Abstract
Excessive mucus secretion is the most prominent feature of pseudomyxoma peritonei (PMP), which often leads to significant increase in abdominal circumference, intractable abdominal pain, progressive intestinal obstruction, abdominal organ adhesions, and cachexia. Excessive mucus secretion is also the main cause of death. Cytoreductive surgery (CRS) combined with hyperthermic intraperitoneal chemotherapy (HIPEC) is the recommended treatment for PMP. However, recurrence is frequently observed even after CRS and HIPEC, presenting similar clinical manifestations. Mucin 2 (MUC2) is the main type of mucin in PMP and plays a key role in the progressive sclerosis of mucus. To comprehensively demonstrate the biosynthetic process and molecular features of MUC2 and to provide new directions for the development of PMP mucolytic strategies, this review systematically summarizes the molecular biology of MUC2, including MUC2 gene structure, transcription, translation, post-translational modification, tertiary structure, and factors regulating mucus viscoelasticity. The results show that MUC2 is a highly glycosylated protein, with glycan accounts for 80% to 90% of the dry weight. The assembly pattern of MUC2 is highly complicated, presenting a bead-like filament. Salt concentration, pH, mucin concentration and trefoil factor family may contribute to the increase in mucus viscoelasticity and sclerosis, which could be used to develop drugs to soften or even dissolve mucus in the future.
Keywords: pseudomyxoma peritonei, MUC2, biological synthesis, post-translational modification, clinicopathologic correlation
Introduction
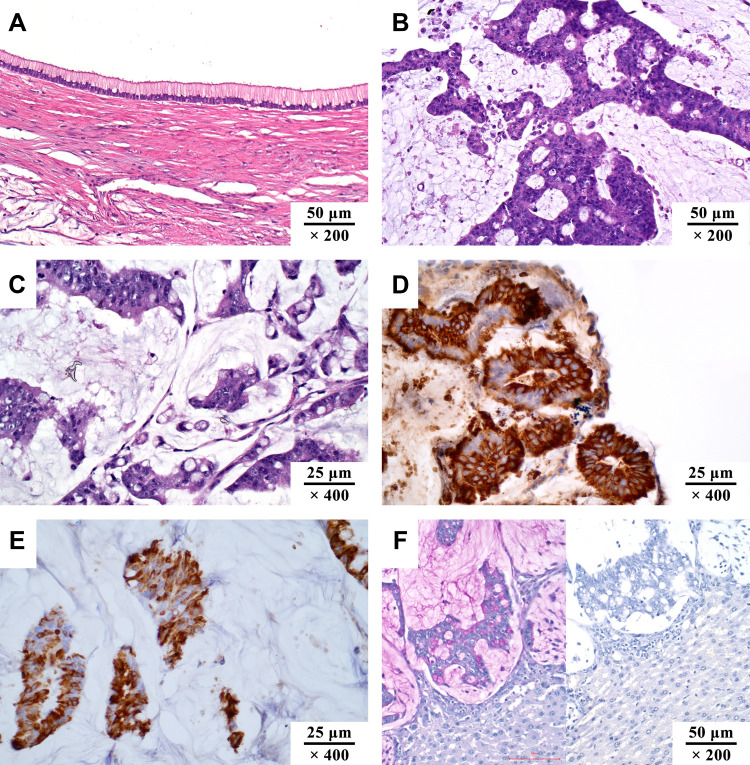
Pseudomyxoma peritonei (PMP) is a clinical syndrome of malignant tumors, which is caused by a primary mucinous tumor that breaks through the primary organ, following spread and implantation in the peritoneum. Typical clinical manifestations include refractory abdominal distension, intractable abdominal pain, progressive intestinal obstruction, and organ adhesion,1–3 which are results of the continuous accumulation of mucus and mucus sclerosis. The mucus sclerosis is the key pathological mechanism in the development of PMP, and the main cause of death.4,5 Among the secreted mucins, mucin 2 (MUC2), MUC5B, and MUC5AC are the major components6 (Figure 1A–F), with the ratio of 2.25:1.54:1.0 in soft mucus, 1:1:1 in intermediate mucus, and 3:2:1 in hard mucus, respectively. Furthermore, MUC2 is the type of mucin with the highest positive rate detected by immunohistochemistry.7 Currently, dissolving mucus intraperitoneally is one of the promising directions to alleviate the accumulation of large amounts of mucus in PMP.8–10 However, the basic knowledge of molecular biology for MUC2 lacks a systematic summary. Therefore, this review aims to summarize the biological synthesis process and structure of MUC2, so as to provide necessary theoretical knowledge for the development of PMP mucolytic treatments.
Figure 1.
The pathological manifestations of pseudomyxoma peritonei. (A) low-grade mucinous carcinoma peritonei (HE, ×200); (B) high-grade mucinous carcinoma peritonei (HE, ×200); (C) high-grade mucinous carcinoma peritonei with signet ring cells (HE, ×400); (D) MUC2 positive (IHC, ×400; MUC2, clone MRQ-18, catalog number ZM-0392); (E) MUC5AC positive (IHC, ×400; MUC5AC, clone MRQ-19, catalog number ZM-0395); (F) PAS staining of mucus, pink area in the left image (HE, ×200). The staining turned negative when incubated with salivary amylase. Unpublished data from the authors’ group.
MUC2 Gene
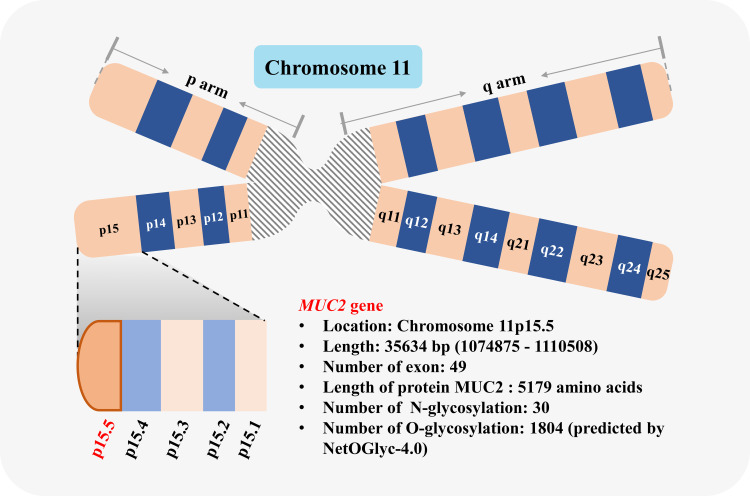
The MUC2 gene is located on chromosome 11p15.5 (Chromosome 11: 1074875-1110508) (Figure 2), with a length of a total of 35,634 bp (NCBI reference sequence: NC_000011.10); the mRNA consists of 49 exons and a total of 16,050 bp (NCBI reference sequence: NM_002457.4). MUC2 is a protein-coding gene and is responsible for encoding MUC2. The MUC2 regulatory sequence is located in the GC-rich region downstream of the open reading frame, or in the coding sequence (CDS), and contains a variety of repetitive and conserved sequences:11,12 ① TATA box, located in −32/-25 bp upstream of the transcription start site; ② CACCC box, located in the 5ʹ-flanking sequence of MUC2 gene. The CACCC box may be a promoter necessary for the transcription of MUC2, located in −88/-80 bp from the transcription start point; ③ Sp1-like targeting sequence, located in the distal region of the 5ʹ-flanking sequence; ④ The fourth promoter is located in −228/-171 bp, which may be related to cell-type specificity.
Figure 2.
Schematic diagram of MUC2 gene. MUC2 gene is located in chromosome 11p15.5.
The factors regulating MUC2 gene expression mainly include three aspects:11,12 ① transcription factor or transcription factor-binding sites, such as short-chain fatty acids, homeobox domain (Cdx), CCCTC-binding factor (CTCF), GATA family, and Sp1 family, and so on; ② DNA methylation and histone modification; ③ p53 may be one of the transcription factors of MUC2 gene.
Biosynthesis of MUC2
Transcription and Translation
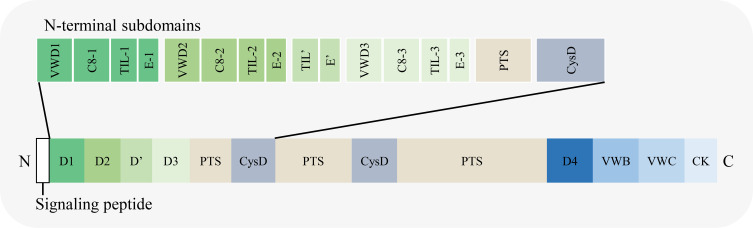
The length of MUC2 protein is 5179 aa (UniProt database, https://www.uniprot.org/; screening keywords, “name: muc2 AND reviewed: yes AND organism: ‘Homo sapiens (Human) [9606]’”, entry identifier: Q02817). The amino acid chain can be divided into three parts13–16 (Figure 3): ① N-terminal, including four domains named D1, D2, D’, and D3. Among them, D1 can be further divided into four subdomains, VWD1, C8-1, TIL1, and E1. D2 is further divided into four subdomains: VWD2, C8-2, TIL2, and E2. D’ is further divided into two subdomains, TIL’ and E’. D3 is further divided into four subdomains: VWD3, C8-3, TIL3, and E3; ② the middle region consists of PTS repeat sequence, CYS domain, PTS repeat sequence, and CYS domain from N-terminal to C-terminal. The PTS repeat sequence consists of 23 amino acid residues (PTTTPITTTTTVTPTPTPTGTQT); ③ C-terminal, including D4, VWB, VWC, and CK domain.
Figure 3.
Schematic diagram of the domains in MUC2 amino acid chain.
Post-Translational Modification
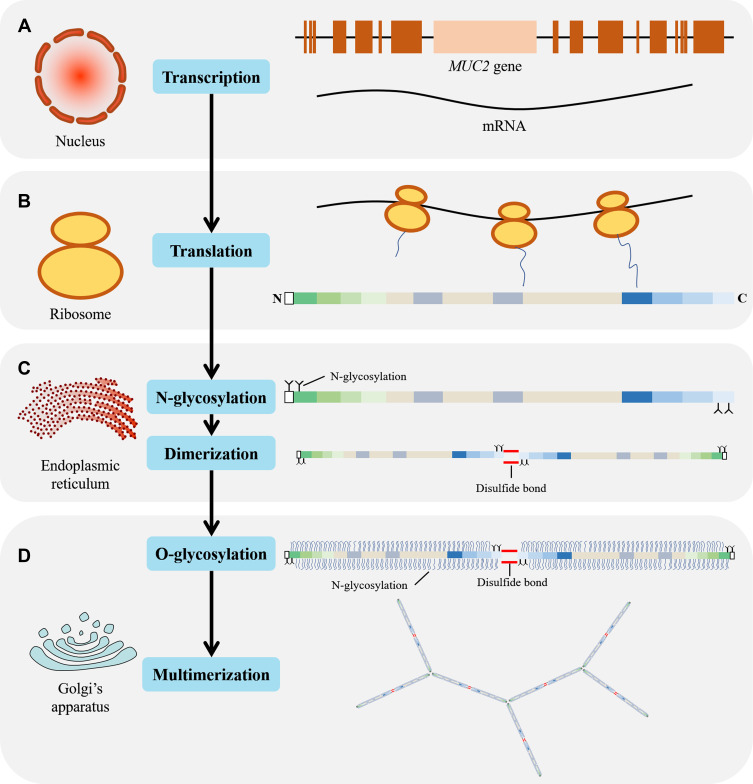
After translation (Figure 4A and B), the polypeptide chain is transported into the endoplasmic reticulum and N-glycosylated, following dimerization through disulfide bond in the CK domain of C-terminal15,17,18 (Figure 4C). The N-glycosylated dimer is then transported to the Golgi apparatus and multimerized through D domains in the N-terminal.15,19 MUC2 is a highly glycosylated protein, with the glycan content accounting for 80% to 90% of the dry weight.20 The types of glycosylation include O-glycosylation, N-glycosylation, and C-mannosylation. Among these, whether C-mannosylation exists in MUC2 is still controversial.21
Figure 4.
Schematic diagram of MUC2 transcription, translation, and post-translational modification. (A) MUC2 mRNA synthesis in the nucleus; (B) In the ribosome, mRNA is translated to form a polypeptide chain with a length of 5179 amino acid residues; (C) In the endoplasmic reticulum, the N-terminus and C-terminus of the peptide chain were N-glycosylated and dimerized through a disulfide bond; (D) In the Golgi apparatus, the dimerized MUC2 are further O-glycosylated and multimerized via disulfide bond.
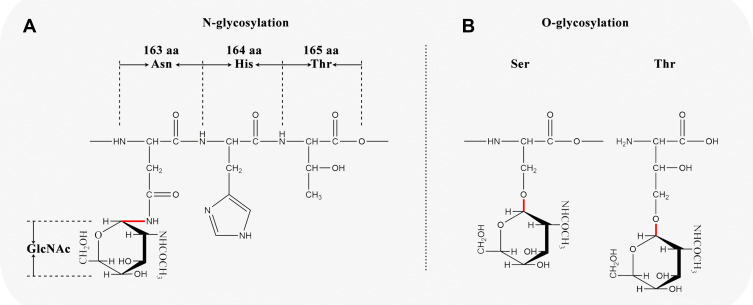
N-Glycosylation
By adding N-acetylglucosamine covalently to the amide nitrogen of Asn in a specific sequence, Asn-X-Ser/Thr, N-glycosylation is performed in the rough endoplasmic reticulum17,22–24 (Figures 4C and 5A). The X amino acid can be any amino acid other than proline. There are 30 N-glycosylation sites in the MUC2 amino acid chain according to UniProt database (entry identifier: Q02817), located in the 163th, 423th, 670th, 770th, 894th, 1139th, 1154th, 1215th, 1230th, 1246th, 1787th, 1820th, 4339th, 4351th, 4362th, 4373th, 4422th, 4438th, 4502th, 4616th, 4627th, 4752th, 4787th, 4881th, 4888th, 4955th, 4970th, 5019th, 5038th, and 5069th amino acids.
Figure 5.
Schematic diagram of N-glycosylation and O-glycosylation sites in MUC2. (A) N-glycosylation; (B) O-glycosylation.
O-Glycosylation
The O-glycosylation is mainly performed in the Golgi apparatus (Figure 4D), with amino acids glycosylated mainly in the PTS repeat sequence17,25,26 (Figure 5B). It is reported that O-glycosylation protects mucosa by preventing the mucin backbone from proteases in the gastrointestinal tract.27 However, the pattern of O-glycosylation is complicated, and no consensus O-glycosylation amino acid sequence motif has been identified.17 To get a general view on the O-glycosylation in MUC2, we use NetOGlyc-4.0 (https://services.healthtech.dtu.dk/service.php?NetOGlyc-4.0) to predict potential O-glycosylation sites. The results showed that there are 1804 amino acid sites that may be O-glycosylated in MUC2, of which Ser accounted for 6.3% (113/1804) and Thr accounted for 93.7% (1691/1804).
Structure
Three-dimensional (3D) structure of MUC2 was searched in the major international protein databases, including PDBe database (https://www.ebi.ac.uk/pdbe/), PDBJ database (https://pdbj.org/), and RCSB PDB database (https://www.rcsb.org/). However, no 3D structure of full length of 5179 amino acids was found, most of which are peptide chains of varying lengths: ① MUC2 D3 domain (6RBF), 856–1240 aa;28 ② MUC2 21–1397 aa (7A5O, 6TM2);13 ③ MUC2 CysD1 domain (6TM6), 1300–1396 aa;13 ④ Tri-O-GalNAc glycosylated mucin sequence (2LHW) based on MUC2 glycoprotein tandem repeat sequence, ACE-PTTTPLK-NH2;29 ⑤ Di-O-GalNAc glycosylated mucin sequence based on the tandem repeat of MUC2 glycoprotein (2LHX, 2LHY, 2LHZ), ACE-PTTTPLK-NH2;29 ⑥ Mono-O-GalNAc glycosylated mucin sequence based on tandem repeat sequence of MUC2 glycoprotein (2LI0, 2LI1, 2LI2), ACE-PTTTPLK-NH2;29 ⑦ Mucin sequence based on MUC2 glycoprotein tandem repeat sequence (2LHV), ACE-PTTTPLK-NH2.29
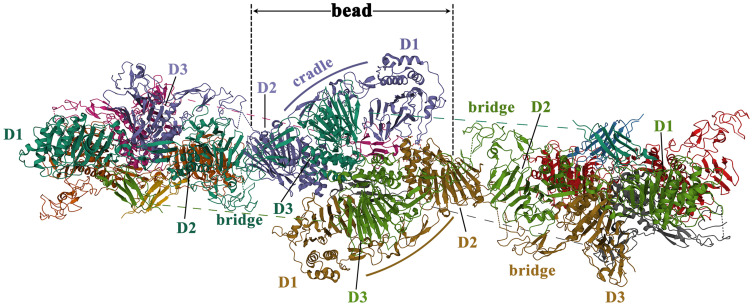
To illustrate the mechanism of MUC2 mucin polymer assembly, Javitt et al13,28 produced and analyzed a peptide chain of about 1400 amino acid residues in the head region of MUC2, namely the D1D2D3CysD1 domain (Figure 3). The assembly of the MUC2 head region presented the following 5 characteristics: ① MUC2 protomers (Figure 7A in reference 29) assemble through disulfide bond (Figure 7B in reference 29) to form bead-like filaments (pH 5.4–6.2, 37°C). CysD1 domain plays a key role in the assembly of MUC2. It was hypothesized that the heads of two CTCK-linked polypeptides linked non-covalently by a reciprocal exchange of D3 assemblies (Figure 7C-7D in reference 29). In the last step of MUC2 production, the non-covalent interactions were released and the bead was disassembled, resulting in expanded polymers (Figure 7E in reference 29); ② Each bead in the filament consists of 8 components (Figure 6): a D1 domain, a D2 domain, and a cradle from the first MUC2; a D3 domain from the second MUC2; another D3 domain from the third MUC2; a D1 domain, a D2 domain, and a cradle from the fourth MUC2; ③ The TIL1-E1 subdomain between the D1 domain and the D2 domain forms a cradle structure (Figure 6). And TIL2-E2-TIL’-E’ between D2 and D3 forms a bridge-like structure (Figure 6). The arc formed by the PTS domain is connected to the D3 domain; ④ Two adjacent dimers are connected by a disulfide bond between the D3 domains (C1088-C1088, C1130-C1130).28 The disulfide bond between two dimers is the result of the formation of bead-like filaments, while not a necessary condition for the formation of filaments. Besides, disulfide bonds may become more sensitive to pH. At pH=5.7, histidine/arginine side-chain clusters (His1128, His1133, and Arg1166) are formed near the Cys1130 disulfide bond at the interface of the D3 in two dimers. The pH-sensitive amino acids on the surface of these polymers are highly conserved among mucins and between mucins and VWF; ⑤ The result of the coarse-grained simulations of the PTS-rich tail and dimerized carboxy termini affixed to the head filament scaffold showed that no topological or steric barriers exist for full-length MUC2 to form similar bead-like filament. However, due to the lack of assembly intermediates of filament-like structures in mucin polymers, it is currently hard to compare the differences between these mimic structures and physiological mucin polymers.
Figure 6.
The schematic diagram of the quaternary structure of the MUC2 head. Reproduced from the RCSB PDB database (https://www.rcsb.org/), PDB ID: 7A5O. Sehnal D, Rose AS, Koča J, Burley SK, Velankar S. Mol*: towards a common library and tools for web molecular graphics. Proceedings of the workshop on molecular graphics and visual analysis of molecular data. Brno, Czech Republic: Eurographics Association; 2018:29–33.30
It should be particularly emphasized that the bead-like filament described by Javitt et al only contains the head region of MUC2, not a multimer formed by the head-to-head and tail-to-tail connection of the full-length MUC2 protein. However, this structure may represent a dense multimeric scaffold formed at similar pH ranges in the Golgi apparatus and secretory vesicles.
Factors Regulating Mucus Viscoelasticity
Progressive mucus sclerosis is one of the important manifestations during the progression of PMP. Mucus sclerosis-related factors may include: ① Cellular factors: higher tumor cellularity or fibrous connective tissue hyperplasia; ② Mucus factors: the increase in mucus viscoelasticity due to the influence of the abdominal environment. The factors regulating the viscoelasticity of mucus mainly include the following four aspects.
1. Salt concentration. Ionic strength may be one of the important factors affecting the viscoelasticity of mucus.20 Under physiological conditions, due to the negative charge of glycoproteins and strong electrostatic repulsion at moderate salt concentrations, the PTS domain of MUC5B forms a free coiled structure.31 Wagner et al32 demonstrated that the viscoelasticity of MUC5AC increased as the salt concentration increased, which may be caused by affecting the salt bridge and reducing the degree of mucin entanglement. The underlying mechanism could be that extracellular HCO3− can better isolate intracellular Ca2+, which is a necessary condition for muco-edema and viscoelasticity.
2. pH. pH is closely related to the viscoelasticity of mucus.20 When pH ≥ 4, conformation of gastric mucin is random coil-like. When the pH < 4, it transforms into an anisotropic, extended conformation. At pH 2–4, mucus changes from a solution-like state to gel-like state as a result of the conformational change driven by Glu and/or Asp residues (pKa≈4), the breaking of salt bridge, and the exposure of CYS domain.20
3. Mucin concentration. When the mucin concentration is high, the formed gel can maintain a weak gel state even under the influence of chaotropic agents, which destroy weak-binding effects (such as hydrogen bonds, electrostatic interactions, hydrophobic chains). When mucin concentration is more than 10 mg/mL, inter-mucin association is found. When the mucin concentration is more than 20–30 mg/mL, it becomes a pure viscous solution. At moderate mucin concentration and low pH, mucin supplementary reversible interactions could increase mucus viscosity.33,34
4. Other factors. Trefoil factor family (TFF) may increase mucus viscosity.35 Besides, it is reported that the elimination of N-glycosylation could downregulate mucin secretion.36
The Pathophysiological Role and Therapeutic Significance of MUC2 in the Progression of PMP
PMP is a malignant tumor syndrome with an indolent course. Clinically, intestinal obstruction and organ adhesion resulting from mucus progressive sclerosis are the main causes of death, with rare cases of organ infiltration-related death. Therefore, mucus progressive sclerosis is the core pathological mechanism for the development of PMP. During the transformation from colloid to semi-solid and finally to solid status, the physical and chemical composition and properties are greatly changed:7 ① In terms of physical properties, the turbidity increases by more than 270%, and the kinematic viscosity increases by more than 40%; ② In terms of chemical composition, the absolute amount of protein, glucose, lipid, sulfhydryl bond, disulfide bond, etc., in mucus is reduced, but the content of sialic acid increases by more than 230%; ③ The ratio of MUC2:MUC5B:MUC5AC is 2.25:1.54:1.0 in soft mucus, 1:1:1 in semi-hard mucus, and 3:2:1 in hard mucus, respectively.
According to O’connell et al,5 MUC2 contributed greatly to the formation of highly gelatinized mucus in PMP, as MUC2 is more extensively glycosylated and is larger in volume compared with MUC5AC under equimolar mass. Tumor cells are wrapped by a large amount of mucus, and the ratio of mucus to tumor cells can be as high as 1000:1, which forms a protective system for tumor cells:6 ① facilitate tumor cells motivity, so that they can get easy access to other parts of the abdominal cavity; ② provide a “protective cover” for tumor cells to prevent the immune system from attacking and the diffusion of chemotherapeutic drugs; ③ provide a tumor microenvironment suitable for tumor growth. Clinically, patients ready for aggressive surgery are often found to have widespread tumor intraperitoneally, hard mucus, and severe organ adhesion, which increases the difficulty of the operation and the incidence of adverse events. Therefore, softening or even dissolving mucus before surgery could reduce the degree of organ adhesion to a certain extent and reduce the risk of surgery.
Conclusions
PMP is a kind of malignant tumor syndrome with milder aggressive behavior, and it often presents an indolent course in clinical practice. Intestinal obstruction and organ adhesions due to progressive mucus scleroses are the main causes of death. MUC2 is an important type of mucin in PMP, with a huge molecular weight and highly glycosylated amino acid side chains. Salt concentration, pH value, mucin concentration, and TFF family, are the possible factors increasing the viscosity of mucin. The current diagnosis and treatment strategy for PMP should still be based on cytoreductive surgery combined with hyperthermic intraperitoneal chemotherapy. The dilution, softening, and dissolving of mucus through the cleavage of O-glycosylation and mucin backbone in a more alkaline environment is one of the promising directions to reduce the risk of surgery and improve the prognosis.
Funding Statement
General Program of the National Natural Science Foundation of China, No. 82073376; Beijing Municipal Administration of Hospitals’ Ascent Plan, No. DFL20180701.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Govaerts K, Lurvink RJ, De Hingh IHJT, et al. Appendiceal tumours and pseudomyxoma peritonei: literature review with PSOGI/EURACAN clinical practice guidelines for diagnosis and treatment. Eur J Surg Oncol. 2021;47:11–35. doi: 10.1016/j.ejso.2020.02.012 [DOI] [PubMed] [Google Scholar]
- 2.Carr NJ, Cecil TD, Mohamed F, et al. A consensus for classification and pathologic reporting of pseudomyxoma peritonei and associated appendiceal neoplasia: the results of the Peritoneal Surface Oncology Group International (PSOGI) modified Delphi process. Am J Surg Pathol. 2016;40:14–26. doi: 10.1097/pas.0000000000000535 [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Xu HB, Peng Z, Cui SZ, Wu W. Chinese expert consensus on cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for pseudomyxoma peritonei. Natl Med J China. 2019;99:1527–1535. [Google Scholar]
- 4.O’Connell JT, Hacker CM, Barsky SH. MUC2 is a molecular marker for pseudomyxoma peritonei. Mod Pathol. 2002;15:958–972. doi: 10.1097/01.Mp.0000026617.52466.9f [DOI] [PubMed] [Google Scholar]
- 5.O’Connell JT, Tomlinson JS, Roberts AA, McGonigle KF, Barsky SH. Pseudomyxoma peritonei is a disease of MUC2-expressing goblet cells. Am J Pathol. 2002;161:551–564. doi: 10.1016/s0002-9440(10)64211-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amini A, Masoumi-Moghaddam S, Ehteda A, Morris DL. Secreted mucins in pseudomyxoma peritonei: pathophysiological significance and potential therapeutic prospects. Orphanet J Rare Dis. 2014;9:71. doi: 10.1186/1750-1172-9-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai K, Akhter J, Mekkawy A, Chua TC, Morris DL. Physical and chemical characteristics of mucin secreted by pseudomyxoma peritonei (PMP). Int J Med Sci. 2017;14:18–28. doi: 10.7150/ijms.16422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amini A, Masoumi-Moghaddam S, Ehteda A, Liauw W, Morris DL. Potentiation of chemotherapeutics by bromelain and N-acetylcysteine: sequential and combination therapy of gastrointestinal cancer cells. Am J Cancer Res. 2016;6:350–369. [PMC free article] [PubMed] [Google Scholar]
- 9.Amini A, Masoumi-Moghaddam S, Ehteda A, Liauw W, Morris DL. Depletion of mucin in mucin-producing human gastrointestinal carcinoma: results from in vitro and in vivo studies with bromelain and N-acetylcysteine. Oncotarget. 2015;6:33329–33344. doi: 10.18632/oncotarget.5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillai K, Akhter J, Chua TC, Morris DL. A formulation for in situ lysis of mucin secreted in pseudomyxoma peritonei. Int J Cancer. 2014;134:478–486. doi: 10.1002/ijc.28380 [DOI] [PubMed] [Google Scholar]
- 11.Yamashita MSA, Melo EO. Mucin 2 (MUC2) promoter characterization: an overview. Cell Tissue Res. 2018;374:455–463. doi: 10.1007/s00441-018-2916-9 [DOI] [PubMed] [Google Scholar]
- 12.Van Seuningen I, Pigny P, Perrais M, Porchet N, Aubert JP. Transcriptional regulation of the 11p15 mucin genes. Towards new biological tools in human therapy, in inflammatory diseases and cancer? Front Biosci. 2001;6:D1216–D1234. [DOI] [PubMed] [Google Scholar]
- 13.Javitt G, Khmelnitsky L, Albert L, et al. Assembly mechanism of mucin and von Willebrand factor polymers. Cell. 2020;183:717–729.e716. doi: 10.1016/j.cell.2020.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson HE, Ambort D, Bäckström M, et al. Intestinal MUC2 mucin supramolecular topology by packing and release resting on D3 domain assembly. J Mol Biol. 2014;426:2567–2579. doi: 10.1016/j.jmb.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Vilar J, Hill RL. The structure and assembly of secreted mucins. J Biol Chem. 1999;274:31751–31754. doi: 10.1074/jbc.274.45.31751 [DOI] [PubMed] [Google Scholar]
- 16.Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639–649. doi: 10.1038/nri.2016.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bansil R, Turner BS. The biology of mucus: composition, synthesis and organization. Adv Drug Deliv Rev. 2018;124:3–15. doi: 10.1016/j.addr.2017.09.023 [DOI] [PubMed] [Google Scholar]
- 18.Asker N, Axelsson MA, Olofsson SO, Hansson GC. Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J Biol Chem. 1998;273:18857–18863. doi: 10.1074/jbc.273.30.18857 [DOI] [PubMed] [Google Scholar]
- 19.Bansil R, Turner BS. Mucin structure, aggregation, physiological functions and biomedical applications. Curr Opin Colloid Interface Sci. 2006;11:164–170. doi: 10.1016/j.cocis.2005.11.001 [DOI] [Google Scholar]
- 20.Demouveaux B, Gouyer V, Gottrand F, Narita T, Desseyn J-L. Gel-forming mucin interactome drives mucus viscoelasticity. Adv Colloid Interface Sci. 2018;252:69–82. doi: 10.1016/j.cis.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 21.Zanetta JP, Pons A, Richet C, et al. Quantitative gas chromatography/mass spectrometry determination of C-mannosylation of tryptophan residues in glycoproteins. Anal Biochem. 2004;329:199–206. doi: 10.1016/j.ab.2004.02.033 [DOI] [PubMed] [Google Scholar]
- 22.Chavan M, Lennarz W. The molecular basis of coupling of translocation and N-glycosylation. Trends Biochem Sci. 2006;31:17–20. doi: 10.1016/j.tibs.2005.11.010 [DOI] [PubMed] [Google Scholar]
- 23.Yan A, Lennarz WJ. Unraveling the mechanism of protein N-glycosylation. J Biol Chem. 2005;280:3121–3124. doi: 10.1074/jbc.R400036200 [DOI] [PubMed] [Google Scholar]
- 24.Yamashita K, Hara-Kuge S, Ohkura T. Intracellular lectins associated with N-linked glycoprotein traffic. Biochim Biophys Acta. 1999;1473:147–160. doi: 10.1016/s0304-4165(99)00175-0 [DOI] [PubMed] [Google Scholar]
- 25.Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim Biophys Acta. 1999;1473:67–95. doi: 10.1016/s0304-4165(99)00170-1 [DOI] [PubMed] [Google Scholar]
- 26.Silverman HS, Sutton-Smith M, McDermott K, et al. The contribution of tandem repeat number to the O-glycosylation of mucins. Glycobiology. 2003;13:265–277. doi: 10.1093/glycob/cwg028 [DOI] [PubMed] [Google Scholar]
- 27.van der Post S, Subramani DB, Bäckström M, et al. Site-specific O-glycosylation on the MUC2 mucin protein inhibits cleavage by the Porphyromonas gingivalis secreted cysteine protease (RgpB). J Biol Chem. 2013;288:14636–14646. doi: 10.1074/jbc.M113.459479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javitt G, Calvo MLG, Albert L, et al. Intestinal gel-forming mucins polymerize by disulfide-mediated dimerization of D3 domains. J Mol Biol. 2019;431:3740–3752. doi: 10.1016/j.jmb.2019.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borgert A, Heimburg-Molinaro J, Song X, et al. Deciphering structural elements of mucin glycoprotein recognition. ACS Chem Biol. 2012;7:1031–1039. doi: 10.1021/cb300076s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sehnal D, Rose AS, Koča J, Burley SK, Velankar S. Mol*: towards a common library and tools for web molecular graphics. Proceedings of the workshop on molecular graphics and visual analysis of molecular data. Brno, Czech Republic: Eurographics Association; 2018:29–33. [Google Scholar]
- 31.Davies HS, Singh P, Deckert-Gaudig T, et al. Secondary structure and glycosylation of mucus glycoproteins by Raman spectroscopies. Anal Chem. 2016;88:11609–11615. doi: 10.1021/acs.analchem.6b03095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner CE, Turner BS, Rubinstein M, McKinley GH, Ribbeck K. A rheological study of the association and dynamics of MUC5AC gels. Biomacromolecules. 2017;18:3654–3664. doi: 10.1021/acs.biomac.7b00809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao X, Bansil R, Bhaskar KR, et al. pH-dependent conformational change of gastric mucin leads to sol-gel transition. Biophys J. 1999;76:1250–1258. doi: 10.1016/s0006-3495(99)77288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georgiades P, Pudney PD, Thornton DJ, Waigh TA. Particle tracking microrheology of purified gastrointestinal mucins. Biopolymers. 2014;101:366–377. doi: 10.1002/bip.22372 [DOI] [PubMed] [Google Scholar]
- 35.Thim L, Madsen F, Poulsen SS. Effect of trefoil factors on the viscoelastic properties of mucus gels. Eur J Clin Invest. 2002;32:519–527. doi: 10.1046/j.1365-2362.2002.01014.x [DOI] [PubMed] [Google Scholar]
- 36.Bell SL, Khatri IA, Xu G, Forstner JF. Evidence that a peptide corresponding to the rat Muc2 C-terminus undergoes disulphide-mediated dimerization. Eur J Biochem. 1998;253:123–131. doi: 10.1046/j.1432-1327.1998.2530123.x [DOI] [PubMed] [Google Scholar]