Abstract
Standardization and genotype independence of methods used to quantify hepatitis C virus (HCV) RNA in clinical specimens are necessary for accurate assessment of the role of HCV quantitation as a prognostic marker for HCV infection and monitoring of the response to antiviral treatment. Commercially available methods used to measure HCV loads include PCR-based (Roche Monitor) and hybridization-based (Quantiplex bDNA-2) methods. Recently, a new version of the Roche Monitor assay (version 2.0) has become available; it has been modified to achieve more equal quantitation of different HCV genotypes. Consistent with previous reports, Roche Monitor version 1.0 substantially underestimated concentrations of RNA transcripts of types 2b, 3a, 4a, 5a, and 6a and virus loads in individuals infected with genotypes 2 to 6 relative to reference tests. However, version 2.0 achieved equivalent quantitation of each genotype over a narrow quantitative range (103 to 5 × 105 copies of RNA/ml) but significantly underestimated RNA concentrations above this range. The assay showed an equivalent inability to quantify high levels of HCV RNA in plasma samples, and this was responsible for the falsely narrow range of virus loads detected in HCV-infected individuals. In contrast, the Chiron bDNA-2 assay could only measure RNA concentrations in the upper quantitative range (2 × 105 to 5 × 107 copies of RNA/ml) but showed equivalent sensitivity for genotypes 1 to 5; however, concentrations of type 6a RNA transcripts and virus loads in clinical specimens from individuals infected with type 6a were underestimated by a factor of 2 to 4. Differences were observed between PCR- and hybridization-based assays in their relative quantitation of HCV RNA transcripts and HCV genomic RNA, which may cause problems with the use of transcripts for interassay calibration.
Quantitation of hepatitis C virus (HCV) RNA sequences in plasma has been used extensively as a prognostic marker for individuals undergoing treatment with interferon and in the subsequent monitoring of their responses. Several investigators have found significant differences in virus load associated with infection with different genotypes of HCV (1, 2, 10, 11, 13, 14, 18), and it had been argued that this could be one the factors involved in genotype-specific differences in the outcome of interferon therapy, in particular, the reduced responses of individuals infected with type 1b compared to those infected with types 2 and 3 (5, 14, 18). Proper assessment of the effect of HCV genotype on outcome requires a method for the quantitation of viral loads in plasma that is equally sensitive for each genotype.
We have previously investigated the genotype independence of the Chiron bDNA assays (Quantiplex RNA assay bDNA-1) and Quantiplex bDNA 2, Roche Monitor assay version 1.0, and nested PCR at limiting dilution by using samples of genotypes 1, 2, and 3 (8). Significant differences in the efficiency of detection of genotypes 1, 2, and 3 were observed for the bDNA-1 and Roche Monitor assays, whereas the bDNA-2 assay and the nested PCR at limiting dilution were able to quantify the RNA sequences of different genotypes with equal efficiencies. In the current study, we have extended this investigation to a comparison of virus load measurement for genotypes 4 and 6 in the bDNA-2, limiting-dilution PCR, and Roche Monitor version 1.0 assays and retested all of the genotypes using the new version (version 2.0) of the Roche Monitor assay. This assay had been modified by the manufacturer with the express intention of achieving more equal quantitation of different genotypes.
MATERIALS AND METHODS
Samples.
Ninety-six plasma samples from HCV-infected, PCR-positive blood donors (20 samples each of genotypes 1, 2, and 3; 19 of genotype 4; 1 of genotype 5; and 16 of genotype 6) were analyzed for HCV RNA levels. Type 4 samples were from Middle Eastern countries, and type 6 samples were from Hong Kong. Fresh plasma samples were aliquoted to ensure that all of the quantitative assays were carried out on samples that had been frozen and thawed only twice. All samples were tested with the bDNA-2, limiting-dilution PCR, and Roche Monitor version 1.0 and 2.0 assays. Previously reported results for type 1, 2, and 3 samples in the bDNA-2, limiting-dilution PCR, and Roche Monitor version 1.0 assays were included in the current study for comparison, while RNA transcripts were retested. Samples from individuals infected with HCV genotypes 4, 5, and 6 were genotyped by restriction fragment length polymorphism analysis of the 5′ noncoding region (5′NCR) (19). Samples were aliquoted and stored at −40°C and were only frozen and thawed twice prior to testing.
Quantitation of HCV-infected samples.
Three commercial assays for the detection of HCV RNA were used, i.e., Quantiplex HCV RNA assay 2.0 or bDNA-2 and the Roche Monitor assay versions 1.0 and 2.0. These assays were performed in accordance with the manufacturers’ instructions. Unamplified HCV RNA was detected in the bDNA-2 assay by capture onto the solid phase by hybridization to oligonucleotides complementary to the 5′NCR and core regions, followed by detection and signal amplification to labelled probes. All samples were tested with kit lot no. MMM169.
The Roche Monitor assays were based upon reverse transcription and amplification of the HCV RNA with primers from the 5′NCR in the presence of an internal control which shared primers with HCV. Both the competitor and sample DNAs were detected by hybridization to a biotin-labelled probe, followed by incubation with enzymatically labelled streptavidin and detection in a colorimetric assay. Samples were tested with kit lots H0315 and G1722 (version 1.0) and H0526 and H011 (version 2.0).
For the in-house limiting-dilution method (8, 15), viral RNA was extracted from 100 μl of plasma by incubation with proteinase K (1 mg/ml) and sodium dodecyl sulfate (0.55%) in the presence of poly(A) (40 mg/ml) and purified by phenol-chloroform extraction as previously described (3). HCV RNA was reversed transcribed and amplified by nested PCR with primers specific for the 5′NCR. Limiting dilution of cDNA was then carried out by using an assumed efficiency of 5% for the reverse transcription step, as previously established (8, 20).
Sequence analysis.
Sequence analysis of the 5′NCR was performed by cycle sequencing in accordance with the instructions supplied with the Thermo Sequenase kit from Amersham International.
Quantitation of HCV RNA transcripts.
HCV transcripts of all six genotypes were provided by J. Detmer (Bayer Diagnostics, Emeryville, Calif.). The methods used for their synthesis and quantitation have been described elsewhere (4, 7). Briefly, to assess the quality of the preparations, HCV RNA transcripts were electrophoresed on 1.5% formaldehyde gels and scanned on an Ambis 400 radioanalytic imager (Ambis, Inc., San Diego, Calif.). The preparations of the HCV RNA transcripts used contained less than 3% free nucleotides and were composed of at least 80% full-length transcripts. Three independent analytical methods were used to quantify the transcripts. These included phosphate determination (6), measurement of A260, and hyperchromicity analysis (17). Transcripts were quantified by the bDNA-2 and limiting-dilution assays and both Roche Monitor assays.
Statistical analysis.
Differences in the distribution of quantitative values between genotypes were detected by the Mann-Whitney U test. The correlation between assays in the quantitation of HCV RNA was measured by using the Spearman nonparametric test.
RESULTS
Quantitation of HCV by four different assays.
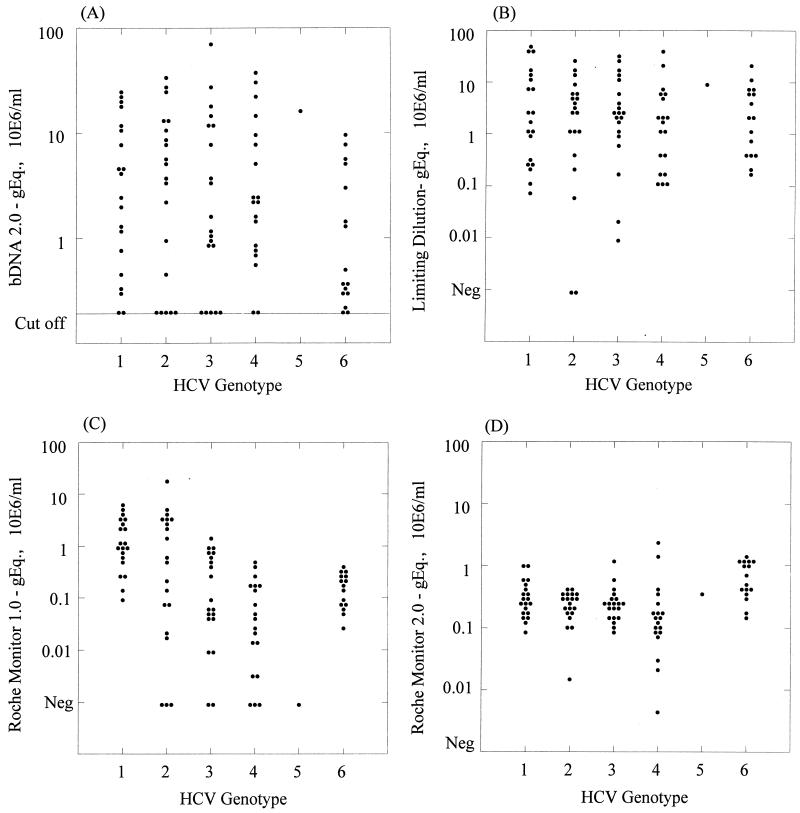
HCV RNA in samples from a total of 96 blood donors infected with genotypes 1 to 6 was quantified by using the bDNA-2 assay in replicate in accordance with the manufacturer’s instructions, once by the Roche Monitor 1.0 and 2.0 assays and in at least eight replicates at limiting dilution by the in-house assay (Table 1). The cutoff sensitivity of the bDNA-2 assay was 0.2 × 106 copies of RNA per ml, whereas the Roche Monitor and limiting-dilution assays had a cutoff sensitivity of approximately 1,000 copies of RNA per ml. Of 35 type 4 and 6 samples, 4 (11.4%) were below the cutoff of the bDNA-2 assay and a different four samples were below the cutoff of the Roche Monitor 1.0 assay, but all samples were positive by the Roche Monitor 2.0 assay. Samples below the cutoff of each assay were assigned the virus load of the cutoff. This approximation did not affect the nonparametric methods used to analyze the data.
TABLE 1.
Quantitation of HCV RNA in plasma samples by four assays
| Genotype (no. of samples) | bDNA-2
|
Limiting dilution
|
Roche Monitor version 1.0
|
Roche Monitor version 2.0
|
||||
|---|---|---|---|---|---|---|---|---|
| Median virus load (range) | Ratioa(P value)b | Median virus load (range) | Ratio (P value) | Median virus load (range) | Ratio (P value) | Median virus load (range) | Ratio (P value) | |
| 1 (20) | 3.145 (<0.2–23.51) | 2.065 (0.074–49.00) | 1.041 (0.100–6.800) | 0.273 (0.089–1.020) | ||||
| 2 (20) | 4.245 (<0.2–32.05) | 1.24 (0.914)c | 2.850 (<0.001–24.00) | 1.38 (0.755)c | 0.555 (<0.001–19.00) | 0.53 (0.208)c | 0.254 (0.015–0.428) | 0.93 (0.387)c |
| 3 (20) | 1.381 (<0.2–71.04) | 0.40 (0.569)c | 2.400 (<0.001–33.00) | 1.16 (0.968)c | 0.076 (<0.001–1.500) | 0.07 (<0.001)d | 0.246 (0.094–1.090) | 0.900 (0.409)c |
| 4 (19) | 2.239 (<0.2–39.11) | 0.66 (0.955)c | 1.840 (0.111–36.80) | 0.89 (0.399)c | 0.038 (<0.001–0.523) | 0.04 (<0.001)d | 0.147 (0.004–2.300) | 0.54 (0.013)d |
| 6 (16) | 0.439 (<0.2–9.622) | 0.131 (0.080)d | 2.320 (0.184–23.20) | 1.12 (0.787)c | 0.184 (0.026–0.382) | 0.18 (<0.001)d | 0.568 (0.134–1.320) | 2.08 (0.005)d |
Ratio of median virus load to that of genotype 1 samples.
Probability of difference in virus load occurring by chance (Mann-Whitney U test).
Not significantly different from genotype 1 samples.
Significantly different (P < 0.05) from genotype 1 samples.
Correlations between the different assays for virus load measurement were obtained. Spearman correlation coefficients ranged from 0.706 for virus loads detected in the bDNA-2 and limiting-dilution assays to 0.351 between Roche Monitor versions 1.0 and 2.0. Higher correlation coefficients were obtained for samples of genotype 1 than for those of other genotypes (0.747 for bDNA-2 with limiting dilution and 0.821 and 0.847 with Roche Monitor versions 1.0 and 2.0, 0.611 and 0.540 for limiting dilution with Roche Monitor versions 1.0 and 2.0, and 0.714 between Roche Monitor versions 1.0 and 2.0). Correlation coefficients ranged from 0.445 to 0.731 (genotype 2), from 0.432 to 0.756 (genotype 3), from 0.246 to 0.756 (genotype 4), and from −0.313 to 0.903 (genotype 6) in pairwise comparisons of the quantitative results of the four assays.
Virus load and genotype.
In three of the four assays, a wide range of virus loads were observed among donors infected with each genotype (Table 1 and Fig. 1). However, the range of virus loads obtained by using Roche Monitor 2.0 was much narrower than that obtained with the other assays. The median values calculated for these distributions differed between different quantitation assays. For example, among the type 1 samples, median values of 3.415, 2.065, 1.041, and 0.273 were observed for the bDNA-2, limiting-dilution, and Roche Monitor version 1.0 and 2.0 methods, respectively, whereas for type 4 samples, medians of 2.239, 1.84, 0.038, and 0.147 were obtained. When using the Mann-Whitney U test for pairwise comparisons between genotypes, we observed no significant difference in virus load between donors infected with types 1, 2, 3, and 4 when samples were analyzed with the bDNA-2 and limiting-dilution assays. However, virus loads of type 6 samples were lower than those of type 1 samples in bDNA-2 (median of 0.439 compared with 3.415), a difference that approached statistical significance (P = 0.08). However, no difference in virus load between type 6 and 1 samples was detected by the limiting-dilution assay (medians of 2.320 and 2.065, respectively; P = 0.787).
FIG. 1.
Range of HCV RNA levels in plasma samples from blood donors infected with genotypes 1 to 6 as detected by the bDNA-2 (A), limiting-dilution (B), Roche Monitor version 1.0 (C), and Roche Monitor version 2.0 (D) quantitative assays. The cutoff value of the bDNA-2 assay (0.2 × 106 copies/ml) is indicated.
Differences in virus loads between genotypes were observed with the Roche Monitor version 1.0 assay. Virus loads of non-type 1 genotypes were significantly lower than those of the type 1 samples (P values were all ≤0.001), apart from the type 2 samples, where the difference did not reach statistical significance. Small differences between genotype 1, 2, and 3 viral loads were observed when the Roche Monitor version 2.0 assay was used. However, type 4 samples showed statistically significantly lower levels of RNA than type 1 samples (median, 0.147; P = 0.013) while type 6 sample RNA levels were higher (median, 0.568; P = 0.005).
The ratios of virus loads individually measured for each sample by the bDNA-2 and Roche Monitor version 1.0 and 2.0 assays to those measured by the limiting-dilution assay were calculated (Table 2). A particularly wide range of ratios were observed with the Roche Monitor version 1.0 assay versus the limiting-dilution assay, with values for types 3, 4, and 6 consistently lower that those obtained for genotype 1 (P < 0.01). In contrast, no significant difference in the ratios obtained for the limiting-dilution assay with the bDNA-2 assay were observed for genotypes 1, 2, 3, and 4 (Table 2). However, the median ratio of genotype 6 samples to type 1 samples was 0.623, significantly lower (P = 0.03) than in the reference test. In contrast, the ratio of Roche Monitor version 2.0 to limiting-dilution assay results for type 6 samples was much higher than those calculated for the other genotypes (2.950). These samples had been tested at a 1-in-10 dilution in the former assay because of the limited sample volume available. Subsequent experiments (see below) suggested that the difference in quantitation between samples of type 6 and those of other genotypes might not have been observed had it been possible to test them undiluted.
TABLE 2.
Efficiency of detection of different genotypes relative to detection by limiting-dilution assay
| Genotype (no. of samples) | Efficiency of detection by:
|
|||||
|---|---|---|---|---|---|---|
| bDNA-2
|
Roche Monitor version 1
|
Roche Monitor version 2
|
||||
| Median (range) | Ratioa(P value)b | Median (range) | Ratio (P value) | Median (range) | Ratio (P value) | |
| 1 (20) | 1.155 (0.092–20.96) | 0.531 (0.043–11.74) | 0.103 (0.006–3.009) | |||
| 2 (20) | 1.401 (0.076–200.0) | 1.210 (0.516)c | 0.307 (0.002–5.758) | 0.578 (0.330)d | 0.096 (0.012–97.00) | 0.932 (1.00)c |
| 3 (20) | 0.943 (0.013–20.00) | 0.816 (0.850)c | 0.050 (0.000–1.000) | 0.094 (<0.001)d | 0.084 (0.008–13.80) | 0.816 (0.745)c |
| 4 (19) | 2.200 (0.075–13.25) | 1.905 (0.126)c | 0.036 (0.000–0.497) | 0.068 (<0.001)d | 0.154 (0.001–1.448) | 1.495 (0.844)c |
| 6 (16) | 0.720 (0.134–1.387) | 0.626 (0.030)d | 0.078 (0.009–0.500) | 0.147 (0.001)d | 0.304 (0.019–6.467) | 2.950 (0.203)c |
Ratio of efficiency of detection to that of genotype 1 samples.
Probability of difference in efficiency of detection between genotypes (Mann-Whitney U test).
Not significantly different from genotype 1 values.
Significantly different from genotype 1 values (P <0.05).
Sequence analysis of the 5′NCRs of types 4 and 6.
Current GenBank entries do not provide sequence information upstream of the 5′NCR primers used in the Roche and limiting-dilution assays. In order to determine if sequence variation within the binding sites of the primers used for amplification could account for the differences in viral load detected in the genotype 4 and 6 samples, we obtained nucleotide sequences by PCR using a novel primer from the extreme 5′ end of the HCV genome in a heminested PCR. No sequence variability in binding sites was detected in either the type 4 or 6 samples by either the Roche or limiting-dilution assays.
Quantitation of HCV RNA transcripts.
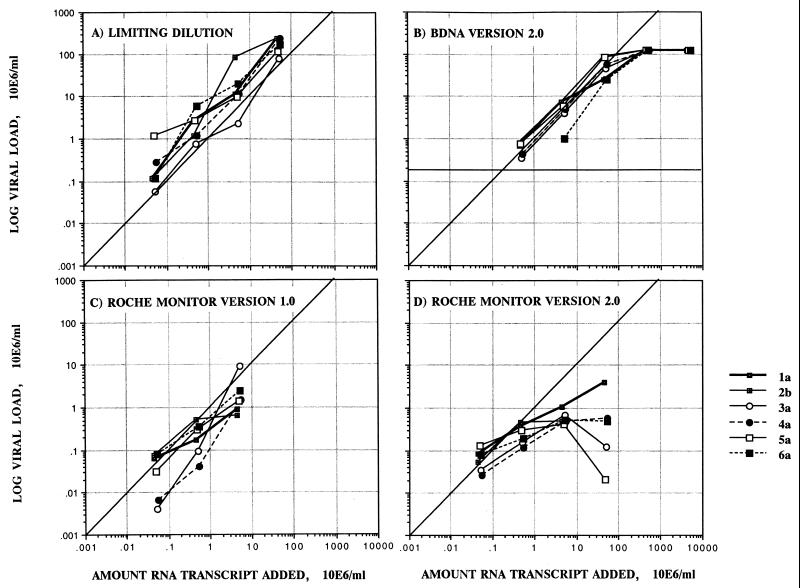
To compare the absolute quantitations of RNA sequences provided by the different assays, four different concentrations of RNA transcripts of HCV genotypes 1 through 6 were analyzed by Roche Monitor test versions 1.0 and 2.0 and the bDNA-2 and limiting-dilution assays (Fig. 2). The results obtained for each sample were deemed valid by the kit instructions, although both Roche Monitor test versions 1.0 and 2.0 produced unexpectedly low quantitation values above concentrations of 106 copies of RNA/ml (Fig. 2) and were unable to accurately quantify samples with greater than 106 HCV RNA copies per ml. In contrast, the bDNA-2 assay was relatively accurate in detecting HCV RNA levels over a range of transcript concentrations of 5 × 105 to 5 × 107 but was unable to detect HCV RNA levels below the stated quantitation limit of 2 × 105 copies/ml. The assay was less sensitive toward genotype 6a transcripts in comparison with the other genotypes. Surprisingly, HCV RNA levels detected with the in-house limiting-dilution assay were consistently higher than the amount of HCV RNA transcript added to the test for all genotypes, with the exception of genotype 3a (within the error margin); these findings contrast with the equivalence of virus loads between this assay and the bDNA-2 assay when plasma samples are tested. This may have occurred because RNA transcripts may be more efficiently reverse transcribed than full-length HCV genomic sequences (see Discussion).
FIG. 2.
Measurement of the absolute efficiencies of quantitative assays by using RNA transcripts of genotypes 1a, 2b, 3a, 4a, 5a, and 6a. The ratio of the amount of HCV RNA detected by each assay to a known amount of transcript added was calculated with increasing amounts of RNA transcript to determine the linear dynamic range of each assay.
Quantitation range of the Roche Monitor version 2.0 assay.
Following the observation of consistent underquantitation of RNA transcripts at concentrations of >106 copies/ml, we investigated whether a similar “saturation” effect influences the quantitation of clinical specimens. Using samples of genotypes 1, 2, and 3, we compared the viral loads detected by Roche Monitor version 2.0 in undiluted samples with those obtained after 1-in-10 dilution prior to testing and where the measured virus loads were multiplied by 10 (Table 3). The values of optical density at 450 nm (OD450) obtained with the Roche Monitor version 2.0 assay of undiluted samples all fell within the operating range of the assay (OD450, ≥0.15 and ≤2.0). However, virus loads in the samples diluted 1 in 10 were consistently higher than those of undiluted samples and were closer to those measured by the bDNA-2 or limiting-dilution assay. (This is consistent with the finding reported in Table 1 for higher virus loads in the type 6 samples.) For example, samples of genotype 1 showed a median viral load ratio of Roche Monitor version 2.0 to limiting dilution of 0.138 when tested undiluted, compared with 0.774 for samples diluted 1 in 10.
TABLE 3.
Comparison of quantitation of HCV RNA in clinical specimens at two dilutions by Roche Monitor version 2.0 with that by other assays
| Genotype (no. of samples) | Median virus load, range (106 RNA copies/ml)
|
||||
|---|---|---|---|---|---|
| bDNA-2 | Limiting dilution | Roche Monitor version 1.0 | Roche Monitor version 2.0
|
||
| Undiluted samples | Samples diluted 1:10 | ||||
| 1 (4) | 6.152 (0.296–18.12) | 4.565 (0.240–7.700) | 2.750 (0.130–4.000) | 0.250 (0.145–0.996) | 1.74 (1.07–4.66) |
| 2 (4) | 11.81 (7.560–13.26) | 6.500 (1.070–24.00) | 2.135 (0.210–4.600) | 0.313 (0.188–0.337) | 1.75 (1.56–3.61) |
| 3 (4) | 9.623 (1.063–18.60) | 4.300 (1.730–17.00) | 0.683 (0.066–1.500) | 0.290 (0.219–0.316) | 1.76 (0.956–2.10) |
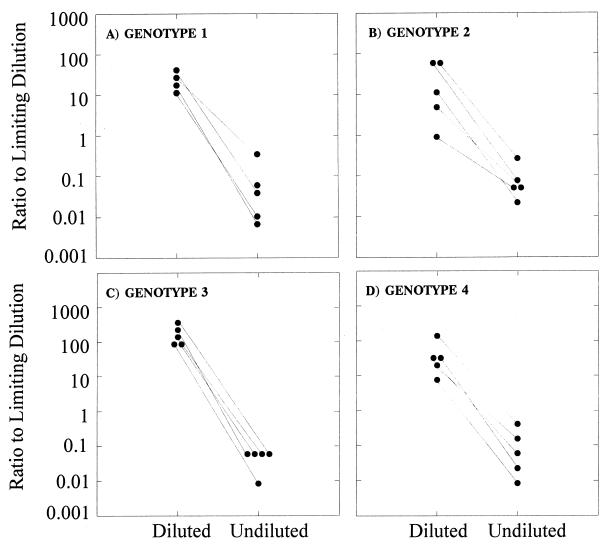
In order to determine if the Roche Monitor assay version 2.0 was operating outside its optimal range, a different set of samples of genotypes 1, 2, 3, and 4 were diluted to an estimated viral load of n × 104 HCV copies per ml according to that calculated by limiting-dilution assay and retested by Roche Monitor version 2.0. The results obtained, once corrected for the dilution factor, were compared to the viral loads obtained from undiluted aliquots of the same samples (Fig. 3). Consistently higher viral loads were obtained when samples were diluted prior to testing for clinical samples with viral titers higher than 105 to 106 HCV copies per ml of plasma.
FIG. 3.
Efficiencies of detection of HCV by the Roche Monitor version 2.0 and limiting-dilution assays for genotypes 1 (A), 2 (B), 3 (C), and 4 (D). Five samples of each genotype were tested undiluted and diluted to n × 104 according to the viral load calculated by limiting dilution to within the dynamic range of the Roche Monitor version 2.0 assay.
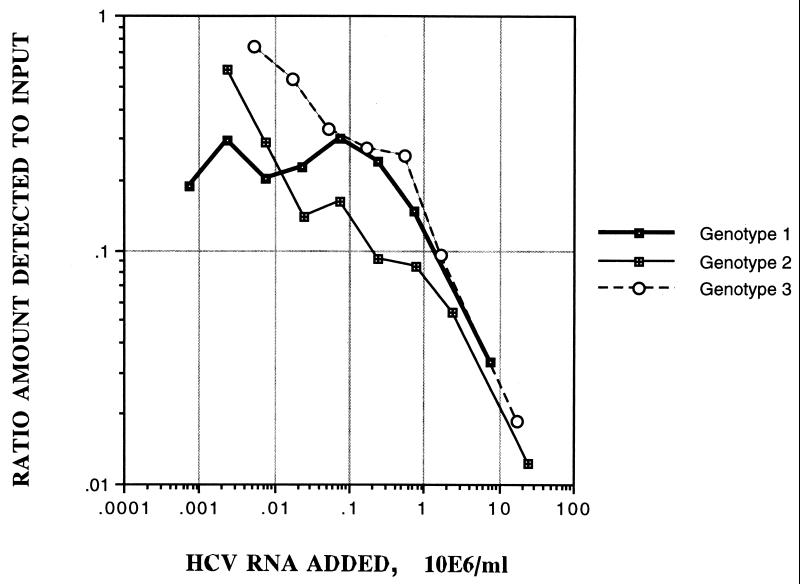
To investigate the difference in quantitation over a wide range of dilutions, three further samples (1 each of genotypes 1 to 3) were serially diluted and retested by the Roche Monitor version 2.0 assay (Fig. 4; Table 4). Plotting of the ratio of the amount of HCV RNA detected by Roche Monitor version 2.0 at various dilutions to the input, determined by limiting dilution, against the amount of HCV RNA added, showed that the measured HCV RNA levels increased in proportion to the dilution. For example, the type 2 sample virus load increased from 2.94 × 105 HCV copies per ml of plasma undiluted to 1.43 × 107 HCV copies per ml at the final dilution (104.0), giving ratios of the amount detected to the amount added of 0.012 and 0.596, respectively. For each of the samples, the Roche Monitor version 2.0 assay measured virus loads comparable to those measured by limiting dilution only at greater-than-100-fold dilutions.
FIG. 4.
Three plasma samples of genotypes 1, 2, and 3 were subjected to serial dilution prior to testing by the Roche Monitor version 2.0 assay. The ratio of the amount detected by the Roche Monitor version 2.0 assay to the amount added, according to limiting dilution (see Table 4), was plotted against the dilution factor.
TABLE 4.
Titration of three plasma samples using Roche Monitor version 2.0
| Input sample (no. of RNA copies/ml)a | Test dilution | Sample
|
Internal control
|
Virus load (no. of RNA copies/ml) | ||
|---|---|---|---|---|---|---|
| Dilution | OD450 | Dilution | OD450 | |||
| Genotype 1 (7.4 × 106) | 1:1 | 1:625 | 0.500 | 1:25 | 0.958 | 2.47 × 105 |
| 1:10 | 1:125 | 0.535 | 1:25 | 0.441 | 1.08 × 106 | |
| 1:31.6 | 1:125 | 0.398 | 1:25 | 0.598 | 1.77 × 106 | |
| 1:100 | 1:125 | 0.191 | 1:25 | 0.550 | 2.24 × 106 | |
| 1:316 | 1:25 | 0.203 | 1:25 | 0.511 | 1.67 × 106 | |
| 1:1,000 | 1:5 | 0.252 | 1:25 | 0.493 | 1.51 × 106 | |
| 1:3,160 | 1:1 | 0.457 | 1:25 | 0.456 | 2.20 × 106 | |
| 1:10,000 | 1:1 | 0.150 | 1:25 | 0.483 | 1.35 × 106 | |
| Genotype 2 (2.4 × 107) | 1:1 | 1:625 | 0.515 | 1:25 | 0.842 | 2.94 × 105 |
| 1:10 | 1:125 | 0.781 | 1:25 | 0.626 | 1.30 × 106 | |
| 1:31.6 | 1:125 | 0.530 | 1:25 | 0.789 | 2.06 × 106 | |
| 1:100 | 1:125 | 0.214 | 1:25 | 0.734 | 2.21 × 106 | |
| 1:31.6 | 1:125 | 0.150 | 1:25 | 0.731 | 3.89 × 106 | |
| 1:1,000 | 1:25 | 0.191 | 1:25 | 0.814 | 3.32 × 106 | |
| 1:3,160 | 1:5 | 0.314 | 1:25 | 0.525 | 6.92 × 106 | |
| 1:10,000 | 1:5 | 0.192 | 1:25 | 0.418 | 1.43 × 107 | |
| Genotype 3 (1.7 × 107) | 1:1 | 1:625 | 0.563 | 1:25 | 0.865 | 3.16 × 105 |
| 1:10 | 1:125 | 0.869 | 1:25 | 0.571 | 1.63 × 106 | |
| 1:31.6 | 1:125 | 0.447 | 1:25 | 0.351 | 4.33 × 106 | |
| 1:100 | 1:125 | 0.270 | 1:25 | 0.506 | 4.68 × 106 | |
| 1:316 | 1:25 | 0.360 | 1:25 | 0.407 | 5.56 × 106 | |
| 1:1,000 | 1:25 | 0.256 | 1:25 | 0.495 | 9.19 × 106 | |
| 1:3,160 | 1:25 | 0.166 | 1:25 | 0.565 | 1.25 × 107 | |
Virus load in test specimens was determined by limiting dilution.
DISCUSSION
This study is an extension of the previous study (8) which used a range of quantitative assays to measure virus load in individuals infected with HCV genotypes 1, 2, and 3. The existence of genotype-specific differences in the sensitivities of these assays, as documented for the original bDNA-1 assay (2, 4, 7) and the Roche Monitor version 1.0 assay (2), affects their quantitative accuracy, making it difficult to properly assess virological and host factors that influence disease severity and response to interferon therapy and interferon-ribavirin combination therapy. For example, underestimation of the virus load in type 2-infected individuals may lead to erroneous attribution of the virus load to their greater response to treatment, even though it has been documented extensively that the median virus loads of type 1- and 2-infected individuals are similar (12, 16, 18) and that (poorly understood) virological differences between the two genotypes contribute additionally and independently of the virus load to the likelihood of an antiviral response.
Among currently available commercial assays for virus load measurement, the Roche Monitor version 1.0 competitive assay shows the greatest differences in sensitivity to genotypes 1 and 2 (times a factor of 9) and 3 (times a factor of 12) (8). In the current study, correction factors of times 53 for type 4 samples and times 15 for type 6 samples might be applied to produce values as calibrated to the limiting-dilution and bDNA-2 assays, as proposed previously for types 2 and 3, although such corrections are unlikely to be accurate for individual samples (8). The new version of the Roche Monitor assay was modified to reduce the differences in sensitivity to non-type 1 genotypes. For low concentrations of RNA transcripts, detection efficiencies were equivalent between genotypes and produced quantitative values corresponding closely to the amount added; these findings stand in contrast in both aspects to those of the previous assays of type 1 to 3 transcripts (8). However, the narrowness of the quantitative range was demonstrated by transcripts and by testing of clinical specimens at different input dilutions (Fig. 3, 4, and 5). Unfortunately for clinical use, measured virus loads from clinical specimens might vary over nearly 2 logs, depending on the test concentration (Fig. 4), with no indication from the test results of whether the virus load was correct or not. For example, undiluted type 2 and 3 samples produced incorrect virus loads of 2.94 × 105 and 3.16 × 105 from OD450 values of 0.515 and 0.563 (internal control values, 0.842 and 0.865) yet they met the manufacturer’s acceptance criteria (Table 4).
Equal sensitivities for all of the genotypes in the Roche Monitor version 2.0 assay appeared to be supported by the equivalence in median virus loads of clinical specimens from individuals infected with genotypes 1 to 4. However, this latter observation is likely to also have been an artifact of the inability to quantify higher virus loads (the median virus load of the type 6a samples tested at a 1:10 dilution was greater than those of the other genotypes). The extremely narrow range of quantitation was the probable reason for the restricted range of virus loads measured in the clinical specimens compared with other assays (Fig. 1) and the poor correlation coefficients with results from other assays. The distribution of virus loads differed principally from the other assays, including the original Roche Monitor assay, in lacking virus load values of greater than 106/ml (Fig. 1).
In contrast to the Roche Monitor version 2.0 assay, the bDNA-2 assay had a range of quantitation that was more appropriate for virus load testing of untreated study subjects, with 85% of the test specimens having virus loads in the linear range of the assay (2 × 105 to 5 × 107 copies/ml, as evaluated by using the RNA transcripts). However, while several studies have documented the equivalence of quantitation of HCV genotypes 1, 2, and 3 (7–9), both transcripts and testing of clinical specimens in the current study revealed a measurable reduction in sensitivity for type 6a sequences compared with the limiting-dilution assay (twofold to eightfold). The extent to which type 6a samples may be underquantified requires further investigation with a larger number of samples.
Finally, and separately from the genotype issue, quantitation of RNA transcripts and HCV genomic RNA in clinical specimens revealed subtle differences in their efficiency of detection by PCR-based and hybridization-based assays. Absolute quantitation with the limiting-dilution assay requires an empirical measurement of the efficiency of reverse transcription in order to convert the measured frequency of cDNA sequences to an RNA concentration. For clinical specimens, we and others have used an efficiency of 5% as a conversion factor for HCV (and human immunodeficiency virus type 1) PCR. In the current study and in our previous study, a 5% efficiency of reverse transcription leads to equivalence of virus loads to those detected by the bDNA-2 assay. However, use of this value for quantitation of RNA transcripts produced consistent overestimation of the RNA concentration (apart from type 3a transcripts). To obtain equivalence to the amount of RNA transcript actually added and to the results of the bDNA-2 assay would necessitate a substantially greater efficiency of reverse transcription (25% for genotypes 1a, 2b, 4a, 5a, and 6a).
Both reverse transcription efficiency and hybridization to the probes used in the bDNA-2 assay may be influenced by the secondary structure of the RNA target. The RNA transcripts used for assay calibration used in this and previous studies (7, 8) were 823 bases in length and may therefore show internal base pairing in this region different from that of full-length sequences. Structural differences that affect the accessibility of internal sequences to hybridization to the probes used in the bDNA-2 assay or to reverse transcription may therefore account for the differences in relative quantitation by PCR- and hybridization-based assays. Potentially, this could prevent the use of subgenomic RNA transcripts for the calibration of assays for the quantitation of HCV genomic RNA in clinical specimens.
In summary, this study describes the equivalent or nearly equivalent quantitation of HCV genotypes by two commercially available assays that operate over quite different ranges of virus loads, which necessarily limits the applications for which the respective assays could be used clinically.
ACKNOWLEDGMENTS
We are very grateful to F. Davidson, Scottish National Blood Transfusion Service; L. Prescott, Department of Medical Microbiology, University of Edinburgh; G. M. Dusheiko, Department of Medicine, Royal Free Hospital, London; and C. K. Lin, Hong Kong Red Cross, Kowloon, for providing samples of genotypes 4 and 6 for quantitative analysis. We are also indebted to J. Detmer and J. Kolberg for providing RNA transcripts of genotypes 1 to 6.
This study was funded in part by a research grant to J.M. and P.S. from Nucleic Acid Systems, Chiron Corporation.
REFERENCES
- 1.Berger A, von Depka Prondzinski M, Doerr H W, Rabenau H, Weber B. Hepatitis C plasma viral load is associated with HCV genotype but not with HIV coinfection. J Med Virol. 1996;48:339–343. doi: 10.1002/(SICI)1096-9071(199604)48:4<339::AID-JMV7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 2.Chan C Y, Lee S D, Hwang S J, Lu R H, Lu C L, Lo K J. Quantitative branched DNA assay and genotyping for hepatitis C virus RNA in Chinese patients with acute and chronic hepatitis C. J Infect Dis. 1995;171:443–446. doi: 10.1093/infdis/171.2.443. [DOI] [PubMed] [Google Scholar]
- 3.Chan S-W, McOmish F, Holmes E C, Dow B, Peutherer J F, Follett E, Yap P L, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 4.Collins M L, Zayati C, Detmer J J, Daly B, Kolberg J A, Cha T A, Irvine B D, Tucker J, Urdea M S. Preparation and characterization of RNA standards for use in quantitative branched DNA hybridization assays. Anal Biochem. 1995;226:120–129. doi: 10.1006/abio.1995.1199. [DOI] [PubMed] [Google Scholar]
- 5.Craxi A, Magrin S, Fabiano C, Linea C, Almasio P. Host and viral features in chromic HCV infection: relevance to interferon responsiveness. Res Virol. 1995;146:273–278. doi: 10.1016/0923-2516(96)80571-1. [DOI] [PubMed] [Google Scholar]
- 6.Daly J A, Ertingshausen G. Direct method for determining inorganic phosphate in serum with the “CentrifiChem.”. Clin Chem. 1972;18:263–265. [PubMed] [Google Scholar]
- 7.Detmer J, Lagier R, Flynn J, Zayati C, Kolberg J, Collins M, Urdea M, Sanchez-Pescador R. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J Clin Microbiol. 1996;34:901–907. doi: 10.1128/jcm.34.4.901-907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins A, Davidson F, Simmonds P. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by Quantiplex HCV RNA assay versions 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J Clin Microbiol. 1997;35:187–192. doi: 10.1128/jcm.35.1.187-192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau J Y N, Simmonds P, Urdea M S. Implications of variations of “conserved” regions of hepatitis C virus genome. Lancet. 1995;346:425–426. doi: 10.1016/s0140-6736(95)92786-7. [DOI] [PubMed] [Google Scholar]
- 10.Lu R H, Hwang S J, Chan C Y, Chang F Y, Lee S D. Quantitative measurement of serum HCV RNA in patients with chronic hepatitis C: comparison between Amplicor HCV Monitor system and branched DNA signal amplification assay. J Clin Lab Anal. 1998;12:121–125. doi: 10.1002/(SICI)1098-2825(1998)12:2<121::AID-JCLA8>3.0.CO;2-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahaney K, Tedeschi V, Maertens G, Dibisceglie A M, Vergalla J, Hoofnagle J H, Sallie R. Genotypic analysis of hepatitis C virus in American patients. Hepatology. 1994;20:1405–1411. doi: 10.1002/hep.1840200605. [DOI] [PubMed] [Google Scholar]
- 12.Martinot Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Lebreton V, Milotova V, Benhamou J P, Erlinger S. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22:1050–1056. [PubMed] [Google Scholar]
- 13.Matsumoto A, Tanaka E, Suzuki T, Ogata H, Kiyosawa K. Viral and host factors that contribute to efficacy of interferon-alpha(2a) therapy in patients with chronic hepatitis C. Dig Dis Sci. 1994;39:1273–1280. doi: 10.1007/BF02093793. [DOI] [PubMed] [Google Scholar]
- 14.Orito E, Mizokami M, Nakano T, Terashima H, Nojiri O, Sakakibara H, Mizuno M, Ogino M, Nakamura M, Matsumoto Y, Miyata K I, Lan J Y N. Serum hepatitis C virus RNA level as a predictor of subsequent response to interferon-alpha therapy in Japanese patients with chronic hepatitis C. J Med Virol. 1994;44:410–414. doi: 10.1002/jmv.1890440418. [DOI] [PubMed] [Google Scholar]
- 15.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Brown A J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith D B, Davidson F, Yap P L, Brown H, Kolberg J, Detmer J, Urdea M, Simmonds P, Mellor J, Neville J, Prescott L, Dow B C, Follett E A C, Skuldamrongpanich T, Tanprasert S, Nuchaprayoon C, Lin C K, Kew M C, Crookes R, Conradie J D, Lin M, Seed C, Deolim G A B, Martins I A, Medgyesi G A, Krusius T, Saeed A A, Teo D, Lee S H. Levels of hepatitis C virus in blood donors infected with different viral genotypes. J Infect Dis. 1996;173:727–730. doi: 10.1093/infdis/173.3.727. [DOI] [PubMed] [Google Scholar]
- 17.Warshaw M M, Tinico I. Optical properties of sixteen dinucleoside phosphates. J Mol Biol. 1966;20:29–38. doi: 10.1016/0022-2836(66)90115-x. [DOI] [PubMed] [Google Scholar]
- 18.Yamada G, Takatani R, Kishi F, Takahashi M, Doi T, Tsuji T, Shin S, Tanno M, Urdea M S, Kolberg J A. Efficacy of interferon alfa therapy in chronic hepatitis C patients depends primarily on hepatitis C virus RNA level. Hepatology. 1995;22:1351–1354. [PubMed] [Google Scholar]
- 19.Zein N N, Rakela J, Persing D H. Genotype-dependent serologic reactivities in patients infected with hepatitis C virus in the United States. Mayo Clin Proc. 1995;70:449–452. doi: 10.4065/70.5.449. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L Q, Simmonds P, Ludlam C A, Leigh Brown A J. Detection, quantitation and sequencing of HIV-1 virus from the plasma of seropositive individuals and from factor VIII concentrates. AIDS. 1991;5:675–681. doi: 10.1097/00002030-199106000-00006. [DOI] [PubMed] [Google Scholar]