Abstract
Background & Aims:
An efficient cell culture system for hepatitis B virus (HBV) is indispensable for research on viral characteristics and anti-viral reagents. Currently, for the HBV infection assay in cell culture, viruses derived from HBV genome-integrated cell lines of HepG2.2.15 or HepAD-38 are commonly used. However, these viruses are not suitable for the evaluation of polymorphism dependent-viral characteristics or resistant mutations against anti-viral reagents. HBV obtained by the transient transfection of the ordinary HBV molecular clone has limited infection efficiencies in cell culture.
Approach & Results:
We found that an 11 amino acid deletion (d11) in the preS1 region enhances the infectivity of cell culture-generated HBV (HBVcc) to sodium taurocholate co-transporting polypeptide-transduced HepG2 (HepG2/NTCP) cells. Infection of HBVcc derived from a d11-introduced genotype C strain (GTC-d11) was approximately 10-fold more efficient than infection of wild-type GTC (GTC-wt), and the number of infected cells was comparable between GTC-d11- and HepG2.2.15-derived viruses when inoculated with the same genome equivalents. A time-dependent increase in pre-genomic RNA and efficient synthesis of covalently closed circular DNA were detected after infection with the GTC-d11 virus. The involvement of d11 in the L-HBs protein in the enhanced infectivity was confirmed by an HBV reporter virus and hepatitis D virus infection system. The binding step of the GTC-d11 virus onto the cell surface was responsible for this efficient infection.
Conclusions:
This system provides a powerful tool for studying the infection and propagation of HBV in cell culture, and also for developing the anti-viral strategy against HBV infection.
Keywords: HBV, genotype, cell culture, NTCP, preS1
INTRODUCTORY STATEMENT
Hepatitis B virus (HBV) infection is a major cause of chronic liver disease, including cirrhosis and hepatocellular carcinoma (1). Although effective vaccines to HBV are available in many countries, the global prevalence of infection of this virus is estimated to be over 290 million (2). HBV has a partially double-stranded 3.2 kb DNA genome and replicates using a reverse transcriptase enzyme that lacks proofreading ability. Therefore, HBV has diverse genetic variability and is classified into at least 8 genotypes: A to H. Some differences in clinical features and outcomes have been reported among these genotypes (3–8). HBV comprises four open reading frames (ORFs) coding for hepatitis B surface antigen (HBsAg), hepatitis B e (HBe) antigen/hepatitis B core (HBc) antigen (HBe/c), hepatitis B polymerase (HBp), and X protein (HBx). HBsAg consists of 3 species: small (S-), middle (M-) and large (L-) HBs proteins. The S-HBs protein contains 266 amino acids, and the M-HBs and L-HBs proteins contain additional regions of preS2 and preS1+preS2, respectively. The S-HBs and L-HBs proteins exist in non-glycosylated and mono-glycosylated forms due to an N-linked glycosylation site at the position of amino acid 146 in the S-HBs region (9). These HBs proteins are known to form the envelope of infectious HBV particles (S-, M- and L-HBs) and empty subviral particles (mainly S- and M-HBs) and play a pivotal role in the infection of this virus. The eradication of chronic HBV infection by the currently available treatments is not expected because HBV covalently closed circular DNA (cccDNA) in hepatocytes cannot be eliminated by these treatments (10). To explore new classes of anti-HBV reagents, an efficient infection and replication system of HBV in cell culture is indispensable. Sodium taurocholate co-transporting polypeptide (NTCP) was discovered as an HBV receptor, and NTCP-transduced HepG2 (HepG2/NTCP) or HuH-7 cells enabled the observation of HBV infection and replication in cell culture (11). In this system, viruses derived from HBV genotype D genome-integrated cell lines of HepG2.2.15 or HepAD-38 are commonly used as inocula (12, 13). However, such viruses are not suitable to investigate effects of strain-specific viral characteristics or resistance-associated polymorphisms on anti-viral reagents. The viruses obtained by the transient transfection of ordinary HBV molecular clones have limited infection efficiencies in cell culture.
In this study, to establish an efficient HBV infection system with cell culture-generated HBV (HBVcc), we explored the regulatory region for infection efficiency in the HBV genome and found that introduction of an 11 amino acid deletion (d11) in the preS1 region confers efficient infectivity.
EXPERIMENTAL PROCEDURES
HBV molecular clones and production of HBVcc
A replication-competent HBV clone with a 1.38-fold genome length was constructed by using the genome sequence of HBV (14). In this study, we used HBV clones of multiple genotypes, as follows: 2 genotype C clones (GTC and GTC2), one genotype A clone (GTA), one genotype B clone (GTB), and one genotype D clone (GTD). The accession numbers are LC488828 (GTA), AB246341 (GTB) (15), AB246345 (GTC) (15), AB246344 (GTC2) (15) and LC488706 (GTD). The usage of the HBV genome sequence from a patient serum sample in this study was approved by the Ethics Committees (the approval number is 377 in National Institute of Infectious Diseases). For the transfection of plasmids of HBV molecular clones, Lipofectamine 3000 Reagent (Thermo Fisher Scientific, Waltham, MA) was used. The HBV DNA titer was measured by real-time PCR with primers and a probe set targeting the HBs region after treatment with DNase (RQ1 RNase-Free DNase, Promega, Madison, WI) (16). The production of HBsAg and HBcrAg was measured by a chemiluminescent enzyme immunoassay analyzer with commercial assay kits (Lumipulse G1200, Fujirebio, Tokyo, Japan) (17–19).
Detailed descriptions of further material and methods are provided in the Supplemental Materials.
RESULTS
Infection of HBVcc into HepG2/NTCP cells
To compare infection efficiencies, we constructed replication-competent HBV molecular clones with a 1.38-fold genome length. We prepared HBV molecular clones of genotype C (GTC) and genotype D (GTD) and generated HBVcc by transient transfection into HepG2 cells. The generated viruses were inoculated into HepG2/NTCP-C4 cells at 500 genome equivalents (GEq) per cell, and the infection efficiencies were compared with HepG2.2.15-derived HBV (genotype D). The inoculation of the wild-type GTC virus (GTC-wt) resulted in several HBc-positive cells at 12 days after inoculation, and the infection efficiency was substantially lower than that of HepG2.2.15-derived HBV (Figure 1A). However, the inoculation of the GTD virus resulted in more HBc-positive cells than the inoculation of the GTC-wt virus. From these observations, we reasoned that the infection efficiency of the GTD virus is higher than that of the GTC virus. We explored GTD-specific features in the HBV genome by analysing an alignment of consensus sequences of GTD and other genotype strains in Hepatitis Virus Database (http://s2as02.genes.nig.ac.jp/db/) and identified an 11 amino acid deletion (d11) in the preS1 region (Figure 1B). We introduced d11 to the GTC molecular clone (GTC-d11) and generated the virus by transient transfection into HepG2 cells. The inoculation of the virus derived from GTC-d11 resulted in a much higher number of infected cells than that of the GTC-wt virus, and the number of infected cells was comparable to that of the infection of HepG2.2.15-derived HBV (Figure 1A). The enhanced infectivity of GTC-d11 was confirmed in PXB cells isolated from urokinase-type plasminogen activator/severe combined immunodeficient (uPA/SCID) mice inoculated with primary human hepatocytes (PhoenixBio, Hiroshima, Japan) (Supplementary Figure S1). The amount of pre-genomic RNA (pgRNA) in the GTC-d11 virus-infected cells exhibited a time-dependent increase similar to that of the infection of HepG2.2.15-derived HBV, whereas this increase was not observed with the infection of the GTC-wt virus (Figure 1C).
Figure 1. Production and infection of HBVcc.
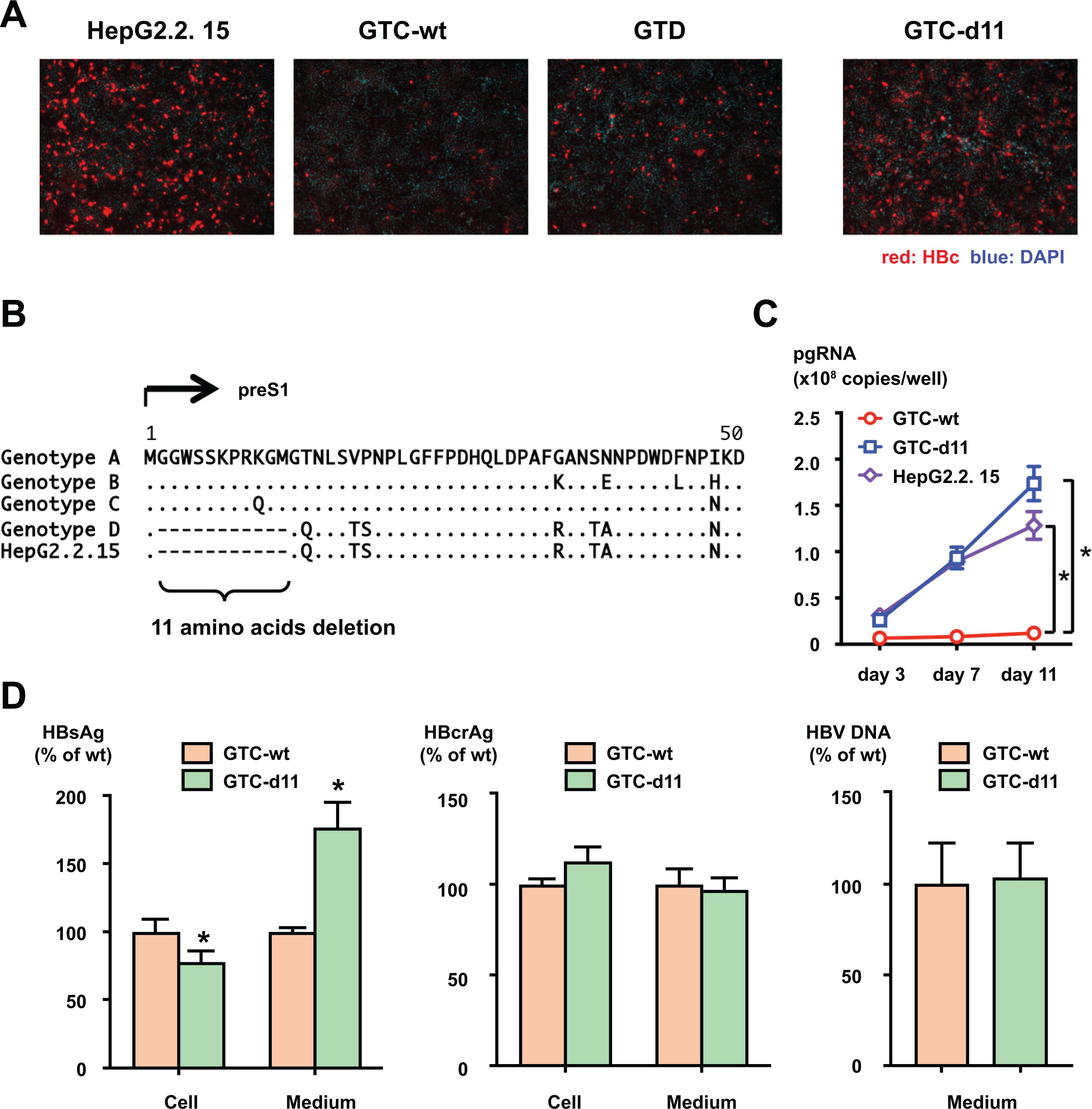
(A) HBVcc of GTC-wt, GTD, and GTC-d11 were infected into HepG2/NTCP-C4 cells at 500 GEq/cell. HBV-positive cells were visualized by staining with an anti-HBc antibody, and nuclei were visualized by DAPI. (B) Alignment of consensus amino acid sequences in the preS1 region of genotype A, B, C, and D clones. The amino acid sequence of HepG2.2.15 is also included. The deletion of 11 amino acids is indicated by the bracket. (C) Production of pgRNA in HBV-infected cells. Total RNA was extracted from GTC-wt-, GTC-d11-, and HepG2.2.15-derived virus-infected cells, and pgRNA was measured by real-time PCR. (D) Production of HBV proteins and HBV DNA in GTC-wt- and GTC-d11-transfected cells. HBsAg and HBcrAg titers in HBV molecular clone-transfected cells and in culture medium were measured. The HBV DNA titer in culture medium was also quantified by real-time PCR after treatment with DNase. The data are indicated as the mean ± standard deviation of the triplicate assay. Asterisk indicates a significant difference (p < 0.05).
To compare the production of HBV proteins in GTC-wt- and GTC-d11-transfected cells, HBsAg and hepatitis B core-related antigen (HBcrAg) were measured in culture media and transfected cells. The amounts of HBcrAg were comparable both in culture media and in transfected cells of GTC-wt- and GTC-d11. However, the amount of HBsAg of GTC-d11-transfected was higher in the culture medium and slightly lower in transfected cells compared with the GTC-wt-transfected, suggesting the efficient secretion of HBsAg including L-HBs such as filaments of GTC-d11 (Figure 1D). HBV DNA levels in culture media were comparable between these clones.
Iodixanol density gradient analysis of HBVcc
To assess features of HBVcc, GTC-wt and GTC-d11 viruses generated in culture media were analyzed by an iodixanol density gradient after purification with a heparin column and adjustment by the amount of HBsAg. In the density gradient of GTC-wt, the titers of HBsAg and HBV DNA peaked at fraction 13, and the titer of HBcrAg peaked at fraction 16 that containing the unenveloped naked-capsids (Figure 2A, upper panel). In the gradient of GTC-d11, these peaks were observed at the same fraction as those in the GTC-wt virus gradient (Figure 2B, upper panel). The infectivity also peaked at the same fraction (fraction 14) in both gradients. When inoculated with the same amount of these fractions, the number of infected cells was substantially higher after inoculation of the GTC-d11 virus than that of the GTC-wt virus, although the HBV DNA titer of the peak fraction of infectivity was higher for the GTC-wt virus than that for the GTC-d11 virus (Figures 2A and 2B, lower panels). This difference was more pronounced when cells were inoculated with the same HBV DNA amount of the viruses (500 GEq/cell), and these infections were inhibited by administration of anti-HBs human immune globulin (HBIG) (Figure 2C). We compared the amounts of HBsAg, HBcrAg, and HBV DNA in the peak fraction of infectivity and found that the contents of HBsAg and HBcrAg were at the same levels when adjusted by HBV DNA titers (Table 1).
Figure 2. Iodixanol density gradient analysis of HBVcc.
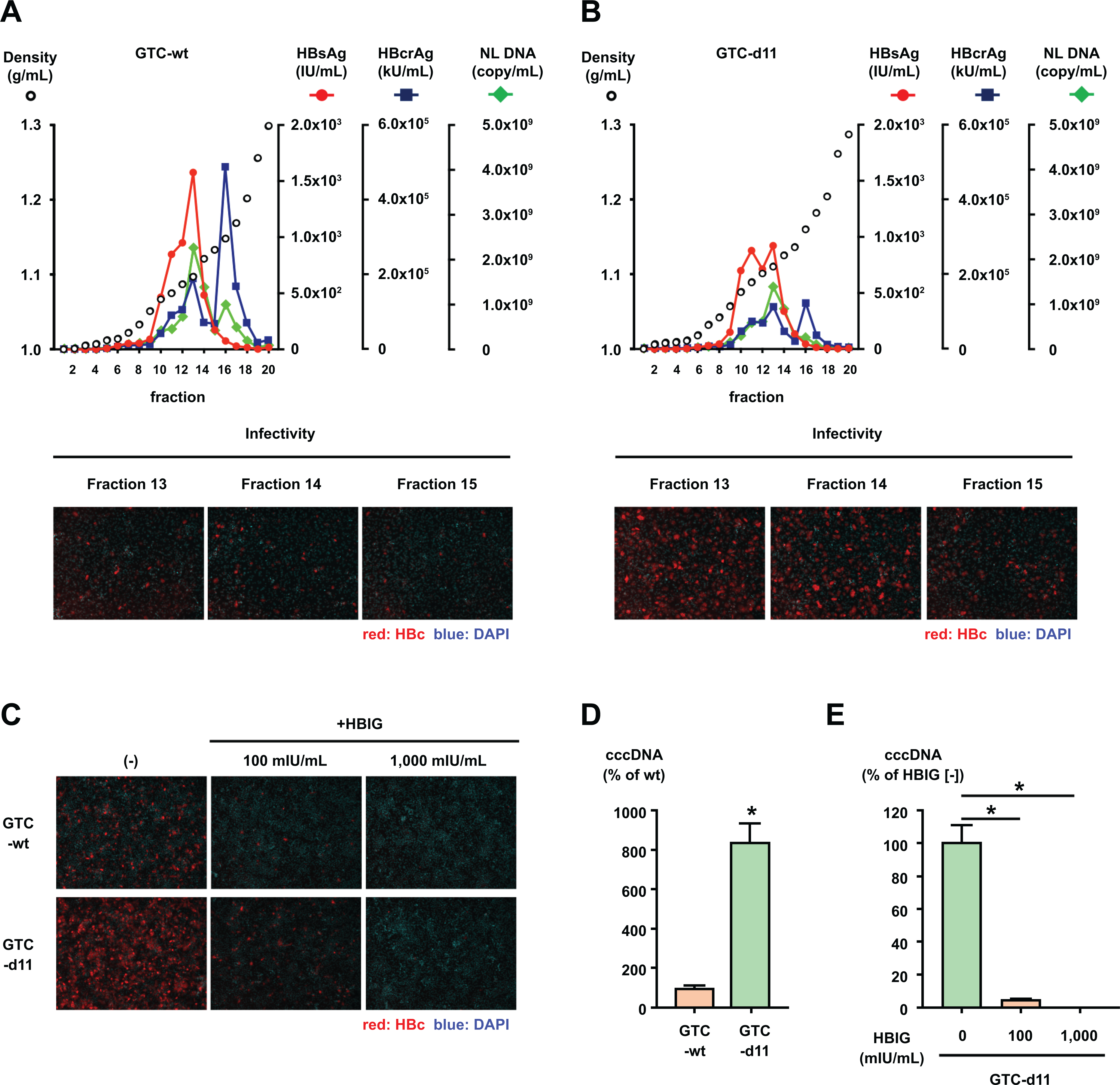
(A, B) HBVcc of GTC-wt and GTC-d11 were purified by a heparin column and analyzed by an iodixanol density gradient. The titers of HBsAg and HBcrAg were measured in each fraction, and the HBV DNA was quantified by real-time PCR after treatment with DNase. The infectivity of HBVcc in the indicated fractions was also evaluated by inoculation of the same volume of fractions onto HepG2/NTCP-C4 cells. (C) The same amount of HBVcc (500 GEq/mL) in fraction 14 of each gradient of GTC-wt and GTC-d11 was mixed with HBIG at the indicated concentrations and inoculated onto HepG2/NTCP-C4 cells. HBV-positive cells were visualized by staining with an anti-HBc antibody, and nuclei were visualized by DAPI. (D) Cell culture-generated viruses of GTC-wt and GTC-d11 were inoculated onto HepG2/NTCP-C4 cells. Synthesized cccDNA in infected cells was extracted by the Hirt extraction method and quantified by cccDNA-specific real-time PCR. (E) The cell culture-generated GTC-d11 virus was treated with HBIG at the indicated concentrations and inoculated onto HepG2/NTCP-C4 cells. The cccDNA in HBV-infected cells was measured by cccDNA-specific real-time PCR after the Hirt extraction method. The data are indicated as the mean ± standard deviation of the triplicate assay. Asterisk indicates a significant difference (p < 0.05).
TABLE 1.
Profiles of HBVcc in Fractions of an Iodixanol Density Gradient
| GTC-wt |
GTC-d11 |
|||
|---|---|---|---|---|
| Fraction 13 | Fraction 14 | Fraction 13 | Fraction 14 | |
| Density (g/mL) | 1.097 | 1.121 | 1.110 | 1.125 |
| HBV DNA (copies/mL) | 2.26 × 109 | 1.38 × 109 | 1.38 × 109 | 8.98 × 108 |
| HBsAg (IU/mL) | 1.58 × 103 | 4.85 × 102 | 9.33 × 102 | 3.38 × 102 |
| HBcrAg (kU/mL) | 1.90 × 105 | 7.17 × 104 | 1.12 × 105 | 4.55 × 104 |
| HBsAg/HBV DNA (IU/copy) | 6.98 × 10−7 | 3.52 × 10−7 | 6.68 × 10−7 | 3 80 × 10−7 |
| HBcrAg/HBV DNA (kU/copy) | 8.40 × 10−5 | 5.22 × 10−5 | 8.08 × 10−5 | 5.12 × 10−5 |
We also evaluated the synthesis of cccDNA in cells infected with these viruses. The amount of synthesized cccDNA in the GTC-d11 virus-infected cells was approximately 8.5-fold higher than that in the GTC-wt virus-infected cells (Figure 2D). The synthesis of cccDNA of the GTC-d11 virus was also inhibited by administration of HBIG (Figure 2E). We confirmed the much amount of core-associated HBV DNA in the GTC-d11 virus-infected cells in comparison with the GTC-wt virus-infected cells by Southern blotting (Supplementary Figure S2).
Effects of d11 on HBV/NL infection
The effects of d11 on infectivity were also assessed in the recombinant HBV reporter virus system of genotype C: HBV/NL (20, 21). This system uses 2 plasmids: an encapsidation competent-plasmid (HBV-NL) encoding NanoLuc luciferase (NL) but no HBe/c and an encapsidation incompetent-plasmid (HBV-dE) encoding all HBV proteins. We modified both plasmids by introducing d11 into the wild-type plasmids (HBV-NL-wt and HBV-dE-wt) and named them HBV-NL-d11 and HBV-dE-d11, respectively (Figure 3A). We generated HBV/NL by transfection of two pairs of plasmids: HBV-NL-wt + HBV-dE-wt or HBV-NL-d11 + HBV-dE-d11. The NL activity after infection of the virus generated by HBV-NL-d11 + HBV-dE-d11 was approximately 8.3-fold higher than that of the virus generated by HBV-NL-wt + HBV-dE-wt (Figure 3B).
Figure 3. d11-associated enhancement of infectivity in an HBV/NL system.
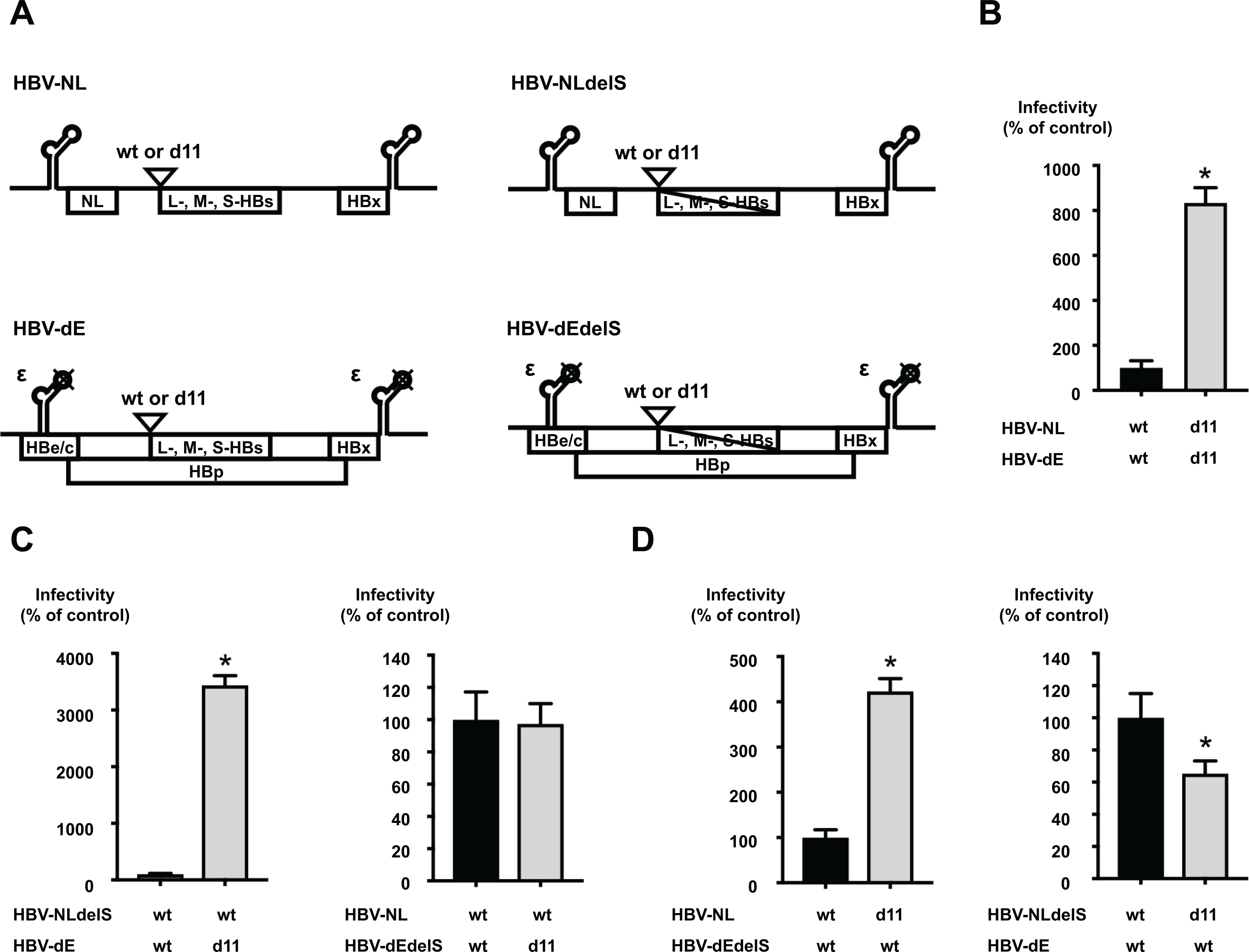
(A) The structure of plasmids for HBV/NL used in this study. The constructs of the encapsidation competent-reporter plasmid (HBV-NL-wt) and the encapsidation incompetent-plasmid (HBV-dE-wt) were prepared. The d11 was introduced to these plasmids and designated HBV-NL-d11 and HBV-dE-d11, respectively. The substitutions of the start codons of L-, M-, and S-HBs was also introduced to these plasmids and generated HBV-NLdelS-wt, HBV-NLdelS-d11, HBV-dEdelS-wt, and HBV-dEdelS-d11. NL; NanoLuc. (B) Infection of reporter HBV generated by transfection of HBV/NL plasmids with or without d11. The generated HBV/NL was quantified after DNase treatment and inoculated onto HepG2/NTCP-18C cells at 3 GEq/cell. After 8 days of incubation, infected cells were lysed, and the NL activities were measured. Infectivity of the virus with d11 is represented as the percentage of infectivity of the virus without d11. (C) Assessment of the effects of d11 in the L-HBs and HBp proteins. The HBV reporter generated by the transfection of the indicated plasmids was inoculated at 50 GEq/cell. The infectivity of the virus with d11 is represented as the percentage of infectivity of the virus without d11. (D) Assessment of the effects of d11 in the L-HBs protein and packaged genome length. The HBV reporter generated by the transfection of the indicated plasmids was inoculated at 50 GEq/cell. The infectivity of the virus with d11 is represented as the percentage of infectivity of the virus without d11. The data are indicated as the mean ± standard deviation of the quintuplicate assay. Asterisk indicates a significant difference (p < 0.05).
The introduction of d11 in the preS1 region affects not only the length of the L-HBs protein but also the length of the HBp protein or the packaged genome. To identify the responsible factor for the d11-associated enhancement of infectivity, we constructed additional plasmids by substituting the all start codons of the L-, M-, and S-HBs proteins in the plasmids of HBV-NL-wt and HBV-dE-wt to delete the expression of all HBs species and named them HBV-NLdelS-wt and HBV-dEdelS-wt, respectively (Figure 3A). The modification of delS was also introduced to the plasmids of HBV-NL-d11 and HBV-dE-d11 and named HBV-NLdelS-d11 and HBV-dEdelS-d11. The infectivity of viruses generated by transfection of two combinations, HBV-NLdelS-wt + HBV-dE-wt and HBV-NLdelS-wt + HBV-dE-d11, was compared. In the combination of HBV-NLdelS-wt + HBV-dE-d11, d11 in the HBV-dE plasmid shortened the length of both L-HBs and HBp, but the length of the packaged genome in the HBV-NLdelS-wt plasmid was intact. The NL activity after infection of the HBV-NLdelS-wt + HBV-dE-d11 virus was approximately 34-fold higher than that after infection of the HBV-NLdelS-wt + HBV-dE-wt virus (Figure 3C, left panel). To see the effects of d11 on the HBp protein only, we also compared the viruses of HBV-NL-wt + HBV-dEdelS-wt and HBV-NL-wt + HBV-dEdelS-d11 and found that the NL activities after infection of these viruses were comparable (Figure 3C, right panel). These data indicate that the L-HBs protein with d11 is responsible for the enhanced infectivity and that the HBp protein with d11 is not associated.
Next, we compared the infectivity of viruses generated by transfection of two combinations: HBV-NL-wt + HBV-dEdelS-wt or HBV-NL-d11 + HBV-dEdelS-wt. In the combination of HBV-NL-d11 + HBV-dEdelS-wt, d11 in the HBV-NL-d11 plasmid shortened the length of the L-HBs protein and the packaged genome, but the length of the HBp protein in the HBV-dEdelS-wt plasmid was intact. The NL activity after infection of the HBV-NL-d11 + HBV-dEdelS-wt virus was approximately 4.2-fold higher than that after infection of the HBV-NL-wt + HBV-dEdelS-wt virus (Figure 3D, left panel). To see the effects of d11 on the genome length only, we also compared the viruses of HBV-NLdelS-wt + HBV-dE-wt and HBV-NLdelS-d11 + HBV-dE-wt and found that the NL activity after the infection of the HBV-NLdelS-d11 + HBV-dE-wt virus was slightly lower than that after infection of the HBV-NLdelS-wt + HBV-dE-wt virus (Figure 3D, right panel). These data suggest that the shortened genome due to d11 slightly reduces infectivity but d11 in the L-HBs protein enhances infectivity. To delineate the responsible residue(s) among 11 amino acids in the preS1 region, we constructed the five deletion mutants, HBV/NL-d2, HBV/NL-d4, HBV/NL-d6, HBV/NL-d8, and HBV/NL-d10, (2aa, 4aa, 6aa, 8aa, and 10aa deletion after the first Methionine of the preS1 region, respectively), and compared the infectivity of HBV/NL generated by transfection with HBV-dEdelS-wt into HepG2 cells. Among these mutant viruses, HBV/NL-d2, HBV/NL-d4, and HBV/NL-d6 exhibited similar or lower infectivity to HBV/NL-wt. HBV/NL-d8 showed the slightly enhanced infectivity, and HBV/NL-d10 had the moderately enhanced infectivity but it was not reached to the level of HBV/NL-d11, indicating that d11 is essential for the fully enhanced infectivity (Supplementary Figure S3).
Iodixanol density gradient analysis of HBV/NL
The viruses generated by transfection of HBV-NL-wt + HBV-dE-wt (HBV/NL-wt) and HBV-NL-d11 + HBV-dE-d11 (HBV/NL-d11) were also analyzed by an iodixanol density gradient after purification with a heparin column. In the density gradient of HBV/NL-wt, as observed in the GTC-wt virus (Figure 2A), HBsAg and HBV DNA peaked at fraction 13, HBcrAg peaked at fraction 16, and the infectivity peaked at fraction 14 (Figure 4A). In the density gradient of HBV/NL-d11, all these parameters peaked at the same fraction as that of HBV/NL-wt (Figure 4B). After inoculation of these peak fractions, the NL activity of the HBV/NL-d11 virus inoculated was 8.8-fold higher than that of the HBV/NL-wt virus inoculated, although the titer of NL DNA was higher in the HBV/NL-wt virus than in the HBV/NL-d11 virus (2.41 × 108 copies/mL vs. 1.13 × 108 copies/mL, respectively). The difference in the infectivity of these viruses was more pronounced when inoculated with the same NL DNA amount of the viruses, and the NL activity was approximately 14.5-fold higher in the HBV/NL-d11 virus-infected cells than in the HBV/NL-wt virus-infected cells (Figure 4C, upper panel). These infections were similarly inhibited by treatment with HBIG or the preS1 peptide (the equivalent of Myrcludex B) in a dose-dependent manner (Figure 4C, lower panel). The effective HBIG concentrations required to inhibit 50% (EC50) of the HBV/NL-wt and HBV/NL-d11 viruses were comparable at 0.122 mIU/mL (95%CI = 0.0750 to 0.199 mIU/mL) and 0.199 mIU/mL (95%CI = 0.104 to 0.382 mIU/mL), respectively. The EC50 values of the HBV/NL-wt and HBV/NL-d11 viruses for the preS1 peptide were also comparable at 38.9 nM (95%CI = 12.0 to 134 nM) and 45.3 nM (95%CI = 17.3 to 121 nM), respectively.
Figure 4. Iodixanol density gradient analysis of HBV/NL with or without d11.
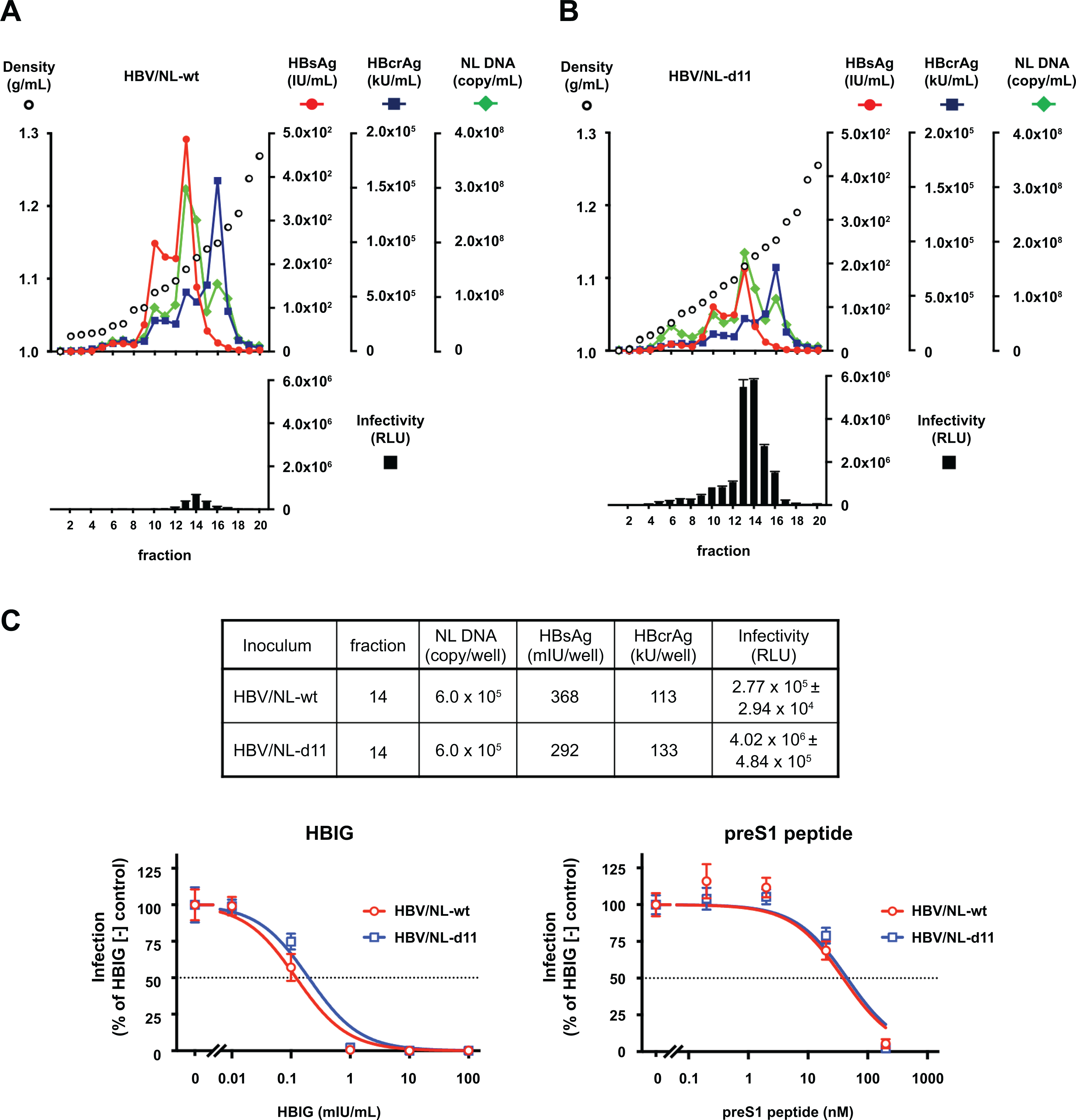
(A, B) HBV/NL generated by transfection of HBV/NL-wt + HBV-dE-wt (HBV/NL-wt) and HBV/NL generated by transfection of HBV/NL-d11 + HBV-dE-d11 (HBV/NL-d11) were analyzed by an iodixanol density gradient. The titers of HBsAg and HBcrAg were measured in each fraction, and the NL DNA was quantified by real-time PCR after treatment with DNase. The infectivity of generated HBV/NL was evaluated by inoculation of the same volume of fractions onto HepG2/NTCP-C18 cells. The infected cells were lysed 8 days after infection, and NL activity was measured. (C) Fraction 14 of each gradient was adjusted by NL DNA and infected into HepG2/NTCP-18C cells. The titers of HBsAg and HBcrAg in inocula and NL activities in infected cells are indicated in the upper panel. Inhibition of the HBV/NL infection by HBIG (lower left) or the preS1 peptide (lower right) was assessed by measuring NL activity at 8 days after infection, and dose-response curves are depicted.
Mechanistic analysis of d11-associated enhancement of HBV infection
To confirm the effect of d11 in the L-HBs protein, we used the hepatitis D virus (HDV) infection system because HDV uses the HBs proteins for its envelope. We prepared cell culture-generated HDV that contained the wild-type L-HBs protein (GTC-wt/HDV) or with the d11-introduced L-HBs protein (GTC-d11/HDV). When inoculated with the same amount of HDV RNA, many more HDAg-positive cells were detected in the GTC-d11/HDV-inoculated cells than in the GTC-wt/HDV-inoculated cells (Figure 5A, left panel). The HDV RNA titer of the GTC-d11/HDV-inoculated cells was approximately 6.0-fold higher than that of the GTC-wt/HDV-inoculated cells.
Figure 5. Mechanistic analysis of d11-associated enhancement in HBV infection.
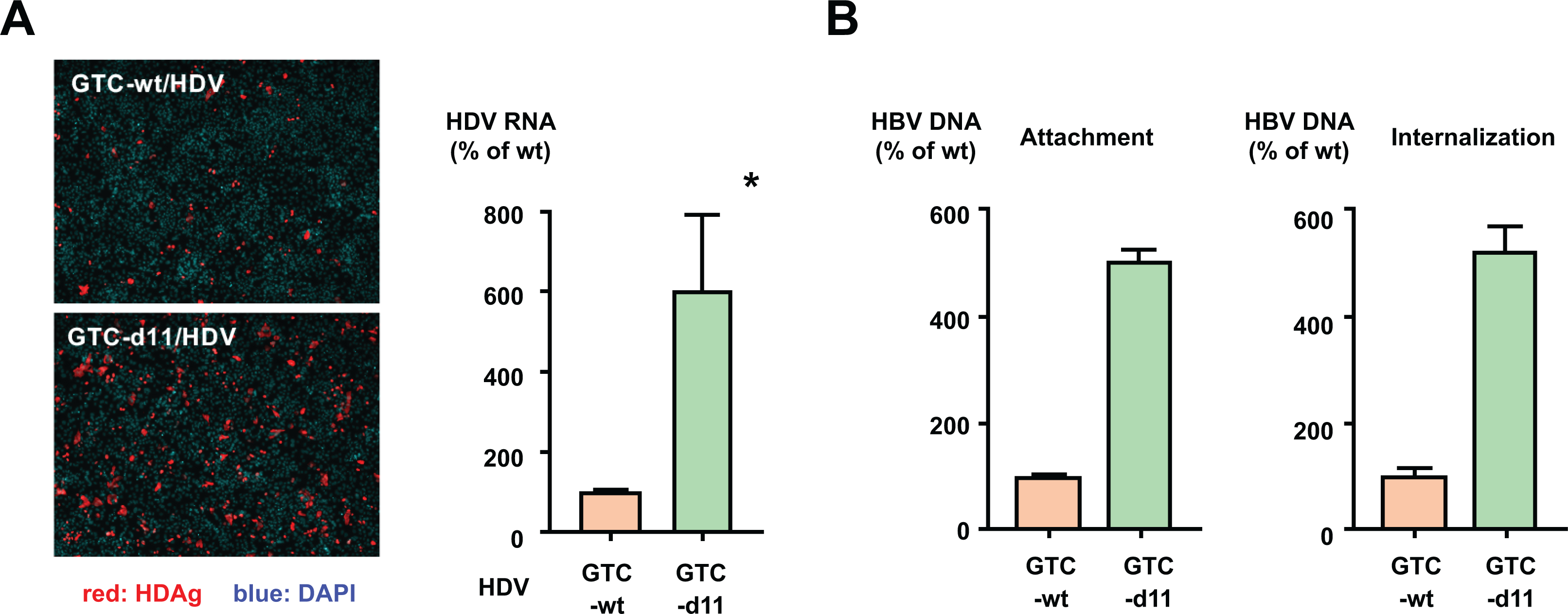
(A) Effects of d11-introduced L-HBs on HDV infection. HDV enveloped with the wild-type L-HBs (GTC-wt/HDV) or with d11-introduced L-HBs (GTC-d11/HDV) was inoculated onto HepG2/NTCP-C4 cells. HDV-infected cells were visualized by staining with an anti-HDAg antibody and counted. Nuclei were visualized by DAPI. The HDV RNA in the infected cells was measured by real-time PCR after treatment with DNase. The data are indicated as the mean ± standard deviation of the triplicate assay. (B) The attachment and internalization of the GTC-wt and GTC-d11 viruses to HepG2/NTCP-C4 cells were analyzed. The purified HBV were attached to the surface of HepG2/NTCP-C4 cells at 4ºC for 3 h without PEG8000, and after that, viruses were internalized by moving the virus-attached cells to 37ºC. The cells were harvested at each step, and HBV DNA was measured by real-time PCR. The data are indicated as the mean ± standard deviation of the triplicate assay. Asterisk indicates a significant difference (p < 0.05).
Then, we evaluated the underlying mechanisms of the L-HBs protein with d11 in the HBV infection steps: attachment or internalization of viruses. After the incubation of HepG2/NTCP-C4 cells with purified HBV at 4ºC, the amount of attached GTC-d11 viruses on the cell surface was approximately 5.1-fold higher than that of attached GTC-wt viruses (Figure 5B, left panel). The amount of internalized GTC-d11 viruses was similarly higher than that of GTC-wt viruses (Figure 5B, right panel). These data indicated that the efficiency of the cell-surface attachment was enhanced in the GTC-d11 viruses compared with that in the GTC-wt viruses. Furthermore, we compared the viral attachment of GTC-wt and GTC-d11 viruses on the original HepG2 cells without NTCP expression and not susceptible to HBV. The amount of attached GTC-d11 viruses on the cell surface was two-fold higher than that of attached GTC-wt viruses (Supplementary Figure S4), indicating the enhanced interaction of GTC-d11 viruses with not only NTCP but also other factors such as HSPG.
To compare the ratios of L-, M- and S-HBs in GTC-wt and GTC-d11 viruses, we analyzed the contents of HBs species by immunoblotting in peak fractions of HBsAg and infectivity of the iodixanol density gradient. The L-HBs and S-HBs proteins were detected as two forms: non-glycosylated and mono-glycosylated. The M-HBs protein was not clearly detected in these viruses. Contrary to expectations, the percentages of L-HBs in all HBs species were comparable between GTC-wt and GTC-d11 viruses, 36.1% vs. 35.8% in fraction 13 and 34.0% vs. 36.1% in fraction 14, respectively (Supplementary Figure S5). However, the ratios of glycosylated to non-glycosylated L-HBs were different between these viruses. In the GTC-wt virus, 33.5% and 42.9% of the total L-HBs proteins were glycosylated in fractions 13 and 14, respectively. In the GTC-d11 virus, 68.7% and 64.7% of the total L-HBs proteins were glycosylated in fractions 13 and 14, respectively. This difference in glycosylation efficiency was not observed in the S-HBs proteins of these viruses. This high percentage of glycosylated L-HBs protein was also observed in the HepG2.2.15-derived virus, which had high infectivity.
Effects of d11 in L-HBs of other HBV strains
To assess the effects of d11 in the L-HBs protein in other HBV strains, we exploited replication-competent HBV molecular clones of genotype A (GTA-wt), genotype B (GTB-wt), and another genotype C (GTC2-wt). d11 was introduced to these molecular clones, and they were named GTA-d11, GTB-d11, and GTC2-d11. By infection of HBVcc of these clones, enhancements in infectivity by d11 were observed. The number of HBV-positive cells by inoculation of d11 viruses was higher than that by inoculation of wild-type viruses (Figure 6A). The amount of pgRNA in the GTA-d11 virus-inoculated cells was 1.7-fold higher than that in GTA-wt inoculated cells (Figure 6B, left panel). Likewise, the amounts of pgRNA in GTB-d11 and GTC2-d11 virus-inoculated cells were 2.4-fold and 11-fold higher than those in GTB-wt and GTC2-wt virus-inoculated cells, respectively (Figure 6B, middle and right panel). These data indicate that the d11-associated enhancement in infectivity is not strain specific or genotype specific.
Figure 6. Enhancement in infectivity by d11 in other HBV genotypes and strains.
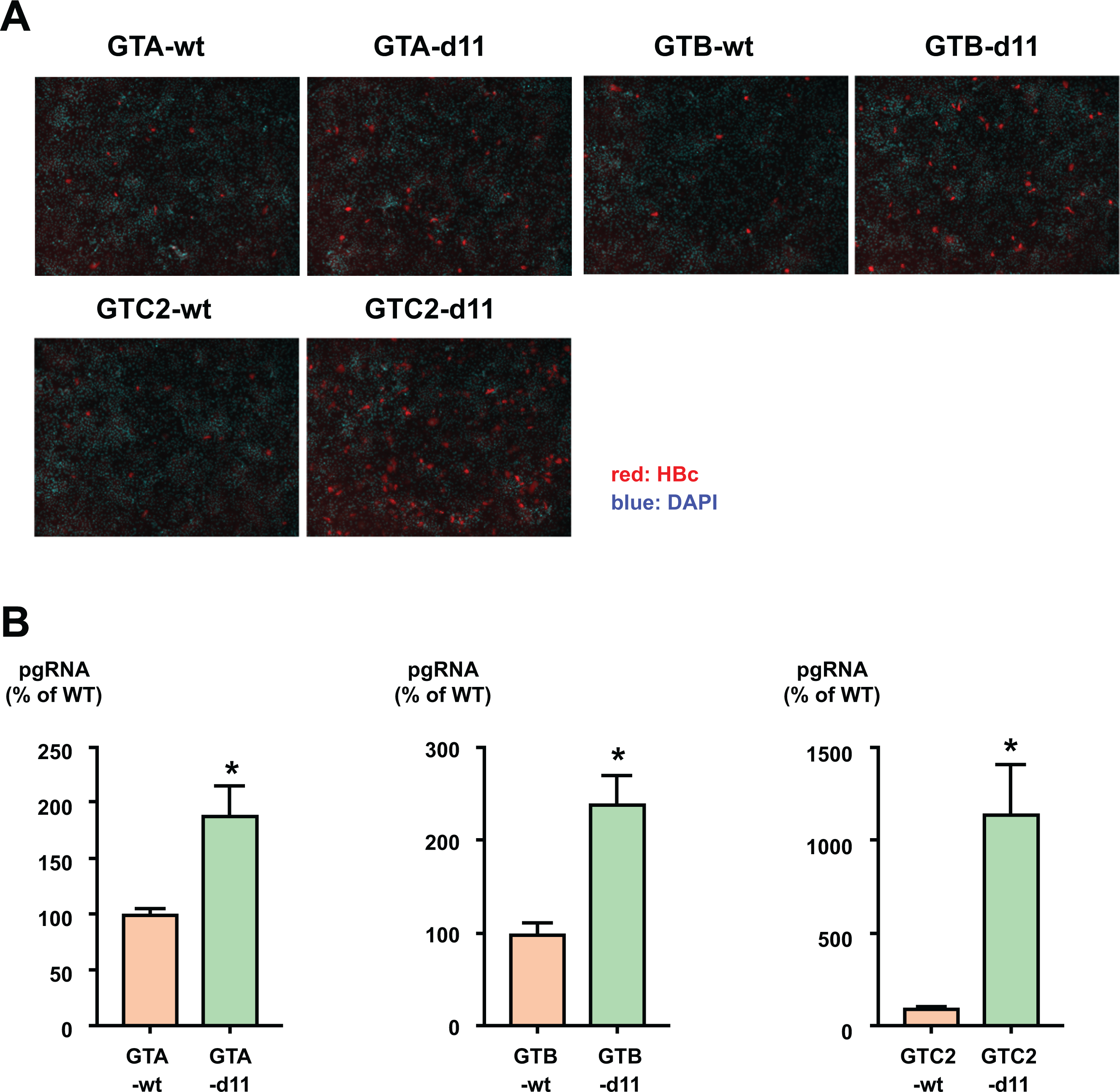
(A) Cell culture-generated viruses of GTA wild type (GTA-wt) or d11 (GTA-d11), GTB wild type (GTB-wt) or d11 (GTB-d11), and another clone of GTC wild type (GTC2-wt) or d11 (GTC2-d11) were inoculated onto HepG2/NTCP-C4 cells at 500 GEq/cell. HBV-positive cells were visualized by staining with an anti-HBc antibody, and nuclei were visualized by DAPI. (B) The production of pgRNA in infected cells. The indicated viruses were infected into HepG2/NTCP-C4 cells at 500 GEq/cell. Total RNA was extracted from infected cells, and pgRNA was measured by real-time PCR. The data are indicated as the mean ± standard deviation of the triplicate assay. Asterisk indicates a significant difference (p < 0.05).
DISCUSSION
HBVcc generated by transient transfection is known to have limited infectivity in cell culture. In this study, we focused on the high infectivity of HBVcc of a GTD strain in comparison with that of a GTC strain. We compared the genome structure of GTD and GTC strains and found the d11 in the preS1 region of GTD. Among the 1,141 strains of genotype D deposited in Hepatitis Virus Database (http://s2as02.genes.nig.ac.jp), 1,058 strains (92.7%) carry d11. This d11 was found not only in genotype D strains but also in HBV strains isolated from non-human primates (Supplementary Figure S6). However, strains with d11 were not clustered in a phylogenetic tree (Supplementary Figure S7). This observation may suggest that d11 has some advantages for expansion beyond the host species. We revealed that a GTD-specific d11 in the preS1 region confers efficient infectivity to strains other than GTD strains and that this modification enables the efficient infection of HBVcc in HepG2/NTCP cells.
Infection of the GTC-d11 virus resulted in many infected cells and a time-dependent increase in intracellular pgRNA, similar to infection of a HepG2.2.15-derived virus. Because the production of HBcrAg after transfection of the GTC-d11 molecular clone was comparable to that after transfection of GTC-wt, the increased number of HBc-positive cells after infection of the GTC-d11 virus is ascribed to an enhancement in the infection step but not enhanced production of HBc proteins in infected cells. To analyze the features of the generated viruses affecting the infection step, we analyzed the profiles of the GTC-wt and GTC-d11 viruses in an iodixanol density gradient. In the density gradient, the peaks of HBV markers were detected at the same fraction. The peak of infectivity was also observed at the same fraction. When adjusted by HBV DNA, the amount of HBsAg or HBcrAg was comparable in the peak fraction of infectivity. These data suggest that the amount of viral component per copy of HBV DNA was almost identical in the peak fraction of infectivity. However, when inoculated with the peak fraction of infectivity, including the same HBV DNA amount, the number of HBV-infected cells was much more abundant by infection with the GTC-d11 virus than by infection with the GTC-wt virus. These data suggest that the enhanced infectivity observed in the GTC-d11 virus is ascribed to the enhanced ability of infection but not to the enhanced production of viral particles. This d11-associated enhancement in infectivity was also confirmed by measuring cccDNA synthesis in infected cells, and the infections of these viruses were similarly inhibited by administration of HBIG, suggesting HBs-dependent infection of these viruses. This d11-associated enhancement in infectivity was also observed in a study with the HBV/NL system. When inoculated with the peak fraction of infectivity, including the same NL DNA amount, the NL activity was 14.5-fold higher in HBV/NL-d11 virus-infected cells than in HBV/NL-wt virus-infected cells. HBV/NL infection was similarly inhibited by HBIG or preS1 peptide administration, exhibiting almost identical EC50 values. Thus, the HBV/NL-d11 infection system provides an assay system with a wide dynamic range for screening anti-viral reagents.
HBV has a partially double-stranded DNA genome and overlapped ORFs. Consequently, d11 in the preS1 region affects not only the length of the L-HBs protein but also the lengths of the HBp protein and the packaged genome. Therefore, we assessed the factors responsible for the enhanced infectivity using the HBV/NL system. In the HBV/NL system, we confirmed the enhanced infectivity due to d11 introduction. In assessment with the d11- or delS-introduced plasmid, a shortened HBp protein and a shortened packaged genome did not enhance the infectivity, but a shortened L-HBs protein was associated with enhanced infectivity. The involvement of the d11-introduced L-HBs protein in the enhanced infectivity was also confirmed in the HDV infection system. HDV enveloped with the d11-introduced L-HBs protein exhibited enhanced infectivity compared with a virus with the wild-type L-HBs protein.
In evaluation of the factors responsible for the d11-associated enhanced infectivity, the attachment of viruses to the cell surface was revealed to be a major affecting step. A larger amount of cell surface-attached viruses was detected in GTC-d11 virus-inoculated cells than in GTC-wt virus-inoculated cells. The efficiencies of internalization of GTC-d11 and GTC-wt viruses seemed to be similar because no additional enhancement was observed. Based on the observation of HBs-associated enhancement of virus attachment to the cell surface, we expected that the ratio of L-HBs among HBs species would be higher in the GTC-d11 virus than in the GTC-wt virus because L-HBs has the NTCP-binding region. We compared the ratio of L-HBs/S-HBs between GTC-d11 and GTC-wt viruses and found no remarkable difference. Surprisingly, the glycosylation efficiency of the L-HBs protein was substantially higher in the GTC-d11 virus than in the GTC-wt virus. Over half of the L-HBs protein was glycosylated in the GTC-d11 virus, although this efficient glycosylation was not observed in the S-HBs protein. Efficient glycosylation of the L-HBs protein was also observed in the HepG2.2.15-derived virus, which is a genotype D strain and has high infectivity in cell culture. Therefore, we reasoned that the glycosylation status of L-HBs may be one of the important factors for the enhanced infectivity, although the glycosylation site is in the S-HBs region. The shortening of the L-HBs protein by the introduction of d11 may alter the tertiary structure of the envelope and affect the status of glycosylation. The interaction between the preS domain of the L-HBs protein and the glycosaminoglycan side chains of cell surface-associated heparan sulfate proteoglycans has been reported to be important at the infection step of HBV (22). The glycosylation status of the viral envelope may affect this interaction. Further study will be needed to investigate the detailed mechanisms of this glycosylation-associated enhancement in HBV infection. The crucial role of the orientation of the preS1 sequence of L-HBs in the infectivity of HBV has been reported (23). This study suggested that the external orientation of the L-HBs occurring during the HBV maturation process mediates virus entry. The increased glycosylation status of the GTC-d11 virus may be associated with this orientation of L-HBs. Further investigation will be needed to address this point.
The HBVcc generated by transient transfection is important to assess the effects of genotype- or strain-specific viral characteristics or resistance-associated substitutions on anti-viral reagents because desired polymorphisms can be introduced in the viral genome. We demonstrated that the infection of the GTC-d11 virus was inhibited by administration of HBIG or the preS1 peptide, similar to infection of the GTC-wt virus, and the EC50 values of these viruses for HBIG or preS1 peptide were almost identical. Currently, interferon and nucleotide analogues are used for the first-line treatment regimen (10). These reagents cannot eliminate HBV, and the discovery of other classes of anti-HBV reagents is desirable. An infection system with highly infectious HBV will be useful for screening novel anti-HBV reagents targeting the infection step and assessment of resistance-associated polymorphisms. In addition, in this system, efficient synthesis of cccDNA was detected after infection with the GTC-d11 virus. Therefore, this system is also available to investigate the host factors associated with the synthesis of cccDNA and to explore anti-HBV reagents targeting cccDNA synthesis.
We found that the enhancement in infectivity by d11 introduction is applicable to clones of other genotypes or strains. However, the levels of enhancement were different among strains. The introduction of d11 to the genotype C strains (GTC and GTC2) enhanced the pgRNA production by approximately 10-fold, and this production level was comparable to that induced by infection with the HepG2.2.15 virus. In the genotype A and B strains, the introduction of d11 enhanced pgRNA production by 1.7-fold and 2.4-fold, respectively. The infectivity of d11-introduced genotype A and B strains did not reach the level of that of the HepG2.2.15 virus. We are not sure whether this insufficient enhancement depends on the genotype or the strain. To establish a more efficient HBV infection system with genotype A and B strains, an investigation to isolate clones of other genotypes that infect more efficiently will be needed.
In conclusion, we established an efficient infection system for HBVcc generated by transient transfection by using d11-introduced HBV molecular clones. This system provides a powerful tool for studying the infection and propagation of HBV in cell culture, and also for developing the anti-viral strategy against HBV infection.
Supplementary Material
Acknowledgements
The authors wish to thank Prof. John Taylor (Fox Chase Cancer Center, Philadelphia, PA) for providing the pSVLD3 plasmid.
Financial support:
This work was supported by grants for Research Programs on Hepatitis (JP19fk0310103 and JP19fk0310120) from the Japan Agency for Medical Research and Development, AMED and by Grant-in-Aid for Scientific Research (C) (JSPS KAKENHI Grant Number 19K08482). The funders had no role in the study design, data collection or interpretation or decision to submit the work for publication.
Abbreviations:
- HBV
hepatitis B virus
- HBVcc
cell culture-generated HBV
- NTCP
sodium taurocholate co-transporting polypeptide
- GT
genotype
- ORF
open reading frames
- HBsAg
hepatitis B surface antigen
- cccDNA
covalently closed circular DNA
- GEq
genome equivalent
- pgRNA
pre-genomic RNA
- HBcrAg
hepatitis B core-related antigen
- HBIG
anti-HBs human immune globulin
- NL
NanoLuc luciferase
- EC50
effective concentrations required to inhibit 50%
- HDV
hepatitis D virus
Footnotes
Competing interests:
All authors declare no competing interests.
REFERENCES
- 1.Liang TJ, Block TM, McMahon BJ, Ghany MG, Urban S, Guo JT, Locarnini S, et al. Present and future therapies of hepatitis B: From discovery to cure. Hepatology 2015;62:1893–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razavi-Shearer D, Gamkrelidze I, Nguyen MH, Chen D-S, Van Damme P, Abbas Z, Abdulla M, et al. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. The Lancet Gastroenterology & Hepatology 2018;3:383–403. [DOI] [PubMed] [Google Scholar]
- 3.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol 1988;69 ( Pt 10):2575–2583. [DOI] [PubMed] [Google Scholar]
- 4.Norder H, Courouce AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 1994;198:489–503. [DOI] [PubMed] [Google Scholar]
- 5.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol 2000;81:67–74. [DOI] [PubMed] [Google Scholar]
- 6.Kurbanov F, Tanaka Y, Mizokami M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res 2010;40:14–30. [DOI] [PubMed] [Google Scholar]
- 7.Yamada N, Shigefuku R, Sugiyama R, Kobayashi M, Ikeda H, Takahashi H, Okuse C, et al. Acute hepatitis B of genotype H resulting in persistent infection. World J Gastroenterol 2014;20:3044–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol 2016;64:S4–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito K, Qin Y, Guarnieri M, Garcia T, Kwei K, Mizokami M, Zhang J, et al. Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J Virol 2010;84:12850–12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia Y, Liang TJ. Development of Direct-acting Antiviral and Host-targeting Agents for Treatment of Hepatitis B Virus Infection. Gastroenterology 2019;156:311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sells MA, Zelent AZ, Shvartsman M, Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J Virol 1988;62:2836–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 1997;41:1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada N, Sugiyama R, Nitta S, Murayama A, Kobayashi M, Okuse C, Suzuki M, et al. Resistance mutations of hepatitis B virus in entecavir-refractory patients. Hepatol Commun 2017;1:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, et al. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology 2006;44:915–924. [DOI] [PubMed] [Google Scholar]
- 16.Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, Tanaka S, et al. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol 1999;37:2899–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa K, Nishikawa H, Enomoto H, Iwata Y, Sakai Y, Ikeda N, Takashima T, et al. Proposed model for the prediction of intrahepatic covalently closed circular DNA level in patients with chronic hepatitis B. Hepatol Res 2019;49:271–283. [DOI] [PubMed] [Google Scholar]
- 18.Murayama A, Momose H, Yamada N, Hoshi Y, Muramatsu M, Wakita T, Ishimaru K, et al. Evaluation of in vitro screening and diagnostic kits for hepatitis B virus infection. J Clin Virol 2019;117:37–42. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki Y, Maekawa S, Komatsu N, Sato M, Tatsumi A, Miura M, Matsuda S, et al. Hepatitis B virus (HBV)-infected patients with low hepatitis B surface antigen and high hepatitis B core-related antigen titers have a high risk of HBV-related hepatocellular carcinoma. Hepatol Res 2019;49:51–63. [DOI] [PubMed] [Google Scholar]
- 20.Nishitsuji H, Ujino S, Shimizu Y, Harada K, Zhang J, Sugiyama M, Mizokami M, et al. Novel reporter system to monitor early stages of the hepatitis B virus life cycle. Cancer Sci 2015;106:1616–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishitsuji H, Harada K, Ujino S, Zhang J, Kohara M, Sugiyama M, Mizokami M, et al. Investigating the hepatitis B virus life cycle using engineered reporter hepatitis B viruses. Cancer Sci 2018;109:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulze A, Gripon P, Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 2007;46:1759–1768. [DOI] [PubMed] [Google Scholar]
- 23.Seitz S, Iancu C, Volz T, Mier W, Dandri M, Urban S, Bartenschlager R. A Slow Maturation Process Renders Hepatitis B Virus Infectious. Cell Host Microbe 2016;20:25–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


