CI = confidence interval.
Decreased HIF-1α gene promoter-specific H3K36Ac enrichment in treatment-naïve OSA patients of cohort 2
Based on the findings that global H3K23Ac and global H3K36Ac were decreased in patients with OSA, we next investigated whether such hypoacetylation occurred specifically at the promoter regions of the HIF-1α and HIF-2α genes. We examined this possibility by ChIP method in PBMC lysates from another cohort (cohort 2) of 28 treatment-naïve OSA patients, and 8 matched PS subjects (Table 1). H3K36Ac enrichment over the HIF-1α (0.52 ± 0.92 versus 1.23 ± 1.25 fold change, adjusted p = 0.006; Fig. 2H) gene promoter region was decreased in the treatment-naïve OSA patients as compared with that in the PS subjects. This specific histone modification was negatively correlated with both AHI (r = − 0.548, p = 0.001, Fig. 2I) and ODI (r = − 0.449, p = 0.006, Fig. 2J), and positively with minimum SaO2 (r = 0.434, p = 0.008, Fig. 2K). These correlations remained statistically significant in multivariate linear regression model 1 or model 2 (Supplementary Tables S5, S6, S7), but did not reach statistical significance in the analysis of separate PS and OSA groups. Additionally, H3K36 Ac enrichment over the HIF-2α gene promoter region (0.49 ± 0.3 versus 1.03 ± 1.06 fold change, adjusted p = 0.024, Supplementary Fig. S4) was decreased in the treatment-naïve OSA patients versus the PS subjects. However, this difference was not normally distributed and did not reach statistical significance in the non-parametric analysis. Moreover, no significant difference in either HIF-1α or HIF-2α gene expression level was noted between case and control groups (Supplementary Fig. S4).
Effects of in vitro IHR on histone modifying enzyme expressions and gene-specific histone modifications in human monocytic THP-1 cells
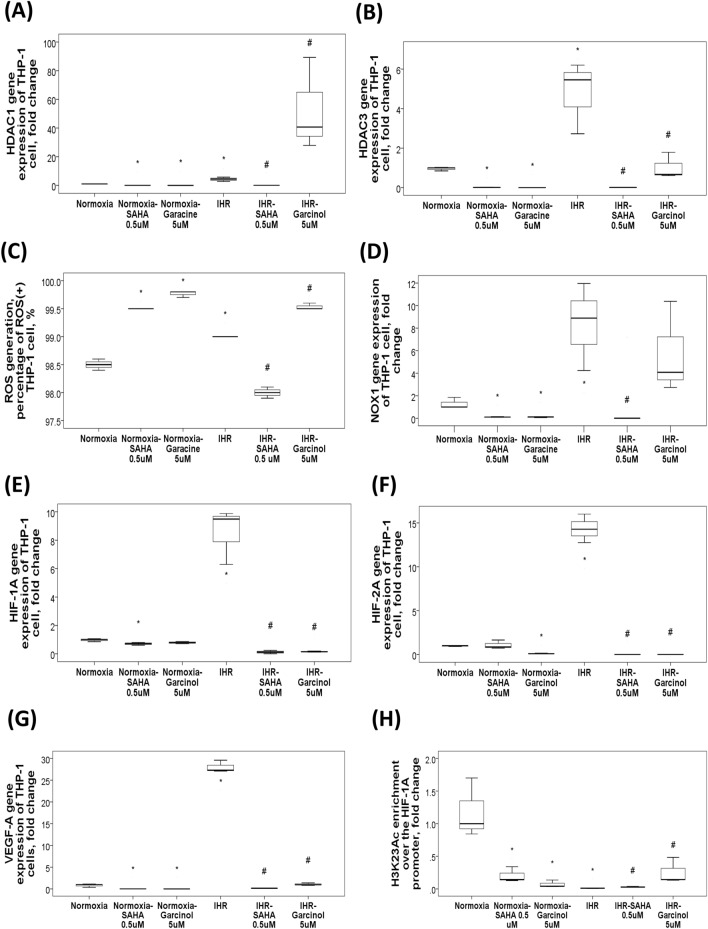
To test the effects of HDAC1 over-expression or under-expression on oxidative stress, THP-1 cells were treated with either suberoylanilide hydroxamic acid (SAHA, HDAC1 inhibitor) or garcinol (HAT inhibitor) under IHR stimuli. Both HDAC1 (Fig. 3A) and HDAC3 (Fig. 3B) gene expressions were increased with 9-day IHR stimuli, while SAHA 0.5 μM treatment resulted in their down-regulations and garcinol 5 μM treatment resulted in their up-regulations under IHR stimuli as compared with IHR alone condition. SAHA reversed IHR-induced elevation of reactive oxygen species (ROS) production (Fig. 3C) in association with normalization of IHR-induced over-expression of the NOX1 gene (Fig. 3D), while garcinol had the opposite effect. Additionally, SAHA reversed IHR-induced up-regulations of the HIF-1α (Fig. 3E), HIF-2α (Fig. 3F), and VEGF-A (Fig. 3G) gene expressions, and IHR-induced H3K23 hypoacetylation over the HIF-1α promoter region (Fig. 3H).
Figure 3.
Changes in histone modifying enzyme expressions and H3K23Ac/H3K36Ac enrichment in response to in vitro intermittent hypoxia with re-oxygenation (IHR) stimuli, and the effect of HDAC1 inhibition. HDAC inhibitor, SAHA, treatment resulted in reversion of IHR-induced up-regulations of the (A) HDAC1, and (B) HDAC3 genes. SAHA treatment reversed (C) IHR-induced over-production of reactive oxygen species (ROS) along with (D) reversion of NADPH oxidase 1 (NOX1) over-expression. Both SAHA (a HDAC1 inhibitor) and garcinol (a HAT inhibitor) treatment reversed IHR-induced up-regulations of the (E) HIF-1α, (F) HIF-2α, and (G) VEGF-A genes, and (H) reversed IHR-induced H3K23 hypoacetylation over the HIF-1α promoter region. *p < 0.05, compared with normoxic conditions by U or Kruskal–Wallis test. **p < 0.01, compared with normoxic condition by U-test. #p < 0.05, compared with IHR condition by Kruskal–Wallis test. HDAC = histone deacetylase; SAHA = suberoylanilide hydroxamic acid; HAT = histone acetyltransferase; HIF = hypoxia inducible factor; VEGF = vascular endothelial growth factor.
Discussion
Epigenetic changes are involved in the switching 'on' and 'off' of many genes, and histone modifications has been regarded as epigenetic gene targets for the regulation of disease-associated cellular changes in hypoxic microenvironment. Little is known about the role of histone modifications in OSA, which is characterized by chronic IHR rather than persistent hypoxia. Therefore, in this study, we sought to identify specific histone modification patterns associated with OSA and its clinical phenotypes. We have proved, for the first time, the relationship between global H3K23/H3K36 hypoacetylation in association with HDAC1 up-regulation and the development of EDS/hypertension phenotype in OSA patients. Specifically, HIF-1α gene promoter-specific H3K36 enrichment was decreased in OSA patients and in response to in vitro IHR stimuli. In contrast, global H3K79 hypermethylation along with KDM6B down-regulation was found to be related to higher incident cardiovascular risk. Additionally, we found that SAHA, a HDAC inhibitor, could reverse IHR-induced ROS over-production through NOX1 in vitro.
H3K36Ac is localized predominantly to the promoters of RNA polymerase II-transcribed genes, a pattern inversely related to that of H3K36 me16. H3K36 acetylation has been shown to increase chromatin accessibility, and promotes DNA double-strand break repair. Furthermore, H3K36 modification is cell cycle regulated with chromatin-bound H3K36me3 peaking in G1 where non-homologous end joining occurs, while H3K36Ac peaking in S/G2 phase when homologous recombination predominates17. The H3K36 hypoacetylation in OSA patients and in response to in vitro short-term IHR stimuli suggest that DNA double-strand break repair may be dampened by chronic IHR in OSA through H3K36 hypoacetylation. The decreased enrichment of H3K36 acetylation over the HIF-1α gene promoter region in OSA patients and in response to IHR stimuli suggest that H3K36Ac may play a specific role in regulating HIF-1α-mediated adaptive responses to hypoxia. However, separate cohorts were used for the global histone and HIF-1α promoter-specific histone enrichment analyses, and this may limit the cause-effect interpretation of the experiments. Moreover, correlations between global H3K23Ac/H3K36Ac expression and metrics such as AHI, arousal index, mean SaO2 were quite weak, so further studies are needed. Finally, many environmental exposures, such as cigarette smoking, may cause specific histone modification change. In the lung tissues of smokers with COPD, the expressions of HDAC2, HDAC5, HDAC8, HDAC7, and HDAC10 were decreased significantly, while H3K23Ac or H3K36Ac has not yet been linked to smoking exposure18. However, we applied multivariate linear regression models to adjust for all possible confounding factors.
In line with our findings that global H3K23 was hypoacetylated in OSA patients, H3K23 acetylation has been reported to be reduced in diabetic mice, and its hypo-acetylation contributed to Drosophila learning defect and depression-like behaviors in rats19–21. Since the co-existence of both H4K16ac and H3K23ac was correlated with higher gene expression levels in a plant study, and lipopolysaccharide can trigger IL-6 production via enhancing recruitment of H3K23 acetylation to IL-6 promoter region, we speculated that the simultaneous hypoacetylation of H3K36 and H3K23 in OSA patients may lead to impaired DNA repair ability under chronic IHR circumstances, and affect specific adaptive gene responses through regulating HIF-1α22,23.
Up-regulation of HDAC1 has been shown to be associated with adverse prognosis of various cancers, and plays critical roles in cellular senescence, aging, myelination, and adult neurogenesis24. In line with our findings that HDAC1 was up-regulated in OSA patients and in response to IHR stimuli, HDAC1 inhibition was reported to protect against hypoxia-induced swelling in cardiomyocytes through enhancing cell stiffness25. Class I HDACs, such as HDAC1 and 3, can enhance HIF-1α stability and HIF-1 transactivation function in persistent hypoxic conditions through binding to the oxygen-dependent degradation domain of HIF-1α26. Although HDAC1 expressions were reduced after CPAP treatment in a small group of OSA patients, the lack of a time control group or sham CPAP device and the lack of CPAP adherence data limit the interpretation of these data. In line with our findings that HDAC3 was down-regulated in OSA patients with high hypoxic load, HDAC3 has been reported to mediate cardioprotection of remifentanil post-conditioning by targeting GSK-3beta in cardiomyocytes in IHR injury27,28. In contrast, HDAC3 could contribute to IHR-induced cell apoptosis of myocardioblasts28. However, the reversion of IHR-induced ROS over-production with SAHA through inhibition of the NOX1 gene indicates that HDAC1/3 inhibition may be a new therapy to reduce oxidative stress in OSA. Several recent studies have shown that HDAC inhibition with SAHA, trichostatin A, or valproic acid, reduced ROS production in experimental diabetes, pulmonary hypertension, and atherosclerosis, through inhibitions of NOX1/2/4/5 expression via epigenetically regulating chromatin accessibility29–32. HDAC inhibitors have displayed clinical efficacy in treating specific tumors33. SAHA inhibits both class I and class II HDACs, but selectively alters transcription of as few as 2–5% of genes34. Further research is required to determine whether HDAC1 inhibition could be used to reduce cardiovascular risk in OSA patients.
H3K79 methylation has been shown to be implicated in several processes of DNA replication and repair of DNA damage35. Moreover, H3K79 methylation, shows a high positive correlation with transcriptional activity36,37. In this study, the global H3K79 hypermethylation in OSA patients with high hsCRP, which has been linked with a high cardiovascular risk15, suggest that H3K79me3 may serve as an endogenous protective mechanism against chronic IHR-induced DNA damage in OSA. Most KDM enzymes are structurally similar to the HIF hydroxylase factor inhibiting HIF-1, suggesting that KDM enzymes may act as molecular oxygen sensors in the cell38. Hypoxia-inducible KDMs have been shown to support the adaptive and adverse gene transcription induced by HIF-1α, and they can also control genome-wide chromatin landscape, especially KDMs which demethylate H3K9 and H3K27 sites39. The down-regulation of KDM6B in OSA patients with high hypoxic load and incident cardiovascular events in this study suggest that long-term oxidative stress-related insults may impair the maintenance of chromatin landscape and provoke cellular senescence and endothelial dysfunction associated with chronic IHR circumstances in OSA. Although previous studies showed that hypoxia either inhibits or induces the enzymatic activity of KMD6B depending on HIF-2α, KDM6B protein levels remained unchanged under hypoxic conditions and decreased upon re-oxygenation40–42. Further study is required to determine whether KDM6B supplementation can reverse H3K79 hypermethylation and ameliorate adverse consequences in severe OSA. One limitation is that this study was conducted using PBMCs; thus, the findings associating KDM6B to future cardiovascular events may not extend to other cell types. Given that a recent U.S. healthcare system–based analysis of mortality and markers of severe morbidity identify sleep apnea as a risk factor for COVID-19 mortality, our results may provide particular insight into immune function with OSA and discover mechanistic pathways underlying COVID-19 morbidity43. Another limitation is that we did not check some histone modifying enzymes known to regulate histone methylation, such as other JmjC-KDM demethylases and G9a methylase44,45.
In conclusions, H3K23/H3K36 hypoacetylation is associated with the development of prevalent hypertension or disease severity in patients with sleep-disordered breathing, probably through up-regulation of HDAC1, while H3K79 hypermethylation is associated with higher risk of incident cardiovascular diseases, probably through down-regulation of KDM6B. HIF-1α and HIF-2α gene promoter-specific H3K36 hypoacetylation may play a role in regulating adaptive gene responses to chronic IHR injury of OSA. HDAC1 inhibition can reduce oxidative stress induced by IHR through inhibition of NOX1. These findings provide further insights into novel epigenetic mechanisms by which chronic IHR leads to adverse consequences in OSA, and opens the possibility of using HDAC1 inhibitor in preventing the morbidities and mortality.
Methods
Subjects
This study was approved by the Institutional Review Board of Chang Gung Memorial Hospital, Taiwan (certificate number: 102-3887B). The study participants were recruited from the sleep center of Kaohsiung Chang Gung Memorial Hospital from January 2014 through December 2017. Written informed consent was obtained from each subject participating in the study aged 20 years or older. All the methods were carried out in accordance with relevant guidelines and regulations. The exclusion criteria included ongoing infections, autoimmune disease, use of immunosuppressive agent in the past 6 months, narcolepsy, severe obesity (body mass index, BMI, ≧35 kg/m2), old age (> 65 year-old), and those with a BMI < 21 kg/m2. OSA (apnea hypopnea index, AHI, ≧5 events/hour) and PS (AHI < 5 events/hour) were diagnosed by full night polysomnography examination at the sleep center of Kaohsiung Chang Gung Memorial Hospital46. The Epworth Sleepiness Scale (ESS) recorded at the examination was used to measure sleep propensity in every study subject47. Prevalent hypertension was defined as systolic blood pressure ≧140 mmHg and/or diastolic blood pressure ≧90 mmHg or medication use at diagnosis of OSA. Incident cardiovascular events (coronary artery disease: angioplasty, coronary artery bypass grafting, myocardial infarction, or angina) or hypertension were defined as those occurred in the following 3 years after the first blood sampling at diagnosis48.
Isolation of PBMC RNA and protein from whole blood samples
PBMCs were isolated from heparinized blood of all study subjects using a two-layer Ficoll-Histopaque density gradient centrifugation (Histopaque 1.077 and 1.119; Sigma Diagnostics, St.Louis, MO) method. Samples were stored in RNAlater® RNA Stabilization Solution (Ambion®) at − 80 °C until analysis. An RNeasy®Plus Mini Kit (Qiagen, Hilden, Germany) was used for isolation of high quality total RNA.
Measurement of histone modifying enzyme gene expressions in the PBMCs using quantitative reverse-transcriptase polymerase chain reaction (RT-PCR)
To determine the expressions of KDM1A, KDM4B, KDM4C, KDM5A-D, KDM6B, DOT1L, HDAC1-7, KAT2A, and KAT6B of the isolated PBMCs, the gene expressions were analyzed using quantitative RT-PCR. The house keeping gene GAPDH was chosen as an endogenous control to normalize the expression data for each gene. All PCR primers (random hexamers) were designed and purchased from Roche according to the company’s protocols (www.roche-applied-science.com), and their sequences are given in Table 2. A total of 300 ng RNA was used for synthesis of first strand cDNA with QuantiTectReverse Transcription Kit (QIAGEN, Germany), and the reverse transcription reaction was added to 5 μl of master mix (QIAGEN, SYBR Green PCR kit; Roche, Germany). The PCR reactions with 45 cycles of amplification were run in a Roche LightCycle 480 QuantiFast R machine. Relative expression levels were calculated using the ΔΔCt method with the median value for the control group as the calibrator49.
Table 2.
Primer sequences for quantitative real-time polymerase chain reactions used in the present study.
| Gene name | Primer sequences | |
|---|---|---|
| KDM1A (LSD1) | Forward | 5′-CATCATGGTGCAAGAAGAGC |
| Reverse | 5′-ATGTGGGAAAGGCAGACAAG | |
| KDM4B (JMJD2B) | Forward | 5′-CTGAAATGCGTGTACTGCCG |
| Reverse | 5′-CGTTGCGGTTCTTGGTGATG | |
| KDM4C (JMJD2C) | Forward | 5′-GCTCGATTTTCCACAGCCTC |
| Reverse | 5′-CAGGGTCGGCCACATATTCA | |
| KDM5B (JAR1D1B) | Forward | 5′-GTACTGTGAAGGACGCACCA |
| Reverse | 5′-TAGCACCGTTTACAGGCTGG | |
| KDM6B (JMJD3) | Forward | 5′-CACCCACTGTGGTCTGTTGT |
| Reverse | 5′-TGTCTCCGCCTCAGTAACAG | |
| HDAC1 | Forward | 5′-CGGTGCTGGACATATGAGAC |
| Reverse | 5′-TGGTCCAAAGTATTCAAAGTAGTCA | |
| HDAC2 | Forward | 5′-CCAGATGTTCTGGCATCCTC |
| Reverse | 5′-ACAGCCCCTGTTGTCCTGT | |
| HDAC3 | Forward | 5′-TGGCATTGACCCATAGCCTG |
| Reverse | 5′-TGCATATTGGTGGGGCTGAC | |
| HDAC4 | Forward | 5′-CGGAAGCATGTGTTTCTGCC |
| Reverse | 5′-TTCTCCATGGAACGGACAGC | |
| HDAC5 | Forward | 5′-TCTTGTCGAAGTCAAAGGAGC |
| Reverse | 5′-GAGGGGAACTCTGGTCCAAAG | |
| HDAC6 | Forward | 5′-TGGCTATTGCATGTTCAACCA |
| Reverse | 5′-GTCGAAGGTGAACTGTGTTCCT | |
| HDAC7 | Forward | 5′-GAACAGTCCATCCCAACAGC |
| Reverse | 5′-GGCTCCTTCCGTCTCCAG | |
| HIF-1α promoter | Forward | 5′-CACCCCCATCTCCTTTCTCT-3′ |
| Reverse | 5′-GGGTTCCTCGAGATCCAATG-3′ | |
| HIF-2α promoter | Forward | 5′-GAAGTGCGGAGGCAGGAG-3′ |
| Reverse | 5′-GGCAGCCCACTTTAAAAACTC-3′ | |
| GAPDH | Forward | 5′-GAAGAGCCAAGGACAGGTAC |
| Reverse | 5′-CAACTTCATCCACGTTCACC | |
KDM Lysine de-methylase, HDAC Histone de-acetylase, LSD1 Lysine-specific demethylase 1, JMJD Jumonji domain-containing proteins, HIF hypoxia-inducible factor, GAPDH Glyceraldehyde 3-phosphate dehydrogenase.
Measurement of global histone modification patterns and histone modifying enzyme protein expressions in the PBMC samples using ELISA method
Protein levels of H3K9Ac, H3K14Ac, H3K23Ac, H3K36Ac, H3K56Ac, H4K16Ac, H3K4me3, H3K9me3, H3K27me3, H3K36me3, H3K79me3, KDM1A, KDM4, KDM5,KDM6B, and HDAC1-4 in the PBMCs were assessed by a commercial ELISA method (Epigentek, USA, and R&D systems, Minneapolis, MN), as described previously50. Briefly, 40 μl dye (Bio-Rad Protein Assay Dye Reagent Concentrate #500-0006) was added to 10 μl bovine serum albumin with serial dilution to generate a standard curve for total protein concentration by measuring values at OD 595 nm. Accordingly, protein lysate equivalent to 20 ng of total protein was used for each protocol in ELISA analysis.
Measurement of gene promoter-specific H3K23Ac and H3K36Ac enrichment with Chromatin immunoprecipitation (ChIP) followed by quantitative RT-PCR
3–5 × 105 PBMCs from another cohort of 8 PS subjects, and 28 treatment-naïve OSA patients were harvested and fixed with 1% formaldehyde at room temperature, followed by glycine to stop the crossing linking reaction. Cells were re-suspended in cell lysis buffer containing 1× Protease Inhibitor Cocktail II (200×, Millipore, USA). Extracts were sonicated using a Standard Sonicator (Diagenode Bioruptor) to achieve chromosome fragment lengths of 200–400 bp. 100 μl sonicated cell extract were diluted in 400 μl ChIP Dilution Buffer (EZ-Magna ChIPTM A/G kit, Millipore, USA), and incubated with 1 μg of anti-H3K23Ac or anti-H3K36Ac (Millipore, USA) at 4 °C for 1 hour while rotating. Two aliquots were reserved as ‘‘controls’’—one incubated without antibody and the other with non-immune Mouse IgG (25 μg, Millipore, USA). Protein A/G magnetic beads were added and incubated at 4 °C overnight while rotating. Cross-linking of the immunoprecipitated chromatin complexes and ‘‘input controls’’ (10% of the total soluble chromatin) were reversed by heating the samples at 62 °C for 4 hours, followed by Proteinase K 1 µL incubation (Millipore, USA). The DNA were purified by spin column and stored at − 20 °C51.
For real-time RT-PCR analysis, amplification of HIF-1α and HIF-2α promoter DNA was performed using the TaqMan reagent kit (PerkinElmer Life Sciences)52. The PCR primers for the two selected gene promoters located from − 300 to + 30 bp of their transcription start site were listed in Table 2. The conditions for the reaction were: 50 °C, 2 min; 94 °C, 10 min, then 95 °C, 20 s; 60 °C, 30 s for 50 cycles. Resulting PCR products were measured and elaborated by the sequence detector ABI GeneAmp PCR System 9700 (PerkinElmer Biosystems). Histone enrichment was compared by fold enrichment method, in which the ChIP signals are divided by the no-antibody signals, representing the ChIP signal as the fold increase in signal relative to the background signal53,54.
In vitro human monocyte cell line culture under IHR stimuli
The human monocytic leukemia cell line THP-1 cells obtained from ATCC (1 × 106 cells/ml) were re-suspended in a 5 cm culture dish containing 5 ml RPMI 1640 medium, and then exposed to normoxia or IHR in a custom-designed, incubation chambers which are attached to an external O2-CO2-N2 hand-driven controller as previously described55. Air-phase set point for IHR consisted of a 25-min hypoxic period (0% O2 and 5% CO2), followed by 35 min of re-oxygenation (21% O2 and 5% CO2), 7 h each day for 9 days56. Control cells were maintained in NOX condition for the same durations. HDAC inhibitor, SAHA (Sigma-Aldrich, Saint Luis, MO, USA), and HAT inhibitor, Garcinol (Sigma-Aldrich, Saint Luis, MO, USA), were used to inhibit or augment HDAC expression, respectively. H2DCFDA (catalog no. D6883; Sigma, USA) was used to measure THP-1 intracellular ROS. Cell-associated mean fluorescent intensity was measured by flow cytometry in FL1 channel at excitation and emission wavelengths of 488 and 535 nm, respectively, using the CytomicsTM FC500 (Beckman Coulter ; USA ). Gene expression levels of the NADPH oxidase NOX1/2/4/5 genes were determined using quantitative RT-PCR method.
Statistical analysis
Continuous values were expressed as mean ± standard deviation (SD). The differences between two groups were analyzed using the Student’s t-test, Mann–Whitney U-test, or X2-test, as appropriate. Subgroup comparisons of continuous variables in the validation cohort were performed using the one-way ANOVA with LSD method for post-hoc tests, followed by multivariate linear regression analysis to adjust for all potential confounding factors (age, BMI, gender, smoking, alcoholism, and co-morbidities), and to obtain adjusted p values. The null hypothesis was rejected at p < 0.05. All analyses were performed using SPSS software version 20.0 (SPSS Corp., Chicago).
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Chang Gung Memorial Hospital, Taiwan (Certificate number: 102-3887B). Written informed consent was obtained from each subject participating in the study.
Supplementary Information
Acknowledgements
The authors acknowledge the technical support provided by the Genomic and Proteomic Core Laboratory, and Internal Medicine Core Facility of the Kaohsiung Chang Gung Memorial Hospital. We wish to thank Ms. Shu-Jun Kong, Ms. Lian-Rong Liu, and Mr. Wei-Zhe Liu for manual scoring of all PSG. We also thank Chih-Yun Lin and the Biostatistics Center, Kaohsiung Chang Gung Memorial Hospital for statistics work. This is not an industry-sponsored study. All the authors declare that they have no potential conflicts of interest or financial support in any for-profit business agency.
Abbreviations
- OSA
Obstructive sleep apnea
- IHR
Intermittent hypoxia with re-oxygenation
- CPAP
Continuous positive airway pressure
- H
Histone
- Ac
Acetylation
- me
Methylation
- HDAC
Histone deacetylase
- KDM
Histone lysine demethylase
- PBMCs
Peripheral blood mononuclear cells
- PS
Primary snoring
- HIF
Hypoxia-inducible factor
- BMI
Body mass index
- AHI
Apnea hypopnea index
- ODI
Oxygen desaturation index
- ESS
Epworth Sleepiness Scale
- RT-PCR
Reverse-transcriptase polymerase chain reaction
- ChIP
Chromatin immunoprecipitation
Author contributions
Y.C.C. performed the statistical analysis and drafted the manuscript. M.C.S., C.H.C., Y.C.C., and H.C.L. enrolled the study participants. P.Y.H., C.C.H., and C.W.L. performed the in vitro experiments. Y.Y.L., C.P.L., and T.Y.W. carried out the gene expression and histone experiments. M.C.L., C.C.H., and M.C.S. conceived of the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Grants (NMRPG8F6071-6073/105-2314-B-182A- 092-MY3/NMRPG8J6131-6133/108-2314-B-182A-129-MY3 to M.C. Lin) from the Ministry of Science and Technology, Taiwan, and by grants from the Chang Gung Memorial Hospital (CMRPG8G0281/8G0282/8J0581/ 8J0582/8J0231/8J0202 to M.C. Lin), Taiwan. The funding body has no role in the design of the study, or collection, analysis, and interpretation of data, or in writing the manuscript.
Data availability
The data that support the findings of this study are available in supplementary information online and from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chang-Chun Hsiao, Email: cchsiao@mail.cgu.edu.tw.
Meng-Chih Lin, Email: linmengchih@hotmail.com.
Mao-Chang Su, Email: maochangsu@yahoo.com.tw.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-00052-9.
References
- 1.Zinchuk AV, Gentry MJ, Concato J, Yaggi HK. Phenotypes in obstructive sleep apnea: A definition, examples and evolution of approaches. Sleep Med. Rev. 2016 doi: 10.1016/j.smrv.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khayat R, Pleister A. Consequences of obstructive sleep apnea: Cardiovascular risk of obstructive sleep apnea and whether continuous positive airway pressure reduces that risk. Sleep Med. Clin. 2016;11:273–286. doi: 10.1016/j.jsmc.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 3.de Lima FF, Mazzotti DR, Tufik S, Bittencourt L. The role inflammatory response genes in obstructive sleep apnea syndrome: A review. Sleep Breath. 2016;20:331–338. doi: 10.1007/s11325-015-1226-7. [DOI] [PubMed] [Google Scholar]
- 4.Weiss P, Kryger M. Positive airway pressure therapy for obstructive sleep apnea. Otolaryngol. Clin. N. Am. 2016;49:1331–1341. doi: 10.1016/j.otc.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Mashaqi S, Gozal D. The impact of obstructive sleep apnea and PAP therapy on all-cause and cardiovascular mortality based on age and gender: Aa literature review. Respir. Investig. 2020;58:7–20. doi: 10.1016/j.resinv.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Ma Q, Zhang L. Epigenetic programming of hypoxic-ischemic encephalopathy in response to fetal hypoxia. Prog. Neurobiol. 2015;124:28–48. doi: 10.1016/j.pneurobio.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YC, et al. Whole genome DNA methylation analysis of obstructive sleep apnea: IL1R2, NPR2, AR, SP140 methylation and clinical phenotype. Sleep. 2016;39:743–755. doi: 10.5665/sleep.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandamme J, et al. H3K23me2 is a new heterochromatic mark in Caenorhabditis elegans. Nucleic Acids Res. 2015;43:9694–9710. doi: 10.1093/nar/gkv1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Husmann D, Gozani O. Histone lysine methyltransferases in biology and disease. Nat. Struct. Mol. Biol. 2019;26:880–889. doi: 10.1038/s41594-019-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack AP, et al. H3K56me3 is a novel, conserved heterochromatic mark that largely but not completely overlaps with H3K9me3 in both regulation and localization. PLoS ONE. 2013;8:e51765. doi: 10.1371/journal.pone.0051765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock RL, Dunne K, Walport LJ, Flashman E, Kawamura A. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics. 2015 doi: 10.2217/epi.15.24. [DOI] [PubMed] [Google Scholar]
- 12.Khangura RK, Bali A, Jaggi AS, Singh N. Histone acetylation and histone deacetylation in neuropathic pain: An unresolved puzzle? Eur. J. Pharmacol. 2017;795:36–42. doi: 10.1016/j.ejphar.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Tsai YP, Wu KJ. Epigenetic regulation of hypoxia-responsive gene expression: Focusing on chromatin and DNA modifications. Int. J. Cancer. 2014;134:249–256. doi: 10.1002/ijc.28190. [DOI] [PubMed] [Google Scholar]
- 14.Barbe F, et al. Effect of continuous positive airway pressure on the incidence of hypertension and cardiovascular events in nonsleepy patients with obstructive sleep apnea: A randomized controlled trial. JAMA. 2012;307:2161–2168. doi: 10.1001/jama.2012.4366. [DOI] [PubMed] [Google Scholar]
- 15.Bouloukaki I, et al. Obstructive sleep apnea syndrome and cardiovascular disease: The influence of C-reactive protein. World J. Exp. Med. 2015;5:77–83. doi: 10.5493/wjem.v5.i2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris SA, et al. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 2007;282:7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pai CC, et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 2014;5:4091. doi: 10.1038/ncomms5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zong D, Liu X, Li J, Ouyang R, Chen P. The role of cigarette smoke-induced epigenetic alterations in inflammation. Epigenet. Chromatin. 2019;12:65. doi: 10.1186/s13072-019-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu P, et al. Liver histone H3 methylation and acetylation may associate with type 2 diabetes development. J. Physiol. Biochem. 2015;71:89–98. doi: 10.1007/s13105-015-0385-0. [DOI] [PubMed] [Google Scholar]
- 20.Li HY, et al. Abnormal modification of histone acetylation involved in depression-like behaviors of rats induced by chronically unpredicted stress. NeuroReport. 2017;28:1054–1060. doi: 10.1097/WNR.0000000000000879. [DOI] [PubMed] [Google Scholar]
- 21.Li KL, et al. Histone acetyltransferase CBP-related H3K23 acetylation contributes to courtship learning in Drosophila. BMC Dev. Biol. 2018;18:20. doi: 10.1186/s12861-018-0179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu L, Chen X, Sanders D, Qian S, Zhong X. High-resolution mapping of H4K16 and H3K23 acetylation reveals conserved and unique distribution patterns in Arabidopsis and rice. Epigenet. Off. J. DNA Methylation Soc. 2015;10:1044–1053. doi: 10.1080/15592294.2015.1104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun D, Wen Q, Wang C. KAT6B promotes LPS-triggered IL-6 production via enhancing recruitment of H3K23 acetylation to IL-6 promoter region. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2017;33:1441–1147. [PubMed] [Google Scholar]
- 24.Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp. Gerontol. 2010;45:279–285. doi: 10.1016/j.exger.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, et al. Histone deacetylase 1 inhibition protects against hypoxia-induced swelling in H9c2 cardiomyocytes through regulating cell stiffness. Circ. J. 2017;82:192–202. doi: 10.1253/circj.CJ-17-0022. [DOI] [PubMed] [Google Scholar]
- 26.Kim SH, et al. Regulation of the HIF-1alpha stability by histone deacetylases. Oncol. Rep. 2007;17:647–651. [PubMed] [Google Scholar]
- 27.Chen M, et al. HDAC3 mediates cardioprotection of remifentanil post-conditioning by targeting GSK-3beta In H9c2 cardiomyocytesin hypoxia/reoxygenation injury. Shock. 2017 doi: 10.1097/SHK.0000000000001008. [DOI] [PubMed] [Google Scholar]
- 28.Chen M, Liu Q, Chen L, Zhang L, Gu E. Remifentanil postconditioning ameliorates histone H3 acetylation modification in H9c2 cardiomyoblasts after hypoxia/reoxygenation via attenuating endoplasmic reticulum stress. Apoptosis Int. J. Progr. Cell Death. 2017;22:662–671. doi: 10.1007/s10495-017-1347-5. [DOI] [PubMed] [Google Scholar]
- 29.Chen F, et al. Inhibition of histone deacetylase reduces transcription of NADPH oxidases and ROS production and ameliorates pulmonary arterial hypertension. Free Radic. Biol. Med. 2016;99:167–178. doi: 10.1016/j.freeradbiomed.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manea SA, et al. Epigenetic regulation of vascular NADPH oxidase expression and reactive oxygen species production by histone deacetylase-dependent mechanisms in experimental diabetes. Redox Biol. 2018;16:332–343. doi: 10.1016/j.redox.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansch N, et al. The enzyme activity of histone deacetylase 8 is modulated by a redox-switch. Redox Biol. 2019;20:60–67. doi: 10.1016/j.redox.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manea SA, et al. Pharmacological inhibition of histone deacetylase reduces NADPH oxidase expression, oxidative stress and the progression of atherosclerotic lesions in hypercholesterolemic apolipoprotein E-deficient mice; potential implications for human atherosclerosis. Redox Biol. 2020;28:101338. doi: 10.1016/j.redox.2019.101338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai Y, Maru Y, Tanaka J. Action mechanisms of histone deacetylase inhibitors in the treatment of hematological malignancies. Cancer Sci. 2016;107:1543–1549. doi: 10.1111/cas.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komatsu N, et al. SAHA, a HDAC inhibitor, has profound anti-growth activity against non-small cell lung cancer cells. Oncol. Rep. 2006;15:187–191. [PubMed] [Google Scholar]
- 35.Wood K, Tellier M, Murphy S. DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules. 2018 doi: 10.3390/biom8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcos-Villar L, et al. Epigenetic control of influenza virus: role of H3K79 methylation in interferon-induced antiviral response. Sci. Rep. 2018;8:1230. doi: 10.1038/s41598-018-19370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farooq Z, Banday S, Pandita TK, Altaf M. The many faces of histone H3K79 methylation. Mutat. Res. Rev. Mutat. Res. 2016;768:46–52. doi: 10.1016/j.mrrev.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhry H, Harris AL. Advances in hypoxia-inducible factor biology. Cell Metab. 2018;27:281–298. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Salminen A, Kaarniranta K, Kauppinen A. Hypoxia-inducible histone lysine demethylases: Impact on the aging process and age-related diseases. Aging Dis. 2016;7:180–200. doi: 10.14336/AD.2015.0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prickaerts P, et al. Hypoxia increases genome-wide bivalent epigenetic marking by specific gain of H3K27me3. Epigenet. Chromatin. 2016;9:46. doi: 10.1186/s13072-016-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon DK, et al. The involvement of histone methylation in osteoblastic differentiation of human periosteum-derived cells cultured in vitro under hypoxic conditions. Cell Biochem. Funct. 2017;35:441–452. doi: 10.1002/cbf.3302. [DOI] [PubMed] [Google Scholar]
- 42.Lee HY, Choi K, Oh H, Park YK, Park H. HIF-1-dependent induction of Jumonji domain-containing protein (JMJD) 3 under hypoxic conditions. Mol. Cells. 2014;37:43–50. doi: 10.14348/molcells.2014.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cade BE, Dashti HS, Hassan SM, Redline S, Karlson EW. Sleep apnea and COVID-19 mortality and hospitalization. Am. J. Respir. Crit. Care Med. 2020;202:1462–1464. doi: 10.1164/rccm.202006-2252LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hancock RL, Dunne K, Walport LJ, Flashman E, Kawamura A. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics. 2015;7:791–811. doi: 10.2217/epi.15.24. [DOI] [PubMed] [Google Scholar]
- 45.Chopra A, Cho WC, Willmore WG, Biggar KK. Hypoxia-inducible lysine methyltransferases: G9a and GLP hypoxic regulation, non-histone substrate modification, and pathological relevance. Front. Genet. 2020;11:579636. doi: 10.3389/fgene.2020.579636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YC, et al. Co-upregulation of Toll-like receptors 2 and 6 on peripheral blood cells in patients with obstructive sleep apnea. Sleep Breath. 2015 doi: 10.1007/s11325-014-1116-4. [DOI] [PubMed] [Google Scholar]
- 47.Hurlston A, et al. The Epworth Sleepiness Scale in service members with sleep disorders. Mil. Med. 2019;184:e701–e707. doi: 10.1093/milmed/usz066. [DOI] [PubMed] [Google Scholar]
- 48.Appleton SL, et al. Hypertension is associated with undiagnosed OSA during rapid eye movement sleep. Chest. 2016;150:495–505. doi: 10.1016/j.chest.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Gabert J, et al. Standardization and quality control studies of 'real-time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia: A Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 50.Chen YC, et al. Genome-wide gene expression array identifies novel genes related to disease severity and excessive daytime sleepiness in patients with obstructive sleep apnea. PLoS ONE. 2017;12:e0176575. doi: 10.1371/journal.pone.0176575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taneyhill LA, Adams MS. Investigating regulatory factors and their DNA binding affinities through real time quantitative PCR (RT-QPCR) and chromatin immunoprecipitation (ChIP) assays. Methods Cell Biol. 2008;87:367–389. doi: 10.1016/S0091-679X(08)00219-7. [DOI] [PubMed] [Google Scholar]
- 52.Chen YC, et al. Histone H3K14 hypoacetylation and H3K27 hypermethylation along with HDAC1 up-regulation and KDM6B down-regulation are associated with active pulmonary tuberculosis disease. Am. J. Transl. Res. 2017;9:1943–1955. [PMC free article] [PubMed] [Google Scholar]
- 53.Bhatnagar S, et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–120. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haring M, et al. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen YC, et al. Co-upregulation of Toll-like receptors 2 and 6 on peripheral blood cells in patients with obstructive sleep apnea. Sleep Breath. 2015;19:873–882. doi: 10.1007/s11325-014-1116-4. [DOI] [PubMed] [Google Scholar]
- 56.Hoffmann MS, Singh P, Wolk R, Narkiewicz K, Somers VK. Obstructive sleep apnea and intermittent hypoxia increase expression of dual specificity phosphatase 1. Atherosclerosis. 2013;231:378–383. doi: 10.1016/j.atherosclerosis.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in supplementary information online and from the corresponding author upon reasonable request.