Abstract
The emergence and persistence of selfish genetic elements is an intrinsic feature of all living systems. Cellular organisms have evolved a plethora of elaborate defense systems that limit the spread of such genetic parasites. CRISPR-Cas are RNA-guided defense systems used by prokaryotes to recognize and destroy foreign nucleic acids. These systems acquire and store fragments of foreign nucleic acids and utilize the stored sequences as guides to recognize and destroy genetic invaders. CRISPR-Cas systems have been extensively studied, as some of them are used in various genome editing technologies. Although Type III CRISPR-Cas systems are among the most common CRISPR-Cas systems, they are also some of the least investigated ones, mostly due to the complexity of their action compared to other CRISPR-Cas system types. Type III effector complexes specifically recognize and cleave RNA molecules. The recognition of the target RNA activates the effector large subunit – the so-called CRISPR polymerase – which cleaves DNA and produces small cyclic oligonucleotides that act as signaling molecules to activate auxiliary effectors, notably non-specific RNases. In this review, we provide a historical overview of the sometimes meandering pathway of the Type III CRISPR research. We also review the current data on the structures and activities of Type III CRISPR-Cas systems components, their biological roles, and evolutionary history. Finally, using structural modeling with AlphaFold2, we show that the archaeal HRAMP signature protein, which heretofore has had no assigned function, is a degenerate relative of Type III CRISPR-Cas signature protein Cas10, suggesting that HRAMP systems have descended from Type III CRISPR-Cas systems or their ancestors.
Keywords: CRISPR-Cas Type III, prokaryotic immunity, RNA-guided defense, cyclic oligonucleotides, CRISPR evolution, HRAMP
INTRODUCTION
All known life forms on Earth share a universal feature – almost all their biological traits are encoded in the nucleic acid sequences that are replicated through a template-based principle. While changes in nucleic acid sequence may have a great influence on biological traits (e.g., protein structures it encodes), these changes usually produce little effect on the chemical and physical properties of nucleic acids themselves, allowing the existence of nucleic acids with every possible sequence. Since an increase in the replication fidelity could be achieved only at a price of increased energy consumption [1], replication of nucleic acids is inevitably error-prone. This necessarily implies an existence of genetic heredity with the variation of biological traits encoded by nucleic acids, which, in turn, provides necessary factors for the evolution by natural selection and/or genetic drift.
Any community of evolving self-replicating living systems inevitably gives rise to genetic parasites and corresponding defense systems [2], so the eternal arms race between selfish elements and their hosts apparently goes back to the origin of life. This competition leads to the development of the anti-parasite defense systems that target different mechanisms involved in the parasite’s life cycle. In their turn, parasites evolve to dodge the defense systems. The hosts are apparently incapable to completely get rid of genetic parasites, since this entails reduction of the horizontal gene transfer essential for the long-term genome stability and evolution [2]. The evolution of defense systems sometimes goes through very peculiar ways, including the shuffling of components of different defense systems and, most strikingly, adoption of components of genetic parasites themselves for the host defense. From this point of view, CRISPR-Cas systems are especially fascinating, as they appear to be bizarre mosaics of “tamed” transposons, toxin–antitoxin systems, and other components of unclear origin. In this review, we describe the structural and mechanistic features of Type III CRISPR-Cas, probably, the most complex defense system of prokaryotes.
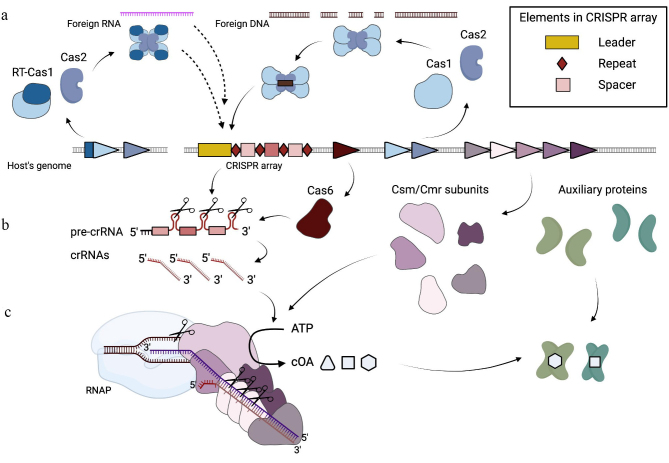
The diversity of CRISPR-Cas loci and the main features of the adaptive immunity mediated by different CRISPR-Cas systems have been already discussed in dozens of reviews (see, for example, [3, 4]). Most CRISPR-Cas loci include CRISPR arrays that consist of two or more repeats separated by unique spacers and adjacent clusters of cas genes. The CRISPR-Cas immune response can be divided into three stages: (a) adaptation, (b) expression, and (c) interference (as shown for Type III CRISPR-Cas systems in Fig. 1). At the adaptation stage, short fragments of DNA are inserted into a CRISPR array, forming a new spacer. Adaptation is mediated by the Cas1-Cas2 integration complex. Although this complex is highly conserved, the exact mechanism of spacer acquisition depends on the CRISPR-Cas system type. At the expression stage, CRISPR arrays are transcribed into pre-CRISPR RNA molecules that are further processed into mature small CRISPR RNAs (crRNAs). The processing stage proceeds via mechanisms depending on the CRISPR-Cas system type. At the interference stage, crRNAs are incorporated into the Cas proteins effector complexes and used as guides for the recognition of foreign nucleic acid sequences (protospacers) that are then destroyed by Cas nucleases. CRISPR-Cas systems have been found in ~90% sequenced archaeal and ~40% eubacterial genomes [5]. Based on the effector complex composition, CRISPR-Cas systems can be divided into two classes. Class 1 includes systems with the multisubunit effector complexes; Class 2 effectors are single multidomain proteins. CRISPR-Cas systems were further subdivided into 6 types and multiple subtypes based on the composition and organization of cas loci: types I, III and IV belong to class 1, and types II, V and VI belong to class 2 [4]. In this review, we focus only on the Type III systems.
Fig. 1.
Type III adaptive immunity. a) Adaptation: insertion of small fragments of invader-derived DNA into the host CRISPR array with the formation of a new spacer-repeat unit. In some systems, the spacers can be acquired from RNA through the activity of the RT domain fused to Cas1. b) Expression: maturation of crRNAs and assembly of effector complexes. c) Interference: triggering of the immune response by specific recognition of foreign RNA.
HISTORY OF TYPE III CRISPR-Cas RESEARCH
Type III CRISPR-Cas systems are widespread in both archaeal and bacterial genomes (34 and 25% of all CRISPR-cas loci, correspondingly) [5]. The genes currently known as Type III cas genes were first identified during the search for conserved gene clusters in the known genomes of hyperthermophilic archaea. At that time, it was hypothesized that these genes belong to previously unknown DNA repair systems [6]. Among the genes present in such clusters, Makarova et al. identified a subset encoding large conserved multidomain proteins. These proteins were shown to contain a domain similar to the Palm domain, a component of various enzymes, such as A, B and Y superfamilies of DNA-dependent DNA polymerases, viral RNA-dependent RNA polymerases, DNA-dependent RNA polymerases of some viruses and mitochondria, reverse transcriptases, and a large group of cyclases and nucleotidyltransferases [7, 8]. On these grounds, these large conserved multidomain proteins were predicted to be polymerases/cyclases. Several families of other protein-coding genes were found to be associated with the predicted polymerase/cyclase genes, but their roles remained unclear. Believing that the discovered loci belonged to a new DNA repair system, Makarova et al. named these proteins RAMPs (Repair-Associated Mysterious Proteins). Besides the polymerase/cyclase-RAMP-encoding loci, Makarova et al. identified another kind of conserved gene clusters that were later recognized as Type I CRISPR-Cas systems. However, the linkage of both kinds of clusters to CRISPR arrays was missed at the time. Few years later, Haft et al. performed an extensive search and classification of protein-coding genes located in the vicinity of CRISPR arrays and delineated the organization of these genes in specific loci [9]. Among the currently recognized CRISPR-Cas types, Haft et al. also described Csm (CRISPR-Cas Subtype Mycobacterium tuberculosis) and Cmr (CRISPR-Cas Module RAMP). Some products of the csm and cmr genes were found to be homologous to each other. Both the csm and cmr loci contain genes for the Palm-domain proteins (csm1 and cmr2) and encode at least two homologous RAMP proteins (csm3 and cmr4). The csm and cmr loci were later designated as Type III-A and Type III-B CRISPR-Cas systems [10]. Haft et al. noticed that the cmr loci never occur as the only CRISPR-Cas system in prokaryotic genomes. Consistently, it was shown that Type III-B CRISPR-Cas systems often lack the adaptation module and therefore must rely on the spacer acquisition machinery of other CRISPR-Cas systems [5].
Type III CRISPR-Cas systems from different organisms were experimentally studied by several independent scientific groups; however, the results of these studies were rather puzzling. Based on the data of in vivo experiments, Marraffini et al. characterized a Type III-A CRISPR-Cas system from Staphylococcus epidermidis as a DNA-targeting system [11]. On the other hand, Type III effector complexes were shown to specifically recognize and cleave RNA targets in vitro [12, 13]. Although we now know that conclusions made based on the in vivo results were incorrect, this work is worth to be discussed in detail, since it is important not only in terms of CRISPR-Cas research but also from a methodological point of view. In their experimental system, Marraffini et al. used a conjugative plasmid and two strains of S. epidermidis. One of these strains lacked CRISPR-Cas systems, while another one harbored a Type III-A system with a spacer matching the sequence of the plasmid-borne nes gene encoding a nickase (a component of conjugational machinery). It was shown that the Type III-A system suppressed the conjugal transfer of the nes-harboring plasmids. Since the spacer matched the coding strand of the nes gene (i.e., the resulting crRNA did not recognize the sense nes transcripts), it was expected that the system would target DNA. Furthermore, the fact that expression of nickase is needed only in the donor strain to initiate the DNA transfer, but not in the recipient strain to maintain the plasmid, also supported the targeting of DNA. The authors considered that the CRISPR array may be transcribed from both strands, thus producing crRNAs targeting the nes transcript, yet no anti-sense CRISPR transcripts were found. Finally, the authors performed an ingenious experiment, disrupting the nes protospacer sequence with a self-splicing intron and showing that such plasmid evaded Type III-A-mediated immunity, providing the strongest argument in favor of DNA targeting. However, they did not consider the possibilities that (i) there is a considerable level of anti-sense nes transcription and/or (ii) Type III immunity is triggered by the recognition of nascent RNA by the effectors. Few years later, the same group established that plasmid protospacers are transcribed in both directions and proved the in vivo specificity of Type III-A systems towards RNA [14].
Despite primary targeting of RNA, Type III CRISPR-Cas systems protect cells from viruses with DNA genomes and interfere with plasmid transformation, as long as viral or plasmid DNA is transcribed with the production of RNA molecules complementary to the crRNA spacers. Transcripts complementary to the protective crRNAs do not have to be essential for the viral or plasmid life cycle/maintenance [14, 15]. The first insights explaining this puzzling observation were obtained from the in vitro characterization of the activities of the Csm and Cmr effector complexes. In addition to the Palm domains, the large subunits of Type III effector complexes (Csm1 and Cmr2 proteins, further designated as Cas10) also contain the HD domains (named after conserved histidine and aspartate residues) [5]. The HD domain proteins are also associated with other CRISPR-Cas systems. For example, in Type I systems, the Cas3 protein destroys target DNA due to the single-strand DNase activity of its HD domain [16]. The binding of target RNA activates the single-strand DNase activity of Cas10 HD domain in both Type III-A [17] and Type III-B [18, 19] effector complexes. Therefore, a model of the co-transcriptional DNA cleavage was proposed to explain the mechanism of Type III immunity. According to this model, when a Type III effector recognizes a nascent transcript, the HD nuclease domain of the Cas10 subunit is activated and cleaves the single-stranded DNA within the transcription bubble (Fig. 2a) [17]. Some support for this model was provided by the in vitro experiments of the Marraffini group [20], although later these results were put to doubt [21]. Be that as it may, mutations of the catalytic residues in the Cas10 nuclease domain do not affect the interference with the plasmid transformation. In contrast, mutations of the catalytic residues of the Cas10 Palm domain significantly attenuated the Type III anti-plasmid immunity [22]. These results clearly imply that the Type III immune response cannot be reduced to the co-transcriptional DNA cleavage only.
Fig. 2.
A model of the co-transcriptional cleavage by Type III effectors and activation of auxiliary nucleases triggered by the target recognition. a) The recognition of target RNA by Type III effector stimulates the activities of HD and Palm domains of the Cas10 subunit. The Palm domain catalyzes the synthesis of cyclic oligoadenylates (cOAs) while the HD domain degrades single-stranded DNA within the transcription bubble. b) cOAs activate the auxiliary nucleases that target DNA or RNA molecules. The activity of the auxiliary effectors is regulated through the degradation of cOAs by ring nucleases or, in some cases, by the auxiliary proteins. c) The avoidance of self-targeting in Type III CRISPR-Cas systems: the complementarity between the target and repeat-derived 5′-tag of crRNA prevents the activation of both the HD and the Palm domains of the Cas10 subunit.
CRISPR-Cas loci often contain genes coding for proteins that are not directly involved in the spacer acquisition, crRNA maturation, or formation of effector complexes. The role of most of these genes (usually referred to as auxiliary) is still poorly understood. Among such genes, there is a family coding for proteins with a specific variant of the Rossmann fold domain (the so-called CRISPR-Cas associated Rossmann Fold, or CARF). In these proteins, CARF domains are frequently linked to various domains with predicted RNase, DNase or DNA-binding activities. The Rossmann fold is a common motif in nucleotide-binding proteins; it was suggested that CARF-domain proteins participate in the CRISPR-Cas mediated immune response by sensing some nucleotide ligands with subsequent activation of their effector nuclease domains [23]. The disruption of the csx1 and csm6 genes, which encode CARF domain proteins and are frequently associated with the Type III cas operons, greatly hinders the ability of Type III CRISPR-Cas systems to interfere with the plasmid transformation [15, 22], providing an additional layer of complexity to the Type III-mediated immunity.
In the Csx1 and Csm6 proteins, the CARF domains are fused with the HEPN (Higher Eukaryotes and Prokaryotes Nucleotide-binding) domains [23]. Proteins with the HEPN domains exhibit the RNase activity and are frequently involved in various defense systems [24]. Some Csx1 and Csm6 proteins cleave single-stranded RNA in vitro through their HEPN domains [25, 26]. Yet, the observed RNase activities are relatively weak, suggesting that the activity of these proteins in the Type III mediated immune response in vivo may be somehow upregulated [25, 26]. This hypothesis was confirmed by the experiments showing that upon recognition of the target RNA, Type III effector complexes convert ATP molecules to a range of cyclic oligoadenylates (cOAs) employing the polymerization activity of the Cas10 Palm domains [27, 28]. These cOAs function as secondary messengers triggering the non-specific RNase activity of the CARF-HEPN proteins (Fig. 2, a and b) [27-29]. The exact role of the non-specific RNase activity of the Csx1/Csm6 proteins is still rather speculative. Inhibition of viral transcription, cell dormancy, or even death were proposed as possible outcomes of the non-specific RNA degradation by the activated Csx1/Csm6 [30,31]. In the latter case, one can envision that inhibition of infection by DNA viruses detected by standard plaque assays, can be achieved even without the activation of the DNase activity of Type III effectors. This scenario, however, is inconsistent with the observations that cells mounting Type III interference against lytic viruses clear the infection and survive [32].
The fact that individual cells mounting the Type III interference remain viable implies that the cOA-mediated activation of cellular RNases must be transient. There are two obvious ways to control the activity of the Csm6/Csx1 nucleases – regulation of the cOAs synthesis or degradation of cOAs. Both mechanisms have been experimentally confirmed. First, the cOA-synthesizing activity of Cas10, which is activated upon the binding of target RNA, is abolished upon the target cleavage [33, 34]. Second, several cOA-degrading nucleases have been characterized. In some organisms, cOAs are degraded by dedicated ring nucleases [35]; other organisms contain CARF-HEPN proteins capable of degrading cOAs that activate them [36, 37]. Interestingly, a highly efficient ring nuclease encoded by a virus infecting Sulfolobus was shown to counter the Type III CRISPR-Cas immunity of the host [38].
Strikingly, the cOA-dependent arm of prokaryotic immunity is similar to one of the pathways of mammalian innate immunity. In the latter case, the presence of double-stranded RNAs in the cytoplasm activates oligoadenylate synthetase (OAS) that converts ATP to 2′-5′ oligoadenylates, which, in turn, activate RNase L. RNase L non-specifically degrades RNA in the cytoplasm [39]. This resemblance turns out to be even more exciting considering that the catalytic core of OAS shares similarity with the Palm domain [40], while the activity of RNase L relies on a distinct variant of the HEPN domain [24]. Recently, the OAS-RNase L pathway has drawn a particular attention, since it was discovered that nucleotide polymorphisms in the locus encoding OAS genes are associated with the COVID-19-induced respiratory failure, suggesting that this pathway is important in the immune response against SARS-Cov-2 [41].
While the role of the cOAs pathway in Type III CRISPR-Cas immunity is relatively clear, the significance of the single-strand DNase activity of the Cas10 HD domain remains obscure. Originally, it was shown that mutations of the Cas10 HD domain catalytic residues have no effect on the Type III-A-mediated interference with the plasmid conjugation in S. epidermidis cells [22] (in this work, the interference was registered as a decreased number of transconjugant colonies). However, it was discovered later that unlike the wild-type system, the Type III-A system with inactivated Cas10 HD domain did not prevent the formation of transconjugant colonies harboring the targeted plasmid, but rather severely retarded their growth, so that the colonies became visible only after a prolonged incubation [31]. Interestingly, when both the HD and the Palm domains were inactivated, no interference was observed and the number, as well as the appearance of the targeting and non-targeting transconjugant colonies were the same [31]. Thus, it appears that both Cas10 domains are needed for the full interference. In fact, the requirement for the cOA-dependent arm of the Type III-A interference may depend on the RNA target abundance. When the transcript recognized by the Type III-A effector is abundant, the activity of the HD domain alone suffices for the full interference; when the target abundance is low, the activities of both the HD and the Palm domain become essential. It is possible that the Type III interference is kinetically controlled: cOA-dependent non-specific RNase inhibits propagation of targeted genetic elements, thus giving sufficient time for their degradation due to the slower activity of the HD domain [31]. This scenario seemingly requires the co-transcriptional DNA cleavage. However, despite the appeal of the co-transcriptional DNA cleavage model, there is little experimental support for it. Co-transcriptional DNA cleavage was observed only in the in vitro experiments described in a single paper [20], in which the double-stranded transcription templates were created by combining a complementary template and the non-template-strand DNA oligos. However, further studies showed that what has been considered as the co-transcriptional DNA cleavage in these experiments could be observed only in the presence of excess of the “targeted”, non-template-strand DNA oligonucleotides over the non-targeted ones, whose sequence was complementary to the RNA transcript [21]. The results suggest that in the experiments of Samai et al. [20], Type III effectors cleaved free single-stranded DNA rather than the DNA within the transcription bubble [21]. As an alternative hypothesis explaining the ability of Type III systems to protect from DNA invaders while leaving alive the cells mounting the interference, it was suggested that the HD domain is specific toward the single-stranded DNA, including replication intermediates of some viruses and plasmids [21]. In summary, while the importance of the Cas10 HD nuclease activity for the Type III interference can be considered as established, the mechanism of its involvement remains uncertain due to the lack of knowledge on its in vivo targets. Recently, it was observed that the Cas10 HD nuclease activity increases the rate of mutagenesis in the cells harboring Type III CRISPR-Cas systems, implying that Cas10 HD could non-specifically target cellular DNA [42].
While the above results suggest that the Type III-mediated interference against DNA invaders requires orchestrated activities of the Cas10 HD and Palm domains along with cOA-activated RNases, confusingly, many Type III systems lack some of these apparently essential components. This implies that such “incomplete” systems rely on/recruit other mechanisms to compensate for the missing components or may function in a completely different way.
An example of such apparently incomplete system is the Type III-B CRISPR-Cas system of Thermus thermophilus, whose Cas10 protein lacks the HD domain [43]. However, since Cas10 possesses an intact Palm domain, it is still capable of activating auxiliary effectors via the cOAs pathway [44]. The Type III-B locus of T. thermophilus encodes a cOA-activated DNA nickase, which might be responsible for the plasmid degradation in the absence of the Cas10 HD DNase activity [45].
In Type III-F CRISPR-Cas systems, the Palm domain of Cas10 is predicted to be inactive due to mutations in the catalytic site. Indeed, the Type III-F loci lack genes coding for the CARF-domain proteins [4]. One can speculate that the Type III-F systems provide at least partial protection against mobile genetic elements by relying on the activity of the Cas10 HD domain only. Consistent with this view, it has been shown that some “complete” Type III CRISPR-Cas systems protect cells from viral infections even when the genes for the auxiliary CARF-domain RNases are deleted from the host genome [30].
Type III-E systems are even more peculiar, as they completely lack the genes coding for the Cas10 subunits. What is even more curious, in these systems, several Cas7 subunits that form the multisubunit crRNA-binding filament in other Type III systems are fused, forming a single large multidomain protein. Type III-E loci lack genes coding for the CARF-domain proteins; but some of them encode putative caspase-like auxiliary effectors [4]. The action mechanisms of these enigmatic systems are still obscure and await future investigations.
Bioinformatically predicted HRAMP systems of Halobacteria provide yet another example of “incomplete” Type III-related loci. HRAMP (named after Halobacterial RAMP) systems lack CRISPR arrays and consist of the so-called HRAMP signature gene, cas7, and cas5 genes, and are often associated with the nucleases containing DEDDy and HNH domains. The function of the HRAMP systems is unknown, although it was proposed that they are dsDNA-targeting immune systems. The HRAMP signature protein does not display any sequence similarity with known protein families, and consequently is considered to be a protein with an unknown function [46]. The recently developed protein structure prediction tool AlphaFold2 allowed us to detect a high structural similarity between the HRAMP signature proteins and Csm1 (modelling was done using ColabFold AlphaFold2_mmseqs2 with default parameters); and the structure homology search was performed using the Dali server [47-49]; the Dali Z-score for the model of the HRAMP signature WP_013440547.1 from Halogeometricum borinquense and the Csm1 from Streptococcus thermophilus (PDB 6IFR-A) is 10.6. It becomes apparent from the model that the HRAMP signature protein possesses an HD domain that is highly similar to the HD domain of Csm1 with the characteristic His-Asp active site (Fig. 3). The HRAMP signature protein is two times smaller than Csm1 and lacks domains responsible for the interaction with Csm2 and Csm4 (proteins present in Type III-A CRISPR-Cas systems and absent from the HRAMP systems). The Palm domain of the HRAMP signature protein is also significantly reduced compared to Csm1 and is likely non-functional. The inability of the HRAMP signature protein to perform cyclic oligonucleotide synthesis is supported by the absence of the HRAMP-associated CARF-coding genes. Yet, the final judgement shall await the results of experimental testing of the HRAMP signature proteins cyclase activity.
Fig. 3.
HRAMP signature protein is a Cas10-related nuclease with the HD domain. Crystal structure of Csm1 from S. thermophilus (left panel) and the AlphaFold2 model of the HRAMP signature protein WP_013440547.1 from Halogeometricum borinquense (right panel) are shown as ribbon diagrams. Conserved structural elements are colored; dissimilar domains are shown in light grey. The positions of the His-Asp active sites are indicated by arrows (positions in Csm1 are experimentally confirmed). Below, Csm1 from S. thermophilus is shown as a part of the Type III-A CRISPR-Cas effector complex.
The relatedness between the HRAMP and Type III CRISPR-Cas systems was first suggested based on a distant similarity of the HRAMP Cas7 proteins with the Type III Cas7 proteins [46]. The structural homology between the HRAMP signature proteins and Type III Cas10 proteins provides further support for the relatedness of these systems. If HRAMP systems had evolved from complete Type III systems, CRISPR arrays and adaptation modules must have been lost during their evolution. However, one could speculate that HRAMP systems have originated from ancestral Type III systems that had existed before the emergence of the CRISPR arrays and adaptation modules. In the latter case, the HRAMP systems may be molecular relics that could shed light on the functions of ancient prototypical Type III systems.
Although the most common auxiliary effectors in Type III systems are cOA-activated CARF-domain RNases of the HEPN family [4], in some cases, Type III systems employ other cOA-sensing effectors. One such unusual Type III-associated auxiliary effector was recently characterized. This cOA-activated Card1 protein from the Type III-A locus of Treponema succinifaciens, exhibits nuclease activity towards both single-stranded DNA and RNA in vitro. However, Card1 activation in S. aureus cells did not produce detectable changes in the transcriptome [50], suggesting that its RNase activity is either inhibited in vivo or is highly specific and does not cause massive transcript degradation. Expression of Card1 compensates for the lack of the csm6 gene in the Type III-A system of S. epidermidis and restores the ability of the cells to resist phage infection. However, cells harboring Cas10 gene with mutated catalytic residues of the HD domain could not clear the infection by a phage even in the presence of Card1 [50], suggesting that its single-strand DNase activity is not sufficient for interference. The clearance of the targeted plasmid from the cells required the activities of both Cas10 HD and Card1, which suggests that the Cas10 HD nuclease activity is specific towards the protospacer-containing DNA [50].
SPACER ACQUISITION IN TYPE III CRISPR-Cas SYSTEMS
Spacer acquisition machinery of almost all CRISPR-Cas systems, including Type III, employs a complex composed of conserved Cas1 and Cas2 proteins (Fig. 1a) [4]. The Cas1-Cas2 integration complexes are able to capture short DNA fragments with further insertion into the CRISPR arrays [51]. The origin of spacer integration intermediates is still rather obscure; however, it was shown for the Type I and II systems that they can be produced via DNA degradation by the RecBCD or AddAB complexes [52, 53] or, in case of so-called primed adaptation in the Type I systems, by the processive Cas3 nuclease [54]. Since the Type III immune response against plasmids and viruses with DNA genomes requires transcription of the targeted protospacer, the mechanisms that allow preferential acquisition of spacers targeting transcriptionally active sites of plasmids or viral genomes should be beneficial for the cell. Indeed, some Type III CRISPR-Cas loci encode genes for the Cas1 proteins fused with the reverse transcriptase (RT) domains, suggesting that spacers could be acquired from RNA molecules (Fig. 1a) [55, 56]. Indeed, at conditions of overexpression of the cas1::RT cas2 adaptation modules, acquisition of spacers derived from RNA, particularly from abundant cellular transcripts was demonstrated [57, 58]. Yet, the spacers were acquired in both orientations with an equal efficiency, which suggests that only half of them would be functional in interference.
The mechanisms responsible for the acquisition of RNA-derived spacers are not fully understood. In vitro, the Cas1::RT-Cas2 complex from Marinomonas mediterranea ligated the 3′ ends of RNA molecules to the 5′ ends of repeats in DNA molecules containing a cognate CRISPR array. The resulting RNA-DNA junction intermediates, once formed, served as substrates for reverse transcription [57]. Interestingly, the RNA molecules could be ligated to either side of the first repeat of the array [57], which is consistent with the observed lack of orientation bias in the acquired spacers. On the other hand, the Cas1::RT-Cas2 complex from Thiomicrospira ligated RNA molecules to the CRISPR-containing DNA fragments in vitro but was unable to convert such intermediates to extended CRISPR arrays, suggesting that in vivo such conversion either requires additional factors, or that the cDNA synthesis must occur before the integration of prespacers into the CRISPR array, which should proceed in both orientations to explain the lack of spacer orientation bias [59].
The presence of the RT domain appears to be a derived feature since the Cas1 part of the Cas1-RT fusion is able to integrate spacers from DNA only [57]. Moreover, most Type III loci encode standard Cas1 proteins [57] without RT domains. In such systems, the adaptation machinery should be directed to DNA and, thus, be indifferent to the protospacer transcription, unless these systems employ other, yet to be described mechanisms allowing to acquire spacers from transcriptionally active sites. The adaptation module in T. thermophilus has a standard cas1-cas2 configuration and no RT domain proteins are encoded in the genome. Yet, an extreme bias in the spacers acquired in the course of infection by a lytic phage was observed: only cells that expressed crRNAs targeting viral transcripts were detected in the infected cultures. However, this bias was due to the purifying selection for protective crRNAs and not caused by the intrinsic biases of the adaptation machinery [32].
THE AVOIDANCE OF SELF-TARGETING IN TYPE III CRISPR-Cas SYSTEMS
CRISPR-Cas effector complexes recognize nucleic acids that are complementary to the spacer part of the bound crRNAs. Since crRNAs are derived from CRISPR arrays, the effectors might target genomic DNA (or, in the case of RNA-targeting systems, the “antisense” CRISPR array transcripts). Such targeting shall be detrimental and thus must be avoided. The immune response mediated by the DNA-targeting CRISPR-Cas systems (Types I, II, and V) requires protospacer adjacent motifs (PAMs) – short (few nucleotides) degenerate sequences that are located in the target DNA near the protospacer but are not present near the spacers in the CRISPR arrays [60]. The effector complexes scan the double-stranded DNA for the PAM sequences, and, upon PAM recognition, initiate the melting of the DNA duplex followed by the formation of the R-loop complex with the complementary targets [61-63]. In the Type III systems, self-targeting is avoided via a completely different mechanism. While Type III effectors recognize and cleave any RNA molecule complementary to the crRNA spacer part, activation of the HD and Palm catalytic domains of the Cas10 subunit requires the absence of complementarity between the target and repeat-derived 5′-tag of crRNA (Fig. 2c) [17, 64, 65]. Strikingly, a similar “tag-antitag” principle of “self versus non-self” discrimination is employed by the Type VI effectors [66, 67]. While being evolutionary and structurally unrelated to each other, both Type III and Type VI effectors target RNA. The recognition of the target triggers the non-specific nuclease activity [68, 69].
Why do DNA-targeting CRISPR-Cas systems require PAMs, whereas RNA-targeting systems rely on different mechanisms? In principle, the tag-antitag strategy should be suitable for discriminative DNA targeting (in fact, the tag-antitag strategy of Type III systems was elaborated when these systems had been believed to target DNA [64]). Yet, only the PAM-dependent mechanisms have evolved, convergently, in the DNA-targeting systems of Types I, II, and V [70]. To complicate the matters, it was suggested that Type III systems are ancestral to all Class 1 systems [70], implying that derived Type I systems have switched from the initial tag-antitag to the PAM-dependent self-avoidance.
The apparent preference for the PAM-dependent mechanisms in the DNA-targeting systems may be explained by the kinetics of target recognition by the effectors. The nitrogenous bases of nucleotides in the single-stranded RNA molecules are exposed, allowing direct interaction with the effector-bound crRNA. In contrast, in double-stranded DNA, complementary interactions with crRNA are impossible, at least initially, and the recognition requires local melting of the DNA duplex, which must dramatically slow down the process of target recognition. The binding energy of the interaction between the effector protein components and the double-stranded PAM sequence may provide the driving force for the initial target melting. In addition, the requirement for a PAM limits the number of available targets by at least an order of magnitude, decreasing the time needed to locate a matching protospacer. One can thus speculate that the primary reason for the use of PAMs is not so much to ensure the self-versus non-self discrimination, but to accelerate the search for matching protospacers in the double-stranded DNA fast enough to provide immunity.
THE ORIGIN OF TYPE III CRISPR-Cas SYSTEMS
The immune response mediated by Type III CRISPR-Cas relies on an interplay between several complex mechanisms, including acquisition of new spacers, maturation of crRNA, assembly of effector complexes, target recognition, synthesis of signaling compounds, activation of auxiliary effectors, and regulation of all aforementioned processes. Such baroque complexity inevitably raises a question about the origin and evolution of Type III systems.
The first glimpses into this puzzle came from the analysis of cas genes decoupled from the CRISPR-Cas systems. Some of the “solo” cas1 genes found outside of the CRISPR-Cas loci [71] were shown to belong to a new family of mobile genetic elements, named casposons, which employ Cas1 proteins for the integration/excision [72, 73]. The mechanism of casposon integration clearly resembles the mechanism of insertion of new spacers into the CRISPR arrays, suggesting that CRISPR-associated Cas1 have originated from a casposon integrase, while CRISPR repeats could be derived from the terminal repeats flanking the casposons [70, 73]. Along with the Cas1 proteins, casposons also encode the Cas4-like proteins [72], which are components of the spacer acquisition machinery in some CRISPR-Cas systems [51].
Another important component of the spacer integration complex is Cas2, a protein homologous to the VapD family nuclease toxins and thus presumably derived from corresponding toxin–antitoxin systems [74]. Although in many cases, Cas2 proteins retain nuclease activity, it is not essential for the spacer integration [75], in which Cas2 plays the structural functions by tethering the Cas1 dimers.
The origins of the effector complexes are less clear. Here, we focus only on a putative evolutionary history of Class 1 CRISPR-Cas systems, which includes Types I, III, and IV. These multisubunit complexes have similar architectures and share several homologous key subunits, suggesting their common origin (Fig. 4). The backbone of the Type I and Type III effectors consists of a crRNA molecule covered by several Cas7 monomers in a complex with small Cas11 subunits. The 5′ end of crRNA is bound to a Cas5 family protein. The large subunits (Cas8 in Type I and Cas10 in Type III) are located in the vicinity of the 5′ end of crRNA [76, 77]. Type I and Type III systems also share a common mechanism of crRNA maturation via the action of Cas6 proteins that recognize the stem-loop structures formed by the repeat sequences in pre-crRNA [78]. Although less is known about the Type IV effectors, it is clear that their backbone is also formed by several Cas7 subunits bound to crRNA [79]; Cas5 and Cas6 homologs are also encoded within the Type IV loci [4]. The key components of Type I and Type III effectors, such as Cas10, Cas5, Cas7, and Cas6, share structural similarity and contain domains with the RRM fold [71, 80]. In addition, Type I Cas11 proteins share structural similarity with the C-terminal domain of Cas10 from the Type III systems [71].
Fig. 4.
Comparison of structures of Type I and Type III effectors; homologous proteins are depicted by matching colors.
Based on these observations, Koonin and Makarova suggested a scenario of the origin of Class 1 CRISPR-Cas systems (Fig. 5). According to this scenario, despite their enormous complexity, Type III systems appear to be ancestral to all known Class 1 systems [4, 70, 71]. It is envisioned that the origin of Class 1 effectors goes back to a putative signaling system that included a Cas10-like polymerase and a CARF-HEPN effector protein that could produce nucleotide-based secondary messengers (likely, cOAs) in response to stress/environmental signals, followed by the activation of the RNase activity of the CARF-HEPN protein [4]. Indeed, loci encoding Cas10-like polymerases fused with the CARF-HEPN domains were detected through bioinformatic analysis [80], however, these systems are not yet characterized. The duplication of the ancestral Cas10-like protein gene followed by the fragmentation could have given rise to the Cas7-like and Cas11-like subunits. Genes encoding Cas5 and Cas6 could have originated through a fusion of two Cas7-like genes, since all these genes share a specific structural motif missing from the Cas10 proteins [70]. The acquisition of the adaptation module components from casposons and toxin–antitoxin systems finally could have given rise to a functional adaptive immune system [70].
Fig. 5.
Proposed scenario of the origin of the Type III CRISPR-Cas systems. Adopted from Koonin et al., 2019 [70].
We can only speculate about the original functions of ancestral Cas10-like-CARF-HEPN signaling systems and stimuli that activated the polymerase/cyclase activity of the Cas10 ancestors. Likewise, the functions of the prototypical Type III systems that must have existed before the acquisition of the adaptation module are unknown. However, distinct variants of Class 1 systems that are not linked to the CRISPR arrays are known [46]. Experimental characterization of these systems may shed light on the functions of ancient Class 1 effectors. Interestingly, it was shown that Type IV effector complexes heterologously expressed in Escherichia coli preferentially associate with small RNAs transcribed from plasmids despite the presence of a transcribed cognate CRISPR array and functional crRNA maturation components [81]. One could envision that ancient prototypical Type III effectors could also bind RNAs derived from mobile genetic elements, employing them as guides for the target recognition in the absence of CRISPR arrays and functioning as a primitive and inefficient immunity system similar to prokaryotic Argonaute proteins [82, 83]. Additionally, the Palm domain of Cas10 shares similarity with the catalytic core of the Thg1 enzyme, an unusual 3′-5′ RNA polymerase essential for the maturation of histidine tRNA [8]. Given this fact and assuming that the RNA-binding Cas7 proteins have originated from the Cas10 RRM domain, one could propose that the ancestor of the present-day Cas10 proteins possessed RNA-binding and RNA polymerase activities and was a part of systems involved in the synthesis, repair, and/or maturation of RNA molecules. Next, the RNA polymerase activity could have specialized/degenerated for the production of signaling compounds. The ligands for the ancestor Cas10 polymerase are unknown, but given the fact that the ancient Cas10 might have possessed the RNA-binding activity, it is possible that such ligands were RNA molecules. While it is hard to envision which RNA molecules activated the immune response in such system, it is noteworthy that many transposable elements, including some casposons, use tRNA genes as targets for their integration [72, 84]. Since Type IV effectors bind small RNAs, including tRNAs [85], one can speculate that before the acquisition of adaptation modules, ancestral Type III systems had recognized the transcripts produced from tRNA genes corrupted by the insertions of mobile genetic elements.
Defense systems that employ sensors producing nucleotide-based messengers to activate the downstream effectors are exceedingly diverse and widespread across all cellular organisms. Beside the described cOA pathway in Type III systems and eukaryotic OAS, one could mention a large family of cGAS/DncV-like nucleotidyltransferases. In animals, cyclic GMP-AMP synthase (cGAS) produces cyclic GMP-AMP in response to the cytosolic DNA. Cyclic GMP-AMP activates the pathway leading to the upregulation of numerous immune response genes [86]. Multiple cGAS homologs have been identified in the genomes of prokaryotes; many of them are associated with the genes for various effectors, including nucleases, phospholipases and transmembrane proteins that comprise so-called cyclic oligonucleotide-based antiphage signaling systems (CBASS) [87-90]. Since signal transduction between the sensors (cyclases) and effectors in such defense systems is mediated by small diffusing molecules, numerous sensor/effector pairs and interconnections between different systems has become possible. Interestingly, some of cGAS/DncV-like cyclases produce cOAs [90]; moreover, some of such cOA-activated CBASS effectors are also employed by the Type III CRISPR-Cas systems [91, 92].
CONCLUSIONS
Although being extremely complex, Type III systems appear to be ancestral to all Class 1 systems. Given the prevalence of Class 1 systems, Type III systems could in fact be the most ancient among all CRISPR-Cas systems. The mechanisms of Type III immunity have remained enigmatic for a long time. The discovery of the cOA pathway solved a big part of the puzzle; however, several aspects of the Type III immunity remain unclear. While the DNase activity of the Cas10 HD domain is essential for the immune response, its targets are still unknown, and we could only speculate about the biological role of this activity. Although auxiliary RNases (Csm6 and Csx1) were characterized as non-specific in in vitro experiments, the data on their in vivo specificity are lacking. There are a number of genes associated with the Type III CRISPR-Cas systems, but only few of such genes have been experimentally characterized. Several types of membrane-associated effectors have not been studied at all [93]. Beside complete Type III systems, simplified RAMP-containing loci were predicted by bioinformatics analysis. Such systems could be intermediates in the evolution of the present-day Type III CRISPR-Cas systems. In particular, the structural homology between the HRAMP signature proteins and the Cas10 proteins of Type III systems discovered by us suggests that the HRAMP systems have either originated from the existing Type III systems or from their ancestors. Finally, the activity and biological functions of the Cas10-CARF-HEPN signaling systems remain to be characterized. To summarize, our current understanding covers (incompletely) only a small part of mechanisms behind the action of the Type III CRISPR-Cas systems and lots of exciting research remains to be done.
Acknowledgments
We would like to thank Svetlana Belukhina for her help during the preparation of the review.
Funding
This work was supported by the grant of the Ministry of Science and Higher Education of the Russian Federation (075-15-2019-1661). M. K. was supported by the grant from the Russian Foundation for Basic Research (20-34-90147); D. A. was supported by the grant from the Russian Science Foundation (19-74-00124); I. F. was supported by the grant from the Russian Science Foundation (21-14-00122) and Grant of the President of the Russian Federation (МК-3793.2021.1.4).
Ethics declarations
The authors declare no conflict of interest. This article does not contain description of studies with the involvement of humans or animal subjects performed by any of the authors.
Contributor Information
Matvey V. Kolesnik, Email: matveykolesnik@gmail.com
Iana Fedorova, Email: femtokot@gmail.com.
Karyna A. Karneyeva, Email: karyna.karneyeva@skoltech.ru
Daria N. Artamonova, Email: daria.vorontsova@skolkovotech.ru
Konstantin V. Severinov, Email: severik@waksman.rutgers.edu
References
- 1.Bennett C. H. Dissipation-error tradeoff in proofreading. Biosystems. 1979;11:85–91. doi: 10.1016/0303-2647(79)90003-0. [DOI] [PubMed] [Google Scholar]
- 2.Koonin E. V., Wolf Y. I., Katsnelson M. I. Inevitability of the emergence and persistence of genetic parasites caused by evolutionary instability of parasite-free states. Biol. Direct. 2017;12:31. doi: 10.1186/s13062-017-0202-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrangou R., Horvath P. A decade of discovery: CRISPR functions and applications. Nat. Microbiol. 2017;2:17092. doi: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 4.Makarova K. S., Wolf Y. I., Iranzo J., Shmakov S. A., Alkhnbashi O. S., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova K. S., Aravind L., Grishin N. V., Rogozin I. B., Koonin E. V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravind L., Koonin E. V. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anantharaman V., Iyer L. M., Aravind L. Presence of a classical RRM-fold palm domain in Thg1-type 3′-5′ nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biol. Direct. 2010;5:43. doi: 10.1186/1745-6150-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova K. S., Haft D. H., Barrangou R., Brouns S. J. J., Charpentier E., et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marraffini L. A., Sontheimer E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale C. R., Zhao P., Olson S., Duff M. O., Graveley B. R., et al. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Rouillon C., Kerou M., Reeks J., Brugger K., et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg G. W., Jiang W., Bikard D., Marraffini L. A. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng L., Garrett R. A., Shah S. A., Peng X., She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 16.Gong B., Shin M., Sun J., Jung C.-H., Bolt E. L., et al. Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3. Proc. Natl. Acad. Sci. USA. 2014;111:16359–16364. doi: 10.1073/pnas.1410806111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazlauskiene M., Tamulaitis G., Kostiuk G., Venclovas Č., Siksnys V. Spatiotemporal control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell. 2016;62:295–306. doi: 10.1016/j.molcel.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 18.Elmore J. R., Sheppard N. F., Ramia N., Deighan T., Li H., et al. Bipartite recognition of target RNAs activates DNA cleavage by the type III-B CRISPR-Cas system. Genes Dev. 2016;30:447–459. doi: 10.1101/gad.272153.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estrella M. A., Kuo F.-T., Bailey S. RNA-activated DNA cleavage by the Type III-B CRISPR-Cas effector complex. Genes Dev. 2016;30:460–470. doi: 10.1101/gad.273722.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samai P., Pyenson N., Jiang W., Goldberg G. W., Hatoum-Aslan A., Marraffini L. A. Co-transcriptional DNA and RNA cleavage during Type III CRISPR-Cas immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T. Y., Liu J.-J., Aditham A. J., Nogales E., Doudna J. A. Target preference of Type III-A CRISPR-Cas complexes at the transcription bubble. Nat. Commun. 2019;10:3001. doi: 10.1038/s41467-019-10780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatoum-Aslan A., Maniv I., Samai P., Marraffini L. A. Genetic characterization of antiplasmid immunity through a type III-A CRISPR-Cas system. J. Bacteriol. 2014;196:310–317. doi: 10.1128/JB.01130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova K. S., Anantharaman V., Grishin N. V., Koonin E. V., Aravind L. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 2014;5:102. doi: 10.3389/fgene.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anantharaman V., Makarova K. S., Burroughs A. M., Koonin E. V., Aravind L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheppard N. F., Glover C. V. C., 3rd, Terns R. M., Terns M. P. The CRISPR-associated Csx1 protein of Pyrococcusfuriosus is an adenosine-specific endoribonuclease. RNA. 2016;22:216–224. doi: 10.1261/rna.039842.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niewoehner O., Jinek M. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA. 2016;22:318–329. doi: 10.1261/rna.054098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niewoehner O., Garcia-Doval C., Rostøl J. T., Berk C., Schwede F., et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548:543–548. doi: 10.1038/nature23467. [DOI] [PubMed] [Google Scholar]
- 28.Kazlauskiene M., Kostiuk G., Venclovas Č., Tamulaitis G., Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357:605–609. doi: 10.1126/science.aao0100. [DOI] [PubMed] [Google Scholar]
- 29.Han W., Stella S., Zhang Y., Guo T., Sulek K., et al. A Type III-B Cmr effector complex catalyzes the synthesis of cyclic oligoadenylate second messengers by cooperative substrate binding. Nucleic Acids Res. 2018;46:10319–10330. doi: 10.1093/nar/gkx1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang W., Samai P., Marraffini L. A. Degradation of phage transcripts by CRISPR-associated RNases enables Type III CRISPR-Cas immunity. Cell. 2016;164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rostøl J. T., Marraffini L. A. Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR-Cas immunity. Nat. Microbiol. 2019;4:656–662. doi: 10.1038/s41564-018-0353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Artamonova D., Karneyeva K., Medvedeva S., Klimuk E., Kolesnik M., et al. Spacer acquisition by Type III CRISPR-Cas system during bacteriophage infection of Thermusthermophilus. Nucleic Acids Res. 2020;48:9787–9803. doi: 10.1093/nar/gkaa685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasef M., Muffly M. C., Beckman A. B., Rowe S. J., Walker F. C., et al. Regulation of cyclic oligoadenylate synthesis by the Cas10–Csm complex. RNA. 2019;25:948–962. doi: 10.1261/rna.070417.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouillon C., Athukoralage J. S., Graham S., Grüschow S., White M. F. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. Elife. 2018;7:e36734. doi: 10.7554/eLife.36734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Athukoralage J. S., Rouillon C., Graham S., Grüschow S., White M. F. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature. 2018;562:277–280. doi: 10.1038/s41586-018-0557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Doval C., Schwede F., Berk C., Rostøl J. T., Niewoehner O., et al. Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat. Commun. 2020;11:1596. doi: 10.1038/s41467-020-15334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Athukoralage J. S., Graham S., Grüschow S., Rouillon C., White M. F. A Type III CRISPR ancillary ribonuclease degrades its cyclic oligoadenylate activator. J. Mol. Biol. 2019;431:2894–2899. doi: 10.1016/j.jmb.2019.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Athukoralage J. S., McMahon S. A., Zhang C., Grüschow S., Graham S., et al. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature. 2020;577:572–575. doi: 10.1038/s41586-019-1909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornung V., Hartmann R., Ablasser A., Hopfner K.-P. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar S. N., Ghosh A., Wang H. W., Sung S. S., Sen G. C. The nature of the catalytic domain of 2′-5′-oligoadenylate synthetases. J. Biol. Chem. 1999;274:25535–25542. doi: 10.1074/jbc.274.36.25535. [DOI] [PubMed] [Google Scholar]
- 41.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A. D., Rawlik K., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 42.Mo C. Y., Mathai J., Rostøl J. T., Varble A., Banh D. V., Marraffini L. A. Type III-A CRISPR immunity promotes mutagenesis of staphylococci. Nature. 2021;592:611–615. doi: 10.1038/s41586-021-03440-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staals R. H. J., Agari Y., Maki-Yonekura S., Zhu Y., Taylor D. W., et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermusthermophilus. Mol. Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steens J. A., Zhu Y., Taylor D. W., Bravo J. P. K., Prinsen S. H. P., et al. SCOPE enables type III CRISPR-Cas diagnostics using flexible targeting and stringent CARF ribonuclease activation. Nat. Commun. 2021;12:5033. doi: 10.1038/s41467-021-25337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMahon S. A., Zhu W., Graham S., Rambo R., White M. F., Gloster T. M. Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate. Nat. Commun. 2020;11:500. doi: 10.1038/s41467-019-14222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makarova K. S., Karamycheva S., Shah S. A., Vestergaard G., Garrett R. A., Koonin E. V. Predicted highly derived class 1 CRISPR-Cas system in Haloarchaea containing diverged Cas5 and Cas7 homologs but no CRISPR array. FEMS Microbiol. Lett. 2019;366:fnz079. doi: 10.1093/femsle/fnz079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You L., Ma J., Wang J., Artamonova D., Wang M., et al. Structure studies of the CRISPR–Csm complex reveal mechanism of co-transcriptional interference. Cell. 2019;176:239–253.e16. doi: 10.1016/j.cell.2018.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mirdita M., Ovchinnikov S., Steinegger M. ColabFold – Making protein folding accessible to all. bioRxiv. 2021 doi: 10.1101/2021.08.15.456425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rostøl J. T., Xie W., Kuryavyi V., Maguin P., Kao K., et al. The Card1 nuclease provides defence during type III CRISPR immunity. Nature. 2021;590:624–629. doi: 10.1038/s41586-021-03206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson S. A., McKenzie R. E., Fagerlund R. D., Kieper S. N., Fineran P. C., Brouns S. J. J. CRISPR-Cas: adapting to change. Science. 2017;356:eaal5056. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 52.Levy A., Goren M. G., Yosef I., Auster O., Manor M., et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Modell J. W., Jiang W., Marraffini L. A. CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature. 2017;544:101–104. doi: 10.1038/nature21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiriaeva A. A., Savitskaya E., Datsenko K. A., Vvedenskaya I. O., Fedorova I., et al. Detection of spacer precursors formed in vivo during primed CRISPR adaptation. Nat. Commun. 2019;10:4603. doi: 10.1038/s41467-019-12417-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toro N., Martínez-Abarca F., Mestre M. R., González-Delgado A. Multiple origins of reverse transcriptases linked to CRISPR-Cas systems. RNA Biol. 2019;16:1486–1493. doi: 10.1080/15476286.2019.1639310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silas S., Makarova K. S., Shmakov S., Páez-Espino D., Mohr G., et al. On the origin of reverse transcriptase-using CRISPR-Cas systems and their hyperdiverse, enigmatic spacer repertoires. MBio. 2017;8:e00897–17. doi: 10.1128/mBio.00897-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silas S., Mohr G., Sidote D. J., Markham L. M., Sanchez-Amat A., et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science. 2016;351:aad4234. doi: 10.1126/science.aad4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.González-Delgado A., Mestre M. R., Martínez-Abarca F., Toro N. Spacer acquisition from RNA mediated by a natural reverse transcriptase-Cas1 fusion protein associated with a type III-D CRISPR-Cas system in Vibriovulnificus. Nucleic Acids Res. 2019;47:10202–10211. doi: 10.1093/nar/gkz746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J. Y., Hoel C. M., Al-Shayeb B., Banfield J. F., Brohawn S. G., Doudna J. A. Structural coordination between active sites of a CRISPR reverse transcriptase–integrase complex. Nat. Commun. 2021;12:2571. doi: 10.1038/s41467-021-22900-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gleditzsch D., Pausch P., Müller-Esparza H., Özcan A., Guo X., et al. PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 2019;16:504–517. doi: 10.1080/15476286.2018.1504546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao Y., Luo M., Hayes R. P., Kim J., Ng S., et al. Structure basis for directional R-loop formation and substrate handover mechanisms in Type I CRISPR-Cas system. Cell. 2017;170:48–60.e11. doi: 10.1016/j.cell.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh D., Mallon J., Poddar A., Wang Y., Tippana R., et al. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a) Proc. Natl. Acad. Sci. USA. 2018;115:5444–5449. doi: 10.1073/pnas.1718686115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marraffini L. A., Sontheimer E. J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamulaitis G., Kazlauskiene M., Manakova E., Venclovas Č., Nwokeoji A. O., et al. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol. Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 66.Liu L., Li X., Ma J., Li Z., You L., et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170:714–726.e10. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 67.Meeske A. J., Marraffini L. A. RNA guide complementarity prevents self-targeting in Type VI CRISPR systems. Mol. Cell. 2018;71:791–801.e3. doi: 10.1016/j.molcel.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abudayyeh O. O., Gootenberg J. S., Konermann S., Joung J., Slaymaker I. M., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meeske A. J., Nakandakari-Higa S., Marraffini L. A. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature. 2019;570:241–245. doi: 10.1038/s41586-019-1257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koonin E. V., Makarova K. S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374:20180087. doi: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Makarova K. S., Wolf Y. I., Koonin E. V. The basic building blocks and evolution of CRISPR-CAS systems. Biochem. Soc. Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krupovic M., Makarova K. S., Forterre P., Prangishvili D., Koonin E. V. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Béguin P., Charpin N., Koonin E. V., Forterre P., Krupovic M. Casposon integration shows strong target site preference and recapitulates protospacer integration by CRISPR-Cas systems. Nucleic Acids Res. 2016;44:10367–10376. doi: 10.1093/nar/gkw821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Makarova K. S., Grishin N. V., Shabalina S. A., Wolf Y. I., Koonin E. V. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nuñez J. K., Kranzusch P. J., Noeske J., Wright A. V., Davies C. W., Doudna J. A. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jackson R. N., Golden S. M., van Erp P. B. G., Carter J., Westra E. R., et al. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichiacoli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor D. W., Zhu Y., Staals R. H. J., Kornfeld J. E., Shinkai A., et al. Structural biology. Structures of the CRISPR–Cmr complex reveal mode of RNA target positioning. Science. 2015;348:581–585. doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Charpentier E., Richter H., van der Oost J., White M. F. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol. Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Özcan A., Pausch P., Linden A., Wulf A., Schühle K., et al. Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. Nat. Microbiol. 2019;4:89–96. doi: 10.1038/s41564-018-0274-8. [DOI] [PubMed] [Google Scholar]
- 80.Burroughs A. M., Zhang D., Schäffer D. E., Iyer L. M., Aravind L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 2015;43:10633–10654. doi: 10.1093/nar/gkv1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Y., Bravo J. P. K., Taylor H. N., Steens J. A., Jackson R. N., et al. Structure of a type IV CRISPR-Cas ribonucleoprotein complex. iScience. 2021;24:102201. doi: 10.1016/j.isci.2021.102201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Swarts D. C., Jore M. M., Westra E. R., Zhu Y., Janssen J. H., et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuzmenko A., Oguienko A., Esyunina D., Yudin D., Petrova M., et al. DNA targeting and interference by a bacterial Argonaute nuclease. Nature. 2020;587:632–637. doi: 10.1038/s41586-020-2605-1. [DOI] [PubMed] [Google Scholar]
- 84.Krupovic M., Béguin P., Koonin E. V. Casposons: mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery. Curr. Opin. Microbiol. 2017;38:36–43. doi: 10.1016/j.mib.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou Y., Bravo J. P. K., Taylor H. N., Steens J., Jackson R. N., et al. Structure of a type IV CRISPR-Cas effector complex. bioRxiv. 2020 doi: 10.1101/2020.07.31.231399. [DOI] [Google Scholar]
- 86.Hopfner K.-P., Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 87.Millman A., Melamed S., Amitai G., Sorek R. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. 2020;5:1608–1615. doi: 10.1038/s41564-020-0777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cohen D., Melamed S., Millman A., Shulman G., Oppenheimer-Shaanan Y., Kacen A., et al. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature. 2019;574:691–695. doi: 10.1038/s41586-019-1605-5. [DOI] [PubMed] [Google Scholar]
- 89.Lowey B., Whiteley A. T., Keszei A. F. A., Morehouse B. R., Mathews I. T., et al. CBASS immunity uses CARF-related effectors to sense 3′-5′- and 2′-5′-linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell. 2020;182:38–49.e17. doi: 10.1016/j.cell.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ye Q., Lau R. K., Mathews I. T., Birkholz E. A., Watrous J. D., et al. HORMA domain proteins and a Trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Mol. Cell. 2020;77:709–722.e7. doi: 10.1016/j.molcel.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lau R. K., Ye Q., Birkholz E. A., Berg K. R., Patel L., et al. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell. 2020;77:723–733.e6. doi: 10.1016/j.molcel.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Malone L. M., Warring S. L., Jackson S. A., Warnecke C., Gardner P. P., et al. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat. Microbiol. 2020;5:48–55. doi: 10.1038/s41564-019-0612-5. [DOI] [PubMed] [Google Scholar]
- 93.Shmakov S. A., Makarova K. S., Wolf Y. I., Severinov K. V., Koonin E. V. Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc. Natl. Acad. Sci. USA. 2018;115:E5307–E5316. doi: 10.1073/pnas.1803440115. [DOI] [PMC free article] [PubMed] [Google Scholar]