Abstract
Maintenance of chromatin structure under the disruptive force of transcription requires cooperation among numerous regulatory factors. Histone post-translational modifications can regulate nucleosome stability and influence the disassembly and reassembly of nucleosomes during transcription elongation. The Paf1 transcription elongation complex, Paf1C, is required for several transcription-coupled histone modifications, including the mono-ubiquitylation of H2B. In Saccharomyces cerevisiae, amino acid substitutions in the Rtf1 subunit of Paf1C greatly diminish H2B ubiquitylation and cause transcription to initiate at a cryptic promoter within the coding region of the FLO8 gene, an indicator of chromatin disruption. In a genetic screen to identify factors that functionally interact with Paf1C, we identified mutations in HDA3, a gene encoding a subunit of the Hda1C histone deacetylase (HDAC), as suppressors of an rtf1 mutation. Absence of Hda1C also suppresses the cryptic initiation phenotype of other mutants defective in H2B ubiquitylation. The genetic interactions between Hda1C and the H2B ubiquitylation pathway appear specific: loss of Hda1C does not suppress the cryptic initiation phenotypes of other chromatin mutants and absence of other HDACs does not suppress the absence of H2B ubiquitylation. Providing further support for an appropriate balance of histone acetylation in regulating cryptic initiation, absence of the Sas3 histone acetyltransferase elevates cryptic initiation in rtf1 mutants. Our data suggest that the H2B ubiquitylation pathway and Hda1C coordinately regulate chromatin structure during transcription elongation and point to a potential role for a HDAC in supporting chromatin accessibility.
Keywords: Hda1 histone deacetylase complex, H2B K123 ubiquitylation, Rtf1, cryptic transcription, Paf1 complex
Introduction
Transcription of eukaryotic genes requires passage of RNA polymerase through a generally repressive chromatin template. Nucleosomes, which consist of approximately 147 base pairs of DNA wrapped around an octamer of histones H2A, H2B, H3 and H4, present a barrier to RNA polymerase II (Pol II) progression (Farnung et al. 2018; Kujirai et al. 2018; Chen et al. 2019). Organisms have evolved numerous mechanisms to overcome this barrier. Variants of histones can replace their canonical counterparts at certain locations in the genome, such as the presence of H2A.Z in exchange for H2A in the nucleosome downstream of the transcription start site (TSS) (Bagchi et al. 2020). Chromatin remodeling factors, such as those in the SWI/SNF, ISWI, INO80 and CHD families, alter the positioning of nucleosomes to regulate DNA accessibility (Tyagi et al. 2016), and the histone chaperones FACT and Spt6 facilitate the reassembly of nucleosomes following Pol II passage (Gurard-Levin et al. 2014; Hammond et al. 2017; Formosa and Winston 2021). The post-translational modification of histones, though, may represent the most diversified means by which nucleosomes can regulate Pol II transcription.
With high selectivity, histone modifying enzymes carry out the covalent modification (e.g. phosphorylation, acetylation, methylation or ubiquitylation) of amino acids within the N- and C-terminal tails or core domains of the histones (Bannister and Kouzarides 2011; Lawrence et al. 2016). At the genomic level, these modifications are deposited in specific patterns and, depending on their functions, are enriched in transcribed or non-transcribed regions. Enzymes that modify histones are hypothesized to generate a “histone code” that is interpreted by effector proteins, which bind with high specificity to appropriately modified histones (Jenuwein and Allis 2001). For example, acetylation of lysines by histone acetyltransferases (HATs) (Lee and Workman 2007) loosens the interactions of histone tails with the DNA backbone (Hong et al. 1993), providing increased DNA accessibility to the transcription machinery and modulating transcription. In addition, acetylated histones can serve as binding sites for other regulatory factors, such as chromatin remodeling factors (Musselman et al. 2012; Marmorstein and Zhou 2014). Histone deacetylases (HDACs) remove the acetyl groups, thus reversing these effects (Seto and Yoshida 2014; Porter and Christianson 2019; Park and Kim 2020).
Crosstalk among histone modifications allows the integration of multiple signals to control transcription. A well-studied example is the dependency of H3 K4 and H3 K79 di- and tri-methylation (me2 and me3) on the mono-ubiquitylation of a conserved lysine in H2B (K123 in Saccharomyces cerevisiae; K120 in H. sapiens) (Worden and Wolberger 2019). H2B K123 mono-ubiquitylation (H2Bub) is catalyzed by the ubiquitin conjugating enzyme Rad6 and the ubiquitin protein-ligase Bre1 (Robzyk et al. 2000; Hwang et al. 2003; Wood et al. 2003a; Fuchs and Oren 2014) and is enriched on the bodies of actively transcribed genes (Minsky et al. 2008; Batta et al. 2011; Van Oss et al. 2016). The Set1 and Dot1 histone methyltransferases that modify H3 K4 and H3 K79, respectively, require the ubiquitin moiety on H2B for activity (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002; Hsu et al. 2019; Worden and Wolberger 2019; Worden et al. 2020). In turn, H3 K4 methylation as well as H3 K36me3 recruit the NuA3 HAT to the genome through its Yng1 and Pdp3 subunits, respectively, thereby controlling the location of histone acetylation by the Sas3 subunit (Taverna et al. 2006; Martin et al. 2017). In other cases, one histone modification may regulate the removal of another modification. For example, H3 K36me2/3 by Set2 activates the Rpd3S deacetylase complex to remove acetyl groups from histones and restore a repressive chromatin structure that prevents initiation of Pol II transcription within protein coding regions (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005; Drouin et al. 2010; Govind et al. 2010; Ruan et al. 2015; Venkatesh and Workman 2015). Thus, the enzymes that modify histones coordinately control chromatin structure and collectively impose or remove barriers to transcription.
In addition to the histone modifiers themselves, transcription factors that associate with Pol II can significantly influence the epigenetic state of chromatin. Previous work on the S. cerevisiae Polymerase-Associated Factor 1 Complex (Paf1C), which is composed of Paf1, Ctr9, Cdc73, Rtf1 and Leo1, has demonstrated a key role for this transcription elongation complex in regulating conserved transcription-coupled histone modifications. Paf1C has not been shown to possess enzymatic activity; however, mutations in genes encoding Paf1C subunits greatly diminish the levels of H2Bub, H3 K4me2/3, H3 K79me2/3 and H3 K36me3 (Krogan et al. 2003; Ng et al. 2003a,b; Wood et al. 2003b; Laribee et al. 2005; Chu et al. 2007; Piro et al. 2012; Van Oss et al. 2016, 2017). We previously identified a small region within the Rtf1 subunit of Paf1C, termed the histone modification domain (HMD), which appears to function as a critical cofactor in the deposition of H2Bub. The HMD is necessary and sufficient to promote H2Bub in vivo, stimulates H2Bub in vitro, and binds directly to Rad6 (Piro et al. 2012; Van Oss et al. 2016). Mutations that disrupt the Rad6-HMD interaction severely reduce levels of H2Bub, H3 K4me2/3 and H3 K79me2/3 and cause phenotypes indicative of disrupted chromatin structure (Warner et al. 2007; Tomson et al. 2011; Van Oss et al. 2016). For example, certain amino acid substitutions within the HMD lead to transcription initiation at a cryptic TSS within the coding region of a Pol II transcribed gene (Tomson et al. 2011; Van Oss et al. 2016), a mutant phenotype that arises when the repressive chromatin state of transcribed regions is disrupted by the loss of histone chaperones, histone modifiers and chromatin remodeling factors (Kaplan et al. 2003; Cheung et al. 2008; Silva et al. 2012). In this study, we exploited the cryptic transcription initiation phenotype of HMD mutants and performed a genetic screen to investigate the functional interactions involving this domain and the histone modification it facilitates, H2Bub. Our results uncovered an interplay between the factors that establish H2Bub during transcription elongation and the Hda1 deacetylase complex (Hda1C) and suggest that Hda1C, directly or indirectly, impacts chromatin accessibility within gene bodies.
Materials and methods
Yeast strains and growth conditions
Saccharomyces cerevisiae strains were grown at 30°C in YPD or SC media (Rose et al. 1990). The SC-His+Gal medium used to monitor the cryptic initiation phenotype at the GAL1p-FLO8-HIS3 reporter contained 2% galactose. To assess defects in telomeric silencing, SC media contained 0.1% 5-fluoroorotic acid (5-FOA; USBiological, F5050). Saccharomyces cerevisiae strains used in this study are listed in Supplementary Table S1. KY strains are isogenic with FY2, a GAL2+ derivative of S288C (Winston et al. 1995). KA strains were derived from crosses originating with strains from a library of histone H3 mutants (Dai et al. 2008). Strains were made by standard methods for genetic crosses or gene replacements (Rose et al. 1990; Vidal et al. 1991). Strains containing a mutation in RTF1 were identified during tetrad analysis by the presence of an HA-tag sequence, using PCR amplification. Strains containing the htb1-K123R allele were identified by restriction enzyme digestion as previously described (Tomson et al. 2011). Strains containing the hda1-H206A replacement were created by delitto perfetto (Storici and Resnick 2006). A portion of the HDA1 gene (+1538 to +1868) was replaced with pCORE using homologous recombination in KY3880 and selecting for Ura+, G418R colonies. The pCORE segment was replaced with a PCR-amplified DNA fragment containing hda1-H206A by selecting for 5FOAR colonies and screening for G418S. The hda1-H206A mutation was created by PCR mutagenesis of a plasmid containing HDA1. Strains containing the phenotypic reporters were derived from genetic crosses.
For phenotypes assessed by replica plating, strains were first purified on YPD. For serial dilution analyses in Figures 2A and 3, strains were grown overnight in liquid YPD, diluted 1:10 and five-fold serial dilutions were pinned (Sigma-Aldrich R2383) to SC Complete, 5FOA, SC-His+Gal or SC-His media. For serial dilution analysis in Figure 7, strains were grown overnight in liquid YPD, diluted to OD600 = 0.8, and 3 µl of five-fold serial dilutions were spotted onto SC-His+Gal or SC Complete media.
Figure 2.
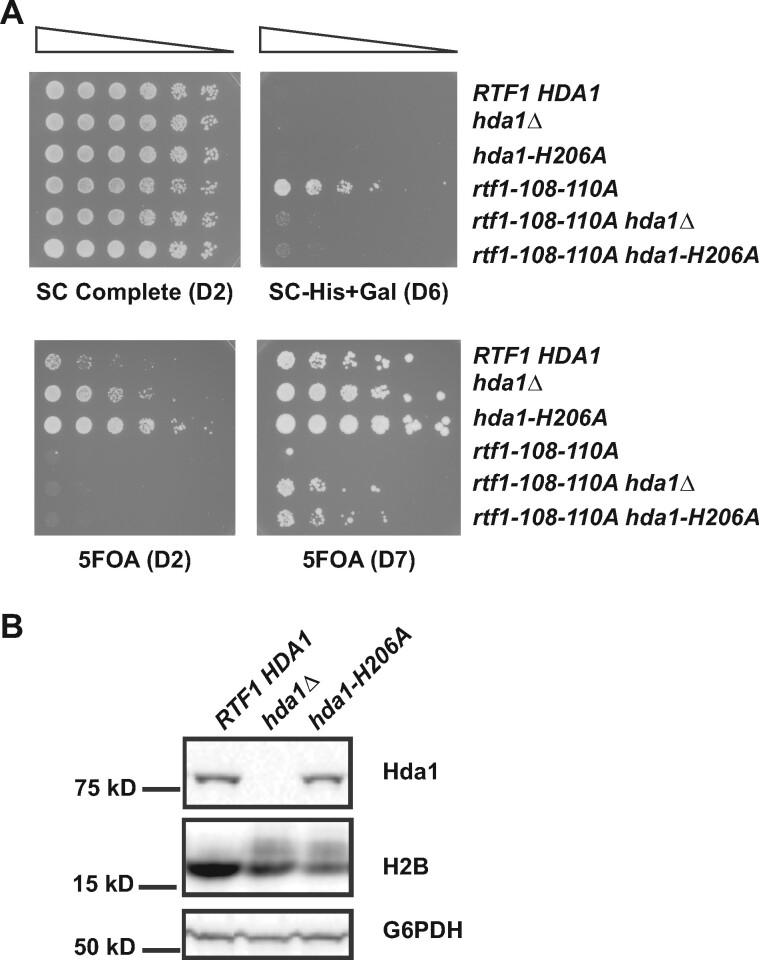
A substitution in the Hda1 catalytic domain suppresses an rtf1 HMD mutant. (A) Cryptic initiation and telomeric silencing phenotypes of the indicated strains containing the GAL1p-FLO8-HIS3 and TELV::URA3 reporters were measured on SC-His+Gal (top panel) and 5FOA-containing media (bottom panel), respectively. (B) Western blot analysis of Hda1 and H2B levels in extracts prepared from a wild type strain, an hda1Δ strain, and an hda1-H206A strain. The following strains were used: KY1523, KY2963, KY3910, KY1494, KY2973 and KY3911
Figure 3.
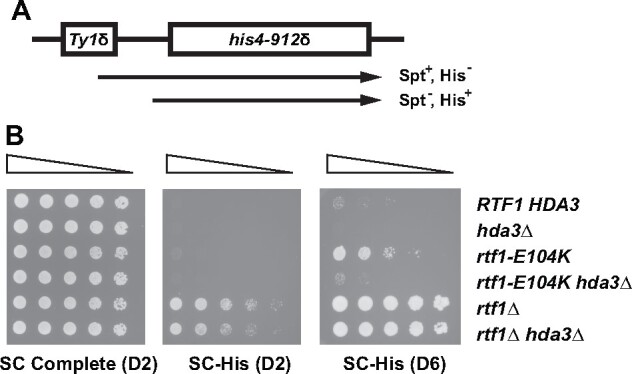
The Spt− phenotype of rtf1-E104K is suppressed by hda3Δ. The Spt− phenotype of rtf1-E104K at the his4-912δ locus (diagrammed at top) was detected on medium lacking histidine (SC-His). Plates incubated for two (D2) or six (D6) days at 30°C are shown. Reduced growth of the rtf1-E104K hda3Δ strain on the SC-His medium indicates suppression of the Spt− phenotype. The following strains were used: KY3191, KY3186, KY1532, KY3178, KY3034 and KY3032.
Figure 7.
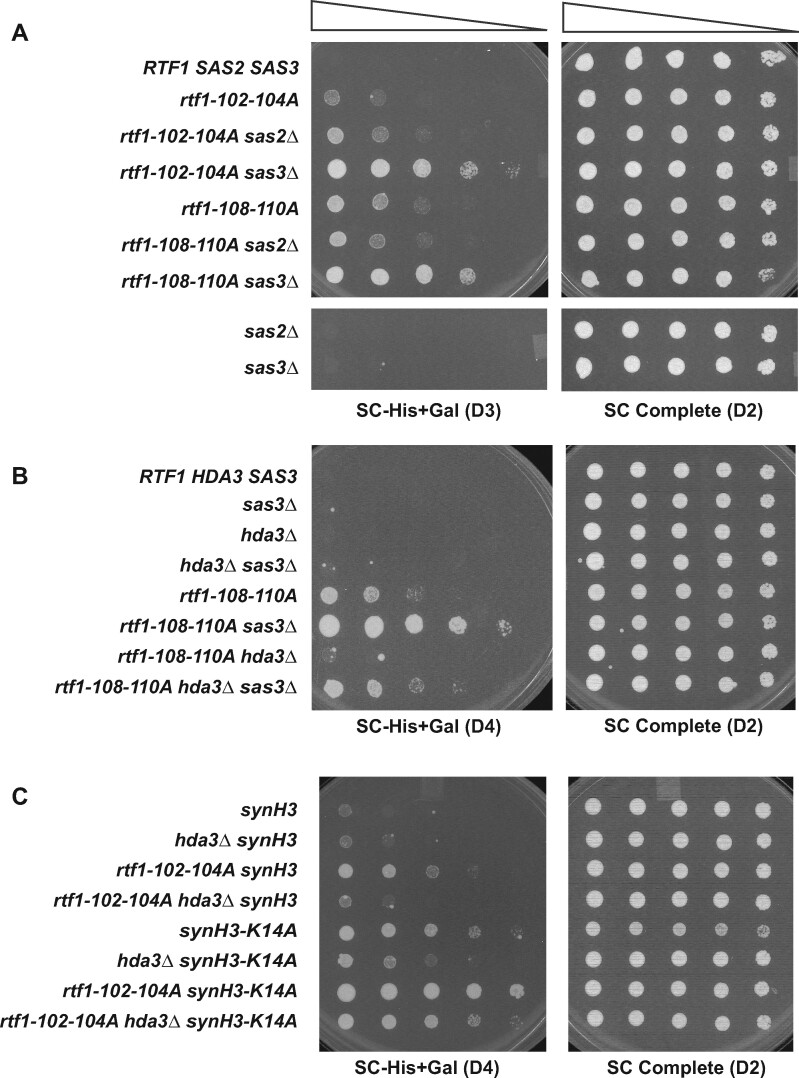
Mutation of SAS3 or H3 K14 enhances the cryptic initiation phenotype of rtf1 HMD mutants. (A, B) Strains with the indicated genotypes were grown as described in the Materials and Methods and serial dilutions were spotted onto SC-His+Gal and SC Complete media and allowed to grow for two (D2), three (D3) or four (D4) days at 30°C before imaging. (C) Strains deleted for HHT1-HHF1 and carrying a synthetic copy of HHT2-HHF2 (synH3), either wild-type or H3 K14A, were used in genetic crosses to create double and triple mutants with rtf1-102-104A and hda3Δ. Strains were grown on the indicated media for two to four days at 30°C. The following strains were used: (A) KY1491, KY2792, KY3366, KY3371, KY1490, KY3369, KY3372, KY3362 and KY3364; (B) KY3423, KY3431, KY3427, KY3438, KY3425, KY3434, KY3429 and KY3441; (C) KA255, KA256, KA257, KA258, KA295, KA296, KA297 and KA298.
Identification of suppressors of an rtf1-108-110A mutation
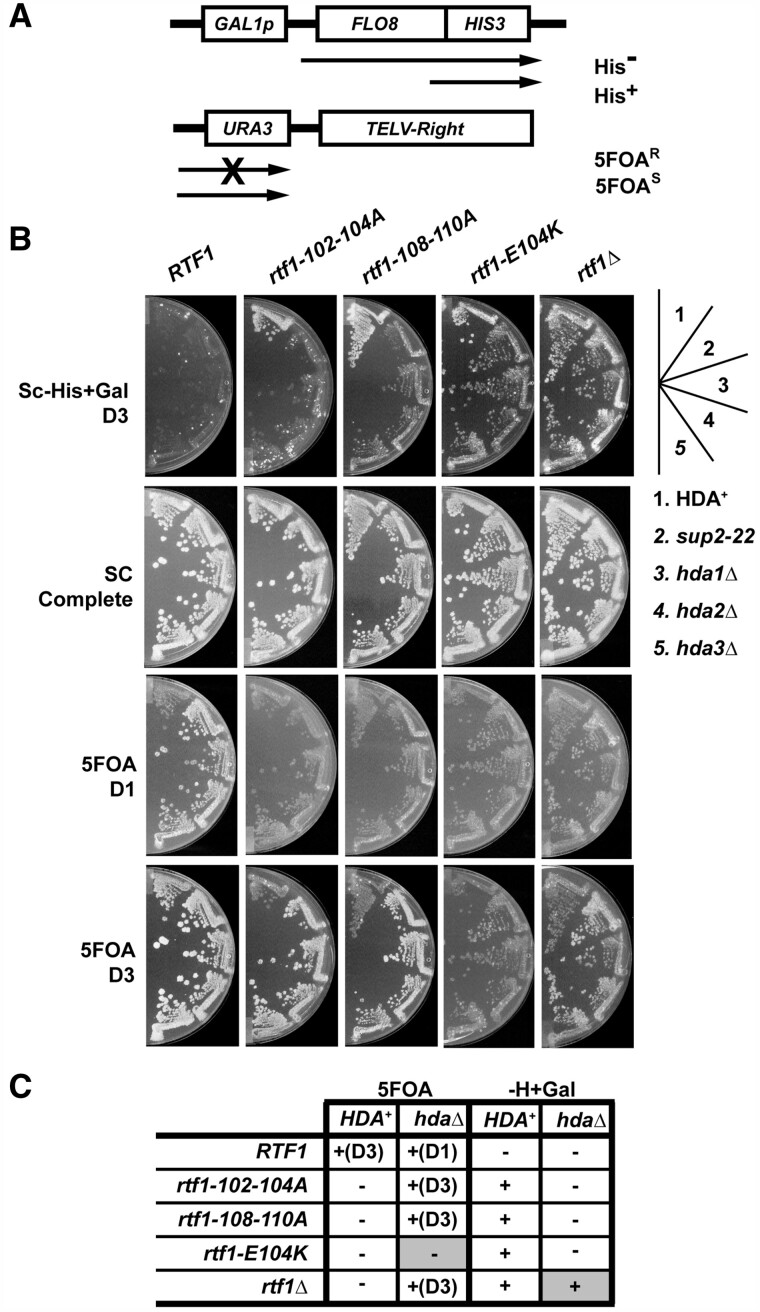
With the original goal of identifying high-copy-number suppressors of the rtf1-108-110A mutation, KY1232 was transformed with a LEU2-marked high-copy-number plasmid library and replica-plated to SC-His-Leu+Gal and SC-Leu + 5FOA media to monitor suppression of the cryptic initiation and telomeric silencing phenotypes, respectively. Using a strategy previously described (Thompson et al. 1993), the pRS425-based (Christianson et al. 1992) library was made in our laboratory from genomic DNA prepared from an rtf1Δ strain (KY957) to avoid recovering RTF1 plasmids that complemented the rtf1-108-110A mutation. Two 5FOA resistant (5FOAR), His- strains were isolated and, surprisingly, retained the suppression phenotypes even after plasmid loss through growth in nonselective conditions, indicating that they harbored chromosomal suppressors of rtf1-108-110A. The two strains, following plasmid loss, were saved as KY1410 and KY1411 and studied further. Diploid yeast from genetic matings with rtf1-108-110A strains and KY1410 and KY1411 showed that the two suppressor mutations were recessive, as the diploids grew on SC-His+Gal media. Additional crosses between KY1410 and KY1411 and with a strain containing RTF1, KY1228, showed that the suppression phenotype of each strain was due to a mutation in a single gene and that the suppressor mutation in each strain was unlinked to the rtf1-108-110A mutation. Haploid derivatives from these crosses were used in genetic matings, which showed that the two suppressor mutations defined a single complementation group. The suppressor mutations in KY1410 and KY1411, named sup2-22 and sup2-23, respectively, behaved similarly in all phenotypic tests.
Several attempts to identify the sup2-22 suppressor mutation by complementation of the cryptic initiation phenotype using plasmid-based yeast genomic DNA libraries were unsuccessful. Two genes that reversed the cryptic initiation phenotype of the rtf1-108-110A sup2-22 strain were HSF1 and ASF1. However, further genetic analysis showed that the suppressor mutation was not in these genes, but rather expression of these genes from a plasmid was likely affecting expression from the cryptic initiation reporter. Specifically, in the FLO8 gene, upstream of where the HIS3 reporter is inserted, we noticed a potential Hsf1 binding site 80 bp upstream of the predicted TATA box for the cryptic transcript, suggesting that overexpression of Hsf1 may drive higher expression of the HIS3 reporter. Further, RTF1 strains that were transformed with an Asf1-expressing plasmid grew on the SC-His+Gal media, suggesting that Asf1 was bypassing the effect of the suppressor. This is consistent with the role of Asf1 in promoting histone exchange and increasing histone acetylation on gene bodies (Hammond et al. 2017; Zhang et al. 2018). Therefore, we performed bulk segregant analysis (Birkeland et al. 2010) followed by whole-genome sequencing to identify the sup2-22 suppressor.
Six tetrads were selected from a cross between KY1232 and KY1498, and genomic DNA was prepared (Gopalakrishnan and Winston 2019) from the two pools of twelve haploid progeny either exhibiting a sup2-22 phenotype or not exhibiting a sup2-22 phenotype. Sequences were obtained from libraries prepared using an NEB Ultra II Library Prep Kit (New England Biolabs # E7103), multiplexed and run on an Illumina MiSeq with a 150-cycle v3 Reagent Kit yielding approximately 7.6 million reads per sample. Raw reads were mapped to the S. cerevisiae genome (S288C version = R64-2-1) (Cherry et al. 2012; Engel et al. 2014) using Bowtie2 (Langmead and Salzberg 2012). SAMtools were used to calculate genotype likelihoods before calling sequence variants using BCFtools (Li et al. 2009; Li 2011). Using VCFtools (Danecek et al. 2011), VCF files for mutant and wild-type segregants were compared and only the mutations found in the suppressor pool were selected from the output text (Ellison et al. 2020). Variants with a quality score > 50 and found within coding regions were retained, leaving three variants. Two of the variants were synonymous (Mtl1 S257S and Flo5 V354V), and a third variant, which created a frameshift (GT to C at +1118), mapped within the HDA3 gene. Sequence variants of interest were further inspected by visualizing the data in the Integrative Genomics Viewer (IGV) from the Broad Institute (Thorvaldsdottir et al. 2013). The mutation at codon 373 of HDA3 is predicted to change seven amino acids in the protein product before a stop codon is encountered. Subsequent direct sequencing of the HDA3 gene in a sup2-23 strain revealed a C to T substitution at nucleotide 592 of the open reading frame, which replaces codon 198 with a stop codon.
Western blot analysis
Western blots to visualize Hda1 and H2Bub were performed using whole-cell extracts prepared using SUTEB buffer as described (Van Oss et al. 2016). H3 methylation was examined using whole-cell extracts prepared using a TCA extraction method (Van Oss et al. 2016). Proteins were run on 15% SDS-polyacrylamide gels. The following antibodies were used: α-Hda1 (Santa Cruz sc-393814; 1:200), α-H2B (Active Motif #39237; 1:3000), α-H2Bub (Cell Signaling #5546; 1:1000), α-G6PDH (Sigma #A9521; 1:20000), α-H3K4Me2 (Millipore #07-030; 1:2000), α-H3K4Me3 (Active Motif #39159; 1:2000), α-H3K79Me2/3 (Abcam #ab2621; 1:1000; note that this antibody recognizes both di- and trimethylated H3K79) and α-H3 antibody (Tomson et al. 2011). Blots were developed using Thermo Scientific West Pico Plus (34580) and images were collected on a BioRad ChemiDoc™ XRS+.
Northern hybridization analysis
Cells were grown at 30°C to OD600 = 0.6 in YP media with 2% galactose. Isolation of RNA and northern analyses were performed as described (Swanson et al. 1991; Shirra and Arndt 1999), except that a 1.5% MOPS-formaldehyde-agarose gel was used. A region from the FLO8 gene (+40 to +671) or the SCR1 gene (−242 to +283) was amplified by PCR and the hybridization probe was made by random priming in the presence of [α-32P-dATP] (Perkin-Elmer BLU512H) using the Klenow fragment (New England Biolabs M0210). Quantification was done using a .tif file generated by phosphorimaging (Amersham Typhoon) and analyzed by Image Lab 6.1 software (Bio-Rad). The SCR1 signal was used for normalization.
Chromatin immunoprecipitation (ChIP) analysis
Cells were grown to 30°C to OD600 = 0.6 in YP media with 2% galactose. With the exception of the sonication step, extracts were prepared as previously described (Shirra et al. 2005). Sonicated chromatin was prepared using a Bioruptor (Diagenode B01060010) with 25 cycles of 30 s on and 30 s off. Immunoprecipitations were performed using 350 µl of extract and 1 µl of 8WG16 antibody (Biolegend 664912). After overnight incubations with primary antibody, 15 µl of Protein A beads (Cytiva 17528001) were added and incubation proceeded at room temperature for 2 h. Beads were washed and the DNA was obtained by reversing the crosslinking, as previously described (Shirra et al. 2005). DNA was purified using the QIAquick PCR Purification Kit (Qiagen #28106). Samples were analyzed by qPCR using a QuantStudio3™ Real-Time PCR System (Thermo Fisher) with SyGreen Blue Mix Lo-ROX (Genesee 17-505B). Input and immunoprecipitated DNA was amplified from the 5' region of the FLO8 gene (+23 to +126; primer efficiency = 1.98) and normalized to DNA amplified near TEL06R (chromosomal coordinates, 269495 to 269598).
Data reproducibility
All growth phenotypes were observed at expected frequencies in genetic crosses. Dilution growth experiments were performed in biological triplicate. Western blots, northern analyses and ChIP experiments were performed in biological duplicate with each biological replicate analyzed twice at minimum. For ChIP experiments, the biological replicates were subjected to immunoprecipitation two independent times. Results shown in Figure 4 were also observed with sup2-22 and hda3Δ strains.
Figure 4.
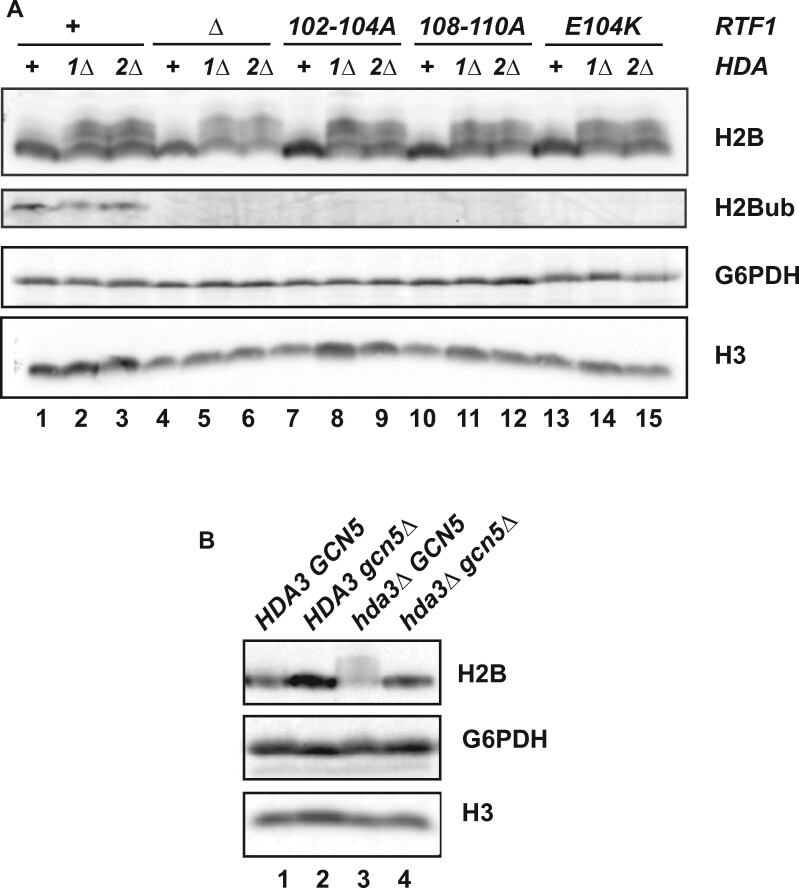
Loss of Hda1C does not restore H2Bub in rtf1 mutants. (A) Western blot analysis of the indicated rtf1 mutant strains in which Hda1C is intact (+) or the HDA1 (1Δ) or HDA2 (2Δ) genes have been deleted. Antibodies against total H2B or H2Bub were used to monitor levels of H2B K123 ubiquitylation. Longer exposures also failed to show a recovery of H2Bub in the hda1Δ and hda2Δ strains. The following strains were used: KY1523, KY2963, KY2934, KY2987, KY2981, KY2983, KY2792, KY2957, KY2965, KY1494, KY2973, KY2974, KY3002, KY2999 and KY3001. (B) Western blot analysis of the indicated strains. The following strains were used: KY433, KY3014, KY2861 and KY3011. In both panels, levels of G6PDH and H3 served as controls.
Results
Loss of Hda1C suppresses three different transcription-associated phenotypes of rtf1 HMD mutants
In S. cerevisiae, the Rtf1 HMD is required for H2Bub through its direct interaction with Rad6 (Van Oss et al. 2016). To identify factors that functionally interact with the HMD, we conducted a genetic screen for suppressors of a previously characterized mutation, rtf1-108-110A, which replaces three consecutive residues within the HMD with alanine and greatly decreases global H2Bub levels in vivo (Tomson et al. 2011). Strains containing the rtf1-108-110A mutation exhibit transcription initiation from a cryptic TSS within the FLO8 open reading frame (Tomson et al. 2011), as assayed by a GAL1p-FLO8-HIS3 reporter construct (Cheung et al. 2008). RTF1 strains carrying this reporter are His- because transcription initiation at the canonical FLO8 TSS gives rise to an mRNA in which the HIS3 coding sequence is out of frame with respect to FLO8. However, in mutants defective in maintaining proper chromatin architecture, such as the rtf1-108-110A mutant, transcription initiation occurs at a cryptic TSS within the FLO8 gene, creating a transcript from which HIS3 is expressed (Figure 1A). We exploited this phenotype to isolate genetic suppressors of rtf1-108-110A with the initial intent of identifying genes that, when overexpressed from a plasmid, would suppress the His+ phenotype of rtf1-108-110A mutants containing the GAL1p-FLO8-HIS3 reporter. Surprisingly, we identified two strains, with mutations in the same complementation group, in which suppression was not dependent on the presence of a plasmid (see Materials and Methods for details). These chromosomal mutations behaved similarly in all phenotypic tests, and we chose the sup2-22 mutation for further analysis. As shown in Figure 1B, the sup2-22 mutation suppressed the cryptic initiation phenotype caused by the rtf1-108-110A mutation and two other rtf1 HMD loss-of-function mutations, rtf1-102-104A and rtf1-E104K (Tomson et al. 2011) (Figure 1B, compare sections 1 and 2 in the rtf1 strains). The sup2-22 mutation did not suppress the cryptic initiation phenotype of an rtf1Δ strain, indicating that suppression is allele-specific.
Figure 1.
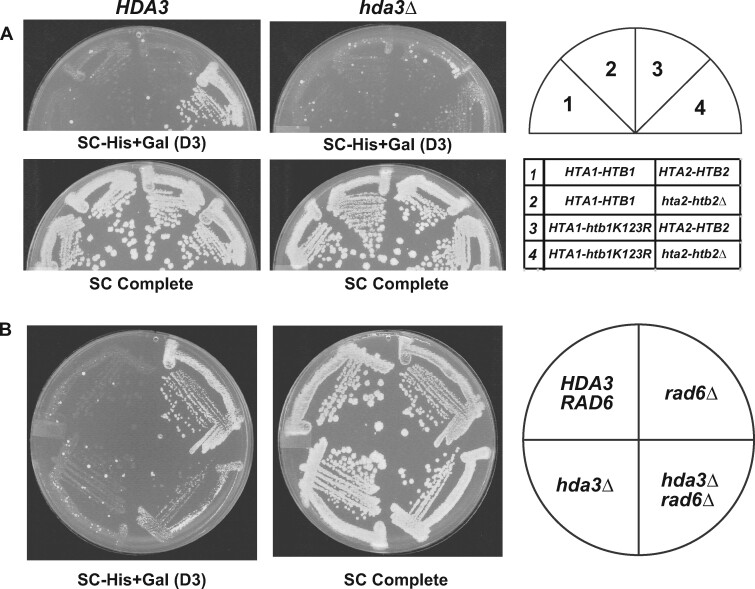
Deletion of any Hda1C subunit suppresses the cryptic initiation and telomeric silencing phenotypes of rtf1 HMD mutants. (A) Schematic diagrams of the cryptic initiation and telomeric silencing reporters used in this study. (B) Suppression of the cryptic initiation and telomeric silencing defects of rtf1 point mutations was assessed by replica plating to SC-His+Gal and 5FOA medium, respectively. SC complete medium was used as a control. Plates were imaged one (D1) or three (D3) days after replica plating and incubating at 30°C. (C) Table summarizing growth phenotypes shown in panel B (+ indicates growth; − indicates lack of growth). Grayed boxes highlight where the phenotype differed from the other rtf1 alleles. The following strains were used: KY1523, KY1496, KY2963, KY2934, KY2868, KY2792, KY1519, KY2957, KY2965, KY2898, KY1494, KY1502, KY2973, KY2974, KY2902, KY1526, KY2855, KY2999, KY3001, KY2867, KY1370, KY1520, KY2981, KY2983 and KY2943.
Bulk segregant analysis followed by whole-genome sequencing identified sup2-22 as a frameshift mutation in the HDA3 gene, which encodes a non-catalytic subunit of the Hda1C HDAC (Lee and Kim 2020). This mutation is predicted to eliminate 283 (out of 655) amino acids from the C-terminus of Hda3. The second, independently derived suppressor mutation identified in our screen, sup2-23, is a nonsense mutation at codon 198 of HDA3. Structural studies indicate that the regions deleted by the sup2-22 and sup2-23 mutations encompass a coiled-coil segment necessary for dimeric interactions between Hda2 and Hda3, which are required for the overall integrity and function of Hda1C (Wu et al. 2001a; Lee et al. 2009, 2021). We asked whether suppression of the rtf1 mutations was specific to our original hda3 mutations or whether loss of any subunit within Hda1C could confer suppression. Complete open reading frame deletions of HDA1, HDA2, or HDA3 were created using one-step gene replacement with the TRP1 gene (Moqtaderi and Geisberg 2013). Removal of any subunit of Hda1C, including the catalytic subunit Hda1, reversed the cryptic initiation phenotype of strains containing the GAL1p-FLO8-HIS3 reporter and rtf1 HMD point mutations (Figure 1B, sections 3 to 5); no differences in suppression were detected among the Hda1C mutants. Consistent with results obtained with the sup2-22 mutation, the cryptic initiation phenotype of strains lacking the entire RTF1 gene was not suppressed by deletion of individual Hda1C subunits. This result suggests that additional regions of Rtf1 or additional proteins that interact with Rtf1 outside of the HMD are necessary to observe the suppression.
Another phenotype displayed by certain rtf1 mutants is the inability to silence transcription near telomeres, as assayed using a TELVR::URA3 reporter (Gottschling et al. 1990; Chu et al. 2007; Tomson et al. 2011; Van Oss et al. 2016). Strains containing this reporter and the wild-type RTF1 gene grow in the presence of 5FOA, a drug that is toxic to cells expressing URA3. Strains, such as rtf1-108-110A mutant, that are defective for silencing of the TELVR::URA3 reporter are 5FOAS. Therefore, we used this reporter as a secondary readout for the suppression of the rtf1 mutations. We originally noted that RTF1 strains with the sup2-22 allele grow better on 5FOA-containing media than wild-type strains (Figure 1B, see growth on 5FOA on day one). Consistent with our identification of sup2-22 as a recessive mutation in HDA3, hda1Δ mutants were previously shown to have increased resistance to 5FOA, using a telomeric reporter (Rundlett et al. 1996; Hang and Smith 2011). The sup2-22, hda1Δ, hda2Δ and hda3Δ mutations suppressed the 5FOA sensitivity of two different rtf1 HMD mutants, rtf1-102-104A and rtf1-108-110A, as well as the rtf1Δ allele. In contrast, the hda mutations did not suppress the 5FOA sensitivity of a different HMD mutant, rtf1-E104K, indicating allele-specific suppression for this phenotype as well (summarized in Figure 1C).
Our results demonstrate that absence of a functional Hda1C suppresses the cryptic initiation and telomeric silencing phenotypes of certain rtf1 HMD mutants. To test if loss of Hda1C catalytic activity could similarly suppress the rtf1-108-110A mutation, we mutated an invariant histidine, H206, in the catalytic center of Hda1 and introduced this hda1-H206A mutation at the endogenous HDA1 locus by allelic replacement (Lee et al. 2021). For both the cryptic initiation and telomeric silencing phenotypes, the hda1-H206A mutation largely phenocopied the hda1Δ mutation with respect to rtf1 suppression (Figure 2A). Western blotting confirmed that the Hda1-H206A mutant protein was expressed at levels similar to wild type (Figure 2B).
In addition to effects on telomeric silencing and cryptic initiation from the internal FLO8 promoter, some mutations that alter the Rtf1 HMD also confer a suppressor-of-Ty (Spt−) phenotype (Warner et al. 2007; Tomson et al. 2011; Van Oss et al. 2016). At the his4-912δ allele, an insertion of a Ty1 δ element in the HIS4 promoter renders cells unable to grow on media lacking histidine due to an upstream shift in transcription initiation (Winston 1992). However, mutations in many genes regulating transcription and chromatin, including RTF1, are able to grow in the absence of histidine because they restore initiation to the native HIS4 TSS and produce a functional HIS4 transcript (Stolinski et al. 1997). This could be considered another case of cryptic transcription since the native promoter is downstream of the promoter provided by the Ty element (Kaplan et al. 2003). Similar to results seen with the GAL1p-FLO8-HIS3 reporter, the Spt− phenotype of rtf1-E104K, but not rtf1Δ, was suppressed by deleting HDA3 (Figure 3).
The identification of loss-of-function mutations in genes encoding an HDAC as suppressors of cryptic initiation was unexpected. Previous work has argued that increased acetylation of histones facilitates cryptic transcription (Rando and Winston 2012; Venkatesh and Workman 2015), likely by relaxing chromatin structure, which could allow spurious pre-initiation complex assembly and the initiation of new transcripts. Thus, mutations that inactivate an HDAC, which would increase histone acetylation, would have been unlikely to reverse the cryptic initiation phenotype of the rtf1 mutant strains. Due to the unanticipated role of Hda1C identified with the GAL1p-FLO8-HIS3 reporter, we focused on the coordination between Hda1C and Rtf1 with respect to cryptic initiation.
Loss of Hda1C does not restore H2Bub in rtf1 strains
The rtf1 alleles studied here are unable to stimulate H2Bub in vivo (Van Oss et al. 2016). One possibility is that suppression of cryptic initiation occurs because loss of Hda1C increases H2Bub levels. However, as shown by western blot analysis, the severe H2Bub defect of the rtf1 mutants was not rescued by deletion of HDA1 or HDA2 (Figure 4A). We also asked if loss of Hda1C recovered H3 methylation marks dependent on a functional Rtf1 HMD and H2Bub (Warner et al. 2007; Piro et al. 2012; Van Oss et al. 2016). While slight changes in global levels of H3 K4me2, H3 K4me3 and H3 K79me2/3 were detected in strains lacking Hda3, we also observed a small increase in total H3 recovered from these strains (Supplementary Figure S1). When normalized to total H3 levels, the changes in H3 K4 or H3 K79 methylation marks were less than two-fold. Coupled with previous results showing that set1Δ and dot1Δ mutants do not express the GAL1p-FLO8-HIS3 reporter (Quan and Hartzog 2010), these observations suggest that suppression of the cryptic initiation phenotypes of the rtf1 mutants is unlikely due to the recovery of H3 K4 or H3 K79 methylation. However, we cannot exclude the possibility that, within a bulk population, a small percentage of rtf1 hda3 double mutant cells have high levels of H3 methylation and these cells are most affected for cryptic initiation.
Strikingly, western blot analysis of total H2B levels showed bands that ran with slower mobility when Hda1C activity was absent, even in RTF1 strains (Figures 2B, 4, A and B). Given the function of Hda1C, we suspected that these bands represented an enrichment of acetylated forms of H2B (Wu et al. 2001a,b) or possibly another modification that is dependent upon H2B acetylation. As the Gcn5 HAT was previously shown to contribute to H2B acetylation (Grant et al. 1997; Suka et al. 2001), we asked if the slower migrating forms of H2B were dependent on Gcn5. Indeed, in gcn5Δ hda3Δ strains, the slower migrating anti-H2B-reactive bands were no longer observed (Figure 4B, lane 4). Interestingly, we did not observe a shift in the mobility of the ubiquitylated form of H2B upon inactivation of Hda1C (Figure 4A), raising the possibility that H2B K123 ubiquitylation inhibits acetylation or an acetylation-dependent modification of H2B.
Loss of H2Bub leads to cryptic initiation dependent on Hda1C
Since the HDA mutations suppress the cryptic initiation phenotype of rtf1 mutants that are severely defective in H2Bub, we assessed the cryptic initiation phenotype of other strains lacking H2Bub and the dependence on Hda1C for this phenotype. We analyzed strains expressing an H2B mutant protein that cannot be ubiquitylated, H2B K123R, as the only form of H2B and a strain lacking Rad6, the ubiquitin conjugase that targets H2B K123. In both cases, when HDA3 is wild type, we observed growth on SC-His+Gal medium, indicating cryptic initiation from the GAL1p-FLO8-HIS3 reporter (Figure 5A, panel 4, and Figure 5B, upper right), identifying a role for H2B K123 in controlling cryptic initiation using this reporter. Consistent with suppression of the rtf1 mutations, hda3Δ also suppresses the cryptic initiation phenotype of htb1-K123R and rad6Δ strains (Figure 5, A and B). Together these results suggest that the role of Hda1C in supporting cryptic initiation in the rtf1 mutants occurs downstream of the effects of losing H2Bub.
Figure 5.
Deletion of HDA3 suppresses defects in the H2Bub pathway. (A) A strain carrying the H2B K123R substitution as the only source of H2B (section 4) exhibits cryptic initiation as measured with the GAL1p-FLO8-HIS3 reporter when HDA3 is present (left), but not when HDA3 has been deleted (right). The following strains were used: KY1523, KY2878, KY2929, KY2876, KY2868, KY2921, KY2924 and KY2910. (B) A strain in which RAD6 has been deleted exhibits cryptic initiation when HDA3 is present (top right quadrant), but not when HDA3 has been deleted (bottom right quadrant). The following strains were used: KY1523, KY2832, KY2918 and KY2868. For both panels, plates were imaged three days after replica plating and incubation at 30°C.
The genetic interactions between Rtf1 and Hda1C are distinct from those involving other HDACs and chromatin regulators
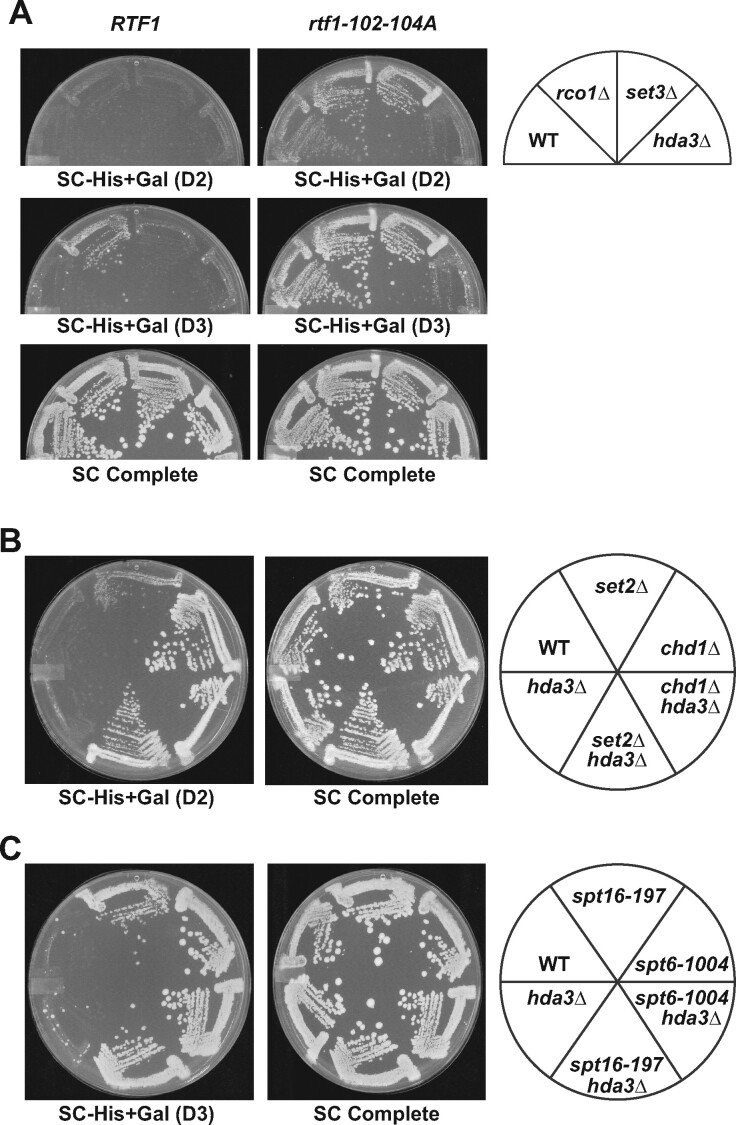
Contrary to our results with Hda1C, previous studies showed that other HDACs suppress cryptic initiation; that is, in their absence, increased levels of cryptic transcription are detected (Cheung et al. 2008; Silva et al. 2012). Given our unexpected result, we asked whether mutations in genes encoding other HDACs, in particular Rpd3S (rco1Δ) (Carrozza et al. 2005; Keogh et al. 2005) or Set3C (set3Δ) (Pijnappel et al. 2001; Kim et al. 2012), would also suppress the transcription defect in rtf1-102-104A strains. By using a deletion of RCO1, we specifically targeted the Rpd3S complex, which has been previously connected to regulation of intragenic cryptic transcription. As expected (Carrozza et al. 2005; Cheung et al. 2008; Silva et al. 2012; McDaniel et al. 2016), a strain lacking the Rpd3S subunit Rco1 and containing the GAL1p-FLO8-HIS3 reporter grew on SC-His+Gal media [Figure 6A, SC-His+Gal (D3)]. In fact, the double mutant, rco1Δ rtf1-102-104A, grew better than either single mutant [Figure 6A, SC-His+Gal (D2)], suggesting the mutations act in different pathways to affect cryptic transcription. In addition, rpd3Δ strains, lacking both the Rpd3S and Rpd3L complexes, showed similar results (Supplementary Figure S2), though strains lacking Rpd3 grow more slowly. Deletion of SET3, which eliminates activity from both the Hos2 and Hst2 HDACs in Set3C, did not lead to a cryptic initiation phenotype as measured by the GAL1p-FLO8-HIS3 reporter, in agreement with previous reports that set3Δ did not elicit transcription from the cryptic promoters in the native FLO8 and STE11 genes (Kim and Buratowski 2009). Furthermore, unlike hda3Δ, set3Δ did not suppress the cryptic initiation phenotype of rtf1-102-104A. Therefore, suppression of cryptic transcription in this rtf1 mutant by hda3Δ is not a common feature of HDAC mutants.
Figure 6.
Genetic interactions between rtf1 mutations and mutations in genes enoding Hda1C subunits are specific. (A) Mutations that disrupt other HDACs do not suppress the cryptic initiation phenotype of an rtf1 HMD mutant. Strains mutated for the Rpd3S (rco1Δ) or Set3C (set3Δ) HDACs and containing either RTF1 or rtf1-102-104A were replica plated to SC-His+Gal or SC Complete media and scanned after two (D2) or three (D3) days of incubation at 30°C. For both the RTF1 and rtf1-102-104A panels, WT indicates the presence of wild-type RCO1, SET3, and HDA3 genes. The following strains were used: KY1523, KY3057, KY3042, KY2868, KY2792, KY3053, KY3038 and KY2898. (B, C) Deletion of HDA3 does not suppress the cryptic initiation phenotypes of strains defective in other chromatin regulators. Strains with the indicated genotypes were replica plated to SC-His+Gal or SC Complete media and scanned after two (D2) or three (D3) days of incubation at 30°C. The following strains were used: (B) KY1523, KY3200, KY3198, KY3199, KY3201 and KY2868 and (C) KY1523, KY3073, KY3084, KY3082, KY3093 and KY2868.
Next, we investigated if the effect of Hda1C was specific to strains defective for H2Bub. Many strains with mutations in genes encoding chromatin and transcription factors show cryptic initiation on the GAL1p-FLO8-HIS3 reporter (Cheung et al. 2008; Quan and Hartzog 2010; Silva et al. 2012). These include strains lacking or mutated in the H3 K36 methyltransferase Set2, the chromatin remodeling enzyme Chd1, and the histone chaperones Spt6 and Spt16. In contrast to rtf1 mutants, hda3Δ did not suppress the cryptic initiation phenotypes of strains lacking Set2 or Chd1 or containing mutations in SPT6 or SPT16 (Figure 6, B and C). In fact, set2Δ hda3Δ and spt16-197 hda3Δ double mutants grew better on the SC-His+Gal medium than set2Δ or spt16-197 single mutant strains. These results show that the genetic interactions we identified between mutants lacking H2Bub and Hda1C are specific. They also suggest that loss of Hda1C is not simply suppressing cryptic initiation by elevating Pol II transcription from the GAL1 promoter and impeding internal initiation events. In support of this, levels of the full-length FLO8 transcript produced from the reporter construct are unchanged by deletion of HDA3 (Supplementary Figure S3, A–C), and Pol II occupancy at the FLO8 5′ region is similar in wild-type strains and hda3Δ mutants (Supplementary Figure S3, A and D).
Sas3 and H3 K14 suppress cryptic initiation in an rtf1 mutant
Since loss of Hda1C suppresses the cryptic initiation phenotype of rtf1 mutants, we hypothesized that deletion of a HAT that works in opposition to Hda1C might exacerbate the cryptic initiation phenotype of the rtf1 mutants and reverse the effect of inactivating Hda1C. Based on our western blot results showing that deletion of GCN5 reversed the H2B mobility shift caused by hda3Δ (Figure 4B), we investigated if Gcn5 was the relevant HAT. Gcn5 is the catalytic subunit of several HAT complexes (Lee and Workman 2007), any of which might be involved in regulating cryptic initiation. If Gcn5 antagonized Hda1C, then the rtf1-102-104A hda3Δ gcn5Δ triple mutant would be expected to phenocopy the rtf1-102-104A single mutant and grow on Sc-His+Gal medium. Instead, the triple mutant was strongly His- (Supplementary Figure S4, right panel, section 4). In addition, on its own, the gcn5Δ mutation suppressed the cryptic initiation phenotype of the rtf1-102-104A mutation (Supplementary Figure S4, right panel, section 3). These results suggest that suppression of rtf1-102-104A by hda3Δ is not easily explained by increased acetylation of a Gcn5-dependent target protein.
Because the rtf1 HMD mutants exhibit telomeric silencing defects and Hda1C also plays a role in silencing, we asked whether deletion of two HATs previously implicated in transcriptional silencing would affect the cryptic initiation phenotype of the rtf1 mutants. SAS2 was first discovered in a screen for genes affecting transcriptional silencing at the mating-type locus and SAS3 was subsequently uncovered by its similarity to SAS2 (Reifsnyder et al. 1996). Sas2 and Sas3 are the catalytic subunits of the HATs SAS and NuA3, respectively (John et al. 2000; Shia et al. 2005). While deletion of SAS2 had little effect, deletion of SAS3 exacerbated the cryptic initiation phenotype of the rtf1 mutants, as indicated by enhanced growth on SC-His+Gal medium (Figure 7A). Interestingly, sas3Δ on its own did not confer a cryptic initiation phenotype. Notably, Sas3 and Hda3 appear to oppose each other because deleting both proteins in the context of rtf1-108-110A gave a cryptic initiation phenotype nearly identical to the rtf1 allele alone (Figure 7B). These results suggest that the acetylation state of one or more shared targets of Hda1C and Sas3 modulates the cryptic initiation phenotype of the rtf1 mutants.
One target of Sas3 is H3 K14 (Taverna et al. 2006). We took advantage of an integrated synthetic histone mutant library (Dai et al. 2008) to create strains in which the only copy of H3 had a lysine-to-alanine substitution at position 14 (synH3-K14A). Unlike sas3Δ strains, the H3 K14A mutant showed cryptic initiation using the GAL1p-FLO8-HIS3 reporter (Figure 7C, compare row 5 to row 1 control). The distinct phenotypes of the sas3Δ and H3 K14A mutants may be due to residual acetylation of H3 K14 in the sas3Δ strain, as previous studies showed that H3 K14 is also acetylated by Gcn5 (Church et al. 2017). However, like the sas3Δ mutation, the H3 K14A substitution strongly enhanced the cryptic initiation phenotype of the rtf1-102-104A mutant (Figure 7C). Deleting HDA3 only partially reversed the rtf1-102-104A synH3-K14A cryptic initiation phenotype. Unlike the rtf1-102-104A hda3Δ synH3 control strain, which carries a synthetic version of wild-type H3, the rtf1-102-104A hda3Δ synH3-K14A remained His+. These results suggest that Sas3 represses the cryptic initiation phenotype of rtf1 mutants and Hda1C opposes Sas3 in this process. One possible target for their action is H3 K14. However, because hda3Δ partially suppresses the cryptic initiation phenotype of the synH3-K14A mutant, other Hda1C targets that control chromatin accessibility must exist.
Discussion
In this study, we discovered a connection between Rtf1, a component of the Paf1 transcription elongation complex necessary for H2Bub, and Hda1C, an HDAC complex, in controlling transcription at three loci that are sensitive to chromatin architecture. Previous studies have shown that deletion of certain HDACs, including Rpd3S or Set3C, causes cryptic initiation (Carrozza et al. 2005; Keogh et al. 2005; Li et al. 2007a,b; Kim et al. 2012). This is consistent with the expectation that increased histone acetylation promotes a permissive chromatin environment. In contrast, our study shows that loss of Hda1C activity suppresses the cryptic initiation phenotype of strains deficient in or completely lacking in H2Bub. The interplay between H2Bub and Hda1C in controlling cryptic transcription initiation appears specific, because deletion of HDA3 did not reduce cryptic initiation in strains lacking several other chromatin-related proteins and mutations in genes encoding other HDACs did not suppress the cryptic initiation phenotype of an rtf1 mutant defective in H2Bub.
We and others have shown that mutations in RTF1 and genes encoding other proteins required for H2Bub cause cryptic initiation within the FLO8 gene (Cheung et al. 2008; Fleming et al. 2008; Tomson et al. 2011; Silva et al. 2012). While H2Bub is required for H3 K4me2/3 and H3 K79me2/3 in vivo, previous studies found that strains lacking Set1 or Dot1, the methyltransferases responsible for H3 K4me and H3 K79me, respectively, did not exhibit cryptic initiation at the GAL1p-FLO8-HIS3 reporter (Quan and Hartzog 2010). This suggests that H2Bub functions independently of these downstream modifications in preventing cryptic initiation. H2Bub has been shown to stabilize nucleosomes, particularly in the middle and ends of genes (Chandrasekharan et al. 2009; Batta et al. 2011), and loss of H2Bub in S. pombe up-regulates noncoding RNAs, particularly antisense transcripts (Pagé et al. 2019; Murawska et al. 2020; Sansó et al. 2020). Recent work using optical tweezers has confirmed the increase in stability of nucleosomes containing H2Bub, and this increase in stability is an energetic barrier to passage by Pol II (Chen et al. 2019). One possibility based on our findings is that Hda1C may contribute to the instability of nucleosomes lacking H2Bub by deacetylating a histone or other target.
Deletion of Hda1C did not suppress the cryptic initiation phenotype of strains completely lacking Rtf1. This suggests that the absence of other functional domains within Rtf1, outside of the HMD, contributes to cryptic initiation in a way that is not suppressed by mutations in genes encoding Hda1C. In addition to the HMD, Rtf1 has separable domains required for its interactions with the Pol II elongation complex, Chd1 and other Paf1C subunits (Simic et al. 2003; Warner et al. 2007; Mayekar et al. 2013; Wier et al. 2013; Vos et al. 2020). The N-terminus of Rtf1 is important for proper recruitment of Chd1 to certain open reading frames (Warner et al. 2007), and we found that deletion of CHD1 causes a cryptic initiation phenotype that cannot be suppressed by hda3Δ. This observation suggests that Chd1 and Hda1C affect cryptic initiation through different pathways or that the effect of Chd1 is upstream of the effect of Hda1C. Therefore, while a complete deletion of Rtf1 can affect transcription and chromatin through several mechanisms, our data suggest that only the effect on H2Bub is suppressed by loss of Hda1C.
What is the target of Hda1C that creates a permissive condition for initiation of transcription within the coding sequence? Cells lacking Hda1C have increased acetylation at several positions on H2B, H3 and H4, including H3 K14 (Carmen et al. 1996; Rundlett et al. 1996; Wu et al. 2001a,b; Yang and Seto 2008; Islam et al. 2011; Ha et al. 2019; Lee and Kim 2020). H3 K14Ac has been previously shown to help maintain proper levels of H3 K4me3 by inhibiting the Jhd2 histone demethylase (Maltby et al. 2012). Furthermore, in a positive feedback loop, methylation of H3 K4, a modification dependent on H2Bub, promotes H3 K14 acetylation by Sas3 as part of the NuA3 HAT complex, through the binding of the Yng1 subunit of NuA3 (Taverna et al. 2006; Martin et al. 2017). Our results demonstrate that, in the context of an rtf1 mutation, the absence of Sas3 or H3 K14 enhances cryptic initiation, supporting the idea that an appropriate level or pattern of histone acetylation marks is needed to prevent cryptic initiation. While Hda1C and Sas3 appear to work in opposition in controlling cryptic initiation, the role of Hda3 in promoting cryptic initiation in rtf1 mutants cannot be solely through deacetylation of H3 K14, as we observed partial suppression of the cryptic initiation phenotype of an H3 K14A mutant by hda3Δ.
The enrichment of Hda1C on the bodies of active genes and its interactions with RNA and Pol II argue for a role in controlling histone acetylation levels during transcription elongation (Govind et al. 2010; Ha et al. 2019; Lee and Kim 2020). Moreover, a recent study showed that Hda1C reduces H4 acetylation within the coding regions of genes undergoing high levels of transcription (Ha et al. 2019; Lee and Kim 2020). With the GAL1p-FLO8-HIS3 reporter used here, high levels of transcription driven by the inducible GAL1 promoter are required to detect cryptic initiation in the rtf1 HMD mutants as has been observed for other mutants (Cheung et al. 2008; Silva et al. 2012). Surprisingly, by a mechanism not understood, hda1Δ cells, with higher levels of H4 acetylation, exhibit higher histone occupancy levels, especially on longer genes where the effect of hda1Δ on H4 acetylation is more evident (Ha et al. 2019). Increasing histone occupancy within gene bodies could be one mechanism by which loss of Hda1C counteracts the disruption in chromatin structure caused by loss of H2Bub.
In summary, our results suggest an interplay between the H2Bub pathway and Hda1C in controlling chromatin accessibility during transcription elongation. Unexpectedly, a functional Hda1C permits cryptic initiation in cells lacking H2Bub. While cryptic initiation is most frequently observed under mutant conditions, it is possible that the dynamic removal of H2Bub during transcription elongation could give rise to cryptic transcripts in wild-type cells, albeit at low levels. In this scenario, the activity of Hda1C could regulate the output of these cryptic transcripts. Whether these transcripts would have biological consequences, as has been reported for some internally initiated transcripts (McKnight et al. 2014; Tamarkin-Ben-Harush et al. 2017), is unclear. Future work will be needed to elucidate the histone or non-histone target(s) through which Hda1C impacts cryptic initiation in opposition to H2Bub and delineate whether these targets are functioning directly or indirectly in this process. Recent technical and computational advances in detecting transcript initiation events within genes bodies (Pelechano et al. 2013; Wei et al. 2019) offer the opportunity to study this and additional examples of crosstalk between epigenetic regulators in maintaining chromatin structure during transcription elongation.
Reagent and data availability
All strains and detailed protocols are available upon request. Sequencing data are available at NCBI as part of the BioProject: PRJNA634539, BioSample accessions: SAMN14999039, SAMN14999040. Supplemental Table S1 and Supplemental Figures S1-S4 are available via figshare: https://doi.org/10.25387/g3.15040245.
Acknowledgments
We are grateful to Kristin Klucevsek for creating the sas2Δ and sas3Δ strains, Daniel Sheidy for technical assistance in performing the genetic screen for rtf1 suppressors, Sarah Tripplehorn for assistance with dilution spot tests, Graham Hatfull and members of his research group for assistance with genomic sequencing, and members of the Arndt, Hainer, Kaplan and Lee laboratories for their insightful comments.
Funding
This research was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided and by the following funding sources: National Institutes of Health (NIH) grants R01 GM052593 and R35 GM141964 to K.M.A., NIH predoctoral fellowship (F31GM129917) to M.A.E., and a Colella Research Fellowship to R.A.K. APC charges for this article were fully paid by the University Library System, University of Pittsburgh.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Bagchi DN, Battenhouse AM, Park D, Iyer VR.. 2020. The histone variant H2A.Z in yeast is almost exclusively incorporated into the +1 nucleosome in the direction of transcription. Nucleic Acids Res. 48:157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T.. 2011. Regulation of chromatin by histone modifications. Cell Res. 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF.. 2011. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 25:2254–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland SR, Jin N, Ozdemir AC, Lyons RH, Weisman LS, et al. 2010. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics. 186:1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, et al. 2002. Trans-histone regulatory pathway in chromatin. Nature. 418:498. [DOI] [PubMed] [Google Scholar]
- Carmen AA, Rundlett SE, Grunstein M.. 1996. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 271:15837–15844. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 123:581–592. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW.. 2009. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA. 106:16686–16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gabizon R, Brown AI, Lee A, Song A, et al. 2019. High-resolution and high-accuracy topographic and transcriptional maps of the nucleosome barrier. eLife. 8:e48281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, et al. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40:D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, et al. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P.. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene. 110:119–122. [DOI] [PubMed] [Google Scholar]
- Chu Y, Simic R, Warner MH, Arndt KM, Prelich G.. 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 26:4646–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church M, Smith KC, Alhussain MM, Pennings S, Fleming AB.. 2017. Sas3 and Ada2(Gcn5)-dependent histone H3 acetylation is required for transcription elongation at the de-repressed FLO1 gene. Nucleic Acids Res. 45:4413–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, et al. 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 134:1066–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, et al. ; 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics. 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, et al. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 277:28368–28371. [DOI] [PubMed] [Google Scholar]
- Drouin S, Laramée L, Jacques P-É, Forest A, Bergeron M, et al. 2010. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 6:e1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison MA, Walker JL, Ropp PJ, Durrant JD, Arndt KM.. 2020. MutantHuntWGS: a pipeline for identifying Saccharomyces cerevisiae mutations. G3 (Bethesda). 10:3009–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, et al. 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda). 4:389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnung L, Vos SM, Cramer P.. 2018. Structure of transcribing RNA polymerase II-nucleosome complex. Nat Commun. 9:5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA.. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 31:57–66. [DOI] [PubMed] [Google Scholar]
- Formosa T, Winston F.. 2021. The role of FACT in managing chromatin: disruption, assembly, or repair? Nucleic Acids Res. 48:11929–11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Oren M.. 2014. Writing and reading H2B monoubiquitylation. Biochim Biophys Acta. 1839:694–701. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan R, Winston F.. 2019. Whole-genome sequencing of yeast cells. Curr Protoc Mol Biol. 128:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA.. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 63:751–762. [DOI] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, et al. 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 39:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640–1650. [DOI] [PubMed] [Google Scholar]
- Gurard-Levin ZA, Quivy J-P, Almouzni G.. 2014. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem. 83:487–517. [DOI] [PubMed] [Google Scholar]
- Ha SD, Ham S, Kim MY, Kim JH, Jang I, et al. 2019. Transcription-dependent targeting of Hda1C to hyperactive genes mediates H4-specific deacetylation in yeast. Nat Commun. 10:4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CM, Strømme CB, Huang H, Patel DJ, Groth A.. 2017. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 18:141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang M, Smith MM.. 2011. Genetic analysis implicates the Set3/Hos2 histone deacetylase in the deposition and remodeling of nucleosomes containing H2A.Z. Genetics. 187:1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM.. 1993. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem. 268:305–314. [PubMed] [Google Scholar]
- Hsu PL, Shi H, Leonen C, Kang J, Chatterjee C, et al. 2019. Structural basis of H2B ubiquitination-dependent H3K4 methylation by COMPASS. Mol Cell. 76:712–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, et al. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 11:261–266. [DOI] [PubMed] [Google Scholar]
- Islam A, Turner EL, Menzel J, Malo ME, Harkness TAA.. 2011. Antagonistic Gcn5-Hda1 interactions revealed by mutations to the Anaphase Promoting Complex in yeast. Cell Div. 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD.. 2001. Translating the histone code. Science. 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- John S, Howe LA, Tafrov ST, Grant PA, Sternglanz R, et al. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K.. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to pol II elongation. Mol Cell. 20:971–978. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F.. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 301:1096–1099. [DOI] [PubMed] [Google Scholar]
- Keogh M-C, Kurdistani SK, Morris SA, Ahn SH, Podolny V, et al. 2005. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 123:593–605. [DOI] [PubMed] [Google Scholar]
- Kim TS, Buratowski S.. 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 137:259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S.. 2012. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 150:1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 11:721–729. [DOI] [PubMed] [Google Scholar]
- Kujirai T, Ehara H, Fujino Y, Shirouzu M, Sekine S, et al. 2018. Structural basis of the nucleosome transition during RNA polymerase II passage. Science. 362:595–598. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, et al. 2005. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol. 15:1487–1493. [DOI] [PubMed] [Google Scholar]
- Lawrence M, Daujat S, Schneider R.. 2016. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 32:42–56. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Bollschweiler D, Schäfer T, Huber R.. 2021. Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies. Sci Adv. 7:eabd4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Kim T.. 2020. Histone H4-specific deacetylation at active coding regions by Hda1C. Mol Cells. 43:841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Maskos K, Huber R.. 2009. Structural and functional studies of the yeast class II Hda1 histone deacetylase complex. J Mol Biol. 391:744–757. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL.. 2007. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 8:284–295. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, et al. 2007a. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 316:1050–1054. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, et al. 2007b. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 21:1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. ; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby VE, Martin BJE, Brind'Amour J, Chruscicki AT, McBurney KL, et al. 2012. Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 109:18505–18510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Zhou M-M.. 2014. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 6:a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BJE, McBurney KL, Maltby VE, Jensen KN, Brind'Amour J, et al. 2017. Histone H3K4 and H3K36 methylation independently recruit the NuA3 histone acetyltransferase in Saccharomyces cerevisiae. Genetics. 205:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayekar MK, Gardner RG, Arndt KM.. 2013. The recruitment of the Saccharomyces cerevisiae Paf1 complex to active genes requires a domain of Rtf1 that directly interacts with the Spt4-Spt5 complex. Mol Cell Biol. 33:3259–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel SL, Fligor JE, Ruan C, Cui H, Bridgers JB, et al. 2016. Combinatorial histone readout by the dual plant homeodomain (PHD) fingers of Rco1 mediates Rpd3S chromatin recruitment and the maintenance of transcriptional fidelity. J Biol Chem. 291:14796–14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K, Liu H, Wang Y.. 2014. Replicative stress induces intragenic transcription of the ASE1 gene that negatively regulates Ase1 activity. Curr Biol. 24:1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, et al. 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 10:483–488. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Geisberg JV.. 2013. Construction of mutant alleles in Saccharomyces cerevisiae without cloning: overview and the Delitto Perfetto method. Curr Protoc Mol Biol. 104:13.10C.1-13.10C.17. [DOI] [PubMed] [Google Scholar]
- Murawska M, Schauer T, Matsuda A, Wilson MD, Pysik T, et al. 2020. The Chaperone FACT and histone H2B ubiquitination maintain S. pombe genome architecture through genic and subtelomeric functions. Mol Cell. 77:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde M-E, Côté J, Kutateladze TG.. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 19:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K.. 2003a. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 278:33625–33628. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K.. 2003b. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 11:709–719. [DOI] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K.. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 277:34655–34657. [DOI] [PubMed] [Google Scholar]
- Pagé V, Chen JJ, Durand-Dubief M, Grabowski D, Oya E, et al. 2019. Histone H2B ubiquitylation regulates histone gene expression by suppressing antisense transcription in fission yeast. Genetics. 213:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Kim JS.. 2020. A short guide to histone deacetylases including recent progress on class II enzymes. Exp Mol Med. 52:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM.. 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 497:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, et al. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro AS, Mayekar MK, Warner MH, Davis CP, Arndt KM.. 2012. Small region of Rtf1 protein can substitute for complete Paf1 complex in facilitating global histone H2B ubiquitylation in yeast. Proc Natl Acad Sci USA. 109:10837–10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter NJ, Christianson DW.. 2019. Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases. Curr Opin Struct Biol. 59:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan TK, Hartzog GA.. 2010. Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics. 184:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Winston F.. 2012. Chromatin and transcription in yeast. Genetics. 190:351–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifsnyder C, Lowell J, Clarke A, Pillus L.. 1996. Yeast SAS silencing genes and human genes associated with AML and HIV–1 Tat interactions are homologous with acetyltransferases. Nat Genet. 14:42–49. [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA.. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 287:501–504. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P.. 1990. Methods in Yeast Genetics—A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Ruan C, Lee CH, Cui H, Li S, Li B.. 2015. Nucleosome contact triggers conformational changes of Rpd3S driving high-affinity H3K36me nucleosome engagement. Cell Rep. 10:204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, et al. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 93:14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansó M, Parua PK, Pinto D, Svensson JP, Pagé V, et al. 2020. Cdk9 and H2Bub1 signal to Clr6-CII/Rpd3S to suppress aberrant antisense transcription. Nucleic Acids Res. 48:7154–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Yoshida M.. 2014. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia WJ, Osada S, Florens L, Swanson SK, Washburn MP, et al. 2005. Characterization of the yeast trimeric-SAS acetyltransferase complex. J Biol Chem. 280:11987–11994. [DOI] [PubMed] [Google Scholar]
- Shirra MK, Arndt KM.. 1999. Evidence for the involvement of the Glc7-Reg1 phosphatase and the Snf1-Snf4 kinase in the regulation of INO1 transcription in Saccharomyces cerevisiae. Genetics. 152:73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirra MK, Rogers SE, Alexander DE, Arndt KM.. 2005. The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics. 169:1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AC, Xu X, Kim HS, Fillingham J, Kislinger T, et al. 2012. The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J Biol Chem. 287:1709–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, et al. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolinski LA, Eisenmann DM, Arndt KM.. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 17:4490–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Resnick MA.. 2006. The Delitto Perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 409:329–345. [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M.. 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 8:473–479. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD.. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 418:104–108. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F.. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 11:3009–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarkin-Ben-Harush A, Vasseur J-J, Debart F, Ulitsky I, Dikstein R.. 2017. Cap-proximal nucleotides via differential eIF4E binding and alternative promoter usage mediate translational response to energy stress. eLife. 6:e21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, et al. 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 24:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Koleske AJ, Chao DM, Young RA.. 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 73:1361–1375. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson BN, Davis CP, Warner MH, Arndt KM.. 2011. Identification of a role for histone H2B ubiquitylation in noncoding RNA 3’-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics. 188:273–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi M, Imam N, Verma K, Patel AK.. 2016. Chromatin remodelers: We are the drivers!! Nucleus. 7:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss SB, Cucinotta CE, Arndt KM.. 2017. Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem Sci. 42:788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oss SB, Shirra MK, Bataille AR, Wier AD, Yen K, et al. 2016. The histone modification domain of Paf1 complex subunit Rtf1 directly stimulates H2B ubiquitylation through an interaction with Rad6. Mol Cell. 64:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL.. 2015. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 16:178–189. [DOI] [PubMed] [Google Scholar]
- Vidal M, Strich R, Esposito RE, Gaber RF.. 1991. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 11:6306–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Linden A, Urlaub H, Cramer P.. 2020. Structure of complete Pol II–DSIF–PAF–SPT6 transcription complex reveals RTF1 allosteric activation. Nat Struct Mol Biol. 27:668–677. [DOI] [PubMed] [Google Scholar]
- Warner MH, Roinick KL, Arndt KM.. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol. 27:6103–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Hennig BP, Wang J, Zhang Y, Piazza I, et al. 2019. Chromatin-sensitive cryptic promoters putatively drive expression of alternative protein isoforms in yeast. Genome Res. 29:1974–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AD, Mayekar MK, Héroux A, Arndt KM, VanDemark AP.. 2013. Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci USA. 110:17290–17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL.. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 11:53–55. [DOI] [PubMed] [Google Scholar]
- Winston F. 1992. 47 Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones, and other transcription factors of yeast. Cold Spring Harbor Monograph Archive 22B. Available at: https://cshmonographs.org/index.php/.
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, et al. 2003a. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 11:267–274. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A.. 2003b. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 278:34739–34742. [DOI] [PubMed] [Google Scholar]
- Worden EJ, Wolberger C.. 2019. Activation and regulation of H2B-Ubiquitin-dependent histone methyltransferases. Curr Opin Struct Biol. 59:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden EJ, Zhang X, Wolberger C.. 2020. Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. eLife. 9:e53199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Carmen AA, Kobayashi R, Suka N, Grunstein M.. 2001a. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc Natl Acad Sci USA. 98:4391–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Suka N, Carlson M, Grunstein M.. 2001b. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell. 7:117–126. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E.. 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 9:206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Serra-Cardona A, Zhou H, Wang M, Yang N, et al. 2018. Multisite substrate recognition in Asf1-dependent acetylation of histone H3 K56 by Rtt109. Cell. 174:818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]