Abstract
The rapid loss of reef-building corals owing to ocean warming is driving the development of interventions such as coral propagation and restoration, selective breeding and assisted gene flow. Many of these interventions target naturally heat-tolerant individuals to boost climate resilience, but the challenges of quickly and reliably quantifying heat tolerance and identifying thermotolerant individuals have hampered implementation. Here, we used coral bleaching automated stress systems to perform rapid, standardized heat tolerance assays on 229 colonies of Acropora cervicornis across six coral nurseries spanning Florida's Coral Reef, USA. Analysis of heat stress dose–response curves for each colony revealed a broad range in thermal tolerance among individuals (approx. 2.5°C range in Fv/Fm ED50), with highly reproducible rankings across independent tests (r = 0.76). Most phenotypic variation occurred within nurseries rather than between them, pointing to a potentially dominant role of fixed genetic effects in setting thermal tolerance and widespread distribution of tolerant individuals throughout the population. The identification of tolerant individuals provides immediately actionable information to optimize nursery and restoration programmes for Florida's threatened staghorn corals. This work further provides a blueprint for future efforts to identify and source thermally tolerant corals for conservation interventions worldwide.
Keywords: Acropora cervicornis, coral reefs, climate change, coral restoration, coral bleaching automated stress system, thermal stress assay
1. Introduction
As warming oceans continue to trigger mass bleaching and mortality of reef-building corals around the world, the fate of reef ecosystems is now tightly linked to the heat tolerance of surviving corals. While immediate reduction of emissions and mitigation of warming is of primary importance to ensure coral reef persistence, biological interventions using naturally heat-tolerant corals may also be necessary to maximize resilience in challenging future environments [1–3].
Interventions that could use heat-tolerant corals include: (i) coral propagation and restoration, where corals are grown in nurseries and outplanted to reefs [4,5]; (ii) selective breeding, where individuals are crossbred to produce new generations of sexual recruits for restoration [6,7]; and (iii) assisted gene flow, in which corals or their gametes are imported to a new population [8–10]. Indeed, the strategic use of heat-tolerant corals as broodstock and source material in these efforts could help boost the abundance of heat-tolerant genotypes in natural and restored coral populations, assuming that heat tolerance is heritable [11–13]. However, a prerequisite for practical implementation of these interventions is to first find and identify heat-tolerant corals, which is itself a major challenge that has traditionally relied on observation during natural bleaching events [14–16], or low-throughput experimental approaches requiring substantial time and resources [17,18]. The difficulty in identifying tolerant corals efficiently, reproducibly or at scale has not only limited their use in restoration programmes, but also hindered general understanding of heat tolerance as a quantitative, empirical trait.
Overcoming these challenges requires a better understanding of the natural variation in coral heat tolerance, the distribution of heat-tolerant genotypes and phenotypes and the degree to which heat tolerance is determined by a coral's genes, symbiotic interactions and environment. There is extensive variation in coral heat tolerance across latitudinal and environmental gradients [19–25], with populations from warmer locations having higher tolerance. However, such differences can also occur over reefal scales of just 10s to 100s of metres [26–28], indicating strong selection across habitats and microenvironments over small spatial scales, and/or an important role for acclimatization [29,30]. Corals' symbioses with Symbiodiniaceae can also play an important role, with thermotolerant symbiont species elevating bleaching thresholds by 1–2°C in corals that can associate with them [14,31,32]. Yet, sometimes even adjacent conspecifics hosting the same symbiont species show dramatic differences in bleaching [16,33], indicating that thermal tolerance varies at the individual level, determined by the genotype of the coral host, its symbionts or their interaction [34]. Through this complexity, there is clear evidence that the heat tolerance of individual corals varies significantly [13,35–39]; however, we still lack robust, quantitative descriptions of this phenotype and the factors underlying its variation.
Recent work has focused on resolving fine-scale, empirical differences in the thermal tolerance of individual coral genotypes [13,18,40], and the development of low-cost, fieldable methods for their large-scale determination. Specifically, sets of portable experimental tanks termed coral bleaching automated stress systems (CBASS; [28]), can facilitate the application of standardized, rapid thermal tolerance assays on corals in remote field settings. Importantly, these rapid, acute assays can recapitulate outcomes of longer-term, ecologically relevant bleaching scenarios [24,41], indicating they can be used to generate meaningful, quantitative and comparable metrics of thermal tolerance. Such efforts are now beginning to be applied to broadly census thermal tolerance in wild coral populations [42], from which the identification of tolerant individuals could inform targeted nursery development [13], and other active conservation interventions.
Here, we used CBASS to census thermal tolerance in highly threatened staghorn corals from coral nurseries in Florida, USA, that together represent the most extensive single-species coral restoration programme in the world [43]. The Caribbean staghorn coral Acropora cervicornis has been decimated regionally since the early 1980s and is a primary focal species in restoration efforts throughout the region, including Florida, where nurseries propagate and outplant tens of thousands of coral fragments to local reefs annually [5,44,45]. We used CBASS on board a research vessel to synoptically and quantitatively compare the thermal tolerances of 229 colonies of A. cervicornis across six coral nurseries from Broward County to the lower Florida Keys (nurseries separated by approx. 250 km, with colonies originating from collection sites spanning approx. 400 km from Broward to the Dry Tortugas). These corals represent an ideal population in which to quantify variation in thermal tolerance owing to coral genotype, because corals are common-gardened in each nursery environment, and associate with the same algal symbiont species (Symbiodinium ‘fitti’) across this gradient [46,47]. These results are critical for optimizing ongoing and future restoration efforts that use heat-tolerant corals, and understanding the evolutionary potential of this threatened species under climate change.
2. Material and methods
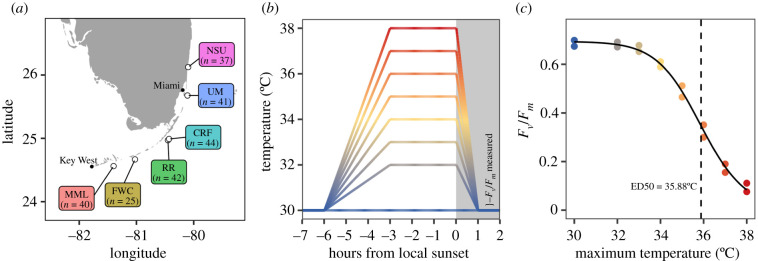
During two ship-based research expeditions in August and October 2020, we measured the thermal tolerance of 229 A. cervicornis colonies from six coral nurseries along Florida's Coral Reef (figure 1a; electronic supplementary material, table S1). The nurseries, from north to south, are operated by Nova Southeastern University (NSU), the University of Miami (UM), the Coral Restoration Foundation (CRF), Reef Renewal (RR), the Florida Fish and Wildlife Conservation Commission (FWC) and Mote Marine Laboratory (MML). Thermal tolerance of nursery corals was measured by CBASS [28], which are portable, field-deployable experimental tanks used to apply rapid, acute heat stress challenges (details below). Fragments of each coral colony were independently exposed to each of eight temperature stress profiles of increasing magnitude (with maximum temperatures between 30 and 38°C; figure 1b) for 7 h, after which maximum photochemical efficiency (Fv/Fm) was measured as an indicator of each fragment's stress response. These data were used to construct a dose–response curve for each colony, from which the effective dose of heat stress required to reduce Fv/Fm by 50% (ED50 value; [41]) was calculated as a metric of each colony's thermal tolerance (figure 1c; details below).
Figure 1.
Study design for census of coral heat tolerance across Florida nurseries. (a) Locations of coral nurseries visited by the R/V Coral Reef II in August and October 2020, and the number of coral individuals tested at each nursery (MML, Mote Marine Laboratory; FWC, Florida Fish and Wildlife Conservation Commission; RR, Reef Renewal; CRF, Coral Restoration Foundation; UM, University of Miami; NSU, Nova Southeastern University; see the electronic supplementary material, table S1 for details). (b) Temperature treatments for CBASS assays. Eight tanks were used to heat one fragment of each coral individual to eight different target temperatures. (c) A dose–response curve modelling Fv/Fm (measured two times per fragment) as a function of maximum treatment temperature was fitted for each coral individual to estimate ED50 (indicated by dashed line) as a metric of thermal tolerance. (Online version in colour.)
(a) . Coral bleaching automated stress systems set-up and assay procedure
CBASS were constructed following the general design of Voolstra et al. [28], using beverage coolers (Coleman 24 Can Party Stacker) partitioned into two independent halves (tanks) with an acrylic divider. Each 8 l tank was equipped with one titanium aquarium heater (Finnex TH-300 W) and two thermoelectric chillers (Nova Tec IceProbe). Seawater was circulated within each tank using a submersible powerhead (SUNSUN JVP 530 GPH), and fresh incoming seawater was supplied to each tank at a rate of approximately 1 ml s−1 (turnover = approx. 2.2 h). Light was provided by LED aquarium lights (Phlizon 165 W) mounted above each cooler and manually adjusted to provide 550 µmol photons m−2 s−1 at the centre of each tank, measured at the depth of the coral fragments with an underwater photosynthetically active radiation sensor (Apogee Instruments). Temperature profiles (figure 1b) were executed by custom controllers (ELEGOO Mega 2560) with temperature sensors (Vktech DS18b20) activating the heaters and/or chillers as needed to achieve prescribed set points. For the tanks heated to 34°C or below on the October field expedition, Inkbird (ITC-308) temperature controllers were used instead.
Temperature profiles consisted of a 30°C baseline temperature, a 3 h ramp up to a prescribed maximum temperature (ranging from 30°C to 38°C), a 3 h hold at the maximum temperature and a 1 h ramp down to 30°C (figure 1b). Profiles were timed such that the end of the maximum temperature hold period coincided with local sunset, and the temperature ramped back down in darkness. After returning to 30°C, Fv/Fm was measured for each coral fragment using a DIVING-PAM-II (Walz, Effeltrich, Germany) chlorophyll fluorometer. Fluorometer settings included: measuring light intensity = 1, measuring light frequency (ML-F) = 4, ML-F high = off, damping = 2, F0 mode = off, saturating pulse intensity = 8 and saturating pulse width = 0.8 s. The gain setting was adjusted as needed to produce an F0 measurement above 100. Two Fv/Fm measurements were taken for each coral fragment (if possible depending on size), from non-overlapping areas of tissue that were facing upward, perpendicular to incident light.
(b) . Coral bleaching automated stress systems deployment and field operations
CBASS were set-up on the deck of the R/V Coral Reef II during field expeditions to coral nurseries operated by NSU (August 2020), MML, FWC, RR, CRF and UM (October 2020; figure 1a; electronic supplementary material, table S1). At each coral nursery, divers collected eight fragments (or ramets; approx. 5 cm each) from available genets of A. cervicornis from the nursery's long-term collection (n = 25–44 genets per nursery; figure 1a). The total number of unique genet × nursery combinations (i.e. ‘colonies’) collected from was 229, comprising up to 172 genets (some genets were present at multiple nurseries from historical sharing of source material (electronic supplementary material, figure S1)). Collected fragments were held prior to experimentation in the ship's 14000 l livewell system, which was continuously filled with fresh oceanic seawater whenever possible, and assayed within 1–2 days of collection.
Approximately 1 h before temperature ramping began, fragments were placed into the experimental tanks, all at 30°C. One fragment of each colony was placed in a consistent position in a coordinate grid arrangement in each of eight tanks. Each tank was set to follow a different temperature profile with maximum target temperatures ranging from 30°C to 38°C in increments of 1°C (in August, 38°C was omitted, while in October, 31°C was omitted; figure 1b). Each tank, therefore, contained one fragment from all of the coral colonies in that run, which ranged from n = 13–22. In August, one system of eight tanks was used, while in October, two systems of eight tanks were used, such that up to 44 colonies could be assayed in a single day.
Thermal stress assays followed the procedures outlined in the previous section, and are depicted in figure 1b. Temperatures in each tank throughout each run were recorded by the Vktech temperature probes, or by HOBO pendants (MX2202) for Inkbird-controlled tanks. HOBO pendants were calibrated against Vktech probes and logger-specific offsets were used to adjust recorded data. At the end of each run, Fv/Fm was measured as described in the previous section, with data collected from each tank in random order.
(c) . Coral bleaching automated stress systems data analysis and ED50 calculations
For each coral fragment, one or two data records were generated that included F0 (background fluorescence), Fm (maximum fluorescence) and Fv/Fm (maximum photochemical efficiency). All data were processed in batches according to the CBASS assay date. For each batch, an initial data pre-filtering step was applied which removed any record with Fv/Fm greater than 0.75, and any record from a high-stress treatment (greater than or equal to 36°C) where both a high Fv/Fm and low F0 indicated a multivariate outlier by Mahalanobis distance. The latter filter was applied to remove data points from severely bleached corals where very low signal resulted in spuriously high Fv/Fm. After pre-filtering, Fv/Fm data were then adjusted to account for the physical position (i.e. coordinate grid location) of each coral genet within each tank: the number of rows and columns from the tank centre were used as linear predictors of Fv/Fm, and residuals were added to the mean for each tank to generate adjusted values. This step was taken to account for attenuation of light towards tank edges that tended to result in higher Fv/Fm.
Dose–response curves (e.g. figure 1c) were then used to model the decline in Fv/Fm as a function of the maximum treatment temperature for each genet. The mean temperature recorded during the 3 h hold period in each tank was used rather than the target temperature because there were some minor deviations from target values, particularly in the Inkbird-controlled tanks. Dose–response curves were fitted as three-parameter log–logistic functions using the drc package [48], with the following constraints on parameter values: maximum Fv/Fm = [0.55,0.72]; slope = [10,120]; ED50 = [30,40]. The lower limit was set equal to zero. Based on initial model fits, additional data filtering was performed to remove points with Cook's distance greater than 4/n (where n is the number of data points for the colony), or with a positive residual exceeding 2 standard deviations. Dose–response curves were then re-fitted with filtered data to generate parameter values (and standard errors) for each colony. ED50 parameter values are interpreted as the quantitative thermal tolerance phenotype for each colony and are the primary metric used in downstream statistical analyses. All Fv /Fm data analysis and dose–response curve fitting is detailed with R code in the accompanying data repository.
(d) . Coral source colony locations and temperature regimes
All A. cervicornis in coral nurseries were originally collected by fragmenting wild source colonies on the reef, with source colony latitude and longitude recorded at the time of collection. For each of these sets of coordinates, we obtained satellite sea surface temperature (SST) data. We used a 5 km resolution SST dataset (Coral Reef Watch CoralTemp V3.1 1985–2012 climatology; https://coralreefwatch.noaa.gov/product/5km/index_5km_sst.php) to calculate maximum monthly mean (MMM) temperatures as the average temperature of the hottest month at each pixel. Source colony latitude, longitude and MMM were then used to test for predictive relationships with thermal tolerance phenotypes (see below).
(e) . Symbiodiniaceae analysis
Small biopsies (approx. 0.5 cm2) were collected from healthy tissue of each coral colony using nail clippers and preserved in DNA buffer. DNA was extracted following a modified organic extraction protocol, and Symbiodiniaceae were quantified using genus-specific qPCR assays targeting Symbiodinium [49] and Durusdinium [50] modified from the methods of [51]. Symbiont to host cell ratios were quantified by normalization to the single-copy CAM locus in A. cervicornis [51] with copy number corrections for Symbiodiniaceae [50] and no fluorescence corrections as PowerUp SYBR Green Master Mix (Applied Biosystems) was used to detect the presence of host and symbiont loci.
(f) . Statistical analysis
The total phenotypic variation in thermal tolerance was visualized as a histogram and kernel density estimate of ED50 values of all coral individuals at all nurseries (figure 2a). From this, we subtracted the variation owing to the environment (i.e. differences between nurseries), by calculating adjusted ED50 values (ED50adj) as the grand mean plus residuals from nursery-specific means (figure 2b). The resulting ED50adj distribution was tested for normality using a Shapiro–Wilk test and summarized in figure 2c. Differences in ED50 among nurseries (figure 2b) were analysed using a Welch ANOVA, after Levene's test indicated unequal variances. Wilcoxon rank-sum tests were used to test for pairwise differences between nurseries, with Bonferroni adjusted p-values and α = 0.01. Relationships between ED50adj and source colony locations, thermal regimes and symbiont abundances were tested using linear models (figure 3). Random samples (n = 10 000) of n genets from each nursery were analysed with respect to the proportion of population ED50adj deciles captured in the sample, and the mean probability they contained greater than or equal to 1 thermotolerant genotype (defined as ED50adj in the 75th percentile or higher across the wider population; figure 4). Finally, we assessed reproducibility of the CBASS-derived ED50 metrics by comparing data for 27 genotypes originating from the same nurseries (n = 21 from MML, n = 4 from FWC, n = 2 from UM) that were also tested in an independent set of CBASS assays in June 2020 (see the electronic supplementary material, Methods). The strength of correlation between each genotype's ED50 values from these two sets of experiments was assessed by both Pearson's and Spearman's rank-order correlation coefficients (figure 5).
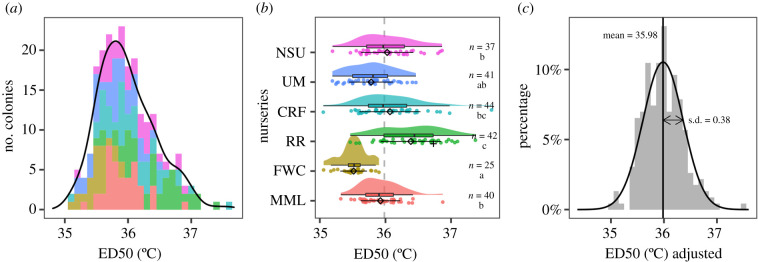
Figure 2.
Variation in coral thermal tolerance. (a) Total phenotypic variation plotted as the number of coral individuals with a given ED50 (bin size = 0.1°C), coloured by nursery as in (b). (b) For each nursery (ordered by latitude; abbreviations as in figure 1), coloured points and probability density functions show ED50 values for each colony, black diamonds and error bars show the mean ± s.d., and boxplots show the median, interquartile range (IQR) and range of values within 1.5 * IQR. The grand mean is shown by the dashed vertical line. Nurseries that do not share a letter have significantly different mean ED50 values (p < 0.01). The black ‘+’ in the RR nursery indicates the one colony dominated by Durusdinium. (c) Adjusted ED50 values, after subtracting variation among nursery environments from total phenotypic variation; used to estimate the genetic (and genotype by environment) component of thermal tolerance. This approximately normal distribution is shown scaled to the per cent of individuals with a given ED50adj (bin size = 0.1°C). (Online version in colour.)
Figure 3.
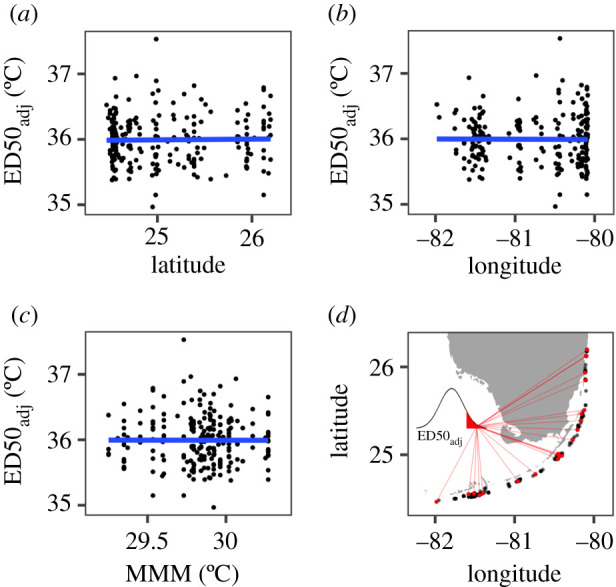
Relationship between thermal tolerance and source colony location and temperature regime. Adjusted ED50 values for nursery corals (controlling for environmental differences among nurseries) are plotted against (a) the original source colony latitude and (b) longitude, and (c) the corresponding maximum monthly mean (MMM) temperatures calculated from satellite SST data with 5 km resolution. Lines represent linear model fits; none of these slopes were significantly different from zero. (d) The most thermally tolerant colonies (top 10% of ED50adj, shown by inset) were sourced from throughout the wider population, as indicated by red line segments and points. All source colonies (except two from Dry Tortugas) are shown by black points along Florida's coral reef. (Online version in colour.)
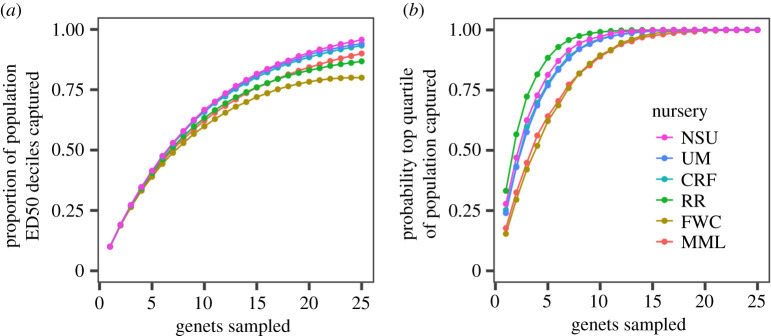
Figure 4.
Thermal tolerance phenotypes captured by randomly selecting genets from each nursery. (a) The proportion of adjusted ED50 deciles for the wider population captured in a random sample of n genets from each nursery. (b) The probability that a random sample of n genets from each nursery contains any of the top 25% most thermally tolerant genets across the wider population. (Online version in colour.)
Figure 5.
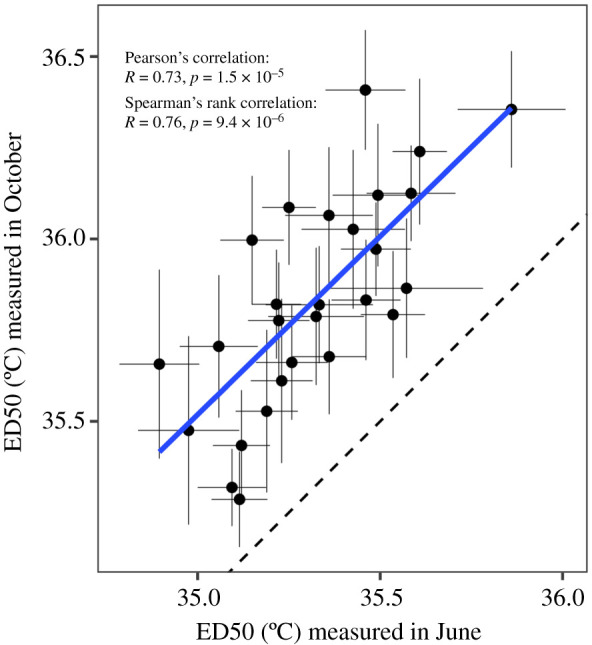
Correlation between independent CBASS runs on the same coral genets sourced from the same nurseries. ED50 values measured in October are plotted against ED50 values measured in June (n = 27 genets). Error bars indicate the fitted parameter standard error. The solid line indicates a linear model fit (slope = 0.98), while the dashed line indicates unity (y = x). (Online version in colour.)
(g) . Data and code availability
All analyses were conducted using R v. 4.0.0 [52] in RStudio v. 1.3.1093 [53]. Figures were created using ggplot2 [54]. All data and code are available on Github (github.com/jrcunning/CBASS_FL_Acer) and archived at Zenodo (doi:10.5281/zenodo.5526941).
3. Results
(a) . Variation in thermal tolerance phenotypes
Thermal tolerance metrics (ED50 values) were generated for 229 coral colonies across the six nurseries, representing up to 172 unique genets (i.e. genetic individuals). Some genets were represented at multiple nurseries (electronic supplementary material, figure S1), either because the source material was directly shared among nurseries, or clonemates were previously identified by single nucleotide polymorphism genotyping, and it is possible that additional unidentified clonemates may exist. Total phenotypic variation in ED50 ranged from 35.05°C to 37.62°C (range = 2.57°C), with a median value of 35.92°C (figure 2a). After removing the effect of nursery environment, the resulting distribution was approximately normal (Shapiro–Wilk W = 0.987; p = 0.038) with a mean of 35.98°C and standard deviation of 0.38°C (figure 2c).
The variance in ED50 was not equal across nurseries (Levene's F5,223 = 4.15, p = 0.0013). Corals from CRF showed the highest and lowest individual values, as well as the highest variation (s.d. = 0.464), while FWC corals showed the lowest variation (s.d. = 0.205). Mean ED50 values also differed significantly across nurseries (Welch's F5,102.39 = 28.30, p < 0.0001). Mean ED50 was highest at RR, intermediate at CRF, NSU, MML and UM, and lowest at FWC (figure 2b), but these differences were not related to the nurseries' MMM temperature. Approximately 37% (ω2 = 0.373) of the total variance in ED50 was accounted for by nursery, and the median pairwise difference between nursery averages was 0.32°C.
After adjusting for these differences between nurseries, we analysed factors that might explain differences within nurseries, where most of the variation occurred. Variation in Symbiodiniaceae genera had no effect, since all colonies analysed (n = 182) hosted exclusively Symbiodinium, except for a single individual from RR that was dominated by Durusdinium (figure 2b). There was no relationship between ED50adj and symbiont to host cell ratios (p = 0.68). We then tested for relationships between ED50adj and the locations of the source colonies on the reef from which nursery fragments were originally collected (electronic supplementary material, figure S2). At this scale, we found no relationship between ED50adj and source colonies’ latitude, longitude or MMM temperature (figure 3a–c). None of these variables, or their interactions, were significant predictors of thermal tolerance. Instead, thermally tolerant phenotypes were distributed throughout Florida's Coral Reef (figure 3d).
To assess the degree to which thermal tolerance phenotypes are genetically fixed or environmentally plastic, we compared ED50adj metrics from 33 coral genets that were present at either two (n = 20), three (n = 12) or four (n = 1) nurseries (electronic supplementary material, figure S3). We found a strong positive correlation in ED50adj for 11 genets maintained at MML and FWC (r = 0.66, p < 0.05), but no correlation among measurements for genets shared between other nurseries (p > 0.05; electronic supplementary material, figure S4).
(b) . Capturing thermal tolerance variation in coral collections
We analysed adjusted ED50 values to ask how many coral genets would need to be randomly drawn from each nursery to (i) capture a majority of the phenotypic variation in the wider population, and (ii) include some of the most thermally tolerant genets in the region. Seven to eight genets from any of the nurseries would encompass, on average, over half of the Florida population's range in ED50adj (figure 4a). To have a high probability (greater than 90%) of capturing at least one tolerant genet relative to the wider population (greater than 75th percentile ED50adj), six to seven genets would need to be sampled from RR, UM, NSU or CRF, or 11 genets from MML and FWC (figure 4b).
(c) . Reproducibility of thermal tolerance phenotypes using coral bleaching automated stress systems
Of the coral genets tested using CBASS in October, 27 of the same genets from the same nurseries were also tested in an independent set of CBASS experiments (see the electronic supplementary material, Methods) in June of the same year, allowing us to test reproducibility. We found a strong linear relationship between the results obtained from the two independent sets of experiments (figure 5), with a Pearson's correlation of 0.73, and Spearman's rank-order correlation of 0.76. Linear regression indicated the slope of the relationship was 0.98, not significantly different from 1. However, ED50 values measured in October were, on average, 0.51°C higher than those measured in June.
4. Discussion
This study represents, to our knowledge, the first large-scale effort to quantify variability in thermal tolerance across a population of the threatened Caribbean staghorn coral, A. cervicornis. Using Fv/Fm ED50 as a standardized metric [41], we found significant variation in thermal tolerance among 229 colonies, ranging from a minimum of 35.05°C to a maximum of 37.62°C. While these temperatures are not representative of the typical temperatures encountered by corals during a natural bleaching event, they do serve as reliable proxies of relative thermal tolerance [28], allowing us to compare differences among individuals. The 2.57°C difference between the least and most thermotolerant individuals found here is comparable to the 2.7°C difference in average ED50 observed between two populations of Stylophora pistillata from the northern and central Red Sea (34.1–36.8°C), also measured using CBASS [24]. The latter populations are separated by approximately 900 km and a MMM temperature gradient of 3.7°C (5 km data product), making it remarkable that the Florida population, spanning just approximately 300 km and only 1°C in MMM, contains an equivalent range in thermal tolerance among individuals. The amount of variation among individuals within other populations is unknown.
Because some of the variation in ED50 we observed may be owing to acclimatization to distinct nursery environments, we used the replicate common-garden nurseries to parse total phenotypic variation into both environmental (i.e. variation between nurseries), and genetic components (i.e. variation within nurseries, which may also include genotype by environment (GxE) effects). Nursery site explained 37% of the total variation in ED50, and the differences between nurseries, representing potential environmental acclimatization effects, were generally small (median pairwise difference = 0.32°C). The average thermal tolerance at each nursery did not correlate with their MMM temperatures, indicating no straightforward thermal acclimatization response across this range. The lowest mean thermal tolerance among nurseries was observed at FWC in the Middle Keys. This could reflect higher nutrient concentrations [55], or some other property of the ‘inimical’ Florida Bay waters that preferentially impact the Middle Keys [56,57] and may reduce thermal tolerance [58,59].
By contrast to the relatively small differences in thermal tolerance between nurseries, approximately 63% of the total variation in thermal tolerance occurred within nurseries, presumably reflecting genetic differences among individuals (although microhabitat, symbiont genotype or technical variability may also be contributing factors). This finding indicates that each nursery already contains some individuals that are relatively thermotolerant compared to the population as a whole. These quantitative thermal tolerance rankings represent immediately actionable information for nursery and restoration programmes designed to increase climate resilience [60]. For example, nurseries might choose to increase propagation and outplanting of more thermotolerant genotypes, because these may have a better chance for long-term survival, and might contribute more heat-tolerant alleles to local gene pools once they reach reproductive maturity. Additionally, selective breeding efforts using nursery corals might focus on crossing the most thermotolerant genets to boost the resilience of sexual recruits used for restoration. Such work will also enable calculation of heritability and breeding values in order to predict the magnitude and speed of generational changes in heat tolerance.
The finding of high variation within nurseries also indicates that tolerant individuals are well distributed throughout the wider population. Indeed, across this broad geographical scale, we found that heat tolerance was not related to the latitude, longitude or the temperature regime (based on satellite-derived SSTs) of the original source colonies from which nursery fragments were initially collected (figure 3). The lack of any spatial or environmental patterns indicates that both thermally tolerant and sensitive genotypes can be found throughout the population, and that tolerant genotypes are not restricted to any particular geographical range or thermal regime within this region. This finding is consistent with Cornwell et al. [42], who also found tolerant individuals distributed across locations and reef types in Palau. If other coral populations are similar, then targeted nursery development or other activities seeking to use tolerant individuals might expect to be able to find and source these individuals locally.
Of course, if corals are moved into a new environment (or sourced from distant locations), it is possible their thermal tolerance might change, such that individuals which are more tolerant in one environment might be less tolerant in another [25,35]. Such GxE interactions could explain why genotypes that were present at multiple nurseries in this study did not always show the same ED50 values or even rank-order in thermal tolerance (electronic supplementary material, figure S3). For example, although the same genets represented at both FWC and MML showed a strong positive correlation in ED50, measurements on other paired genets from other nurseries were uncorrelated (electronic supplementary material, figure S4), which could reflect either technical variability or GxE interactions. Because relatively few genets were shared between nurseries it is hard to generalize from these results. However, reciprocal transplant studies have shown that corals typically maintain similar thermal tolerance even after being moved to a new environment [13,61,62].
Considering the corals within each nursery as collections from the wider population, we can ask how many genets must be collected to capture a wide range of thermotolerance phenotypes, and, in particular, to capture high-tolerance individuals. We found that 7–8 individuals from any nursery encompass most (greater than 50%) of the thermal tolerance range of the wider population and that 6–11 are very likely (greater than 90%) to include at least one individual in the top quartile of thermotolerance across the wider population (figure 4b). These sample sizes broadly fall within the current sampling recommendations for minimum nursery diversity [60], indicating that broad representation of both genetic and phenotypic variation can be captured with similar effort, and further suggesting that phenotypic variation in thermal tolerance is indeed strongly underpinned by genetic variation. If other populations are structured similarly, this information may guide general approaches to the sourcing of corals for selective breeding, or targeted nursery development, without requiring prior knowledge or determination of individual phenotypes.
Direct determination of heat tolerance phenotypes using CBASS holds great potential for advancing knowledge and restoration activities, but it is critical to understand the reliability and limitations of these tools. To test the reproducibility of these thermal stress assays we performed two independent CBASS assays on the same coral genets from the same nurseries, but with different experimental systems, in different locations, and several months apart (June versus October). We found that ED50 values from the two assays were highly correlated, indicating a reproducible tolerance phenotype (figure 5). However, thermal tolerance was, on average, 0.5°C higher in October than June, suggesting seasonal acclimatization to recent summer maximum temperatures (which typically occur in early September). Alternatively, this offset could be owing to differences in experimental systems, emphasizing the importance of rigorous methodological standardization [63] to enable comparison across CBASS studies. Regardless, the highly correlated ED50 values, with a slope of 1 (figure 5), indicate that CBASS can reliably distinguish hierarchies of thermal tolerance among genets and compare differences in the range in thermal tolerance across studies. However, these findings also highlight significant challenges for the merging and/or comparison of CBASS datasets generated from different field locations, experimental systems and/or times of the year. Indeed, direct comparisons between studies might require shared genotypes in order to provide an internal standard. In this context, the synoptic nature of the Florida-wide data presented here is noteworthy.
Quantitative thermal tolerance data are also critical for efforts to identify genomic features associated with this trait. Indeed, genome-wide association studies (GWAS) in corals show promise in identifying loci associated with thermal tolerance but have previously relied on bleaching responses observed in situ in response to natural, heterogeneous stressors [25,64], adding uncertainty to the trait data. The standardized, quantitative trait data provided by CBASS and the Fv/Fm ED50 metric may significantly improve future GWAS, and linking this phenotype with whole-genome sequence data is an important next step in uncovering the genomic determinants of thermal tolerance.
While this census of thermal tolerance across a managed coral population represents a critical step forward in optimizing coral restoration, strategies that rely solely on single phenotypic characters such as thermotolerance ignore the possibility that high-performing genets might suffer from trade-offs, e.g. lower disease resistance [18], slower growth [65] or less rapid lesion healing [66] (but see [37]). Such trade-offs may decrease the overall attractiveness of particular genets and may be particularly important when some traits, such as growth, impact valuable ecosystem services, such as habitat construction or wave attenuation, that may also be desired restoration goals. Consequently, thermotolerance, although a critical factor in determining the long-term survivorship of restored corals, is just one component of a restoration strategy that must include diversity and variation as fundamental criteria to guard against unanticipated threats and challenges [60]. Indeed, because the ultimate goal of coral restoration is to create self-sustaining multi-species communities, ensuring that restoration strategies incorporate diversity while still attempting to bias the next generation of corals in favour of particular traits (whether they be increased thermal tolerance, disease resistance or growth rate), represents a defining challenge. This is true regardless of whether the restoration strategy depends on corals that are derived asexually (nursery rearing and outplanting) or sexually (selective breeding).
In summary, this study reveals considerable variation in thermal tolerance among A. cervicornis genotypes in the world's most extensive reef restoration programme, and this variation is generally reflected at the scale of the individual nursery. Similar censuses of thermal tolerance using CBASS could be extended to neighbouring populations to compare how the variation in heat tolerance in Florida compares to the region, and advance our understanding of the evolutionary potential of coral thermal tolerance under climate change. However, care must be taken to ensure standardized practices to facilitate cross-comparison among studies. In the meantime, our findings suggest that locally available thermally tolerant individuals could be used immediately to help boost the climate resilience of restoration efforts across Florida's Reef.
Supplementary Material
Acknowledgements
We thank J. Delaney and A. Bruckner of FKNMS for permit no. 2020-120. We thank D. Barshis, N. Evensen, and the Shedd Aquarium Fabrication Shop and Teen Learning Laboratory for assistance with CBASS construction, and the captain and crew of the R/V Coral Reef II, A. Kough, S. Kessel, G. Parsons, B. Crown, S. Sousley, B. Williams, A. Ayerza, P. McPherson and H. Wind for field support. We thank E. Kobetz (UM) for approving COVID fieldwork logistics.
Ethics
Florida Keys National Marine Sanctuary Permit no. 2020-120 supported this work.
Data accessibility
All data and code are available on Github (github.com/jrcunning/CBASS_FL_Acer) and archived at Zenodo (doi:10.5281/zenodo.5526941).
Authors' contributions
Funding (R.C., C.D., C.K., J.E.P., A.C.B.), conceptualization (R.C., C.D., C.K., J.E.P., A.C.B.), data curation (R.C., K.E.P., K.J.S., R.F.K., A.D.W., O.M.W.), formal analysis (R.C.), investigation (R.C., K.E.P., K.J.S, R.F.K., A.D.W., O.M.W., W.C.M.), methodology (R.C., K.E.P., K.J.S.), project administration (R.C., K.E.P., K.J.S., A.C.B.), resources (R.C., E.B., M.D.A., D.S.G., G.H., J.L., D.L., K.M., A.L.M., A.M., E.M.M., K.N., B.R., R.v.H.), software (R.C.), supervision (R.C., A.C.B.), validation and visualization (R.C.), writing and editing (all authors).
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
The authors declare they have no conflict of interest regarding this work.
Funding
This work was supported by NSF grant no. OCE-2023705 to R.C., C.K., J.E.P., C.D. and A.C.B. Additional support was generously provided by the Paul M. Angell Family Foundation, Dr Scholl Foundation and the Brunswick Public Foundation.
References
- 1.National Academies of Sciences Engineering, Medicine. 2019. A research review of interventions to increase the persistence and resilience of coral reefs. Washington, DC: The National Academies Press. [Google Scholar]
- 2.Bay LK, et al. 2019. Reef restoration and adaptation program: intervention technical summary. A report provided to the Australian Government by the reef restoration and adaptation program. Townsville, Australia: Australian Institute of Marine Science (AIMS). [Google Scholar]
- 3.Kleypas J, et al. 2021. Designing a blueprint for coral reef survival. Biol. Conserv. 257, 109107. ( 10.1016/j.biocon.2021.109107) [DOI] [Google Scholar]
- 4.Rinkevich B. 2014. Rebuilding coral reefs: does active reef restoration lead to sustainable reefs? Curr. Opin. Environ. Sustain. 7, 28-36. ( 10.1016/j.cosust.2013.11.018) [DOI] [Google Scholar]
- 5.Lirman D, Schopmeyer S. 2016. Ecological solutions to reef degradation: optimizing coral reef restoration in the Caribbean and Western Atlantic. PeerJ 4, e2597. ( 10.7717/peerj.2597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guest JR, Baria MV, Gomez ED, Heyward AJ, Edwards AJ. 2014. Closing the circle: is it feasible to rehabilitate reefs with sexually propagated corals? Coral Reefs 33, 45-55. ( 10.1007/s00338-013-1114-1) [DOI] [Google Scholar]
- 7.Quigley KM, Bay LK, Oppen MJH. 2020. Genome-wide SNP analysis reveals an increase in adaptive genetic variation through selective breeding of coral. Mol. Ecol. 29, 2176-2188. ( 10.1111/mec.15482) [DOI] [PubMed] [Google Scholar]
- 8.van Oppen MJH, Oliver JK, Putnam HM, Gates RD. 2015. Building coral reef resilience through assisted evolution. Proc. Natl Acad. Sci. USA 112, 2307-2313. ( 10.1073/pnas.1422301112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bay RA, Rose NH, Logan CA, Palumbi SR. 2017. Genomic models predict successful coral adaptation if future ocean warming rates are reduced. Sci. Adv. 3, e1701413. ( 10.1126/sciadv.1701413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagedorn M, et al. 2021. Assisted gene flow using cryopreserved sperm in critically endangered coral. Proc. Natl Acad. Sci. USA, 118, e2110559118. ( 10.1073/pnas.2110559118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Oppen MJH, et al. 2017. Shifting paradigms in restoration of the world's coral reefs. Glob. Change Biol. 23, 3437-3448. ( 10.1111/gcb.13647) [DOI] [PubMed] [Google Scholar]
- 12.Lohr KE, Patterson JT. 2017. Intraspecific variation in phenotype among nursery-reared staghorn coral Acropora cervicornis (Lamarck, 1816). J. Exp. Mar. Biol. Ecol. 486, 87-92. ( 10.1016/j.jembe.2016.10.005) [DOI] [Google Scholar]
- 13.Morikawa MK, Palumbi SR. 2019. Using naturally occurring climate resilient corals to construct bleaching-resistant nurseries. Proc. Natl Acad. Sci. USA 116, 10 586-10 591. ( 10.1073/pnas.1721415116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glynn PW, Maté JL, Baker AC, Calderón MO. 2001. Coral bleaching and mortality in Panama and Ecuador during the 1997–1998 El Niño-Southern Oscillation event: spatial/temporal patterns and comparisons with the 1982–1983 event. Bull. Mar. Sci. 69, 79-109. [Google Scholar]
- 15.McClanahan TR. 2004. The relationship between bleaching and mortality of common corals. Mar. Biol. 144, 1239-1245. ( 10.1007/s00227-003-1271-9) [DOI] [Google Scholar]
- 16.Cunning R, Ritson-Williams R, Gates RD. 2016. Patterns of bleaching and recovery of Montipora capitata in Kāne‘ohe Bay, Hawai‘i, USA. Mar. Ecol. Prog. Ser. 551, 131-139. ( 10.3354/meps11733) [DOI] [Google Scholar]
- 17.Edmunds PJ. 1994. Evidence that reef-wide patterns of coral bleaching may be the result of the distribution of bleaching-susceptible clones. Mar. Biol. 121, 137-142. ( 10.1007/BF00349482) [DOI] [Google Scholar]
- 18.Muller EM, Bartels E, Baums IB. 2018. Bleaching causes loss of disease resistance within the threatened coral species Acropora cervicornis. Elife 7, e35066. ( 10.7554/eLife.35066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coles SL, Jokiel PL, Lewis CR.. 1976. Thermal tolerance in tropical versus subtropical Pacific reef corals. Pac. Sci. 30, 159-166. [Google Scholar]
- 20.Berkelmans R. 2002. Time-integrated thermal bleaching thresholds of reefs and their variation on the Great Barrier Reef. Mar. Ecol. Prog. Ser. 229, 73-82. ( 10.3354/meps229073) [DOI] [Google Scholar]
- 21.Ulstrup KE, Berkelmans R, Ralph PJ, van Oppen MJH. 2006. Variation in bleaching sensitivity of two coral species across a latitudinal gradient on the Great Barrier Reef: the role of zooxanthellae. Mar. Ecol. Prog. Ser. 314, 135-148. ( 10.3354/meps314135) [DOI] [Google Scholar]
- 22.Kenkel CD, Goodbody-Gringley G, Caillaud D, Davies SW, Bartels E, Matz MV. 2013. Evidence for a host role in thermotolerance divergence between populations of the mustard hill coral (Porites astreoides) from different reef environments. Mol. Ecol. 22, 4335-4348. ( 10.1111/mec.12391) [DOI] [PubMed] [Google Scholar]
- 23.Dixon GB, Davies SW, Aglyamova GV, Meyer E, Bay LK, Matz MV. 2015. Genomic determinants of coral heat tolerance across latitudes. Science 348, 1460-1462. ( 10.1126/science.1261224) [DOI] [PubMed] [Google Scholar]
- 24.Voolstra CR. 2020. Contrasting heat stress response patterns of coral holobionts across the Red Sea suggest distinct mechanisms of thermal tolerance. Mol. Ecol. 30, 4466-4480. ( 10.1111/mec.16064) [DOI] [PubMed] [Google Scholar]
- 25.Drury C, Lirman D. 2021. Genotype by environment interactions in coral bleaching. Proc. R. Soc. B 288, 20210177. ( 10.1098/rspb.2021.0177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver TA, Palumbi SR. 2011. Do fluctuating temperature environments elevate coral thermal tolerance? Coral Reefs 30, 429-440. ( 10.1007/s00338-011-0721-y) [DOI] [Google Scholar]
- 27.Schoepf V, Stat M, Falter JL, McCulloch MT. 2015. Limits to the thermal tolerance of corals adapted to a highly fluctuating, naturally extreme temperature environment. Sci. Rep. 5, 1-14. ( 10.1038/srep17639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voolstra CR, Buitrago-López C, Perna G, Cárdenas A, Hume BCC, Rädecker N, Barshis DJ. 2020. Standardized short-term acute heat stress assays resolve historical differences in coral thermotolerance across microhabitat reef sites. Glob. Change Biol. 26, 4328-4343. ( 10.1111/gcb.15148) [DOI] [PubMed] [Google Scholar]
- 29.Brown BE, Dunne RP, Goodson MS, Douglas AE. 2002. Experience shapes the susceptibility of a reef coral to bleaching. Coral Reefs 21, 119-126. ( 10.1007/s00338-002-0215-z) [DOI] [Google Scholar]
- 30.Palumbi SR, Barshis DJ, Traylor-Knowles N, Bay RA. 2014. Mechanisms of reef coral resistance to future climate change. Science 344, 895-898. ( 10.1126/science.1251336) [DOI] [PubMed] [Google Scholar]
- 31.Berkelmans R, van Oppen MJH. 2006. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc. R. Soc. B 273, 2305-2312. ( 10.1098/rspb.2006.3567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverstein RN, Cunning R, Baker AC. 2015. Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob. Change Biol. 21, 236-249. ( 10.1111/gcb.12706) [DOI] [PubMed] [Google Scholar]
- 33.Ritson-Williams R, Gates RD. 2020. Coral community resilience to successive years of bleaching in Kāne‘ohe Bay, Hawai‘i. Coral Reefs 39, 757-769. ( 10.1007/s00338-020-01944-4) [DOI] [Google Scholar]
- 34.Baums IB, Devlin-Durante MK, LaJeunesse TC. 2014. New insights into the dynamics between reef corals and their associated dinoflagellate endosymbionts from population genetic studies. Mol. Ecol. 23, 4203-4215. ( 10.1111/mec.12788) [DOI] [PubMed] [Google Scholar]
- 35.Drury C, Manzello D, Lirman D. 2017. Genotype and local environment dynamically influence growth, disturbance response and survivorship in the threatened coral, Acropora cervicornis. PLoS ONE 12, e0174000. ( 10.1371/journal.pone.0174000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirk NL, Howells EJ, Abrego D, Burt JA, Meyer E. 2018. Genomic and transcriptomic signals of thermal tolerance in heat-tolerant corals (Platygyra daedalea) of the Arabian/Persian Gulf. Mol. Ecol. 27, 5180-5194. ( 10.1111/mec.14934) [DOI] [PubMed] [Google Scholar]
- 37.Wright RM, Mera H, Kenkel CD, Nayfa M, Bay LK, Matz MV. 2019. Positive genetic associations among fitness traits support evolvability of a reef-building coral under multiple stressors. Glob. Change Biol. 25, 3294-3304. ( 10.1111/gcb.14764) [DOI] [PubMed] [Google Scholar]
- 38.Kavousi J, Denis V, Sharp V, Reimer JD, Nakamura T, Parkinson JE. 2020. Unique combinations of coral host and algal symbiont genotypes reflect intraspecific variation in heat stress responses among colonies of the reef-building coral, Montipora digitata. Mar. Biol. 167, 1-5. ( 10.1007/s00227-019-3632-z) [DOI] [Google Scholar]
- 39.Drury C. 2020. Resilience in reef-building corals: the ecological and evolutionary importance of the host response to thermal stress. Mol. Ecol. 29, 448-465. ( 10.1111/mec.15337) [DOI] [PubMed] [Google Scholar]
- 40.Parkinson JE, Banaszak AT, Altman NS, LaJeunesse TC, Baums IB. 2015. Intraspecific diversity among partners drives functional variation in coral symbioses. Sci. Rep. 5, 15667. ( 10.1038/srep15667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evensen NR, Fine M, Perna G, Voolstra CR, Barshis DJ. 2021. Remarkably high and consistent tolerance of a Red Sea coral to acute and chronic thermal stress exposures. Limnol. Oceanogr. 66, 1718-1729. ( 10.1002/lno.11715) [DOI] [Google Scholar]
- 42.Cornwell B, Armstrong K, Walker NS, Lippert M, Nestor V, Golbuu Y, Palumbi SR. 2021. Widespread variation in heat tolerance and symbiont load are associated with growth tradeoffs in the coral Acropora hyacinthus in Palau. Elife 10, e64790. ( 10.7554/eLife.64790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young CN, Schopmeyer SA, Lirman D. 2012. A review of reef restoration and coral propagation using the threatened genus Acropora in the Caribbean and Western Atlantic. Bull. Mar. Sci. 88, 1075-1098. ( 10.5343/bms.2011.1143) [DOI] [Google Scholar]
- 44.Schopmeyer SA, et al. 2017. Regional restoration benchmarks for Acropora cervicornis. Coral Reefs 36, 1047-1057. ( 10.1007/s00338-017-1596-3) [DOI] [Google Scholar]
- 45.Moulding AL, Griffin SP, Nemeth MI, Ray EC. 2020. Caribbean Acropora outplanting in U.S. Jurisdiction: 1993–2017. NOAA Technical Memorandum NMFS-SER-10. ( 10.25923/N4TX-1A30) [DOI]
- 46.Baums IB, Johnson ME, Devlin-Durante MK, Miller MW. 2010. Host population genetic structure and zooxanthellae diversity of two reef-building coral species along the Florida Reef Tract and wider Caribbean. Coral Reefs 29, 835-842. ( 10.1007/s00338-010-0645-y) [DOI] [Google Scholar]
- 47.Reich HG, Kitchen SA, Stankiewicz KH, Devlin-Durante M, Fogarty ND, Baums IB. 2021. Genomic variation of an endosymbiotic dinoflagellate (Symbiodinium ‘fitti’) among closely related coral hosts. Mol. Ecol. 30, 3500-3514. ( 10.1111/mec.15952) [DOI] [PubMed] [Google Scholar]
- 48.Ritz C, Baty F, Streibig JC, Gerhard D. 2015. Dose-response analysis using R. PLoS ONE 10, e0146021. ( 10.1371/journal.pone.0146021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter RN. 2017. Environmental controls on the reassembly of Symbiodinium communities in reef corals following perturbation: implications for reef futures under climate change. PhD thesis, University of Miami, Miami, FL. ProQuest Dissertations Publishing. 10283915. [Google Scholar]
- 50.Cunning R, Baker AC. 2012. Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Change 3, 259-262. ( 10.1038/nclimate1711) [DOI] [Google Scholar]
- 51.Palacio-Castro AM. 2019. Abiotic controls on endosymbiotic algal communities and their implications for coral bleaching susceptibility and recovery. PhD thesis, University of Miami, Miami, FL. ProQuest Dissertation Publishing. 13881222. [Google Scholar]
- 52.R Core Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See http://www.r-project.org/. [Google Scholar]
- 53.RStudio Team. 2020. RStudio: integrated development environment for R. Boston, MA: R Studio, PBC. See http://www.rstudio.com.
- 54.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Berlin, Germany: Springer. [Google Scholar]
- 55.Szmant AM, Forrester A. 1996. Water column and sediment nitrogen and phosphorus distribution patterns in the Florida Keys, USA. Coral Reefs 15, 21-41. ( 10.1007/BF01626075) [DOI] [Google Scholar]
- 56.Ginsburg RN, Shinn EA. 1964. Distribution of the reef-building community in Florida and the Bahamas. AAPG Bull. 48, 527-527. ( 10.1306/bc743c69-16be-11d7-8645000102c1865d) [DOI] [Google Scholar]
- 57.Manzello DP, Enochs IC, Kolodziej G, Carlton R. 2015. Coral growth patterns of Montastraea cavernosa and Porites astreoides in the Florida Keys: the importance of thermal stress and inimical waters. J. Exp. Mar. Biol. Ecol. 471, 198-207. ( 10.1016/j.jembe.2015.06.010) [DOI] [Google Scholar]
- 58.Wiedenmann J, D'Angelo C, Smith EG, Hunt AN, Legiret F-E, Postle AD, Achterberg EP. 2013. Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Change 3, 160-164. ( 10.1038/nclimate1661) [DOI] [Google Scholar]
- 59.Vega Thurber RL, Burkepile DE, Fuchs C, Shantz AA, McMinds R, Zaneveld JR. 2014. Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Change Biol. 20, 544-554. ( 10.1111/gcb.12450) [DOI] [PubMed] [Google Scholar]
- 60.Baums IB, et al. 2019. Considerations for maximizing the adaptive potential of restored coral populations in the western Atlantic. Ecol. Appl. 29, e01978. ( 10.1002/eap.1978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schoepf V, Carrion SA, Pfeifer SM, Naugle M, Dugal L, Bruyn J, McCulloch MT. 2019. Stress-resistant corals may not acclimatize to ocean warming but maintain heat tolerance under cooler temperatures. Nat. Commun. 10, 4031. ( 10.1038/s41467-019-12065-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barott KL, et al. 2021. Coral bleaching response is unaltered following acclimatization to reefs with distinct environmental conditions. Proc. Natl Acad. Sci. USA 118, e2025435118. ( 10.1073/pnas.2025435118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grottoli AG, et al. 2021. Increasing comparability among coral bleaching experiments. Ecol. Appl. 31, e02262. ( 10.1002/eap.2262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fuller ZL, et al. 2020. Population genetics of the coral Acropora millepora: toward genomic prediction of bleaching. Science 369, eaba4674. ( 10.1126/science.aba4674) [DOI] [PubMed] [Google Scholar]
- 65.Bay RA, Palumbi SR. 2017. Transcriptome predictors of coral survival and growth in a highly variable environment. Ecol. Evol. 7, 4794-4803. ( 10.1002/ece3.2685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaufman ML, Watkins E, van Hooidonk R, Baker AC, Lirman D. 2021. Thermal history influences lesion recovery of the threatened Caribbean staghorn coral Acropora cervicornis under heat stress. Coral Reefs 40, 289-293. ( 10.1007/s00338-020-02025-2) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyses were conducted using R v. 4.0.0 [52] in RStudio v. 1.3.1093 [53]. Figures were created using ggplot2 [54]. All data and code are available on Github (github.com/jrcunning/CBASS_FL_Acer) and archived at Zenodo (doi:10.5281/zenodo.5526941).
All data and code are available on Github (github.com/jrcunning/CBASS_FL_Acer) and archived at Zenodo (doi:10.5281/zenodo.5526941).