Abstract
The coronavirus disease 2019 (COVID-19) pandemic requires the continued development of safe, long-lasting, and efficacious vaccines for preventive responses to major outbreaks around the world, and especially in isolated and developing countries. To combat severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), we characterize a temperature-stable vaccine candidate (TOH-Vac1) that uses a replication-competent, attenuated vaccinia virus as a vector to express a membrane-tethered spike receptor binding domain (RBD) antigen. We evaluate the effects of dose escalation and administration routes on vaccine safety, efficacy, and immunogenicity in animal models. Our vaccine induces high levels of SARS-CoV-2 neutralizing antibodies and favorable T cell responses, while maintaining an optimal safety profile in mice and cynomolgus macaques. We demonstrate robust immune responses and protective immunity against SARS-CoV-2 variants after only a single dose. Together, these findings support further development of our novel and versatile vaccine platform as an alternative or complementary approach to current vaccines.
Keywords: vaccine, vaccinia virus, SARS-CoV-2, RBD, single dose, immune response
Graphical abstract
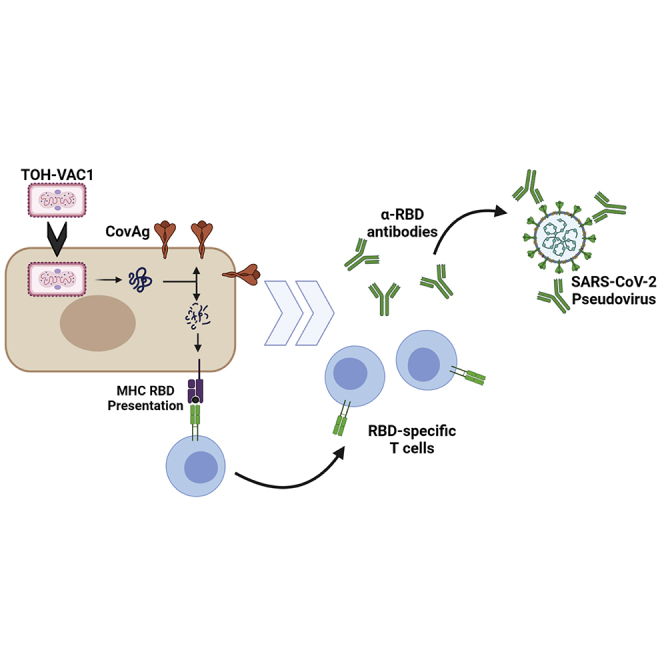
To combat SARS-CoV-2, we characterize a novel vaccine candidate (TOH-Vac1) that uses a replication-competent, attenuated vaccinia virus as a vector to express a membrane-tethered spike receptor binding domain antigen. We evaluate the effects of dose escalation and administration routes on vaccine safety, efficacy, and immunogenicity in mice and cynomolgus macaques.
Introduction
A large, sustained community outbreak of severe acute pneumonia due to a novel coronavirus (COVID-19) was first recognized with household transmission in late 2019 in Wuhan, China. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) had likely emerged from a zoonosis, and onward person-to-person transmission.1 Variant strains have emerged in and spread from the United Kingdom (alpha), South Africa (beta), the United States (epsilon), Brazil (gamma), and India (delta), with variable changes in infectiousness, immune evasion, and pathogenicity (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html). As of September 2021, 216 million people have been infected with SARS-CoV-2 and over 4.5 million have died of COVID-19.2 This devastating pandemic has resulted in the unprecedented rapid development of several new, safe, and efficacious vaccines employing different technology platforms.3 The platforms in furthest clinical development, with regulatory emergency use authorization or full Food and Drug Administration approval, include different mRNA-based vaccines4,5 and nonreplicating adenovirus-vectored vaccines.6,7 The immunodominant target of the humoral response8,9, the spike (S) protein, emerged as the central immunogen focus for SARS-CoV-2 vaccine design.4, 5, 6, 7,10 The S glycoprotein is a large homotrimeric complex that mediates binding and entry of SARS-CoV-2 into host cells. It is divided into two subunits, S1 and S2, which control binding and fusion, respectively. Within the S1 subunit is the receptor binding domain (RBD) that mediates viral attachment to host cells by interacting with the host receptor, angiotensin-converting enzyme 2 (ACE2).11, 12, 13 Since neutralization of this RBD-ACE2 interaction is capable of blocking virus infection,14, 15, 16, 17 the RBD is a popular target for drug development and vaccination strategies.
Despite the incredible progress made over the past year, there are still numerous challenges facing COVID vaccine development.18, 19, 20, 21 At this point, it remains unclear whether the currently authorized vaccines elicit long-term immunogenicity. In addition, multidose vaccines can create financial burdens and additional challenges for manufacturing and administration, which can cause delays between doses that may yet have unforeseen consequences on timeliness, degree, and duration of protection. Special requirements for storage and handling, especially with mRNA vaccines that require −80°C freezers, have also created challenges for delivery to remote and impoverished communities that lack prerequisite equipment.22 Altogether, this creates a demand for new vaccines with increased stability and performance.
Although an estimated 190 candidates are currently undergoing preclinical development and over 90 are in early clinical development,23 few are attempting to immunize with replicating attenuated Vaccinia virus vectors.24 Previous work has demonstrated that Vaccinia virus (VACV) can be engineered to express heterologous antigens and can be strategically attenuated to act as a safe vaccine vector.25,26 VACV has an excellent safety profile, in large part due to its historical use as the vaccine in the worldwide campaign to eradicate smallpox.27 In the context of vaccine development against SARS-CoV, modified Vaccinia Ankara (MVA), a highly attenuated VACV strain incapable of productive infection,28 expressing the spike glycoprotein was shown to be effective in inducing a neutralizing antibody response.29, 30, 31, 32 Recent studies have explored the use of MVA as delivery vector for SARS-CoV-2 antigens.18,19,33, 34, 35
While replication-incompetent viruses such as MVA are typically favored as vaccine vectors because of their safety, replicating viruses offer several additional advantages for vaccine delivery. They have increased persistence at the immunization site, which locally prolongs antigen expression; they are often more effective at stimulating T cell responses, and they are relatively easy to manufacture for large populations. Replicating VACV strains have been used previously for vaccination campaigns against smallpox with adverse effects occurring only in a very small population of individuals. Since then, many attenuations have been discovered that reduce the health risks of VACV strains even further.36, 37, 38, 39, 40, 41 In our lab, we have engineered oncolytic VACV variants used to treat immunocompromised cancer patients without safety concerns.36, 37, 38,42
In this study, we evaluated the use of a replication-competent attenuated Tiantan (TT) strain of VACV as vector for a COVID-19 vaccine. With its role as the initial point of contact between SARS-CoV-2 and the host cellular receptor ACE2, the RBD was selected as the primary immunogen for our vaccine.11, 12, 13 While there are other potential mechanisms to neutralize virus entry through targeting of S, direct inhibition of the RBD-ACE2 interaction is one of the best characterized methods for blocking SARS-CoV-2 infection.43, 44, 45, 46, 47 In the full S trimer, the RBD transitions between two discrete conformations: an “up” state, which is capable of interacting with ACE2, and a “down” state in which the receptor is inactive and shielded from neutralizing antibodies.20 It is still not fully clear what controls the conformation of the RBD in the S complex, but it is known that mutations in both the S1 and S2 subunits can change these two conformations and influence antibody binding.6 Thus, exposure of the RBD antigen for antibody generation in full-length S constructs is dependent on a conformational selection process that is controlled by other domains within S. In contrast, an isolated RBD vaccine exposes epitopes that when targeted can neutralize interactions with host cells. We therefore engineered a chimeric RBD construct fused to the transmembrane region of S (henceforth termed CovAg) to expose essential epitopes for neutralizing antibody generation (Figure 1A). Membrane-tethered expression of RBD was used, as opposed to a secreted form, as membrane anchoring has been found to increase immunogenicity of spike-based mRNA vaccine.48 We found that one individual dose of our strategically attenuated TT CovAg vaccine is sufficient to induce strong and long-lasting RBD-targeted humoral and T cell immune responses in vaccinated mice and cynomolgus macaques. The TT-based viral vector presented in this paper, TOH-VAC1, holds great potential as an additional vaccine platform in the ongoing fight against COVID-19.
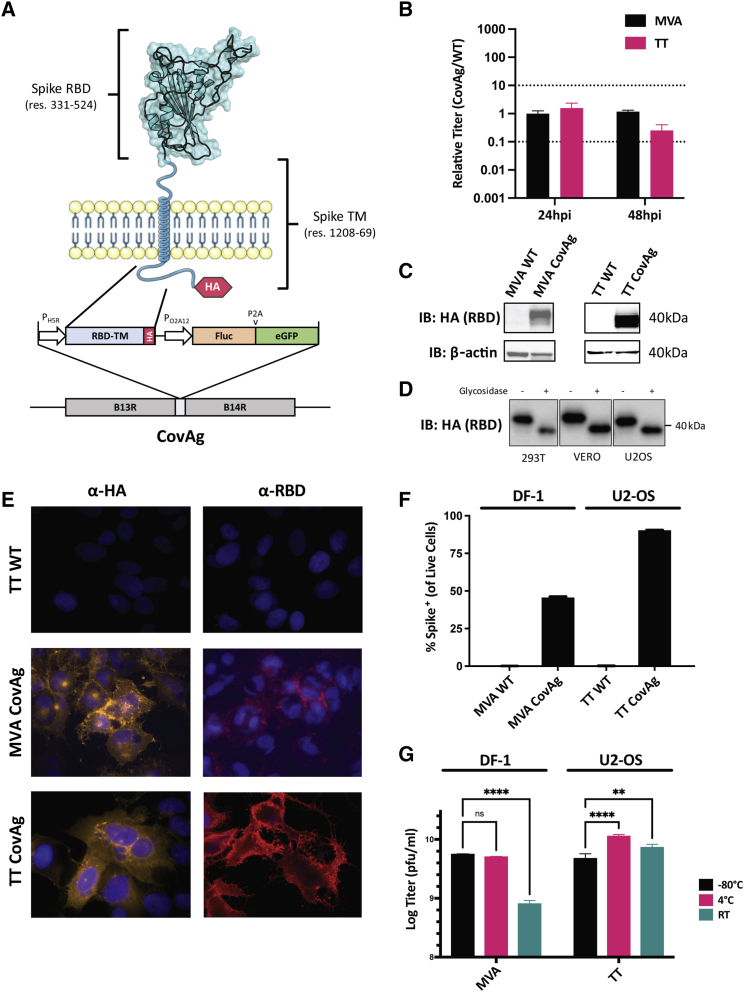
Figure 1.
Design and validation of CovAg vaccine
(A) Schematic of CovAg (spanning residues 331 to 524 and 1,208 to 1,269) antigen49 and its insertion site in the Vaccinia genome. The RBD-TM was designed with a C-terminal HA-tag for detection and is expressed from the Vaccinia H5R promoter (early/late). It was inserted at the B14R locus along with firefly luciferase and GFP for in vivo and in vitro detection, respectively. (B) Comparison of titers from cells infected with MVA CovAg or TT CovAg. Data are shown as CovAg titers relative to WT (n = 3, mean ± SD). Dashed lines reflect 10-fold increase or decrease in points. (C) Immunoblot of CovAg expressed from U2OS cells infected MVA and TT as probed with HA antibody. (D) Immunoblot of CovAg from infected U2OS cells after undergoing treatment with glycosidase to illustrate RBD glycosylation (uncropped western can be found in Figure S2). (E) Immunofluorescence of MVA/TT CovAg constructs with α-HA or α-RBD. For HA antibody samples, cells were permeabilized with 0.2% Triton X-100, whereas RBD samples were left unpermeabilized. (F) Quantification of RBD expressed on the surface of live cells infected with MVA- or TT- CovAg, by flow cytometry (n = 3, mean ± SD). (G) Temperature stability of MVA and TT backbones as probed by plaque assay after storage at −80°C, 4°C, or room temperature (RT) for 7 days (n = 3, log-transformed titer means ± SD; two-way ANOVA with Sidak's correction for multiple comparisons; alpha threshold = 0.05).
Results
Characterization of poxvirus vaccine vectors
The RBD-based CovAg immunogen (Figure 1A, materials and methods) was encoded into two poxvirus strains, TT and MVA, to compare humoral and cellular immunities from both a replicating and nonreplicating viral vector, respectively. In both strains, CovAg was inserted into the B14R locus under the control of a robust early/late promoter (H5R) (Figure 1A).50 For detection and purification of the virus, a GFP-firefly luciferase reporter cassette under the control of a synthetic early/late promoter (O2A12) was incorporated following the CovAg gene (Figure 1A). Recombinant TT and MVA vaccine vectors were generated through homologous recombination and purified by selection of GFP-positive plaques. Virus purity was confirmed via PCR analyses and Nanopore deep sequencing (Figure S1).
In order to compare viral kinetics of the antigen-encoding viruses with the parental wild-type (WT) strains, cells were infected at an MOI of 1 and titers were quantified at 24 and 48 h postinfection (hpi). Standard plaque assays revealed that MVA CovAg replicated similarly to the WT virus in DF-1 chicken embryo fibroblast cells (Figure 1B). Similar titers were observed for TT CovAg and TT WT in U2-OS cells (Figure 1B). Taken together, these results suggest CovAg expression does not significantly impair replication of either MVA or TT viral vectors.
Expression of CovAg from MVA and TT vectors was assessed via immunoblotting with an HA antibody (Figure 1C). Since RBD glycosylation is integral for maintenance of antigenic conformation and generation of neutralizing antibodies against SARS-CoV-2,39,40,51,52 glycosylation of the RBD was assayed by immunoblot following treatment with glycosidase (Figures 1D and S2). The deglycosylated RBD had increased electrophoretic mobility, consistent with the loss of two essential N-linked glycosylation sites at positions N331, N343.52,53 Expression and localization of CovAg was assessed by immunofluorescence (Figure 1E) and flow cytometry (Figure 1F). Together, these data confirmed that CovAg is translocated to the plasma membrane with the RBD exposed on the extracellular surface, and suggest that the poxvirus vectors mediate expression of CovAg in an antigenically relevant conformation.
We investigated the short-term temperature stability of MVA and TT vaccine vectors, which has previously been demonstrated to be stable long-term under a range of temperature conditions.54, 55, 56 Both viruses were stored at −80°C, 4°C, and room temperature for 1 week and then active vaccine particles were quantified by plaque assay. Under these conditions, there was no significant drop in titer TT vectors, but replication of MVA was impaired by storage at room temperature (Figure 1G).
Comparison of MVA and TT viral vaccine vectors
To assess and compare the safety profiles of our MVA and TT CovAg vaccines, BALB/c mice were injected intraperitoneally (i.p.) at varying doses, and their individual body weights were monitored over time, as an indicator of mouse health. At the doses used for vaccination within this study, neither TT nor MVA caused any significant loss average weight (Figures S3A and S3B), and no signs indicative of pathogenic infection were observed. We further investigated the safety of TT by injecting BALB/c intracranially (i.c.) at a dose of 1 × 106 plaque-forming units (pfu), as previously performed.57,58 All mice from these injections survived with no observable signs of cognitive impairment or neurotoxicity (Figure S3C). As a positive control, we observed that all mice injected i.c. with WT vesicular stomatitis virus (VSV), a virus with well-known neurotoxicity59, 60, 61, died within 4 days. In addition, we did not detect any significant difference in mouse body weight between intranasal (i.n.) administration of TT WT or TT CovAg (Figure S3D).
To determine which vector (MVA or TT) is more effective at stimulating immune responses against the CovAg immunogen, BALB/c mice were vaccinated with each virus via i.p., i.n., or subcutaneous (s.c.) injections. Antibody responses were measured by ELISA against recombinant RBD and the antibody titer was calculated by fitting of dilution curves (Figures 2A and 2B, materials and methods). For i.n. and i.p. administration routes, TT CovAg was significantly more effective than MVA CovAg at stimulating humoral responses against the RBD. On average, the antibody titer for TT CovAg was three to four orders of magnitude greater than those of MVA CovAg. In contrast, the antibody titers did not significantly differ between vaccines administered s.c, but the antibody titers induced by TT CovAg s.c. administration were also three to four orders of magnitude lower than i.p. and i.n. administration (Figure 2B).
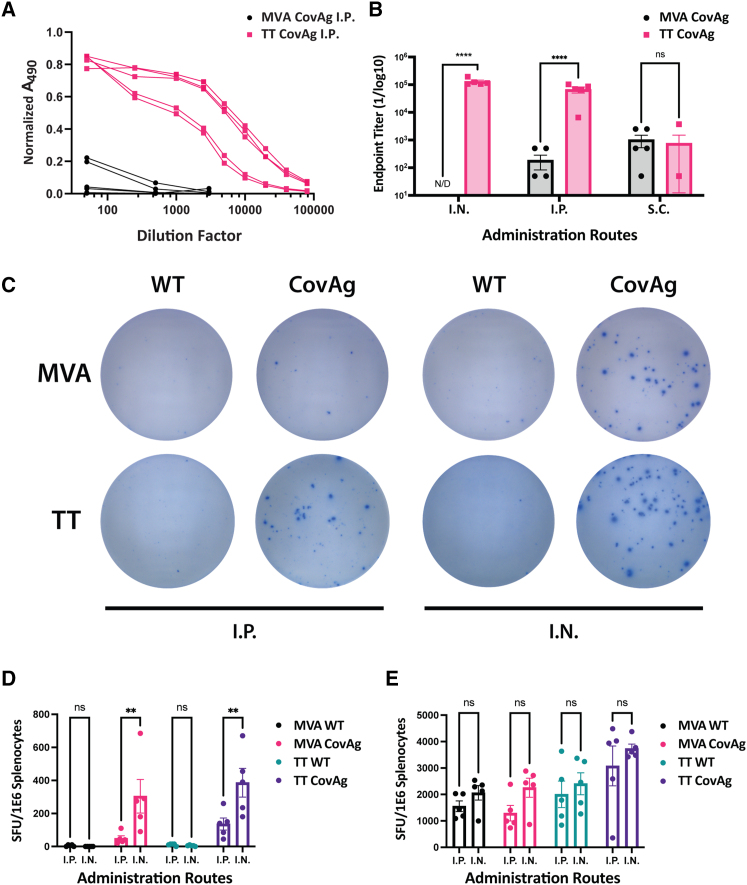
Figure 2.
Comparison of MVA and TT CovAg vaccines
(A) Normalized ELISA absorbance versus serum dilution factors for mice vaccinated i.p. with MVA CovAg (black) or TT CovAg (pink) (n = 5 per group, biological replicates shown). (B) Endpoint titer from RBD ELISAs for MVA and TT vaccines inoculated via different administration routes (n = 5, mean ± SEM; two-way ANOVA with Sidak's correction for multiple comparisons; alpha threshold = 0.05). N/D indicates values were not detectable. (C) Representative wells from IFN-γ ELISPOT experiment for MVA and TT. (D) Spot-forming units (SFU) of MVA and TT vaccines stimulated with spike S1 peptide pool. (E) SFUs of MVA and TT vaccines stimulated with Vaccinia E3 and F2 peptides to examine T cell responses against Vaccinia backbone (n = 5, mean ± SEM; two-way ANOVA with Sidak's correction for multiple comparisons; alpha threshold = 0.05).
MVA and TT vectors were further compared for their ability to stimulate RBD-specific T cell responses via interferonγ (IFN-γ) enzyme-linked immunosorbent spot (ELISPOT) assays. Both the MVA CovAg and TT CovAg vaccines induced T cell responses against RBD epitopes by both i.p. and i.n. administration routes (Figures 2C and 2D). In addition, the T cell responses were more robust for both vectors when administered i.n., and TT had stronger responses compared with MVA. Interestingly, before commercially available peptide pools were available, we identified an immunodominant RBD epitope for BALB/c mice that had elevated responses for TT CovAg, similar to the commercial S1 peptide pool, as compared with DMSO controls (Figures S4A–S4C, Table S1). The responses against this single peptide were weaker for MVA CovAg than TT CovAg (Figures S4A–S4C). Last, we examined T cell responses against the VACV backbone using two well-established T cell epitopes.62 Both MVA and TT had similar responses to these peptides and we did not observe any difference between i.p. and i.n. administration routes (Figure 2E).
Overall, the comparative analyses of antibody and T cell responses for TT CovAg and MVA CovAg showed that TT was a more suitable vaccine platform. Not only did TT CovAg generate antibody titers that were three to four orders of magnitude greater than those generated by MVA (Figure 2B), but ELISPOT results were on average 1.5 to 2.5 times higher with TT CovAg (Figure 2D). Of note, this was accomplished with a TT dose that was 10-fold less than MVA, and no toxicity was observed. Given that TT outperformed MVA through these initial in vivo experiments and given the relative challenge of manufacturing MVA compared with TT, we decided to focus on the TT CovAg platform for further investigation into its suitability as a COVID-19 vaccine.
TT is a potent single-dose vaccine that offers longstanding protection
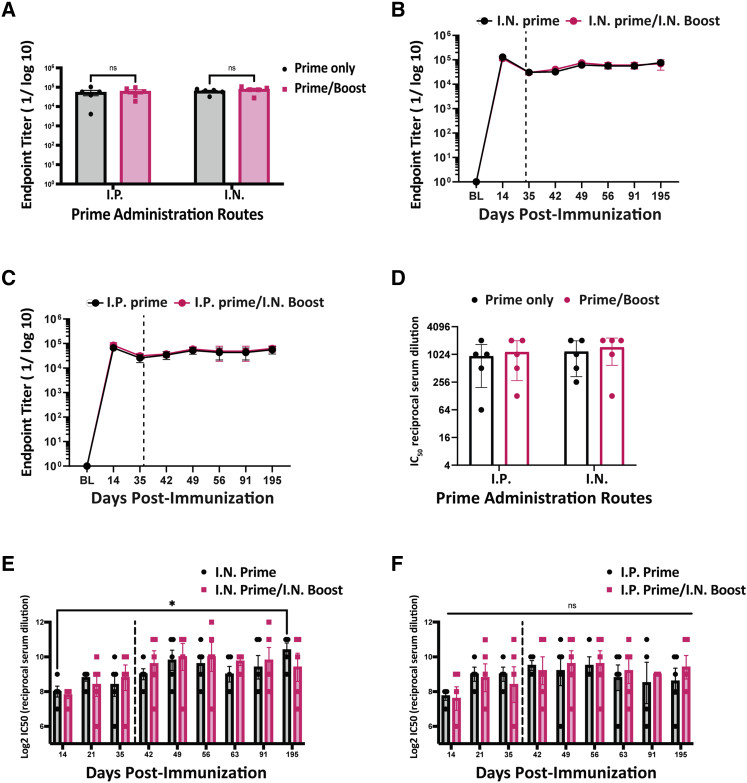
To determine whether our TT platform could be further enhanced, we designed a homologous prime/boost strategy for our vaccine. BALB/c mice were initially inoculated i.p. and i.n. and ELISAs were performed every week to evaluate RBD antibody responses. Upon detection of a slight decrease in antibody titer (around day 35), mice were boosted i.n. with a second dose of the same vaccine. At day 49, the prime/boost responses were compared for each route of administration, and we observed no significant increase in antibody titers after the boost dose (Figure 3A). We continued measuring antibody titers for more than 6 months after the initial immunization, and we showed sustained total antibody titers for both prime only and prime:boost vaccination regimens (Figures 3B and 3C). We also quantified the neutralizing capacity of antibodies produced with and without the boosting dose using a replication-competent VSV pseudotyped with the SARS-CoV-2 spike. This VSV-based surrogate system has been previously demonstrated to resemble both SARS-CoV-2 entry and neutralizing antibody sensitivity.63 Consistent with the ELISA data, antibodies generated over a 6-month period from a single-dose TT CovAg vaccination resulted in robust neutralization without drop in titer over time (Figures 3D–3F). In addition, there was no noticeable difference in the neutralizing response of mice receiving either one or two doses of the vaccine. Last, we showed high antibody titer in mice that were immunized through i.m. administration of TT CovAg, but not TT WT control (Figure S4D).
Figure 3.
Analyses of humoral responses induced by TT CovAg vaccine in BALB/c mice
(A) Endpoint titer of TT CovAg vaccinated mice with and without boosts for various routes of administration. Data were acquired at day 49, 14 days after the boost injection date (n = 5, mean ± SEM; two-way ANOVA with Sidak's correction for multiple comparisons; alpha threshold = 0.05). (B and C) Antibody endpoint titers over 195 days for i.n. (B) or i.p. primed (C) mice. The vertical dashed line depicts the boost date (n = 5, mean ± SEM). (D) VSV-S neutralization IC50 values for samples described in (A). (E and F) VSV-S neutralization IC50 values over 195 days for prime only and prime/boost vaccinations given either i.p. (E) or i.n. (F) (n = 5, Log2 mean ± SEM; ∗p value < 0.05 relative to D14; two-way ANOVA with Dunnet's correction for multiple comparison; alpha threshold = 0.05).
Development of attenuated TT vaccine with improved safety
Given the compelling immunological responses observed in mice vaccinated with TT CovAg, we evaluated its potential as a vaccine candidate in nonhuman primates. Since mice are more resistant to VACV compared with primates, there was concern that TT might pose additional safety risks at the administered dose. To address that potential concern, we further attenuated the VACV backbone by inserting mCherry into the A56R gene of the TT CovAg vaccine to disrupt its function (TOH-VAC1). A56R was selected based on prior studies that showed its deletion significantly attenuated the lethality of the virus in vivo.57 CovAg expression was unaffected by the A56R deletion (Figure S5A). To evaluate the safety benefit of TOH-VAC1, immunodeficient nude mice were injected intravenously (i.v.) at a dose of 1 × 107 pfu. While there was only a slight benefit observed for TOH-VAC1 over the original TT CovAg in terms of relative weight loss (Figure S5B), there was substantial reduction in the number and severity of pox lesions on the skin (Figure S6). Mice treated with TOH-VAC1 or the original TT CovAg showed decreased weight loss compared with TT WT, as a measure of mouse health, and increased survival due to fewer pox lesions. These results are consistent with average weight of mice remaining the same after i.n. administration of TOH-VAC1 or TT CovAg (Figure S3D).
An interesting feature of TT vector with A56R deletion is its enhanced syncytium formation capacity (Figure S5C). To determine whether the attenuated toxicity or syncytia formation could impact the immune response against the CovAg antigen, BALB/c mice were immunized and tested for neutralizing antibodies and T cell responses (Figures S5D and S5E). The titer of neutralizing antibodies and the T cell response against RBD peptides were not significantly impacted by the A56R deletion. Therefore, the novel TOH-VAC1 construct offers a vaccine platform safer than the parental TT vector and without any loss in immunogenicity.
Investigation of TOH-VAC1 efficacy in cynomolgus macaques
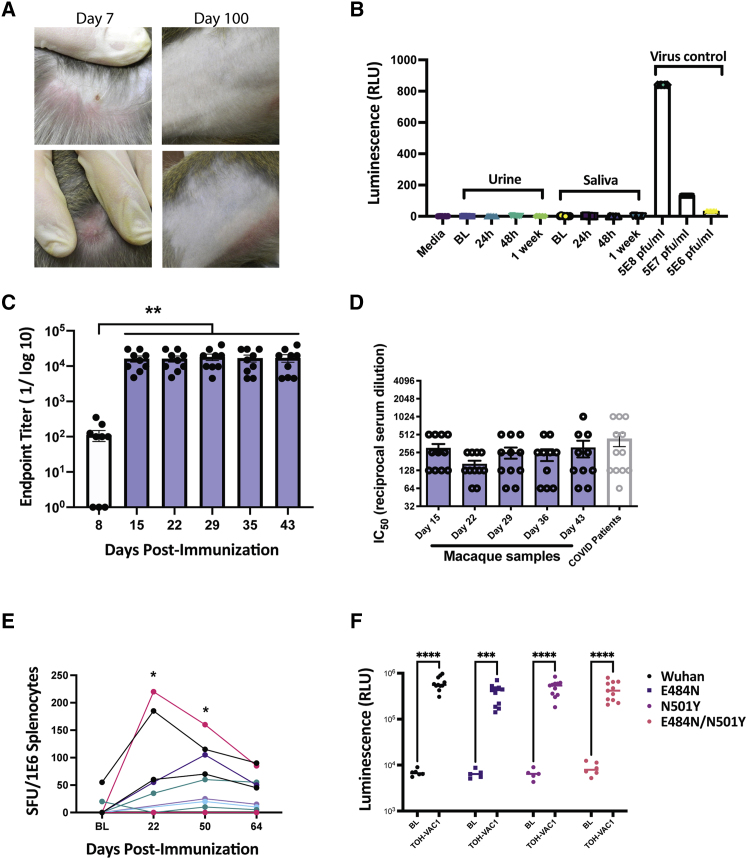
Cynomolgus macaques were immunized i.m. with TOH-VAC1 at a dose of 1 × 107 pfu. With exception of some macaques developing minor swelling or lesions at the injection site, which eventually made a full recovery (Figure 4A), there were no adverse side effects from the TOH-VAC1 vaccine. Average body weight and temperature remained unchanged (Figures S7A and S7B). In addition, virus shedding was not observed in either macaque saliva or urine after immunization, as indicated (Figures 4B and S7C and Tables S3–S5). Antibody responses were measured by ELISA for 43 days following vaccination, and from day 15 onward, we observed a robust and consistent response (Figure 4C). Similarly, the neutralization response against spike-pseudotyped VSV was consistent over the 43 days, with values similar to patients that had previously tested positive for COVID-19 (Wuhan strain) (Figure 4D). RBD-specific T cell responses were also measured from the peripheral blood mononuclear cells (PBMCs) of macaques immunized with TOH-VAC1 (Figure 4E). ELISPOTs were performed on samples collected 22, 50, and 64 days postimmunization to test the strength and duration of memory T cell responses against RBD peptides. The strongest responses were observed on day 22 for two macaques that had more than 150 spot-forming units per million splenocytes. However, five macaques continued to exhibit RBD-specific T cell responses up to 64 days following vaccination. In addition, the T cell responses in three macaques were higher at day 50 compared with day 22. Together, these results demonstrate the long-lasting protective capability of the TOH-VAC1 vaccine.
Figure 4.
Investigation of TT RBD-TM efficacy in cynomolgus macaques
(A) Images of injection sites for TOH-VAC1 after 7 and 100 days. (B) Measurement of viral shedding in macaque urine and saliva. U2OS cells were mixed with either macaque urine, saliva, or pure TOH-VAC1 (virus control) and virus presence was assayed based on firefly luciferase activity. Baseline (BL) samples were sera sample taken from macaques before vaccination. (C) Endpoint antibody titer for RBD-specific antibodies from macaques vaccinated i.m. with TOH-VAC1. Baseline values were subtracted from all other timepoints (n = 11, mean ± SEM; ∗∗p < 0.01 relative to D8; one-way ANOVA with Dunnet's correction for multiple comparison, alpha threshold = 0.05). (D) VSV-S neutralization assay results from macaque samples post immunization (vaccinated i.m. with TOH-VAC1), and results from sera of SARS-CoV-2 (Wuhan strain)-positive patients. No observable neutralization was observed from sera prior to vaccination (macaques n = 11, mean ± SEM; human COVID-19 patients n = 12, mean ± SEM). (E) ELISPOT data from macaque PBMCs stimulated with S1 peptide pool. Each curve depicts results from a single macaque immunized with TOH-VAC1. (F) Seroconversion assay for the RBD of SARS-CoV-2 and three of its variants tested against macaque sera 28 days post immunization (n = 11, mean shown; ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 relative to BL; two-way ANOVA with Sidak's correction for multiple comparisons, alpha threshold = 0.05). BL samples were sera samples taken from macaques before vaccination.
To address potential concerns about the increase in resistance to spike-based vaccines observed for SARS-CoV-2 variants,64, 65, 66 we developed a new biosensor assay (Figures 4F and S8), based on our previous work,12,51 to test the binding of antibodies produced from TOH-VAC1 vaccination against common RBD variants that have emerged around the world. Using this assay, we tested the antibodies produced in macaques against the WT (Wuhan sequence) and three RBD variants, including the N501Y mutation found in the Alpha variant and the E484K mutation found in the Beta and Gamma variants. Both of these variants can reduce virus sensitivity to immune sera from vaccinated individuals and when combined together, resistance to pre-existing antibodies is even further increased.64, 65, 66 However, with our biosensor assay, we found that sera from macaques immunized with TOH-VAC1 interacted with both single and double mutants (Figure 4F).
Discussion
As the COVID-19 pandemic continues with high rates around the world and as new variants emerge with resistance to current vaccines, there maintains an immediate need for the development of accessible vaccines that elicit long-lasting protective immunity. Here, we developed a viral vector-based COVID-19 vaccine with the MVA and TT strains of VACV that express the RBD of the SARS-CoV-2 S protein tethered, via its transmembrane region, to the plasma membrane. Both VACV vectors have been successfully used as vaccines in the past. TT was used as a vaccine in China during the campaign to eradicate smallpox,27 and more recently, has seen new developments as a vaccine for hepatitis B virus67 and HIV.68 On the other hand, MVA has been tested as a vaccine vector for many diseases, due its well-characterized safety and immunogenicity.41,69, 70, 71, 72, 73, 74, 75 Both vaccines are stable over a range of temperatures, which ensures easy storage and distribution,55,56 and VACV can also be freeze-dried and stored for extended periods at 25°C without significant impact on immunogenicity.54
In our study, the comparative analysis of humoral and cellular immune responses between TT and MVA demonstrated that TT is far more effective at stimulating neutralizing antibody production and alloreactive T cell responses against the SARS-CoV-2 RBD (Figure 2). Antibody levels were three to four orders of magnitude lower for MVA immunized mice compared with TT and T cell responses were 2-fold lower. The use of an attenuated nonreplicating vector has several advantages from a safety standpoint, but its inability to replicate in vivo also decreases the quantity and duration of immunogen expression, which consequently leads to reduced immunity. With the replication-competent TT backbone, our vaccine was capable of stimulating robust humoral and cellular immunity with only a single dose, whereas MVA-based SARS-CoV-2 vaccines require two doses to achieve strong immunity.18,19,33, 34, 35 Antibody titers from our vaccine were also similar to those reported for the mRNA and Adenovirus vector-based vaccines currently in use.4,5,73,76
In mice, immunization with a second dose of TT CovAg did not further improve the levels of RBD-specific neutralizing antibodies. In macaques, a single dose of TOH-VAC1 was sufficient to generate neutralizing antibodies titers that are similar to those from former symptomatic COVID-19 patients. As for safety concerns with the use of a replicating vector, we have shown in this study that TT is sufficiently safe for immunization of both mice and macaques. However, if any concerns arise, it is possible to further attenuate TT using deletions like our A56R knockout.57,58,77 In contrast, it is much more challenging to reverse engineer MVA to become more immunogenic or more persistent for prolonging immunogen expression.
Based on our results, it is clear there are many inherent advantages to using a replicating poxvirus vector for a SARS-CoV-2 vaccine. It produces stronger immune responses with only a single injection at a 10-fold lower dose than its nonreplicating MVA counterpart. Biomanufacturing of MVA is also more challenging, as it requires growth in specialized cells such as primary chicken embryonic fibroblasts,18 as opposed to TT, which can be produced using many common immortalized cell lines and at very high yields. Therefore, it will be faster to produce more doses and improve global accessibility to vaccination with our TT vaccine than with a similar MVA-based vaccine platform. Since many nations are also facing challenges obtaining enough vaccines to provide two doses in a timely manner, this single-dose TT vaccine could improve global accessibility to vaccination. This will be essential for getting COVID-19 rates under control, especially with new, highly infectious variants.
Anti-vector immunity can pose a challenge to many viral vector vaccines. However, for VACV, pre-existing immunity fades over time, and can be evaded through the use of mucosal routes of administration.78, 79, 80 There is also existing evidence that VACV-based vaccines suffer little to no impairment in protective efficacy when administered to individuals who have received prior vaccination against smallpox.78,81, 82, 83 These have been met with successful boosting of the immune response, and in at least one case providing increased viral control in the previously vaccinated population.84 There is also an additional advantage in providing a vaccine that provides immunity against both SARS-CoV-2 and smallpox. Although smallpox has been eradicated on the world stage, there are still potential concerns that it could be used for bioterrorism.85
A key factor for a vaccine to provide long-lasting immunity against SARS-CoV-2 and protection against novel SARS-CoV-2 variants will be the generation of T cell responses against highly conserved immunodominant antigens. Despite being a key target for neutralizing antibodies, the RBD, which comprises the main component of the CovAg immunogen used in this study, is not considered an immunodominant antigen.21,86,87 However, our vaccine stimulates robust T cell immunity against the RBD, which is likely attributed to using a replicating viral vector for delivery. Replicating viral vectors such as TT can often stimulate more effective cellular immunity relative to nonreplicating vectors because they achieve higher and longer expression of the target immunogen and act as stronger adjuvants for the immune system.22,86, 87, 88 TOH-VAC1 is therefore a promising backbone for the next generation of vaccination strategies implementing immunodominant T cell antigens.
The large coding capacity of VACV could permit encoding of multiple SARS-CoV-2 antigens, which could serve to both increase T cell immunity against the virus and reduce the chance of novel variants arising that are resistant to the vaccine. The S protein, which has been the target of available vaccines,4,5,73 is prone to mutations that have resulted in increased infectivity and/or vaccine resistance.64 Currently, the four SARS-CoV-2 variants of concern and four variants of interest that have been classified by the World Health Organization all contain one or more mutations within the S protein. In light of this, COVID-19 vaccine development has begun to incorporate other targets such as the nucleocapsid (N), membrane (M), and envelope (E) proteins.89 The N protein is a particularly promising target since it is highly conserved in sequence and structure among coronaviruses, has lower rates of mutations compared with S, and is highly immunogenic,89 driving T cell responses. In addition, a multi-antigen SARS-CoV-2 vaccine may be able to replace S-based vaccines and alleviate potential concerns about vaccine-induced thrombotic thrombocytopenia.90, 91, 92
As the emergence of new SARS-CoV-2 variants continues to increase, it is necessary to assess the risk of vaccine resistance.64 Here, we developed a novel biosensor assay using a split luciferase system, that measures antibody interactions with RBD mutants. The assay can be rapidly performed on samples from vaccinated or infected individuals to reveal whether their existing antibodies are capable of binding mutant RBD and conferring some protection against emerging SARS-CoV-2 variants. Using this assay, we demonstrated that the antibodies generated from our TOH-VAC1 vaccine recognize four RBD variants.
Here, we report the development of a replicating VACV-based vaccine, TOH-VAC1, that expresses the RBD of SARS-CoV-2. An RBD-based MVA vaccine has been generated previously by Liu et al., but antibody responses for it were much weaker than its full-length S counterparts.19 This was likely due to the removal of key T cell epitopes elsewhere in S that were capable of aiding humoral immunity. However, the study also found that the RBD vaccine was more effective than full-length S constructs in generating neutralizing antibodies when used as a booster.5 In our case, we may have been able to overcome the weaker humoral and T cell responses from an RBD antigen with the use of TT, a replicating poxvirus vector. This does open possibilities for heterologous prime-boost strategies with other viral and subunit vaccines incorporating RBD or full-length S as an antigen. In many places, there are currently long waits between doses for approved vaccines due to shortages and issues with manufacturing. TOH-VAC1 provides a potential solution as a single, low dose of replicating viral vaccine that is easy to manufacture and store. Clinical testing will be important to determine the efficacy of this vaccine in humans, but TOH-VAC1 promises to be a valuable tool in the fight against COVID-19.
Materials and methods
Cell lines and viruses
All cell lines were purchased from the American Type Culture Collections (Manassas, VA). VSV-S/GFP was a kind gift from Dr. Sean Whelan (Washington University School of Medicine).63 The Vaccinia TT strain was a gift from Dr. David Evans.93 MVA (VR-1508) was purchased from the ATCC.
Construct design
The TOH-VAC1 construct consists of the RBD of the SARS-CoV-2 spike protein (amino acid residues 331 to 524) fused to its transmembrane (TM) domain (residues 1,208 to 1,270) via a 3x GGGGS linker (the accession number used for spike sequences is MW070087.1). Upstream of the RBD coding sequence is a murine interleukin-12 signal peptide followed by a GGSGGG linker. At the C-terminal end of the TM region is an HA-Tag for convenient detection. The CovAg construct is expressed by the Vaccinia early/late H5R promoter (GenBank accession number: LR877630.1). For recombinant virus selection and tracking of viral growth, genes encoding firefly luciferase and enhanced GFP (eGFP) were incorporated under a separate early/late promoter (composed of O2L and A12L promoters) separated by a P2A sequence. The entire construct noted above was flanked by homology arms for B13R and B14R loci in Vaccinia. The DNA sequence for the described construct is found in Table S1.
Mouse experiments
Six- to 8-week-old female BALB/c or nude mice (The Jackson Laboratory, Bar Harbor, ME) were obtained for studies. All experiments were approved by the University of Ottawa animal care and veterinary services (MEe-2258-R5, or OHRIe-3340-A), including periodic saph bleeds for serum collection.
ELISA
Nunc Maxisorp 96-well flat-bottom plates were coated overnight at 4°C with 125 ng of RBD per well (prepared in-house [see supplemental materials and methods]). The following day, the RBD solution was removed, the plates were washed three times with PBS-Tween (0.1% Tween 20) and blocked with 3% skim milk solution for 1 h. Mouse sera from vaccinated mice were then serially diluted in 1% skim milk and added to the plates to incubate for 2 h at room temperature. In addition, a positive and negative control was added to each plate composed of a monoclonal RBD antibody (1 μg/mL; Cat No: MBS434247, Anti-RBD Domain [SARS-CoV-2 spike], monoclonal antibody, MyBioSource, CA, USA) and a pool of sera taken from mice prior to vaccination. Following the 2-h incubation, plates were washed with PBS-Tween and incubated with anti-mouse immunoglobulin (Ig)G conjugated to horseradish peroxidase (HRP) (1:3000; Cat No: 314930, Goat anti-Mouse IgG [H + L] Secondary Antibody, HRP, Invitrogen) for 1 h at room temperature. Plates were then developed using SigmaFast OPD solution and measured at 490 nm using a BioTek microplate reader.
All experimental absorbance readings were normalized relative to the blank and the positive control (monoclonal RBD antibody at 1 μg/mL) and fit using a quadratic binding polynomial assuming 1:1 binding. The fitting was performed using a Monte Carlo simulation with the nonlinear curve-fitting tool in QtGrace. The reciprocal antibody titer (LDF) was determined by interpolating the dilution factor that intersected with a minimum detection threshold defined by 10x the standard deviation of the responses from mice vaccinated with TT WT, or to a fixed value of 0.025 (whichever was larger).
ELISPOTs
BALB/c mice were inoculated either intranasally or intraperitoneally with 1 × 107 pfu MVA or 1 × 106 pfu TT vaccines. Seven days after injection, mice were euthanized, and spleens were harvested for IFN-γ ELISPOT assays. Splenocytes were isolated and incubated at a density of 2 × 105 cells/well on murine IFN-γ Single-Color ELISPOT plates (ImmunoSpot) with either 1 μM of PepTivator SARS-CoV-2 Prot S1 pool (Miltenyi Biotec) or 10 μM of the CYGVSPTKL peptide (CanPeptide). The CYG peptide was identified via a primary screen of previously discovered SARS-CoV-1 T cell epitopes as a potent SARS-CoV-2 T cell epitope for BALB/c mice (summary of peptides is found in Table S2). In addition, DMSO was utilized as a negative control, while peptides from VacV F2/E362 (SPGAAGYDL/VGPSNSPTF; Genscript) were used as positive controls for VACV, respectively. Splenocytes were stimulated with peptides for 20 h and then the ELISPOT was performed according to the manufacturer's protocol. Plates were imaged and spots were counted using an ImmunoSpot Analyzer.
Pseudovirus neutralization assay
Vero E6 cells were seeded in 96-well plates such that there were 40,000 cells per well at the time of infection. Serum was first diluted in a separate 96-well at a 1:10 dilution in serum-free DMEM and a serial 1 in 2 dilution series was performed. VSV pseudotyped with the SARS-CoV-2 spike glycoprotein63 and co-encoded with eGFP was then added to the serum in an equal volume of serum-free DMEM for a final dilution of 2000 pfu per well and incubated for 1 h at 37°C. After 1 h, media on the cell was replaced with 60 μL of the virus/serum and incubated for 1 h at 37°C. Wells were then topped up with carboxymethylcellulose (CMC) in DMEM (supplemented with 10% fetal bovine serum) for a final concentration of 3% CMC and incubated 24 h at 34°C. GFP foci were imaged and counted using a Cellomics ArrayScan VTI HCS Reader.
Cynomolgus macaque study
Our study with 11 non-naïve cynomolgus macaques was performed at the Institute national de la recherche scientifique (Laval, Québec). The macaques were previously used in an unrelated study involving inoculation with oncolytic Maraba virus but had no prior exposure to any SARS-CoV-2 antigens. Pre-study data (body weight, detailed clinical exam, complete blood count, and liver function) were used for randomization. Each single-dose immunization (i.m.) with TOH-VAC1 at 1 × 107 pfu was followed by observation for 4 h postadministration. Weekly body weight measurements and detailed clinical observations were recorded on bleed-day or the day before administration. Daily cage-side observations were performed (e.g., animal behavior, food consumption, estrous cycle bleeding, feces appearance). Images of the injection sites were acquired after vaccination and at the end of the study to monitor for potential pox lesions. Complete blood count and liver function analyses were conducted (subcontracted by the Centre National de Biologie Experimentale), as well as ELISA (serum) and Nabs (serum), and blood draw every 7 days. Blood draw (9–10 mL) for ELISpot day 1 (d1), pre-boost, post boost, and end of study (ELISA, Nabs, and serum analysis performed by Dr. Lapointe's laboratory). Urine samples and saliva throat-swab (in PBS) samples were collected and stored at −80°C on day −1 pre-vaccination (baseline), then 24 h, 48 h, and 1 week post vaccination to detect potential virus shedding.
Viral shedding luciferase assay
U-2 OS cells were seeded at 1 × 104 cells/well in a 96-well white-bottom plate. The following day, 100 μL of urine or saliva samples or media alone was added to each well with 100 μL of media supplemented with antibiotics to prevent contamination. After 72 h, additional cells were infected with TOH-VAC1, which expresses firefly luciferase, at indicated pfu/mL (positive controls). At 6 hpi, Renilla substrate was added to each well and readout was performed by a BioTek plate reader.
Screening PCR for viral shedding of TOH-Vac1
To determine if macaques vaccinated with TOH-Vac1 shed viral genomes, a PCR amplifying short sequences of two different genomic loci was performed on throat swaps taken on d1 and d8 post vaccination. The genomic loci amplified are B8R and D10R. Successful detection of parts of the viral genome results in the amplification of a 556 base pairs (bp) for B8R and 548 bp for D10R, respectively (Tables S3–S5). Throat swabs were boiled at 98°C for 3 min to inactivate virus and release viral genomes; 2 μL of each sample were used per PCR reaction. As a positive control, U2-OS cells were infected at an MOI of 0.1 with Vaccinia TT WT. The cell culture supernatant was collected 48 hpi, diluted 1/10 with sterile PBS, and boiled at 98°C for 3 min; 2 μL of the positive control were used per PCR reaction. The same amount of water was used as a negative control. After PCR, the complete reaction volume was analyzed on a 1.2% (w/v) agarose gel, stained with RedSafe Nucleic Acid Staining Solution (200,00x); 0.5 μg Thermo Scientific GeneRuler 1 kb Plus DNA Ladder was used as reference.
Seroconversion biosensor assay
Recently we have developed several biosensors for detection of SARS-CoV-2 seroconversion.12,51 Here, we updated our biosensor by fusing nanoluciferase to the C terminus of the RBD via a flexible linker. An IgK signal peptide was inserted at the beginning of the RBD sequence to secrete the fusion protein from cells. All RBD mutations were generated by site-directed mutagenesis. The construct was cloned into pcDNA3.1 and transfected into HEK293T cells. After 2 days, the supernatant was collected and filtered to use for the seroconversion assay. Luciferase activity was measured from the supernatants to adjust the relative concentrations of RBD used for the seroconversion assay. For the seroconversion assay against different variants of SARS-CoV-2, 20 μL of magnetic beads conjugated with protein G were combined with 50 μL of nanoluciferase-conjugated RBD and 5 μL of serum from vaccinated macaques. After 15-min incubation on a shaker at 25°C, the beads were collected using a magnet and washed with PBS three times to remove excess nanoluciferase. The next step was followed by adding 50 μL of diluted nanoluciferase substrate to it. A Synergy microplate reader (BioTek, Winooski, VT, USA) was used to measure luminescence.
Statistical analyses
All graphs and statistical analyses were performed using GraphPad Prism v9. Means of more than two groups were compared by 1-way ANOVA using Tukey correction for multiple comparisons. Two-way ANOVA with Sidak correction for multiple comparisons was used to compare the means of more than two groups split on two independent variables. Dunnet's correction for multiple comparisons was used when comparing the mean of more than two groups against a single baseline or control group. Alpha levels for all statistical tests were set at a threshold of 0.05. Normal distribution of datasets was assessed using D'Agostino & Pearson omnibus K2 normality test and Shapiro-Wilk normality test. All biological replicates are indicated by n value in figure legends. Error bars represent standard deviation (SD) or standard error of the mean (SEM) as indicated in figure legends. For all statistical tests, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns = not significant.
Additional methods can be found in supplemental information section.
Acknowledgments
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was possible by the generous support from the Ottawa Hospital Foundation, the Thistledown Foundation, the Terry Fox Research Institute, and the Canadian Cancer Society. This work was also funded by a FastGrant for COVID-19 Science to C.S.I., J.C.B., and D.J.M. and a grant from the Canadian Institutes of Health Research (#448323) to R.A, J.A., D.W.C., R.L., D.J.M., J.C.B., and C.S.I. T.A. is funded by a CIHR Banting Fellowship. R.S. is funded by a CIHR postdoctoral fellowship. M.J.F.C. is funded by the Taggart-Parkes Fellowship. T.R.J. is funded by a CIHR Frederick Banting and Charles Best Canada and Ontario Graduate Scholarship. Z.T. is funded by an NSERC CGS-D3 and an Ontario Graduate Scholarship. A.S. is supported by NSERC Graduate Scholarships. R.S., M.J.F.C., N.M., J.P., T.R.J., Z.T., J.W., R.R., A.S., S.T., E.B., and J.D. received funding support from MITACS CanPRIME Accelerate fellowships. D.W.C. and J.B.A. are supported in part by a salary award from the Faculty and Department of Medicine, University of Ottawa. The graphical abstract was generated using BioRender.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.10.008.
Contributor Information
John C. Bell, Email: jbell@ohri.ca.
Carolina S. Ilkow, Email: cilkow@uottawa.ca.
Supplemental information
References
- 1.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/
- 3.Organización Mundial de la Salud (2021). Status of COVID-19 Vaccines within WHO EUL/PQ evaluation process (20 January 2021). 2. https://extranet.who.int/pqweb/sites/default/files/documents/Status_COVID_VAX_20Oct2021.pdf
- 4.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., et al. Interim results of a phase 1–2a trial of Ad26.COV2.S covid-19 vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A.J., Cornaby C., Bartelt L., Weiss S., Park Y., et al. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown E.E.F., Rezaei R., Jamieson T.R., Dave J., Martin N.T., Singaravelu R., Crupi M.J.F., Boulton S., Tucker S., Duong J., et al. Characterization of critical determinants of ACE2–SARS CoV-2 RBD interaction. Int. J. Mol. Sci. 2021;22:2268. doi: 10.3390/ijms22052268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimball K.J., Numnum T.M., Kirby T.O., Zamboni W.C., Estes J.M., Barnes M.N., Matei D.E., Koch K.M., Alvarez R.D. A phase I study of lapatinib in combination with carboplatin in women with platinum sensitive recurrent ovarian carcinoma. Gynecol. Oncol. 2008;111:95–101. doi: 10.1016/j.ygyno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W., Derclaye S., Vincent S.P., Soumillion P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C., Ryu D.-K., Lee J., Kim Y.-I., Seo J.-M., Kim Y.-G., Jeong J.-H., Kim M., Kim J.-I., Kim P., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:1–10. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min L., Sun Q. Antibodies and vaccines target RBD of SARS-CoV-2. Front. Mol. Biosci. 2021;8:671633. doi: 10.3389/fmolb.2021.671633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Arriaza J., Garaigorta U., Pérez P., Lázaro-Frías A., Zamora C., Gastaminza P., del Fresno C., Casasnovas J.M., Sorzano C.Ó.S., Sancho D., et al. COVID-19 vaccine candidates based on modified vaccinia virus Ankara expressing the SARS-CoV-2 spike protein induce robust T- and B-cell immune responses and full efficacy in mice. J. Virol. 2021;95:2260–2280. doi: 10.1128/JVI.02260-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R., Americo J.L., Cotter C.A., Earl P.L., Erez N., Peng C., Moss B. One or two injections of MVA-vectored vaccine shields hACE2 transgenic mice from SARS-CoV-2 upper and lower respiratory tract infection. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2026785118. e2026785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., Kopp M., Li D., Parks R., Hsu A.L., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 22.Shomuradova A.S., Vagida M.S., Sheetikov S.A., Zornikova K.V., Kiryukhin D., Titov A., Peshkova I.O., Khmelevskaya A., Dianov D.V., Malasheva M., et al. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53:1245–1257.e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Draftand landscape. https://extranet.who.int/pqweb/sites/default/files/documents/Status_COVID_VAX_20Oct2021.pdf
- 24.Harbour J.C., Lyski Z.L., Schell J.B., Thomas A., Messer W.B., Slifka M.K., Nolz J.C. Cellular and humoral immune responses in mice immunized with vaccinia virus expressing the SARS-CoV-2 spike protein. J. Immunol. 2021;206:2596–2604. doi: 10.4049/jimmunol.2100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutter G., Moss B. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. U S A. 1992;89:10847–10851. doi: 10.1073/pnas.89.22.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutter G., Wyatt L.S., Foley P.L., Bennink J.R., Moss B. A recombinant vector derived from the host range-restricted and highly attenuated MVA strain of vaccinia virus stimulates protective immunity in mice to influenza virus. Vaccine. 1994;12:1032–1040. doi: 10.1016/0264-410x(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 27.Lu B., Yu W., Huang X., Wang H., Liu L., Chen Z. Mucosal immunization induces a higher level of lasting neutralizing antibody response in mice by a replication-competent smallpox vaccine: vaccinia Tiantan strain. J. Biomed. Biotechnol. 2011;2011:970424. doi: 10.1155/2011/970424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer H., Sutter G., Mayr A. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 1991;72:1031–1038. doi: 10.1099/0022-1317-72-5-1031. [DOI] [PubMed] [Google Scholar]
- 29.Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U S A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F., Wei Q., He T., Yu W., Yu J., et al. Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 2005;79:2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song F., Fux R., Provacia L.B., Volz A., Eickmann M., Becker S., Osterhaus A.D.M.E., Haagmans B.L., Sutter G. Middle East respiratory syndrome coronavirus spike protein delivered by modified vaccinia virus Ankara efficiently induces virus-neutralizing antibodies. J. Virol. 2013;87:11950–11954. doi: 10.1128/JVI.01672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ba L., Yi C.E., Zhang L., Ho D.D., Chen Z. Heterologous MVA-S prime Ad5-S boost regimen induces high and persistent levels of neutralizing antibody response against SARS coronavirus. Appl. Microbiol. Biotechnol. 2007;76:1131–1136. doi: 10.1007/s00253-007-1073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Routhu N.K., Cheedarla N., Gangadhara S., Bollimpelli V.S., Boddapati A.K., Shiferaw A., Rahman S.A., Sahoo A., Edara V.V., Lai L., et al. A modified vaccinia Ankara vector-based vaccine protects macaques from SARS-CoV-2 infection, immune pathology, and dysfunction in the lungs. Immunity. 2021;54:542–556.e9. doi: 10.1016/j.immuni.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiuppesi F., d’Alincourt Salazar M., Contreras H., Nguyen V.H., Martinez J., Park Y., Nguyen J., Kha M., Iniguez A., Zhou Q., et al. Development of a multi-antigenic SARS-CoV-2 vaccine candidate using a synthetic poxvirus platform. Nat. Commun. 2020;11:6121. doi: 10.1038/s41467-020-19819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandrasekar S.S., Phanse Y., Hildebrand R.E., Hanafy M., Wu C.W., Hansen C.H., Osorio J.E., Suresh M., Talaat A.M. Localized and systemic immune responses against sars-cov-2 following mucosal immunization. Vaccines. 2021;9:1–17. doi: 10.3390/vaccines9020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J.H., Rivière I., Gonen M., Wang X., Sénéchal B., Curran K.J., Sauter C., Wang Y., Santomasso B., Mead E., et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breitbach C.J., Arulanandam R., De Silva N., Thorne S.H., Patt R., Daneshmand M., Moon A., Ilkow C., Burke J., Hwang T.H., et al. Oncolytic vaccinia virus disrupts tumor-associated vasculature in humans. Cancer Res. 2013;73:1265–1275. doi: 10.1158/0008-5472.CAN-12-2687. [DOI] [PubMed] [Google Scholar]
- 38.Pelin A., Foloppe J., Petryk J., Singaravelu R., Hussein M., Gossart F., Jennings V.A., Stubbert L.J., Foster M., Storbeck C., et al. Deletion of apoptosis inhibitor F1L in vaccinia virus increases safety and oncolysis for cancer therapy. Mol. Ther. Oncolytics. 2019;14:246–252. doi: 10.1016/j.omto.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson R., Edwards R., Mansouri K., Janowska K., Stalls V., Kopp M., Haynes B., Acharya P. Glycans on the SARS-CoV-2 spike control the receptor binding domain conformation. bioRxiv. 2020 2020.06.26.173765. [Google Scholar]
- 40.Bouwman K.M., Tomris I., Turner H.L., van der Woude R., Shamorkina T.M., Bosman G.P., Rockx B., Herfst S., Snijder J., Haagmans B.L., et al. Multimerization- and glycosylation-dependent receptor binding of SARS-CoV-2 spike proteins. PLoS Pathog. 2021;17:e1009282. doi: 10.1371/journal.ppat.1009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samreen B., Tao S., Tischer K., Adler H., Drexler I. ORF6 and ORF61 expressing MVA vaccines impair early but not late latency in murine gammaherpesvirus MHV-68 infection. Front. Immunol. 2019;10:2984. doi: 10.3389/fimmu.2019.02984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breitbach C., Bell J.C., Hwang T.-H., Kirn D., Burke J. The emerging therapeutic potential of the oncolytic immunotherapeutic Pexa-Vec (JX-594) Oncolytic Virother. 2015;4:25. doi: 10.2147/OV.S59640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi C., Sun X., Ye J., Ding L., Liu M., Yang Z., Lu X., Zhang Y., Ma L., Gu W., et al. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020;17:621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X., Wang Y.L., Wu J., Qi J., Zeng Z., Wan Q., Chen Z., Manandhar P., Cavener V.S., Boyle N.R., et al. Neutralizing aptamers block S/RBD-ACE2 interactions and prevent host cell infection. Angew. Chem. Int. Ed. 2021;60:10273–10278. doi: 10.1002/anie.202100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiaojie S., Yu L., Lei Y., Guang Y., Min Q. Neutralizing antibodies targeting SARS-CoV-2 spike protein. Stem Cell Res. 2021;50:102125. doi: 10.1016/j.scr.2020.102125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah M., Ahmad B., Choi S., Goo Woo H. Mutations in the SARS-CoV-2 spike RBD are responsible for stronger ACE2 binding and poor anti-SARS-CoV mAbs cross-neutralization. Comput. Struct. Biotechnol. J. 2020;18:3402–3414. doi: 10.1016/j.csbj.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang S., He Y., Lu H., Siddiqui P., Zhou Y. Induce highly potent neutralizing antibodies conformation-dependent epitopes that protein contains multiple respiratory syndrome coronavirus spike receptor-binding domain of severe acute. J. Immunol. 2021;174:4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 48.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 50.Antoine G., Scheiflinger F., Dorner F., Falkner F.G. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 51.Azad T., Singaravelu R., Taha Z., Jamieson T.R., Boulton S., Crupi M.J.F., Martin N.T., Brown E.E.F., Poutou J., Ghahremani M., et al. Nanoluciferase complementation-based bioreporter reveals the importance of N-linked glycosylation of SARS-CoV-2 S for viral entry. Mol. Ther. 2021;29:1984–2000. doi: 10.1016/j.ymthe.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Consortium, A.A Structural and functional comparison of SARS-CoV-2-spike receptor binding domain produced in Pichia pastoris and mammalian cells. Sci. Rep. 2020;10:21779. doi: 10.1038/s41598-020-78711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y., Liao Q., Chen T., Zhang Y., Yuan W., Xu J., Zhang X. Freeze-drying formulations increased the adenovirus and poxvirus vaccine storage times and antigen stabilities. Virol. Sin. 2020;36:365–372. doi: 10.1007/s12250-020-00250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newman F.K., Frey S.E., Blevins T.P., Yan L., Belshe R.B. Stability of undiluted and diluted vaccinia-virus vaccine. Dryvax. J. Infect. Dis. 2003;187:1319–1322. doi: 10.1086/374564. [DOI] [PubMed] [Google Scholar]
- 56.Essbauer S., Meyer H., Porsch-Özcürümez M., Pfeffer M. Long-lasting stability of vaccinia virus (orthopoxvirus) in food and environmental samples. Zoonoses Public Health. 2007;54:118–124. doi: 10.1111/j.1863-2378.2007.01035.x. [DOI] [PubMed] [Google Scholar]
- 57.Dehaven, B.C., Gupta, K., and Isaacs, S.N.. The vaccinia virus A56 protein: a multifunctional transmembrane glycoprotein that anchors two secreted viral proteins. J. Gen. Virol.;92(Pt 9):1971-1980. [DOI] [PMC free article] [PubMed]
- 58.Lee M.S., Roos J.M., McGuigan L.C., Smith K.A., Cormier N., Cohen L.K., Roberts B.E., Payne L.G. Molecular attenuation of vaccinia virus: mutant generation and animal characterization. J. Virol. 1992;66:2617–2630. doi: 10.1128/jvi.66.5.2617-2630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson J.E., Nasar F., Coleman J.W., Price R.E., Javadian A., Draper K., Lee M., Reilly P.A., Clarke D.K., Hendry R.M., et al. Neurovirulence properties of recombinant vesicular stomatitis virus vectors in non-human primates. Virology. 2007;360:36–49. doi: 10.1016/j.virol.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wollmann G., Paglino J.C., Maloney P.R., Ahmadi S.A., van den Pol A.N. Attenuation of vesicular stomatitis virus infection of brain using antiviral drugs and an adeno-associated virus-interferon vector. Virology. 2015;475:1–14. doi: 10.1016/j.virol.2014.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Pol A.N., Dalton K.P., Rose J.K. Relative neurotropism of a recombinant rhabdovirus expressing a green fluorescent envelope glycoprotein. J. Virol. 2002;76:1309–1327. doi: 10.1128/JVI.76.3.1309-1327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flesch I.E.A., Woo W.-P., Wang Y., Panchanathan V., Wong Y.-C., La Gruta N.L., Cukalac T., Tscharke D.C. Altered CD8 + T cell immunodominance after vaccinia virus infection and the naive repertoire in inbred and F 1 mice. J. Immunol. 2010;184:45–55. doi: 10.4049/jimmunol.0900999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta R.K. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat. Rev. Immunol. 2021;21:340–341. doi: 10.1038/s41577-021-00556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma network for genomic surveillance in South Africa∗, COMMIT-KZN team. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng Y., Chuai X., Chen P., Chen H., Wang W., Ruan L., Li W., Tan W. Recombinant vaccinia vector-based vaccine (Tiantan) boosting a novel HBV subunit vaccine induced more robust and lasting immunity in rhesus macaques. Vaccine. 2017;35:3347–3353. doi: 10.1016/j.vaccine.2017.04.059. [DOI] [PubMed] [Google Scholar]
- 68.Liu Q., Li Y., Luo Z., Yang G., Liu Y., Liu Y., Sun M., Dai J., Li Q., Qin C., et al. HIV-1 vaccines based on replication-competent Tiantan vaccinia protected Chinese rhesus macaques from simian HIV infection. AIDS. 2015;29:649–658. doi: 10.1097/QAD.0000000000000595. [DOI] [PubMed] [Google Scholar]
- 69.Joachim A., Msafiri F., Onkar S., Munseri P., Aboud S., Lyamuya E.F., Bakari M., Billings E., Robb M.L., Wahren B., et al. Frequent and durable anti-hiv envelope viv2 igg responses induced by hiv-1 dna priming and hiv-mva boosting in healthy tanzanian volunteers. Vaccines. 2020;8:1–13. doi: 10.3390/vaccines8040681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Volkmann A., Williamson A.L., Weidenthaler H., Meyer T.P.H., Robertson J.S., Excler J.L., Condit R.C., Evans E., Smith E.R., Kim D., et al. The Brighton Collaboration standardized template for collection of key information for risk/benefit assessment of a Modified Vaccinia Ankara (MVA) vaccine platform. Vaccine. 2020;39:3067–3080. doi: 10.1016/j.vaccine.2020.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morelli M.P., Del Medico Zajac M.P., Pellegrini J.M., Amiano N.O., Tateosian N.L., Calamante G., Gherardi M.M., García V.E. IL-12 DNA displays efficient adjuvant effects improving immunogenicity of Ag85A in DNA prime/MVA boost immunizations. Front. Cell. Infect. Microbiol. 2020;10:581812. doi: 10.3389/fcimb.2020.581812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koch T., Dahlke C., Fathi A., Kupke A., Krähling V., Okba N.M.A., Halwe S., Rohde C., Eickmann M., Volz A., et al. Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: an open-label, phase 1 trial. Lancet Infect. Dis. 2020;20:827–838. doi: 10.1016/S1473-3099(20)30248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Logunov D.Y., Dolzhikova I.V., Shcheblyakov D.V., Tukhvatulin A.I., Zubkova O.V., Dzharullaeva A.S., Kovyrshina A.V., Lubenets N.L., Grousova D.M., Erokhova A.S., et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pittman P.R., Hahn M., Lee H.S., Koca C., Samy N., Schmidt D., Hornung J., Weidenthaler H., Heery C.R., Meyer T.P.H., et al. Phase 3 efficacy trial of modified vaccinia Ankara as a vaccine against smallpox. N. Engl. J. Med. 2019;381:1897–1908. doi: 10.1056/NEJMoa1817307. [DOI] [PubMed] [Google Scholar]
- 75.Hu W.G., Steigerwald R., Kalla M., Volkmann A., Noll D., Nagata L.P. Protective efficacy of monovalent and trivalent recombinant MVA-based vaccines against three encephalitic alphaviruses. Vaccine. 2018;36:5194–5203. doi: 10.1016/j.vaccine.2018.06.064. [DOI] [PubMed] [Google Scholar]
- 76.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mejías-Pérez E., Carreño-Fuentes L., Esteban M. Development of a safe and effective vaccinia virus oncolytic vector WR-Δ4 with a set of gene deletions on several viral pathways. Mol. Ther. Oncolytics. 2018;8:27–40. doi: 10.1016/j.omto.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slike B.M., Creegan M., Marovich M., Ngauy V. Humoral immunity to primary smallpox vaccination: impact of childhood versus adult immunization on vaccinia vector vaccine development in military populations. PLoS One. 2017;12:e0169247. doi: 10.1371/journal.pone.0169247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belyakov I.M., Moss B., Strober W., Berzofsky J.A. Mucosal vaccination overcomes the barrier to recombinant vaccinia immunization caused by preexisting poxvirus immunity. Proc. Natl. Acad. Sci. U S A. 1999;96:4512. doi: 10.1073/pnas.96.8.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang X., Lu B., Yu W., Fang Q., Liu L., Zhuang K., Shen T., Wang H., Tian P., Zhang L., et al. A novel replication-competent vaccinia vector MVTT is superior to MVA for inducing high levels of neutralizing antibody via mucosal vaccination. PLoS One. 2009;4:e4180. doi: 10.1371/journal.pone.0004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Altenburg A.F., van Trierum S.E., de Bruin E., de Meulder D., van de Sandt C.E., van der Klis F.R.M., Fouchier R.A.M., Koopmans M.P.G., Rimmelzwaan G.F., de Vries R.D. Effects of pre-existing orthopoxvirus-specific immunity on the performance of Modified Vaccinia virus Ankara-based influenza vaccines. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-24820-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramírez J.C., Gherardi M.M., Rodríguez D., Esteban M. Attenuated modified vaccinia virus Ankara can Be used as an immunizing agent under conditions of preexisting immunity to the vector. J. Virol. 2000;74:7651–7655. doi: 10.1128/jvi.74.16.7651-7655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goepfert P.A., Elizaga M.L., Sato A., Qin L., Cardinali M., Hay C.M., Hural J., DeRosa S.C., DeFawe O.D., Tomaras G.D., et al. Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J. Infect. Dis. 2011;203:610–619. doi: 10.1093/infdis/jiq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kannanganat S., Nigam P., Velu V., Earl P.L., Lai L., Chennareddi L., Lawson B., Wilson R.L., Montefiori D.C., Kozlowski P.A., et al. Preexisting vaccinia virus immunity decreases SIV-specific cellular immunity but does not diminish humoral immunity and efficacy of a DNA/MVA vaccine. J. Immunol. 2010;185:7262–7273. doi: 10.4049/jimmunol.1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lane J.M., Summer L. In: Bioterrorism Infect. Agents A New Dilemma 21st Century. Fong I.W., Alibek K., editors. Springer; 2009. Smallpox as a weapon for bioterrorism; p. 147. [Google Scholar]
- 86.Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Amaya Hernandez S.P., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 2021;6:eabf7550. doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robert-Guroff M. Replicating and non-replicating viral vectors for vaccine development. Curr. Opin. Biotechnol. 2007;18:546–556. doi: 10.1016/j.copbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J. Virol. 2020;94:e00647-20. doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang S., Liu Y., Wang X., Yang L., Li H., Wang Y., Liu M., Zhao X., Xie Y., Yang Y., et al. SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J. Hematol. Oncol. 2020;13:1–22. doi: 10.1186/s13045-020-00954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grobbelaar L.M., Venter C., Vlok M., Ngoepe M., Laubscher G.J., Lourens P.J., Steenkamp J., Kell D.B., Pretorius E. SARS-CoV-2 spike protein S1 induces fibrin(ogen) resistant to fibrinolysis: implications for microclot formation in COVID-19. Biosci. Rep. 2021;41 doi: 10.1042/BSR20210611. BSR20210611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cines D.B., Bussel J.B. SARS-CoV-2 vaccine–induced immune thrombotic thrombocytopenia. N. Engl. J. Med. 2021;384:2254–2256. doi: 10.1056/NEJMe2106315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qin L., Evans D.H. Genome scale patterns of recombination between coinfecting vaccinia viruses. J. Virol. 2014;88:5277–5286. doi: 10.1128/JVI.00022-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.