Abstract
Human papillomavirus (HPV) can be detected by amplification of viral DNA. A novel PCR primer set generating a short PCR fragment (SPF PCR) was used for amplification of a fragment of only 65 bp from the L1 region and permitted ultrasensitive detection of a broad spectrum of HPV genotypes. The intra- and intertypic sequence variations of the 22-bp interprimer region of this amplimer were studied. Among 238 HPV sequences from GenBank and clinical specimens, HPV genotypes were correctly identified based on the 22-bp sequence in 232 cases (97.2%). Genotype-specific probes for HPV genotypes 6, 11, 16, 18, 31, 33 to 35, 39, 40, 42 to 45, 51 to 54, 56, 58, 59, 66, 68, 70, and 74 were selected, and a reverse hybridization line probe assay (LiPA) (the INNO-LiPA HPV prototype research assay) was developed. This LiPA permits the use of amplimers generated by the SPF as well as the MY 09/11 primers. The assay was evaluated with a total of 1,354 clinical specimens, comprising cervical scrapes (classifications ranging from normal cytology to severe dyskaryosis) and formalin-fixed, paraffin-embedded cervical carcinoma samples. LiPA results were highly concordant with sequence analysis of the SPF amplimer, genotype-specific PCR, and sequence analysis of amplimers generated by MY 09/11 primers. The sensitivity of the SPF primers was higher than that of the GP5+/6+ primers over a broad range of HPV types, especially when multiple HPV genotypes were present. In conclusion, the SPF LiPA method allows extremely sensitive detection of HPV DNA as well as reliable identification of HPV genotypes in both cervical smears and paraffin-embedded materials.
Human papillomaviruses (HPV) constitute a group of viruses associated with benign and malignant lesions of cutaneous and mucosal epithelia. So far, more than 100 different HPV genotypes have been identified, of which approximately 40 have been detected in the anogenital area (7, 34). These include several HPV genotypes that are known to be present in cervical carcinomas and in precursor lesions (5, 10), such as HPV type 16 (HPV-16) and HPV-18. Such genotypes are defined as high-risk genotypes and are associated with a comparatively high risk for invasive disease. In contrast, other genotypes (e.g., HPV-6 and -11) are considered low-risk genotypes, since they are not associated with the development of cervical carcinoma. The clinical classification of HPV types into either high- or low-risk groups is still not completely clear. According to zur Hausen (34), HPV-16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -66, and -69 belong to the class of high-risk types. HPV-59 and -68 can probably also be considered high-risk types (15, 32). Formal classification of HPV is performed by phylogenetic analysis of an ∼450-bp sequence from the L1 region of the genome (7, 34). By definition, HPV genotypes show a sequence dissimilarity of more than 10% in this 450-bp sequence.
Since HPV still cannot be cultured efficiently and the clinical performance of serological assays is still poor, diagnosis of HPV infection is based almost entirely on molecular tools, including liquid hybridization (e.g., Hybrid Capture; Digene Diagnostics, Silver Spring, Md.) (9), Southern and dot blot hybridization with HPV type-specific probes (14, 19), type-specific PCR (3, 31), and general-primer PCR (11, 13, 26, 30). Type-specific PCR permits detection of only a limited number of genotypes and requires the use of multiple PCRs. Therefore, several general or consensus PCR primers have been developed to detect a broad spectrum of HPV genotypes. The majority of large studies to date have been performed with the MY 09/11 (13) and the GP5+/6+ (11) primer sets. A disadvantage of these general primer systems is the relatively large size of the PCR fragment, especially in samples that yield poorly amplifiable DNA, such as formalin-fixed, paraffin-embedded materials (3, 17, 21). Various methods have been described for identification of HPV genotypes after amplification with general or consensus PCR primers. Besides sequence analysis, restriction fragment length polymorphism analysis of PCR amplimers has been described (4). Differentiation between potentially high-risk and low-risk HPV genotypes can also be achieved by hybridization of amplified HPV DNA with type-specific probes, using different test formats, such as dot blot hybridization (13), microtiter plate hybridization (8), or a line blot method (12, 15). Some of these typing systems have several drawbacks, such as the need for multiple hybridization reactions and the limited sensitivity for detection of multiple HPV genotypes in a single sample.
By definition, triage of patients with atypical squamous cells of undetermined significance (ASCUS) is difficult by cytological methods. Identification of high-risk HPV genotypes may permit selection of those patients who are at increased risk for disease and therefore may provide additional clinical value (9, 10, 16, 23). An crucial requirement for this approach is that HPV DNA testing and identification of high-risk HPV genotypes should be highly sensitive and specific, to achieve maximum negative predictive value for development of cervical intraepithelial neoplasia (CIN) (6).
Recently, we have developed novel general short PCR fragment (SPF) primers that amplify a fragment of only 65 bp, permitting highly sensitive, broad-spectrum detection of HPV DNA (17). To extend our knowledge of the natural history and clinical relevance of HPV infections, we developed and clinically evaluated a rapid typing assay for the simultaneous identification of 25 HPV genotypes after highly sensitive, broad-spectrum PCR amplification, using the novel SPF primer set.
MATERIALS AND METHODS
Clinical materials.
Group 1 contained cervical scrapes and biopsies (n = 104) obtained from patients visiting the gynecology outpatient clinics of community hospitals in Delft and the Slotervaart Hospital in Amsterdam, The Netherlands. The patients had been referred to the hospitals for treatment of CIN lesions. Smear preparations were examined and classified according to the Bethesda system (20).
Group 2 (n = 488) and group 3 (n = 278) comprised cervical scrapes obtained from women who were treated in the gynecology outpatient clinics of community hospitals in Delft and Amsterdam, The Netherlands, in 1997 and 1998, respectively. Patients had a history of colposcopy or treatment of dysplasia by loop electrosection of the transformation zone. Cervical scrapes were obtained with a cervix brush and were suspended and transported in 1.5 ml of phosphate-buffered saline (pH 7.2) at room temperature. These samples were all classified as having normal cytology or ASCUS.
Group 4 consisted of a total of 304 cervical scrapes obtained from women who were tested for the presence of HPV between 1988 and 1993 in the Netherlands. These patients had either one cervical smear showing severe dyskaryosis (n = 153) or two cervical smears showing mild or moderate dyskaryosis (n = 151). The interval between two abnormal smears was 1 year or less (6).
Group 5 comprised 180 patients with cervical carcinoma. The cervical biopsies were obtained from women visiting the Russian Cancer Center in Moscow between 1988 and 1994 (29). The specimens had been fixed in formalin, embedded in paraffin, histologically examined after hematoxylin-eosin staining, and classified as squamous cell carcinoma (n = 129) or adenocarcinoma (n = 51).
DNA isolation.
DNA was isolated from scrapes as described previously (17). Briefly, scrapes were resuspended in 1.5 ml of phosphate-buffered saline (pH 7.2). The cell suspension was vigorously shaken, and 120 μl was treated with 40 μl of proteinase K (200 μg/ml) in 3% Triton X-100 for 1 h at 37°C. The proteinase was inactivated by incubation at 95°C for 10 min. Subsequently, 10 μl of the supernatant was used in a PCR.
DNA was isolated from a single 10-μm section of formalin-fixed, paraffin-embedded tissue by treatment with proteinase K (1 mg/ml) in a volume of 250 μl at 56°C for 16 h. Proteinase K was inactivated at 95°C for 10 min, and 10 μl was used directly for PCR.
Plasmids.
Plasmids containing HPV genomic DNA were kindly provided by E.-M. de Villiers, Heidelberg, Germany (HPV-6b, -11, -13, -16, -18, -40, -45, -51, and -53); R. Ostrow, Minneapolis, Minn. (HPV-26); A. Lorincz, Silver Spring, Md. (HPV-31, -35, -43, -44, -56, -61, and -64); T. Matsukura, Tokyo, Japan (HPV-58, -59, -62, -67, and -69); and G. Orth, Paris, France (HPV-30, -33, -34, -39, -42, -52, -54, -55, -66, -68, -70, and -74).
PCR.
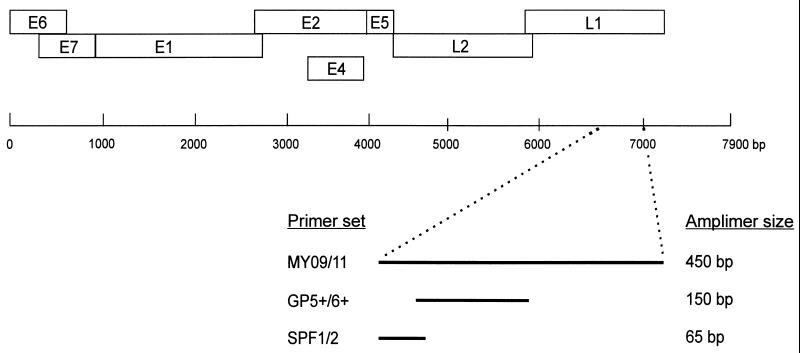
The development of the SPF1/2 PCR primers has been described earlier (17). In addition to this SPF1/2 primer set (comprising six primers) we have further optimized the system. The current version (prototype research kit INNO-LiPA HPV SPF10) comprises four additional primers. The universal primer sets MY 09/11 (13) and GP5+/6+ (11, 14, 15), as well as the type-specific primer sets for HPV-16, -18, -31, and -33 (3), were used exactly as described previously (Fig. 1).
FIG. 1.
Schematic representation of the locations of the different general primer sets (MY 09/11, GP5+/6+, and SPF) on the HPV genome. The circular HPV DNA genome is represented by a single line, and boxes show the positions of the various early (E) and late (L) genes. Within the L1 region, the positions of the amplification targets as well as the expected amplimer sizes for each of the primer sets are indicated.
SPF10 PCR was performed in a final reaction volume of 50 μl containing 10 μl of the isolated DNA, 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 2.0 mM MgCl2, 0.1% Triton X-100, 0.01% gelatin, 200 μM each deoxynucleoside triphosphate, 15 pmol of each of the forward and reverse primers, and 1.5 U of AmpliTaq Gold (Perkin-Elmer). The PCR conditions were as follows: activation of AmpliTaq Gold for 9 min at 94°C was followed by 40 cycles of 30 s at 94°C, 45 s at 52°C, and 45 s at 72°C, with a final extension of 5 min at 72°C. Each experiment was performed with separate positive and several negative PCR controls.
Amplification of isolated DNA was checked with β-globin PCR primers PC03 and PC04 (24).
Sequence analysis.
For sequence analysis of SPF and MY 09/11 amplimers, fragments were excised from 2 and 1% low-melting-temperature TAE agarose gels, respectively, and directly sequenced with the AmpliCycle sequencing kit (Perkin Elmer) and one of the PCR primers. For adequate sequencing of the 65-bp SPF amplimer, the concentration of dideoxyribonucleosides was increased in each of the four termination reaction mixtures by adding 2 μl of 500 μM ddA, 200 μM ddC, 60 μM ddG, and 200 μM ddT, respectively. The sequence products from SPF and MY 09/11 amplimers were separated on 15 and 6% polyacrylamide gels, respectively, by using the Alf-express system (Pharmacia, Uppsala, Sweden). Sequences were read manually from the electrophoresis pattern. The DNA sequences were analyzed with the PC/Gene software and the basic local alignment search tool (1) at the National Center for Biotechnology Information (20a).
HPV sequences from GenBank.
HPV sequences with the following accession numbers were obtained from GenBank and classified according to the Los Alamos database (18a) (sequences marked with an asterisk were used as references for the corresponding HPV genotypes): HPV-6, X00203*, S73503, and L41216; HPV-7, X74463* and M96300; HPV-11, M14119* and U55993; HPV-13, X62843* and M69076; HPV-16, K02718*, U89348, AF003031, M96285, AF043286, U34165, U34167 to U34176, U34179, U34181, U34183, U34185, U34187 to U34189, U341893, U37217, AF003027, AF001600, and AF043287; HPV-18, X05015*, U45889 to U45891, and U89349; HPV-18var, U45892 to U45894 and M96287; HPV-30, X74474* and M96279; HPV-31, J04353* and U37410; HPV-32, X74475* and M96291; HPV-33: M12732*, U45895 to U45897, and A12360; HPV-34, X74476* and M96292; HPV-35, M74117*, X74477, and U45898; HPV-39, M62849* and U45899 to U45905; HPV-40, X74478* and M96293; HPV-44, U31788* and U12493; HPV-45, X74479*, U45906 to 45916, and M96294; HPV-51, M62877* and U45917; HPV-52, X74481*, U45718 to U45923, and M96297; HPV-53, X74482* and M96298; HPV-54, U37488* and U125601; HPV-55, U31791 and U12494; HPV-56, X74483* and M96299; HPV-57, X55965* and U37537; HPV-58, D90400* and U45924 to U45929; HPV-59, X77858*, U45930 to U45933, and U12496; HPV-61, U31793*, U12499, and U12500; HPV-66, U31794* and U12498; HPV-67, D21208* and U12492; HPV-68, M73258* and U45934; HPV-70, U21941* and U22461; and HPV-72, X94164* and U12485.
Reverse hybridization by the LiPA.
Genotype-specific probes were used to develop a line probe assay (LiPA), the INNO-LiPA HPV prototype research genotyping assay. The oligonucleotide probes were enzymatically provided with a poly(dT) tail. Subsequently, probes were immobilized as parallel lines on nitrocellulose membrane strips. the top line contains a positive control biotinylated DNA. The HPV LiPA is performed essentially as described earlier for the hepatitis C virus INNO-LiPA (28). Briefly, 10 μl of PCR product, containing biotin moieties at the 5′ ends of the primers, was denatured by adding 10 μl of NaOH solution. After 10 min, a LiPA strip was put into the tray. Two milliliters of prewarmed (37°C) hybridization buffer (3× SSC [1× SSC is 15 mM Na-citrate and 150 mM NaCl], 0.1% sodium dodecyl sulfate) was added and incubated at 50 ± 0.5°C for 1 h. All incubations and washing steps were performed automatically in an Auto-LiPA. The strips were washed twice for 30 s and once for 30 min at 50°C with 2 ml of hybridization solution. Following this stringent wash, the strips were incubated with 2 ml of alkaline phosphatase-streptavidin conjugate for 30 min at room temperature. Strips were washed twice with 2 ml of rinse solution and once with 2 ml of substrate buffer. Two milliliters of substrate (5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium) was added and incubated for 30 min at room temperature. The reaction was stopped by aspiration of the substrate solution and addition of 2 ml of distilled water. After drying, the strip results were interpreted by eye by comparing the hybridization pattern to standard type-specific templates. The HPV type was determined by eye according to the following criteria.
In most cases the probe name is directly linked to the HPV type (e.g., a purple color on probe line 16 indicates the presence of HPV-16). Probes c31, c56, and c68 are secondary probes, which are of interest only when there is a positive hybridization with the probe line just above (31/40/58, 56/74, or 68/45). These c probes were developed in order to discriminate between infections with single HPV genotypes and mixed infections. HPV-40, -58, -74, and -45 are also identified separately by independent probe lines. A true infection with HPV-31 yields positive hybridization with probe line 31/40/58 as well as c31. The c probes c31, c56, and c68 will also react with other HPV types, which have completely matching sequences. Probe c31 also reacts with amplimers from types 33 and 54.
Since the LiPA does not contain a separate probe for HPV-54, a sample containing type 54 will react exclusively with probe c31. Similarly, probe c56 reacts with type 58 amplimers. Therefore, amplimers of type 58 will show reactivity with probes 30/40/58, c56, and 58. Probe c68 is also reactive with amplimers from types 18 and 39. HPV-74 is identified by the probes 56/74 and 74. In summary, HPV genotypes 6, 11, 16, 18, 34, 35, 39, 42 to 44, 51 to 54, 59, 66, and 70 are recognized by hybridization to a single probe line, whereas HPV types 18, 31, 33, 39, 40, 45, 56, 58, 68, and 74 yield a specific hybridization pattern on the LiPA.
Statistical analyses.
Data were analyzed by using the Fisher exact test or McNemar’s test.
RESULTS
Development of SPF PCR primers.
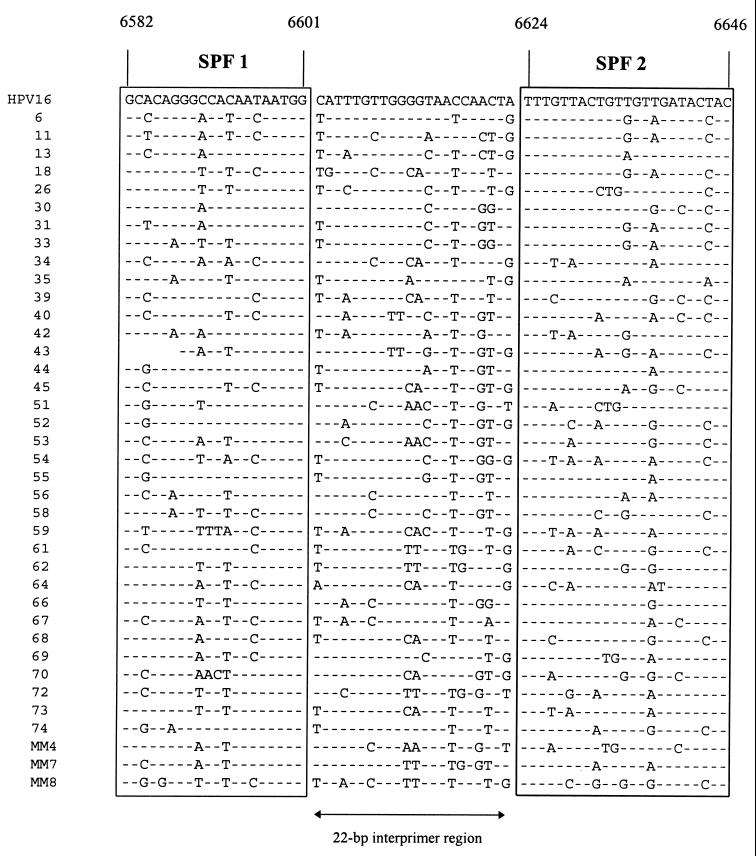
Complete or partial HPV sequences were obtained from GenBank and used for alignment of L1 region sequences. As described earlier (17), the L1 region is relatively well conserved, and several general PCR primers in this part of the HPV genome were developed (Fig. 1). Recently, we have selected a novel set of PCR primers in this region which amplify a fragment of only 65 bp (17). These primers, designated SPF, allowed extremely sensitive amplification from a broad spectrum of HPV genotypes. The present study aimed at using the sequence variation within the amplified fragment of 65 bp for identification of specific HPV genotypes. The SPF primers flank an interprimer region of 22 bp. The alignment of the complete 65-bp fragments amplified by SPF primers from the L1 regions of different mucosal HPV genotypes is shown in Fig. 2. Apparently, all HPV genotypes except genotypes 68 and 73 have a unique SPF interprimer sequence.
FIG. 2.
Alignment of 65-bp nucleotide sequences from the L1 regions of a total of 39 HPV genotypes. The target sequences for the SPF primers are boxed and flank the 22-bp interprimer region, as indicated. Positions are according to the HPV-16 sequence PPH16 (GenBank accession no. K02718).
Inter- and intratypic sequence conservation in the SPF 22-bp interprimer region.
To investigate whether the observed sequence variation among and between HPV genotypes is consistent, a total of 134 sequences from 31 mucosal HPV genotypes, obtained from GenBank, were analyzed (Table 1). For each genotype, the full-length genomic sequence was used as a reference (see Materials and Methods). In 8 (5.9%) of the 134 sequences studied, the interprimer sequences were not completely conserved compared to their respective reference sequences. Four of the nine HPV-18 sequences showed identical single-base-pair mismatches to the reference sequence, and this variant was provisionally designated HPV-18var. One of the two HPV-57 sequences contained a single mismatch, and this sequence had been formally classified as HPV-57b (33). Thus, in these five cases the correct HPV type can be recognized by analysis of the 22-bp sequences. In contrast, one of the HPV-11 sequences showed five mismatches compared to the reference sequence. Among 28 HPV-16 sequences, one sequence contained a single mismatch to the reference sequence. One of the HPV-54 sequences showed two mismatches to the reference. Taken together, analysis of the 22-bp sequences resulted in identification of the correct HPV in 131 (97.8%) of the 134 cases.
TABLE 1.
Conservation of the 22-bp SPF interprimer sequences among mucosal HPV genotypes for 134 HPV sequences obtained from GenBanka
| HPV genotypeb | No. of sequences analyzed | No. of variant 22-bp sequences (no. of mismatches) |
|---|---|---|
| 6 | 3 | 1 (2) |
| 7 | 2 | |
| 11 | 2 | 1 (5) |
| 13 | 2 | |
| 16 | 28 | 1 (1) |
| 18 | 9 | 4 (2)c |
| 30 | 2 | |
| 31 | 2 | |
| 32 | 2 | |
| 33 | 5 | |
| 34 | 2 | |
| 35 | 3 | |
| 39 | 8 | |
| 40 | 2 | |
| 44 | 2 | |
| 45 | 13 | |
| 51 | 2 | |
| 52 | 8 | |
| 53 | 2 | |
| 54 | 2 | 1 (2)d |
| 55 | 2 | |
| 56 | 2 | |
| 57 | 2 | 1 (1)e |
| 58 | 7 | |
| 59 | 6 | |
| 61 | 3 | |
| 66 | 2 | |
| 67 | 2 | |
| 68 | 3 | |
| 70 | 2 | |
| 72 | 2 | |
| Total | 134 | 8 |
Sequence accession numbers are given in Materials and Methods.
For each HPV type, a full-length HPV genomic sequence (see Materials and Methods) was used as a reference.
These four aberrant sequences of genotype 18 were identical and were provisionally designated variant HPV-18var.
This sequence showed two mismatches to the reference HPV-54 sequence and did not match any other HPV type.
This sequence was formally classified as HPV-57b (33).
Since the number of L1 region sequences available in GenBank was limited for several HPV genotypes, a total of 104 clinical isolates were also analyzed. HPV DNA was amplified from these samples by the MY 09/11 as well as the SPF primers. Identification of HPV genotypes was based on sequence analysis of the MY 09/11 amplimer. SPF amplimers were also analyzed, and the HPV genotypes were deduced from the 22-bp SPF interprimer sequences. The results are shown in Table 2. Among the 104 isolates studied, MY 09/11- and SPF-based typing results were initially discordant in 7 cases (5.1%). These discordant cases were further analyzed. Among 19 isolates identified as HPV-16 by the MY 09/11 sequence, SPF analysis showed the presence of HPV-31 in 1 isolate. Type-specific PCR revealed the presence of both HPV-16 and -31 in this isolate. Similarly, in one sample containing HPV-18, SPF PCR detected HPV-16, and the presence of both HPV-16 and -18 was confirmed by type-specific PCR. In the isolate containing HPV-51, sequence analysis of the SPF fragment revealed a single mismatch to the HPV-51 reference sequence. Two of the five samples with HPV-56 yielded discordant results. In one case, the 22-bp sequence showed a single mismatch to the HPV-56 reference sequence, whereas the other case showed the presence of HPV-45. One case of HPV-58 was mistyped as HPV-56 by the SPF system. In the sample containing HPV-73, the SPF system detected HPV-53. Since type-specific primers are not available for HPV-51, -53, -56, -58, and -73, not all discordant results could be analyzed by type-specific PCR. Thus, the HPV types identified from the 22-bp SPF interprimer sequences were initially concordant with MY 09/11 sequences in 99 (95.2%) of the 104 cases studied. Several discordant results were suspected to be due to the presence of multiple HPV types, and this was further analyzed by reverse hybridization (see below). These results show that sequence variation of the SPF amplimer can be used to identify a broad range of HPV genotypes.
TABLE 2.
Identification of HPV genotypes by direct sequencing of MY 09/11 and SPF amplimers and by SPF LiPA in 104 clinical samples (group 1)
| HPV type based on MY 09/11 sequence (n) | No. of isolates for which SPF sequence-based typinga was concordant with MY 09/11 typing | Type(s) determined by the following method when SPF-based typing was initially discordant with MY 09/11 sequence:
|
||
|---|---|---|---|---|
| SPF sequence | Type-specific PCRb | MY 09/11 and SPF LiPA | ||
| 6 (12) | 12 | |||
| 11 (2) | 2 | |||
| 16 (19) | 18 | 31 | 16, 31 | 16, 31 |
| 18 (8) | 7 | 16 | 16, 18 | 16, 18 |
| 30 (1) | 1 | |||
| 31 (9) | 9 | |||
| 33 (7) | 7 | |||
| 35 (6) | 6 | |||
| 39 (1) | 1 | |||
| 45 (3) | 3 | |||
| 51 (1) | 0 | Unknownc | 51 | |
| 52 (9) | 9 | |||
| 53 (1) | 1 | |||
| 55 (1) | 1 | |||
| 56 (5) | 3 | Unknownd | 56 | |
| 45 | 45, 52, 56 | |||
| 57 (1) | 1 | |||
| 58 (6) | 5 | 56 | 56 | |
| 62 (1) | 1 | |||
| 66 (4) | 4 | |||
| 67 (2) | 2 | |||
| 68 (1) | 1 | |||
| 73 (1) | 0 | 53 | 53, 68/73 | |
| 74 (1) | 1 | |||
| MM4 (1) | 1 | |||
| MM7 (1) | 1 | |||
| Total (104) | 97 | 7 | 2 | 7 |
HPV genotype based on sequence of the 22-bp SPF interprimer region.
Available for HPV-16, -18, -31, and -33, according to Baay et al. (3).
The 22-bp sequence from this sample showed a single mismatch to HPV-51.
The 22-bp sequence from this sample showed a single mismatch to HPV-56.
Development of a reverse hybridization LiPA.
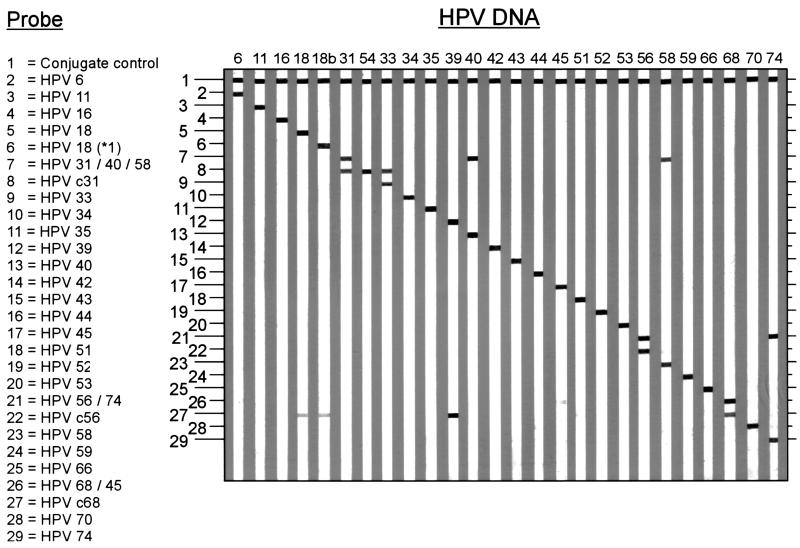
To identify HPV genotypes by hybridization, specific probes were deduced from the SPF sequence alignments (Fig. 2) and used to develop the INNO-LiPA HPV prototype research genotyping assay. Since HPV genotypes often differ by only a single nucleotide in the 22-bp interprimer sequences, well-controlled hybridization conditions are necessary. Therefore, a reverse hybridization LiPA was developed (27), allowing the simultaneous identification of multiple HPV genotypes in a single hybridization step. All probes were optimized to ensure that hybridization occurs only between completely matching sequences. The outline and representative examples of the HPV LiPA are shown in Fig. 3.
FIG. 3.
Outline and representative examples of the INNO-LiPA HPV genotyping assay. The positions of the control line and the 28 specific probes for each of the 25 HPV genotypes are shown at the left. The number above each strip indicates the HPV genotype for which DNA was amplified by SPF primers. The precise interpretation of the hybridization patterns is described in detail in Materials and Methods.
Most of the HPV genotypes are recognized by hybridization to a single type-specific probe. However, a number of HPV genotypes, i.e., 6, 31, 33, 40, 45, 56, 58, 68, and 74, hybridize to more than one probe and can be directly recognized by a specific hybridization pattern on the strip.
To assess the efficacy and reliability of the LiPA, HPV sequences were amplified by SPF primers from a total of 34 plasmids containing complete or partial HPV genomic sequences. The relevant part of the L1 region was amplified from these plasmids and analyzed by direct sequencing. SPF LiPA yielded the expected HPV genotyping results in all cases, and these were completely concordant with sequence data, indicating the high specificity of the reverse hybridization assay.
To determine the efficacy of the SPF system to detect multiple HPV genotypes, mixtures of HPV-11 and HPV-18 target DNAs were tested. A fixed quantity of HPV-11 was combined with increasing concentrations of HPV-18. Conversely, a fixed quantity of HPV-18 DNA was combined with various concentrations of HPV-11. All mixtures were amplified by the SPF primers and tested in the LiPA. The results indicated that the SPF system permits detection of two different HPV genotypes, even if one type is present in a 1,000-fold excess over the other type (data not shown). In contrast, it appears that sequence analysis permitted type-specific detection of both types up to only a 10-fold excess of one genotype over the other.
The amplification target of the SPF primers is located within the target region for the MY 09/11 primer set. Therefore, SPF as well as MY 09/11 amplimers can be used for analysis in the LiPA, provided that PCR primers are biotinylated. SPF and MY 09/11 amplimers from the 104 clinical samples of patient group 1 had already been analyzed by direct sequencing as described above and shown in Table 2. Subsequently, these amplimers were also tested by LiPA. In one HPV-16 isolate, the presence of HPV-16 and -31, as also determined by type-specific PCR, was confirmed, indicating infection with multiple HPV types in this case. Similarly, the presence of both HPV-16 and -18 in the single discordant case of HPV-18 was confirmed. Since the single mismatch was not included in the type-specific probe for these genotypes on the LiPA strip, the HPV-51 isolate and one of the HPV-56 isolates that showed a mismatch to their reference sequences were both correctly typed by the LiPA. In the remaining discordant case of HPV-56, a mixture of HPV-45, -52, and -56 was found. By using the SPF system, one of the HPV-58 isolates was identified as HPV-56 by sequencing as well as by LiPA. The sequence of the MY 09/11 amplimer classified this isolate as HPV-58, but the 22-bp SPF interprimer region in this amplimer was completely homologous to that of HPV-56 (Fig. 2). In the 22-bp SPF interprimer region, the HPV-58 genome differs by only two nucleotides from HPV-56. Therefore, this isolate can be considered as being mistyped by SPF due to intratypic sequence variation of HPV-58. Finally, LiPA analysis of the sample containing HPV-73 revealed the presence of a mixture of HPV-53 and HPV-68 or -73.
Taken together, and including the cases with multiple HPV genotypes, the LiPA results were in agreement with the sequence analysis of the SPF and MY 09/11 fragments, as well as type-specific PCR, in 103 (99%) of the 104 cases.
Evaluation of the HPV LiPA with clinical samples.
To assess the performance of the HPV LiPA system, various groups of clinical specimens were investigated. All clinical samples yielded a β-globin-specific amplimer, confirming the presence of amplifiable DNA. A total of 488 cervical scrapes from patient group 2, classified as having normal cytology or ASCUS, were tested by SPF PCR, as well as with the GP5+/6+ primers. SPF PCR detected HPV DNA in 117 (24%), whereas GP5+/6+ detected HPV-DNA in only 77 (15.7%), of the cases. SPF PCR was repeated for cases that were exclusively positive by SPF to confirm the presence of HPV DNA. The GP5+/6+-positive samples were all positive by SPF PCR. To confirm the specificity of the SPF primer set, the resulting amplimers were subjected to sequence analysis, and the results are shown in Table 3. Based on 100% identity to reference sequences, the 22-bp sequences could be assigned to known HPV genotypes in 93 (79.4%) of the SPF-positive cases, and multiple HPV types were detected in 12 samples (10.2%). GP5+/6+ PCR did not detect HPV DNA in 40 (34.2%) of the 117 SPF-positive cases, and these GP5+/6+-negative cases were distributed over multiple HPV genotypes, suggesting a higher sensitivity of SPF over a broad range of HPV genotypes.
TABLE 3.
Detection of different HPV genotypes by SPF and GP5+/6+ PCR among 488 cervical scrapes from group 2 (1997) and 278 cervical scrapes from group 3 (1998)
| HPV genotype based on analysis of SPF fragments | Group 2
|
Group 3
|
||
|---|---|---|---|---|
| No. SPF positivea | No. GP5+/6+ PCR positive | No. SPF positiveb | No. GP5+/6+ PCR positive | |
| 6 | 6 | 6 | 2 | 1 |
| 11 | 4 | 3 | ||
| 16 | 18 | 12 | 15 | 12 |
| 18 | 4 | 2 | 3 | 3 |
| 31 | 9 | 6 | 6 | 2 |
| 33 | 6 | 4 | 4 | 3 |
| 35 | 1 | 1 | 1 | |
| 39 | 1 | |||
| 42 | 2 | 2 | ||
| 44 | 1 | |||
| 45 | 4 | 4 | 1 | 1 |
| 51 | 1 | 1 | ||
| 52 | 3 | 1 | 2 | 1 |
| 53 | 1 | 2 | 1 | |
| 54 | 1 | |||
| 55 | 1 | |||
| 56 | 3 | 2 | ||
| 58 | 3 | 2 | 1 | |
| 59 | 1 | 1 | ||
| 61 | 2 | 1 | ||
| 62 | 2 | 2 | ||
| 66 | 8 | 2 | 4 | 4 |
| 67 | 2 | 1 | ||
| 68/73 | 1 | |||
| 69 | 1 | 1 | ||
| 70 | 1 | 1 | 1 | 1 |
| 74 | 1 | 1 | 2 | 1 |
| MM7 | 1 | |||
| Unknown | 24 | 18 | 8c | 7 |
| Multiple | 12 | 9 | 10 | 9 |
| Total | 117 | 77 (65.8%) | 70 | 52 (74.3%) |
HPV genotypes were based on sequence analysis of the 22-bp SPF interprimer region.
HPV genotypes were based on SPF LiPA.
Two isolates contained HPV-67 and two contained MM7 as determined by sequence analysis, but these could not be recognized with the current version of the LiPA.
A total of 278 cervical scrapes in patient group 3, also classified as having normal cytology or ASCUS, were investigated by SPF and GP5+/6+ PCR. These results are also shown in Table 3. SPF PCR yielded positive results in 70 samples (25.2%), whereas the use of GP5+/6+ resulted in only 52 positive cases (18.7%). SPF LiPA detected multiple HPV types in 10 (14.2%) of the SPF-positive samples. The cases that remained undetected by GP5+/6+ but were SPF positive were distributed over various HPV types. In 8 (11.4%) of the 70 SPF-positive samples, the HPV type could not be assigned.
Altogether, a total of 766 cervical scrapes (groups 2 and 3), were analyzed, of which 697 (90.1%) had normal cytology and 69 (8.9%) were classified as ASCUS. Among the cases with normal cytology, SPF PCR detected 157 (22.5%), whereas GP5+/6+ detected only 101 (14.4%), HPV-positive samples, and this difference was highly significant (P < 0.001). Among the 69 cases classified as ASCUS, SPF primers detected more HPV-positive samples (30/69 = 43.5%) than the GP5+/6+ primers (26/69 = 37.7%), but this difference was not statistically significant.
Among all SPF-positive samples, 8 (53.3%) of 15 samples containing HPV-31 were GP5+/6+ negative. Similarly, 6 (50%) of the 12 HPV-66 cases remained undetected by GP5+/6+ (Table 3). These findings indicate a significantly lower sensitivity of the latter primer set for these particular HPV types. Sequences from 32 (17.1%) of the 187 SPF-positive samples could not be assigned to any known HPV genotype. Of these 32 samples 26 were tested with the MY 09/11 primers, and 8 were HPV positive. All interprimer sequences were exactly 22 bp in length and were similar to HPV sequences, but they showed between one and five mismatches to any known HPV genotype. Among the 155 samples containing a known HPV genotype, 32 (20.6%) were infected by an HPV with a low-risk genotype (HPV-6, -11, -42, -44, -53, -54, -55, -61, -62, -67, -70, -74, or -MM7).
The fourth group comprised 304 selected cervical smears with mild to moderate (n = 151) or severe (n = 153) dyskaryosis. A total of 299 (98.4%) of the 304 samples were positive by SPF PCR. HPV genotypes were determined by SPF LiPA, and the results are shown in Table 4. Of these, 147 (97.3%) of the 151 cases with mild or moderate dyskaryosis were SPF positive, and 95 of these contained a single HPV genotype. In 2 (1.4%) of the 147 SPF-positive samples, the sequences could not be assigned to a known HPV type, and these will require further characterization. High-risk HPV genotypes (HPV-16, -18, -31, -33, -35, -45, -51, -52, -56, -58, and -66) were present in 95.2% of the cases. Similarly, 152 (99.3%) of the 153 scrapes diagnosed with severe dyskaryosis were HPV positive. HPV genotypes could be assigned in 151 (99.3%) of these samples, and all were classified as high-risk types.
TABLE 4.
Detection of HPV genotypes among 304 cervical smears with mild or moderate dyskaryosis or severe dyskaryosis (group 4)
| HPV genotype based on SPF LiPA | No. of smears with:
|
|
|---|---|---|
| Mild or moderate dyskaryosis (n = 151) | Severe dyskaryosis (n = 153) | |
| 11 | 2 | |
| 16 | 41 | 56 |
| 18 | 9 | 9 |
| 31 | 16 | 14 |
| 33 | 5 | 9 |
| 35 | 3 | |
| 45 | 2 | |
| 51 | 4 | |
| 52 | 3 | 2 |
| 53 | 1 | |
| 56 | 5 | 1 |
| 58 | 3 | 4 |
| 66 | 2 | |
| Unknown | 2 | 1 |
| Multiple | ||
| Double low riska | 2 | |
| Double high riskb | 36 | 41 |
| Triple high riskc | 12 | 9 |
| Quadruple high riskd | 2 | 3 |
| HPV negative | 4 | 1 |
Both samples contained HPV-6 and -11.
Samples containing two HPV genotypes, at least one of which is classified as high risk.
Samples containing three HPV genotypes, at least one of which is classified as high risk.
Samples containing four HPV genotypes, at least one of which is classified as high risk.
Altogether, the SPF sequences could not be assigned to a known HPV genotype in only 3 (1%) of the 304 cases. A total of 105 (34.5%) of the 304 samples contained more than one HPV genotype, and in 103 (98.1%) of these, at least one high-risk genotype was detected. In 21 (6.9%) of the samples, three HPV genotypes were detected, and in 5 (1.6%) cases, four different HPV types were observed.
Group 5 contained 180 formalin-fixed, paraffin-embedded cervical carcinoma samples, comprising 51 adenocarcinomas and 129 squamous cell carcinomas. SPF PCR yielded positive results in all cases. HPV genotypes were identified by sequence analysis and LiPA, and only high-risk HPV types were found, as shown in Table 5. HPV-16 and -18 were found in more than 70% of both groups. The relative prevalence of HPV-18 (17.6%) among the adenocarcinomas was higher than that among the squamous cell carcinomas (6.2%), and this difference was statistically significant (P = 0.027). The number of samples containing other high-risk HPV types was too small to analyze differences between the two groups of carcinomas.
TABLE 5.
HPV genotypes among 180 cervical carcinomas as determined by sequence and LiPA analysis of SPF PCR fragments
| HPV genotype | No. of carcinomas
|
||
|---|---|---|---|
| Total | Adenocarcinoma | Squamous cell carcinoma | |
| 16 | 116 | 31 | 85 |
| 18 | 17 | 9 | 8 |
| 31 | 7 | 2 | 5 |
| 33 | 2 | 2 | |
| 35 | 3 | 1 | 2 |
| 45 | 14 | 3 | 11 |
| 52 | 2 | 2 | |
| 56 | 4 | 4 | |
| 58 | 3 | 3 | |
| 68/73 | 1 | 1 | |
| Unknown | 3 | 1 | 2 |
| Multiple | 8 | 3 | 5 |
| Total | 180 | 51 (28.3%) | 129 (71.7%) |
DISCUSSION
Diagnosis of HPV infection relies on the detection of the viral DNA. The L1 region of the HPV genome has been used for the development of general PCR primer sets, such as MY 09/11, and GP5+/6+ (11, 13). Recently, we have developed a novel SPF primer set, amplifying a fragment of only 65 bp from this L1 region (17). We evaluated the use of these SPF primers and investigated whether sequence variation in the 22-bp SPF interprimer region permits identification of HPV genotypes. Although this 22-bp sequence does not permit formal classification of HPV genotypes, it showed consistent intertypic sequence variation. The inter- and intratypic sequence variation was assessed by analysis of HPV sequences of the L1 region. Altogether, analysis of the 22-bp fragment, as compared to the sequence of the larger MY 09/11 fragment, resulted in reliable genotype identification for 232 (97.5%) of the 238 HPV sequences from GenBank and from clinical materials (Tables 1 and 2). Thus, the intratypic sequence variation among isolates of the same HPV genotype appeared to be limited in this small part of the genome, and the intertypic sequence variation is sufficiently representative for identification of specific HPV genotypes.
Based on the consistent sequence heterogeneity of the SPF amplimer, genotype-specific probes were selected, and a reverse hybridization LiPA was developed. The current version of the INNO-LiPA HPV prototype research genotyping assay comprises the SPF primer set and a reverse hybridization strip containing 28 probes for the detection of 25 different HPV genotypes. The performance of the LiPA was first investigated by analysis of SPF amplimers obtained from HPV DNA-containing plasmids. All HPV genotypes were correctly identified, indicating the high specificity of the SPF LiPA. Also, the LiPA is extremely sensitive for the simultaneous detection of multiple HPV types, even if one type is present in great excess over the other.
Evaluation of the SPF LiPA system was performed with multiple groups of clinical specimens, including cervical smears and cervical carcinomas. Among cervical scrapes with normal cytology or ASCUS, the prevalences of HPV-positive cases, as well as the distributions of genotypes, were highly similar. In these specimens, SPF PCR appears to be significantly more sensitive than GP5+/6+, especially among the cases with normal cytology. This finding confirmed earlier observations (17) and may be explained by the lower concentration of HPV in samples with normal cytology. Although data about the natural history of HPV infections are limited, the viral load is presumably low during the first phase of infection but may increase over time, in parallel with the development of cytological aberrations (25). Since most cervical smears are classified as normal or ASCUS, the use of an extremely sensitive method to diagnose HPV infection is crucial to achieve a maximal negative predictive value for the development of HPV-associated cervical carcinoma. Therefore, the use of the SPF system may have important clinical implications for triage and follow-up of patients with ASCUS and even for patients with normal cytology.
It is not completely clear whether certain HPV genotypes should be considered high-risk types. High-risk HPV types have been frequently detected in cervical and anogenital cancers (18, 34). However, this definition also depends on the accuracy of the HPV detection methods. PCR-based assays can be hampered, especially in archival smears or paraffin-embedded materials, in which the DNA is often damaged. The SPF primers permit efficient detection of HPV DNA in these materials. In the present study, SPF PCR detected HPV DNA in all cervical carcinoma samples, and the HPV genotypes in these samples were classified as high-risk types. HPV-16 and -18 are present in more than 70% of these carcinomas, confirming the importance of these high-risk genotypes. The prevalence of HPV-18 appeared to be significantly higher among adenocarcinoma cases than among squamous cell carcinoma cases, and this is in agreement with earlier observations (2, 19).
The prevalence of multiple HPV infections differed considerably between the clinical groups. Among cases with normal cytology or ASCUS, multiple HPV types were found in 11.8% of the HPV-positive cases, whereas among cases with mild or moderate dyskaryosis, multiple HPV types were detected in 34.5% of the cases. Among the carcinoma samples, only 4.4% contained more than one HPV type, which may, e.g., be due to an underlying CIN lesion in the biopsy taken.
The SPF primers detect a very broad range of HPV genotypes with apparently equal sensitivity. In contrast, other general primers appear to have lower sensitivities for certain HPV genotypes (12, 17, 22), as exemplified by the limited sensitivity of the GP5+/6+ primers for HPV-31 and HPV-66 (Table 3). The differential sensitivities for certain HPV genotypes can be a crucial factor for the detection of multiple HPV genotypes, since this may result in a considerable underestimation of the true prevalence of multiple HPV infections. Both the amplification of a broad range of HPV genotypes by the SPF primers and the sensitive detection of specific genotypes by LiPA play an important role in the overall sensitivity of the SPF system.
The present study also showed that sequence analysis of PCR products does not accurately reflect the presence of multiple HPV types but reveals only the sequence of the most prevalent HPV type(s). In contrast, the reverse hybridization method permits simultaneous and accurate detection of multiple HPV types with much higher sensitivity.
In conclusion, the SPF system can play an important role in further studies of the epidemiology of HPV infections as well as the relationships between cervical carcinoma and specific HPV genotypes.
ACKNOWLEDGMENTS
We thank Semyon Petrov and Frank Smedts for providing the carcinoma biopsies and histological analysis. Els Stet is acknowledged for preparation of LiPA strips. We also thank Jannie Baars for her assistance with cytological examinations and Ron Berkhout for his contributions to the initial sequence analyses.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arends M J, Donaldson Y K, Duvall E, Wyllie A H, Bird C C. HPV in full thickness cervical biopsies: high prevalence in CIN 2 and CIN 3 detected by a sensitive PCR method. J Pathol. 1991;165:301–309. doi: 10.1002/path.1711650405. [DOI] [PubMed] [Google Scholar]
- 3.Baay M F, Quint W G, Koudstaal J, Hollema H, Duk J M, Burger M P, Stolz E, Herbrink P. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J Clin Microbiol. 1996;34:745–747. doi: 10.1128/jcm.34.3.745-747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard H U, Chan S Y, Manos M M, Ong C K, Villa L L, Delius H, Peyton C L, Bauer H M, Wheeler C M. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis. 1994;170:1077–1085. doi: 10.1093/infdis/170.5.1077. . (Erratum, 173:516, 1996.) [DOI] [PubMed] [Google Scholar]
- 5.Bosch F X, Manos M M, Munoz N, Sherman M, Jansen A M, Peto J, Schiffman M H, Moreno V, Kurman R, Shah K V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 6.Burger M P, Hollema H, Pieters W J, Quint W G. Predictive value of human papillomavirus type for histological diagnosis of women with cervical cytological abnormalities. BMJ. 1995;310:94–95. doi: 10.1136/bmj.310.6972.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan S Y, Delius H, Halpern A L, Bernard H U. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clavel C, Rihet S, Masure M, Chypre C, Boulanger J C, Quereux C, Birembaut P. DNA-EIA to detect high and low risk HPV genotypes in cervical lesions with E6/E7 primer mediated multiplex PCR. J Clin Pathol. 1998;51:38–43. doi: 10.1136/jcp.51.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox J T, Lorincz A T, Schiffman M H, Sherman M E, Cullen A, Kurman R J. Human papillomavirus testing by hybrid capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 1995;172:946–954. doi: 10.1016/0002-9378(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 10.Cuzick J, Szarewski A, Terry G, Ho L, Hanby A, Maddox P, Anderson M, Kocjan G, Steele S T, Guillebaud J. Human papillomavirus testing in primary cervical screening. Lancet. 1995;345:1533–1536. doi: 10.1016/s0140-6736(95)91086-7. [DOI] [PubMed] [Google Scholar]
- 11.de Roda Husman A M, Walboomers J M, van den Brule A J, Meijer C J, Snijders P J. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76:1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 12.Gravitt P E, Peyton C L, Apple R J, Wheeler C M. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36:3020–3027. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildesheim A, Schiffman M H, Gravitt P E, Glass A G, Greer C E, Zhang T, Scott D R, Rush B B, Lawler P, Sherman M E, Kurman R J, Manos M M. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs M V, de Roda Husman A M, van den Brule A J, Snijders P J, Meijer C J, Walboomers J M. Group-specific differentiation between high- and low-risk human papillomavirus genotypes by general primer-mediated PCR and two cocktails of oligonucleotide probes. J Clin Microbiol. 1995;33:901–905. doi: 10.1128/jcm.33.4.901-905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs M V, Snijders P J, van den Brule A J, Helmerhorst T J, Meijer C J, Walboomers J M. A general primer GP5+/6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol. 1997;35:791–795. doi: 10.1128/jcm.35.3.791-795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins A, Kristiansen B E, Ask E, Oskarsen B, Kristiansen E, Lindqvist B, Trope C, Kjorstad K. Detection of genital papillomavirus types by polymerase chain reaction using common primers. APMIS. 1991;99:667–673. doi: 10.1111/j.1699-0463.1991.tb01244.x. [DOI] [PubMed] [Google Scholar]
- 17.Kleter B, van Doorn L J, ter Schegget J, Schrauwen L, van Krimpen C, Burger M P, ter Harmsel B, Quint W G V. A novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153:1731–1739. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorincz A T, Reid R, Jenson A B, Greenberg M D, Lancaster W, Kurman R J. Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet Gynecol. 1992;79:328–337. doi: 10.1097/00006250-199203000-00002. [DOI] [PubMed] [Google Scholar]
- 18a.Los Alamos National Laboratory. 23 October 1997, revision date. Los Alamos database. [Online.] http://hpv-web.lanl.gov. [2 June 1999, last date accessed.]
- 19.Low S H, Thong T W, Ho T H, Lee Y S, Morita T, Singh M, Yap E H, Chan Y C. Prevalence of human papillomavirus types 16 and 18 in cervical carcinomas: a study by dot and Southern blot hybridization and the polymerase chain reaction. Jpn J Cancer Res. 1990;81:1118–1123. doi: 10.1111/j.1349-7006.1990.tb02522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundberg G D. The 1988 Bethesda system for reporting cervical/vaginal cytological diagnosis. JAMA. 1989;262:931–934. [PubMed] [Google Scholar]
- 20a.National Center for Biotechnology Information. 13 April 1999, revision date. [Online.] http://www.ncbi.nlm.nih.gov. [2 June 1999, last date accessed.]
- 21.Ohara Y, Honma M, Iwasaki Y. Sensitivity of the polymerase chain reaction for detecting human T-cell leukemia virus type I sequences in paraffin-embedded tissue. Effect of unbuffered formalin fixation. J Virol Methods. 1992;37:83–88. doi: 10.1016/0166-0934(92)90022-6. [DOI] [PubMed] [Google Scholar]
- 22.Qu W, Jiang G, Cruz Y, Chang C J, Ho G Y, Klein R S, Burk R D. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/6+ primer systems. J Clin Microbiol. 1997;35:1304–1310. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richart R M. Screening. The next century. Cancer. 1995;76:1919–1927. doi: 10.1002/1097-0142(19951115)76:10+<1919::aid-cncr2820761308>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 24.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 25.Smits H L, Bollen L J, Tjong-A-Hung S P, Vonk J, van der Velden J, ten Kate F J, Kaan J A, Mol B W, ter Schegget J. Intermethod variation in detection of human papillomavirus DNA in cervical smears. J Clin Microbiol. 1995;33:2631–2636. doi: 10.1128/jcm.33.10.2631-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snijders P J, van den Brule A J, Schrijnemakers H F, Snow G, Meijer C J, Walboomers J M. The use of general primers in the polymerase chain reaction permits the detection of a broad spectrum of human papillomavirus genotypes. J Gen Virol. 1990;71:173–181. doi: 10.1099/0022-1317-71-1-173. [DOI] [PubMed] [Google Scholar]
- 27.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vandenborght B, van Heuverswyn H, Maertens G. Typing of HCV isolates and characterization of new (sub)types using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 28.Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996;34:2259–2266. doi: 10.1128/jcm.34.9.2259-2266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ter Harmsel B, van Muyden R, Smedts F, Hermans J, Kuijpers J, Raikhlin N, Petrov S, Ledebev A, Ramaekers F, Trimbos B. The significance of cell type and tumor growth markers in the prognosis of unscreened cervical cancer patients. Int J Gyn Cancer. 1998;8:336–344. [Google Scholar]
- 30.Tieben L M, ter Schegget J, Minnaar R P, Bouwes Bavinck J, Berkhout R J, Vermeer B J, Jebbink M F, Smits H L. Detection of cutaneous and genital HPV types in clinical samples by PCR using consensus primers. J Virol Methods. 1993;42:265–279. doi: 10.1016/0166-0934(93)90038-s. [DOI] [PubMed] [Google Scholar]
- 31.van den Brule A J, Claas E C, Du Maine M, Melchers W J, Helmerhorst T, Quint W G, Lindeman J, Meijer C J, Walboomers J M. Use of anticontamination primers in the polymerase chain reaction for the detection of human papilloma virus genotypes in cervical scrapes and biopsies. J Med Virol. 1989;29:20–27. doi: 10.1002/jmv.1890290105. [DOI] [PubMed] [Google Scholar]
- 32.van Ranst M A, Kaplan J B, Burk R D. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J Gen Virol. 1992;73:2653–2660. doi: 10.1099/0022-1317-73-10-2653. [DOI] [PubMed] [Google Scholar]
- 33.Wu T C, Trujillo J M, Kashima H K, Mounts P. Association of human papillomavirus with nasal neoplasia. Lancet. 1993;341:522–524. doi: 10.1016/0140-6736(93)90280-t. [DOI] [PubMed] [Google Scholar]
- 34.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]