Abstract
Insm1, Neurod1, and Pax6 are essential for the formation and function of pancreatic endocrine cells. Here, we report comparative immunohistochemical, transcriptomic, functional enrichment, and RNA splicing analyses of these genes using gene knock-out mice. Quantitative immunohistochemical analysis confirmed that elimination of each of these three factors variably impairs the proliferation, survival, and differentiation of endocrine cells. Transcriptomic analysis revealed that each factor contributes uniquely to the transcriptome although their effects were overlapping. Functional enrichment analysis revealed that genes downregulated by the elimination of Insm1, Neurod1, and Pax6 are commonly involved in mRNA metabolism, chromatin organization, secretion, and cell cycle regulation, and upregulated genes are associated with protein degradation, autophagy, and apoptotic process. Elimination of Insm1, Neurod1, and Pax6 impaired expression of many RNA-binding proteins thereby altering RNA splicing events, including for Syt14 and Snap25, two genes required for insulin secretion. All three factors are necessary for normal splicing of Syt14, and both Insm1 and Pax6 are necessary for the processing of Snap25. Collectively, these data provide new insights into how Insm1, Neurod1, and Pax6 contribute to the formation of functional pancreatic endocrine cells.
Keywords: pancreas, endocrine progenitor cells, gene expression, alternative RNA splicing
Introduction
During pancreas development, the formation of islets of Langerhans requires formation of an endocrine cell-specific gene regulatory network (GRN). Prior studies have identified multiple transcription factors (TFs) that contribute to endocrine cell development (Arda et al. 2013; Osipovich et al. 2021). Among these, Neurog3 plays an especially important role. By activating a downstream cascade of pro-endocrine TFs, Neurog3 initiates formation of the endocrine cell-specific GRN (Gu et al. 2002; Ejarque et al. 2013; Bechard et al. 2016). In its absence, the fate of pancreatic pre-endocrine cells is redirected toward the ductal lineage and no endocrine cells are formed (Gradwohl et al. 2000; Schwitzgebel et al. 2000; Wang et al. 2010; Osipovich et al. 2021). In contrast, when other pro-endocrine TFs downstream of Neurog3 are eliminated, including Insm1, Neurod1, and Pax6, pancreatic endocrine-like cells are formed that exhibit marked defects in proliferation and/or hormone expression, suggesting that each of these factors, in turn, regulates a subnetwork of genes (Naya et al. 1997; Sander et al. 1997; St-Onge et al. 1997; Huang et al. 2000; Heller et al. 2004; Gierl et al. 2006; Mellitzer et al. 2006; Osipovich et al. 2014).
TFs activate target genes by directly binding DNA in promoter regions, and ChIP-Seq has been widely used to identify genome-wide TF binding sites. Previous ChIP-Seq studies using cell lines have identified many binding sites for Insm1, Neurod1, and Pax6. Analysis of Insm1 and Neurod1 binding sites in a pancreatic β-cell line derived from adult islets has shown that Insm1, Neurod1, and Foxa2 frequently co-bind to the same genomic regions. However, these analyses have also revealed that only 32% of genes in adult β-cells that are regulated by Insm1 have nearby Insm1 binding (Jia et al. 2015). Similarly, while both Pax6 and Neurod1 often bind to enhancers that are active in insulinoma cells, the binding of both factors occurs at only 40% of such binding sites (Lizio et al. 2015). Furthermore, while there is a large degree of overlap in the binding of Neurod1 and Pax6, Pax6 exhibits both activating and repressive functions in mouse and human β-cell lines (Swisa et al. 2017).
RNA processing is also a vitally important process that is regulated during development. There are hundreds of different RNA-binding proteins (RBPs) that affect polyadenylation, stabilization, and localization dynamics of mRNA. Some of these proteins also introduce nucleotide modifications and cause differential splicing of introns and exons (Licatalosi and Darnell 2010; Wickramasinghe and Venkitaraman 2016; Manning and Cooper 2017; Carazo et al. 2019). Alternative RNA splicing significantly enhances transcriptome diversity, and it is important for cell differentiation (Fiszbein and Kornblihtt 2017). Indeed, Singer et al. (2019) have recently reported that the alternative splicing of Pax6, which requires recruitment of RBPs by the lncRNA Pauper, alters both its transcriptional activity and DNA binding specificity. It has also recently been shown in islets from individuals with type 2 diabetes that many RBPs are dysregulated and that this may impair correct RNA splicing (Jeffery et al. 2019).
In this study, we performed immunohistochemical and bulk transcriptomic analyses to directly compare the effects of eliminating Insm1, Neurod1, and Pax6 during endocrine cell development. Our findings indicate that these three TFs individually and coordinately regulate the expression of common and unique sets of genes necessary for the proliferation and function of pancreatic endocrine cells. Additionally, we find that Insm1, Neurod1, and Pax6 differentially affect the splicing of genes, thereby adding complexity to pancreatic endocrine cell proteomes.
Materials and methods
Mice and genotyping
Insm1tm1.1Mgn (Insm1GFPCre) (Osipovich et al. 2014) and Neurod1tm1Jle (Neurod1LacZ) (Miyata et al. 1999) mice were maintained in a CD-1 background. Pax6tm2Pgr (Pax6flox) mice were obtained from Jackson Laboratory and kept in a C57Bl6/J background (Ashery-Padan et al. 2000; Sun et al. 2008). All animal experimentation was performed under the Vanderbilt University Institutional Animal Care and Use Committee’s oversight. The Insm1GFPCre allele expresses a GFP-Cre fusion protein that contains a nuclear localization sequence and were genotyped as previously described (Osipovich et al. 2014). Pax6flox mice contain loxP sites flanking the initiation codon in exon 4 and exon 6 (the paired domain), requiring Cre activity for excision and were genotyped using the primer pairs 5′-CCTAACAGAGCCCCGTATTC (forward) and 5′-GCCCAACAGTCCAGAGAAAG (reverse). In Neurod1LacZ mice, Neurod1 coding sequences were replaced with a LacZ reporter (Miyata et al. 1999) and therefore intrinsically lack Neurod1 expression. To detect wild-type Neurod1 and Neurod1LacZ we used either 5′-ACCATGCACTCTGTACGCATT (forward) or 5′-GAGAACTGAGACACTCATCTG (forward) in combination with 5′-AAACGCCGAGTTAAAGCCATC (reverse), respectively.
Timed matings and tissue collection
Embryos were isolated from timed matings where noon of the day that vaginal plugs were observed was designated E0.5. Embryonic day (E) 15.5 animals were dissected into ice-cold PBS and visually genotyped for the presence of InsmGFPCre allele based on green fluorescence. For PCR genotyping, embryonic tissues were digested at 55°C in 100 µl of PCR lysis buffer (1× PCR buffer, 0.1% Triton X-100, 100 µg/ml Proteinase K), inactivated at 90°C for 15 min and used for PCR. Whole pancreas was dissected and either processed for fluorescence-activated cell sorting (FACS) or frozen in OCT for immunostaining analyses.
Immunofluorescence
Pancreata were routinely fixed in 2% paraformaldehyde for 2 h at room temperature, incubated in 30% sucrose in PBS overnight on a shaker, then mounted using Tissue Tek O.C.T. compound and stored at −80°C. Immunofluorescence staining of frozen tissue sections was performed as previously described (Burlison et al. 2008). Ten μm tissue sections were stained using anti-GFP (1:500, ThermoFisher, #A10262), insulin (1:1000, Invitrogen, #PA1-26938), glucagon (1:100, Milipore, #AB932), somatostatin (1:1000, Linco), pancreatic polypeptide (1:1000, Linco), and/or Ki67 (1:500, ThermoFisher, #RM-9106-S1) antibodies, and ProLong Gold anti-fade reagent with DAPI (Life Technologies, #P36941) used to mount coverslips. TUNEL (TMR red) assays were performed following the manufacturer’s protocol (ROCHE). Respective images were obtained via confocal microscopy using an LSM 510 Meta microscope at 20× magnification. The image processing software, ImageJ (National Institute of Health), was used to manually identify and quantify fluorescently-positive cells (Schneider et al. 2012). The percentage of GFP-positive cells expressing islet hormones were calculated. Three different biological replicates from each genotype and at least 500 cells were analyzed for each measurement.
Fluorescence-activated cell sorting
Pancreatic cells were dispersed into a single-cell suspension by incubation for 5 min at 37°C in Accumax (Sigma) containing 50 µg/ml DNase I. Reactions were quenched by the addition of sorting buffer [FACS staining buffer (R&D Systems), DNAse (1:1000)]. Cells were then filtered through a 35 µm nylon mesh into a FACS tube (Corning), washed once with FACS staining buffer, centrifuged at 1100 rpm for 3 min, then re-suspended in 500 µl of FACS sorting buffer (Osipovich et al. 2014). 7-aminoactinomycin (7-AAD) was added at a concentration of 1:1000 immediately before sorting using either a Benton Dickenson FACS Aria-II or Aria-III instrument to distinguish between live and dead cells. GFP-positive, 7-AAD negative cells were collected directly into Trizol LS (Invitrogen) containing 40 µg/ml of mussel glycogen (Roche/Sigma). An average of 6000 cells was obtained per embryo sample.
RNA isolation, library construction, and RNA-Seq
Three replicate samples for each genotype were collected, and total RNA was isolated using Trizol LS then treated with DNase I (Life Technologies). RNA was column-purified using the RNA Clean and Concentrator Kit (Zymo Research) and previously published protocols (Choi et al. 2012). RNA integrity was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), and samples with an RNA integrity number (RIN) of 7.0 or greater were used for RNA-Seq. All samples were amplified using the SMART-Seq Ultra Low Input RNA Kit (TAKARA/Clontech) at 10 cycles, except for the Neurog3GFP/+ and Neurog3GFP/GFP samples, which were prepared using the Ovation RNA-Seq System V2 (NuGEN). cDNA was prepared using the Low Input Library Kit (Clontech) and sequenced using either an Illumina HiSeq3000 genome analyzer to obtain paired-end, 75-bp reads, or Illumina NovaSeq6000 to get paired-end, 100-bp reads. RNA samples were sequenced to an average depth of ∼5.0 × 107 reads. RNA-Seq data have been deposited in ArrayExpress (accession no. E-MTAB-10262).
Bioinformatic analysis of RNA-Seq
FastQ files were routinely processed then sanitized using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (last accessed 2021-08-31) and TrimGalore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) (last accessed 2021-08-31), respectively. Individual reads were aligned to the mouse genome (mm-10) using Spliced Transcripts Alignment to a Reference (STAR) software (Dobin et al. 2013). Pairwise differential gene expression analyses were performed using HTSeq (Anders et al. 2015) to obtain read counts, and DESeq2 was used (Love et al. 2014) to quantify and display the differences. Only genes that exceeded an adjusted P-value ≤0.05 were included in the analysis. Gene ontology (GO) analyses were performed using the online Database for Annotation, Visualization, and Integrated Discovery (DAVID v6.8) (Huang da et al. 2009a,b). Neurog3GFP/+ and Neurog3GFP/GFP samples were separately analyzed as described (Osipovich et al. 2021).
Bioinformatic analysis of ChIP-Seq
ChIP-Seq datasets for INSM1 and NEUROD1 binding sites were obtained from ArrayExpress (E-GEOD-54046). Both datasets were obtained using an insulinoma cell line propagated from RIP1-Tag2 transgenic mice (Jia, et al. 2015). Snakemake (5.2.4), a workflow management system, was used to manage the data processing. FastQC (0.11.7) returned a quality score report on each sample’s sequences before and after Trimgalore (0.6.5) discarded sequences with a read length shorter than 20 bp were discarded and trimmed read ends that do not meet the threshold of 20. Trimmed sequences were used by the RNA-Seq aligner Bowtie2 (2.3.5) to align against the mouse genome (gencode 17; GRCm38). On average, 95% of reads were mapped to the reference per sample. Samtools (1.9) converted the SAM files into BAM files and filtered out alignments with a lower MAPQ value than 10, and sort alignments by leftmost coordinates and by read name. Macs2 (2.2.6) was used to perform peak calling on the samples using default parameters for bandwidth and effective genome size. The resulting BED files were modified by bedtools (2.26.0) for pile-up visualization via IGV. Unmodified BED files were reported as very high-quality data by phantompeakqualtools (1.2.2).
Differential splice variant analysis
DSV analysis was performed using the previously described RNA-Seq datasets from control and knock-out (KO) embryos at E15.5. Snakemake (5.2.4), a workflow management system, was used to call five programs necessary to process the samples. FastQC (0.11.8) returned a quality score report on each sample’s sequences before and after Trimgalore (0.5.0) trimmed the sequences. Trimmed paired-end sequences were passed to STAR (2.6.0), an RNA-seq aligner, to align against the mouse genome (gencode 17; GRCm38), and gene counts for each sample were returned. The average uniquely mapped reads count per sample was 52M (89%). QoRTs (Quality of RNA-Seq toolset) converts the gene counts format for analysis in JunctionSeq. Lastly, Multiqc (1.7) compiled the final summary output from the other jobs into a single report. Gene counts for each sample were passed to R (3.5.3) to do a quality control assessment before analysis. Quality control assessment included analyzing a box plot of normalized sample counts, principal component analysis (PCA) plot, sample to sample heatmap clustering, and a density plot of the normalized counts. All samples passed the quality control assessment. Each sample’s QoRTS formatted gene counts were run through JunctionSeq (1.12.1) to determine any differential gene expression through alternative splicing between the KO and control groups.
Sashimi plots and exon junction visualization
Plots of read coverage across exons and corresponding sashimi plots were generated using the Integrative Genomics Viewer (IGV) platform (Robinson et al. 2011). Indexed BAM files from each KO dataset were uploaded directly to IGV for analysis, and mm10 was used as the reference genome. All tracks are set to the same read count scale for a given locus.
Results
An integrated analysis of Insm1, Neurod1, and Pax6, three Neurog3-dependent pro-endocrine TFs
Comparison of the gene expression profiles of purified Neurog3GFP/+ and Neurog3GFP/GFP (Neurog3 KO) cells from E15.5 embryos by bulk RNA-Seq (Osipovich et al. 2021) revealed a marked reduction in the expression of pro-endocrine TFs Insm1, Neurod1, and Pax6(Supplementary Figure S1A). The marked dysregulation of these genes provides further evidence that each lies downstream of Neurog3 in a Neurog3-driven endocrine cell-specific gene regulatory network (Supplementary Figure S1B) essential for the formation and function of pancreatic endocrine cells (Naya et al. 1997; St-Onge et al. 1997; Gierl et al. 2006; Mellitzer et al. 2006).
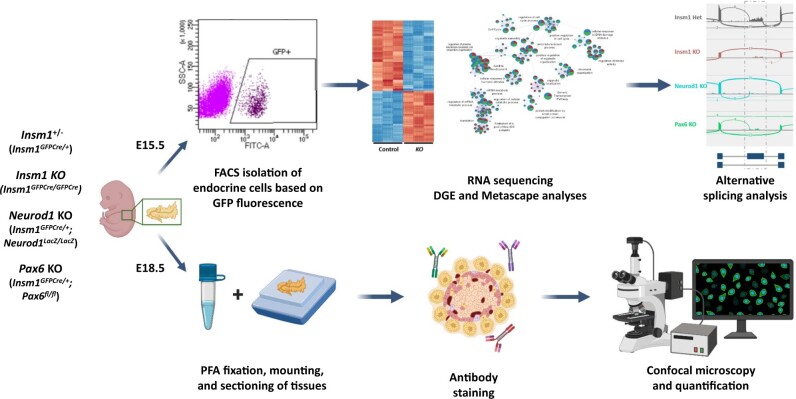
To systematically assess and compare the independent effects of Insm1, Neurod1, and Pax6 in developing endocrine cells, we intercrossed mice containing Insm1GFPCre, Neurod1LacZ, and Pax6flox alleles. From timed matings, we obtained embryos that were heterozygous for Insm1 (Insm1+/−) for use as controls, embryos that were globally deficient in Insm1 (Insm1 KO) or Neurod1 (Neurod1 KO), and embryos that lacked Pax6 specifically in Insm1-expressing cells (Pax6 KO) (Figure 1). Our strategy took advantage of the expression profile of Insm1, specifically that it is expressed in developing endocrine cells, that it is maintained in all endocrine cell types through adulthood, and that it is absent in exocrine pancreas. Green fluorescence produced by the Insm1GFPCre allele enabled us to purify endocrine cells by FACS from E15.5 embryos for RNA-Seq analysis and to accurately identify pancreatic endocrine cells by immunohistochemistry in tissue samples from E18.5 embryos (Figure 1). The findings were then comparatively analyzed.
Figure 1.
An integrated approach for analyzing the roles of Insm1, Neurod1, and Pax6 in pancreatic endocrine cell development. Control and KO embryos were collected at either E15.5 or E18.5. GFP-positive cells from E15.5 pancreata were FACS sorted and collected for RNA-Seq, followed by differential expression, Metascape, and differential splicing analyses. Pancreata from E18.5 embryos were fixed in PFA, sections stained for nuclear GFP signal, endocrine hormones, Ki67 and TUNEL reporter, and marked cells were subsequently quantified. Schematic was created with BioRender.com.
Mice lacking Insm1, Neurod1, or Pax6 exhibit defects in endocrine cell differentiation, proliferation, and apoptosis
Immunofluorescence staining and morphometric analysis of pancreatic tissues from Insm1+/− (control) and Insm1 KO, Neurod1 KO, and Pax6 KO embryos at E18.5 was done by co-staining and quantifying the percentage of pancreatic hormone-positive cells, cell proliferation marker Ki67-positive, or apoptotic TUNEL-positive cells per total number of GFP-positive endocrine cells.
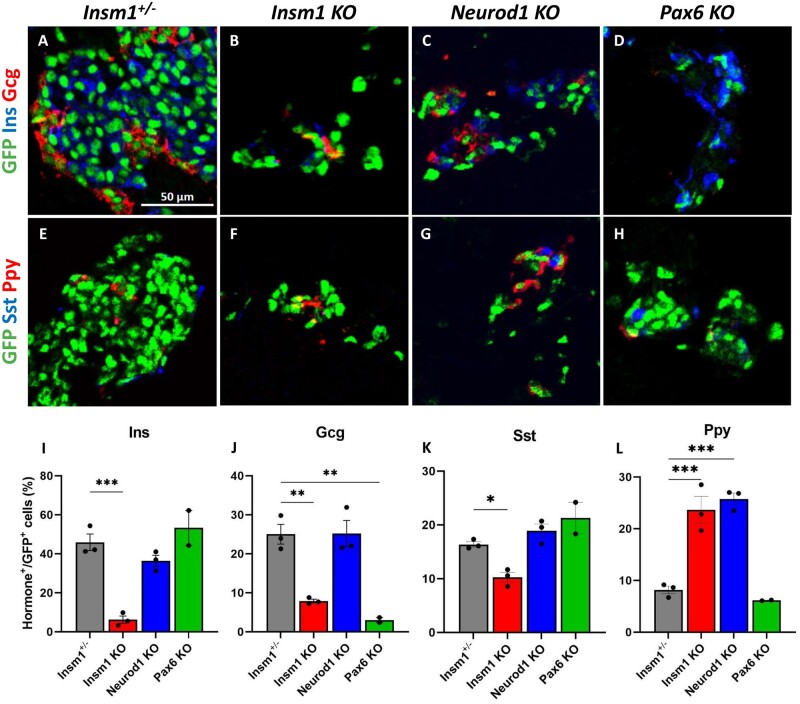
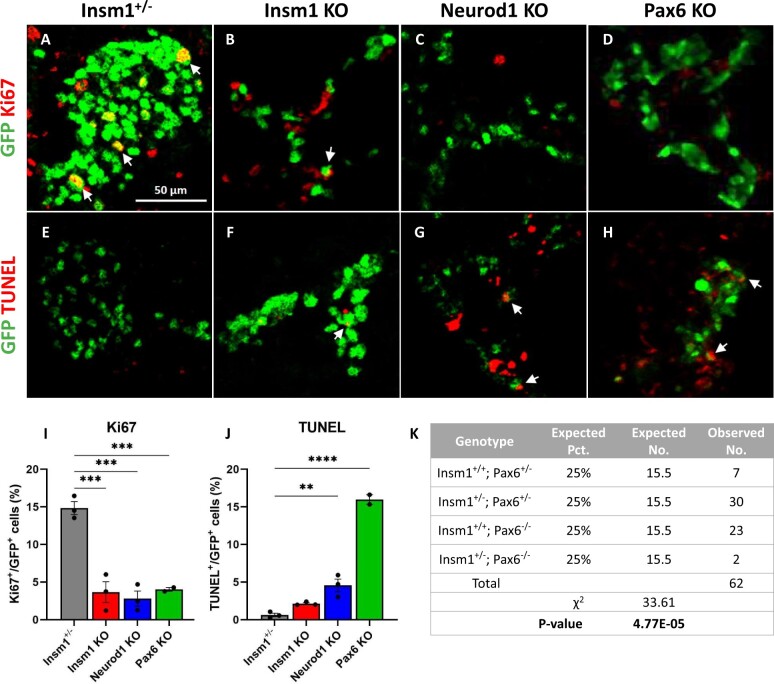
Similar to prior reports, embryos that lack Insm1 exhibit both a disruption in endocrine cell organization and a reduced number of endocrine cells (Gierl et al. 2006; Osipovich et al. 2014). Insm1 KO pancreata were again seen to have a lower number of insulin-, glucagon-, and somatostatin-expressing endocrine cells, and an increase in pancreatic polypeptide-expressing cells, indicating profound defects in endocrine differentiation and lineage specification (Figure 2, B, F, and I–L). Immunostaining with Ki67 revealed reduced proliferation of the Insm1 KO endocrine cells (3.6% vs. 15% of control cells) (Figure 3, B, F, I, and J), whereas the number of apoptotic cells between the control and KO cells was unchanged.
As in previous studies, Neurod1 KO embryos also exhibited a lower overall number of endocrine cells, disrupted islet organization, and increased percentage of pancreatic polypeptide-expressing endocrine cells increased (Figure 2, C, G, and I–L). The Neurod1 KO embryos had fewer Ki67-positive endocrine cells (2.8% vs. 15% in the controls) and a marked increase in TUNEL-positive cells (4.5% vs. 0.6% in control animals) (Figure 3, C, G, I, and J), which is also similar to prior reports (Naya et al. 1997; Romer et al. 2019).
Similarly, analysis of the Pax6 KO embryos revealed disorganized endocrine islets and a reduced overall number of endocrine cells. However, in these mice, there was a marked decrease in the number of glucagon-positive cells (Figure 2, D and J), although the numbers of other hormone-expressing GFP-positive cells were not significantly affected (Figure 2, H, K, and L). There was also a considerable reduction in the number of Ki67-positive cells in the Pax6 KO animals (4% vs. 15% in the controls) (Figure 3, D and I) and an increase in the number of TUNEL-positive endocrine cells (Figure 3, H and J). Notably, we had difficulty obtaining Insm1GFPCre/+, Pax6 KO embryos at E18.5, and only succeeded in obtaining two embryos of the sought-after genotype out of 62 embryos genotyped (P = 4.77E−05 by chi-squared test), indicating that the haploinsufficiency of Insm1 in the setting of a Pax6 KO impairs embryonic survival compared to the effects of a Pax6 KO alone (Figure 3K).
Figure 2.
Impaired differentiation of pancreatic endocrine cells in Insm1, Neurod1, and Pax6 KO embryos. (A–D) Immunofluorescence labeling of pancreata from E18.5 Insm1GFP-expressing embryos using antibodies against GFP (green) that marks pre-endocrine cells, and pancreatic hormones insulin (Ins, blue), and glucagon (Gcg, red). Compared with Insm1+/− mice (A), mice lacking Insm1 (B), Neurod1 (C), and Pax6 (D) exhibit a decrease in total number of endocrine cells, altered endocrine cell morphology and numbers of hormone expressing cells. (E–H) Immunofluorescence labeling of pancreata from E18.5 embryos using antibodies against GFP (green), pancreatic polypeptide (Ppy, red), and somatostatin (Sst, blue) shows altered numbers of hormone expressing cells in Insm1 (F), Neurod1 (G), and Pax6 (H) KO mice. (I–L) Quantification of a percentage of hormone-positive cells among GFP-positive endocrine cells demonstrates defects in differentiation of cells positive for hormones: insulin (Ins) (I), glucagon (Gcg) (J), somatostatin (Sst) (K), and pancreatic polypeptide (Ppy) (L) in Insm1, Neurod1, and Pax6 KO embryonic pancreata in comparison with Insm1+/−. Error bars indicate SEM (n = 3); P-values were determined by one-way ANOVA test. Asterisks indicate P-values of *<0.05, **<0.01, and ***<0.001. Scale bars: 50 μm.
Figure 3.
Decreased proliferation of endocrine cells in Insm1, Neurod1, and Pax6 KOs, and increased apoptosis in Neurod1 and Pax6 KO embryos. (A–D) Immunofluorescence labeling of pancreatic tissues from E18.5 Insm1GFP-expressing embryos using antibodies against GFP (green) that marks all pre-endocrine cells, and antibodies against cell proliferation marker Ki67 (red). Compared to controls (A), mice lacking Insm1 (B), Neurod1 (C), and Pax6 (D) exhibit a decrease in the number of Ki67-positive endocrine cells. Arrows indicate cells co-expressing GFP and Ki67. (E–H) Immunofluorescence labeling of pancreatic tissues from E18.5 embryos with antibodies against GFP (green) and TUNEL assay (red) marking positive apoptotic events. Compared with Insm1+/− mice (E), mice lacking Insm1 (F), Neurod1 (G), and Pax6 (H) exhibit an increase in the number of endocrine cells positive for both TUNEL and GFP. Arrows indicate TUNEL positive cells co-expressing GFP. (I) Quantification of a percentage of Ki67-positive cells among GFP-positive cells demonstrates a proliferation defect in endocrine cells from Insm1, Neurod1, and Pax6 KO pancreata at E18.5. (J) Quantification of a percentage of TUNEL-positive cells among GFP-positive cells demonstrates increased apoptosis in endocrine cells in Neurod1 and Pax6 KO pancreata at E18.5. Error bars indicate SEM (n = 3); P-values were determined by one-way ANOVA test. Asterisks indicate P-values of *<0.05, **<0.01, and ***<0.001. Scale bars: 50 μm.
Together, these analyses indicate that mice lacking Insm1, Neurod1, or Pax6 have abnormal islet morphologies, a reduced number of endocrine cells, and varying defects in differentiation toward hormone-expressing cells. Moreover, all three KOs exhibited decreased endocrine cell proliferation rates, with both the Neurod1 and Pax6 KOs also showing increased apoptosis.
Pancreatic pre-endocrine cells lacking Insm1, Neurod1, and Pax6 have distinct transcriptional profiles
Next, to better understand how each factor contributes to the pro-endocrine cell transcriptome, we performed bulk RNA-Seq on FACS-purified cell populations isolated from E15.5 embryos. Cells were isolated at this earlier time point to minimize variance in gene expression caused by maturation toward specific endocrine subtypes. Hierarchical clustering and PCA of the 12 RNA-Seq samples revealed tight clustering by genotype, indicating the low variance between samples and that each condition analyzed has a unique transcriptional profile (Supplementary Figure S2, A and B).
Dysregulated expression of genes involved in hormone secretion, RNA metabolism and processing, and cell development in Insm1 KO endocrine cells
We previously reported the gene expression profile of both Insm1+/− and Insm1 KO mice at E15.5 using a legacy RNA amplification and sequencing method (Osipovich et al. 2014). To accurately compare the transcriptomes of the three TF KOs in this study, we collected new Insm1 KO datasets using the methods, reagents, and instrumentation described herein. A comparison of the two different Insm1+/− and Insm1 KO datasets indicated that they clustered first by amplification method (Nugen vs. Clontech) and secondarily by genotype (Insm1+/− control vs. Insm1 KO), consistent with a method-dependent batch effect. However, a linear regression analysis of the log2-fold changes of differentially expressed genes from the old and new datasets showed strong correlation (R2 = 0.85) (Supplementary Figure S3, A–C).
Analysis of the new Insm1+/− and Insm1 KO datasets by DESeq revealed a total of 4,694 differentially expressed genes, 2,265 were categorized as downregulated genes (DRGs) and 2429 upregulated genes (URGs) in the KO compared to control embryos (P-value ≤ 0.05) (Supplementary Figures S2C, S4, A and B and Table S1). Among the top DRGs were numerous TFs known to control endocrine cell development, including both Sox9 (McDonald et al. 2012) and Mnx1, which is necessary for endocrine cell fate allocation and in later stages for maintaining the β-cell fate (Sund et al. 2001; Wang et al. 2002; Pan et al. 2015). Also, among the DRGs were the TF Bhlhe23, the histone modifiers Hist1h2bf and Histh2bb, and genes involved in the trafficking and secretion of insulin (Avp, Chgb) (Supplementary Figure S4C). GO and pathway enrichment analysis results from DRGs revealed enrichment in genes involved in cell projection morphogenesis (Bdnf, Brsk2, Camk2b, Etv4), cellular response to hormone stimulus (Adra2a, Gcgr, Insr, Kcnc2), hormone secretion (StxbKp5l, Stx1a, Cacna1d), and mRNA metabolic process (Srsf2, Hnrnpc, Celf4, Celf6) (Supplementary Figure S4, D and F and Table S2).
Among the top URGs, there were many genes necessary for the key functions of mature islet cells. Examples include Kcnc4 (Kv3.4), a voltage-gated potassium channel expressed in β- and δ-cells; and Gpr17, an orphan G protein-coupled receptor known to inhibit neural cell maturation (Gopel et al. 2000; Jacobson and Philipson 2007; Chen et al. 2009). GO and pathway enrichment analyses identified an enrichment in genes involved in the regulation of cellular component organization (Adck1, Arap1), Notch and Wnt signaling pathways (Dll1, Notch1/2, Wnt7b, Frzb, Frz8), and the negative regulation of cellular development (Aatk, Dlx1, Rest, Kctd11) (Supplementary Figure S4, E and F and Table S2). These results indicate that Insm1 stimulates many genes involved in hormone secretion and the function of mature endocrine cells, while repressing genes associated with Notch and Wnt signaling.
Dysregulated expression of genes involved in chromatin organization, cell proliferation, and mitochondrial function in Neurod1 KO endocrine cells
Comparing the Neurod1 KO and control datasets revealed a different set of 4613 dysregulated genes (2131 DRGs and 2482 URGs) (P-value ≤ 0.05) (Supplementary Figures S2D, S5, A and B and Table S1). The top DRGs included many factors known to impact islet cell function, including Prlr, G6pc2, Hspa1a, and Hspa1b (Supplementary Figure S5C). Transcriptional co-regulators (Runx1t1, Myt1l), cell cycle regulation (Cdkn1a, Cdc14b), and chromatin organization (Ctcf, Kat2b) were enriched in DRGs in Neurod1 KO (Supplementary Figure S5D and Table S2).
The top URGs in this dataset included the NF-κB signaling regulators (Tifab, Ccdc3), extracellular matrix factors (Emilin2, Col19a1), and the TRAIL receptor Tnfsrf26 (Supplementary Figure S5C). URGs were enriched in such functional categories as cellular respiration (Coq10a, Uqcc3), mitochondrial organization (Mfn2, Meif1, Meif2, Plekhf1), and autophagy (Atg7, Atg13, Tbc1d17, Vps11) (Supplementary Figure S5D and Table S2). Moreover, genes involved in regulation of cell death were also upregulated (P2rx1, P2rx7, C6). Identification of these enriched GO terms and pathways for the Neurod1 KO mice parallels the changes we and others have observed in islet morphology, including reduced endocrine cell proliferation and increased cell death (Naya et al. 1997; Romer et al. 2019). These findings indicate that the loss of Neurod1 causes a transcriptional response marked by the upregulation of genes involved in NF-κB signaling and energy metabolism, and the downregulation of other pancreatic TFs and genes associated with cell cycle regulation.
Dysregulated expression of genes involved in cell cycle regulation, developmental growth, and apoptosis in Pax6 KO endocrine cells
Comparison of the Pax6 KO embryos and controls revealed a third set of 5770 genes (2756 DRGs and 3014 URGs) that were Pax6 dependent (P-value ≤ 0.05) (Supplementary Figures S2E, S6, A and B and Table S1). Among the top URGs were Prlr, G6pc2, Gcg, Ptgdr, and Ccdc3 (Supplementary Figure S6C). Inspection of the enriched GO terms and pathways for Pax6-dependent DRGs and URGs suggests a critical role for Pax6 in stimulating cell growth and cell cycle, and regulating endoplasmic reticulum (ER) stress and apoptosis in developing endocrine cells (Supplementary Figures S6, D and E and Table S2). Specifically, many genes involved in chromatin organization (Ash1l, p300, Mettl4), cell cycle regulation (Ccnd1, Cdc27, Cdkn1b, Chek1), and developmental cell growth (Fgf9, Igf1r, Tgfb2) were downregulated in Pax6 KO animals (Supplementary Figure S6, D and F). Consistent with the diminished number of α-cells observed at E18.5, there was a marked reduction in Gcg expression, as well as dysregulation of other α-cell-specific genes Brn4 and Pcsk2. GO terms and pathways enriched in URGs include cellular component organization (Elmo1/2, Lima1, Myo7a), response to ER stress (Atf6b, Traf2), and the apoptotic signaling pathway (Casp2, Casp9, Madd) (Supplementary Figure S6, E and F and TableS2). Analysis of dysregulated genes in Pax6 KO animals shows the downregulation of other TFs, cell cycle regulators and genes important for cellular growth, and the upregulation of apoptotic signaling and genes involved in the response to ER stress.
Comparison of the gene sets dysregulated in the Insm1, Neurod1, and Pax6 KO endocrine cells
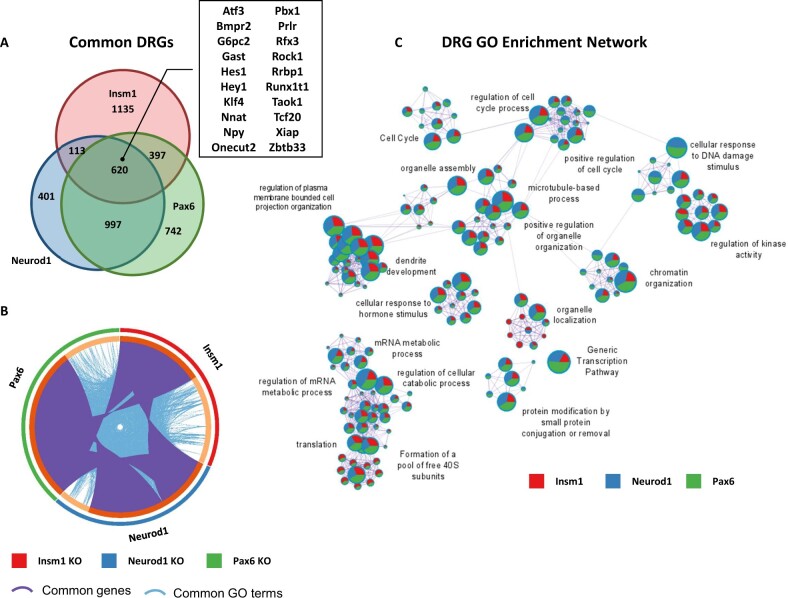
To further assess the similarities and differences in the genes affected by each gene KO, we started by comparing the gene lists. We found that of the 8729 total dysregulated genes (based on adj. P-value ≤ 0.05) across all three KOs, 4719 (54%) were affected by more than one TF (Figures 4A and 5A). Additionally, 578 (6.6%) of genes were inversely regulated across TFs. For example, Mnx1 is upregulated in Pax6- and Neurod1 KO datasets and downregulated in Insm1 KO samples, conversely Rest is downregulated in Pax6- and Neurod1 KO samples and upregulated in Insm1 KO data. Similarly, Fev is upregulated in the absence of Pax6, unchanged in Neurod1 KO and downregulated in Insm1 KO datasets (Supplementary Table S1). This finding indicates that while Insm1, Neurod1, and Pax6 commonly regulate a sizeable fraction of genes, each TF has unique effects on the transcriptome. To quantify these findings and reflect the similarity of the different groups, we determined the percentage of shared genes relative to the total number of dysregulated genes. This analysis revealed a range of 2.6–22.6% similarity between different groups, with the highest similarity between Neurod1 and Pax6 DRGs (22.6%).
Figure 4.
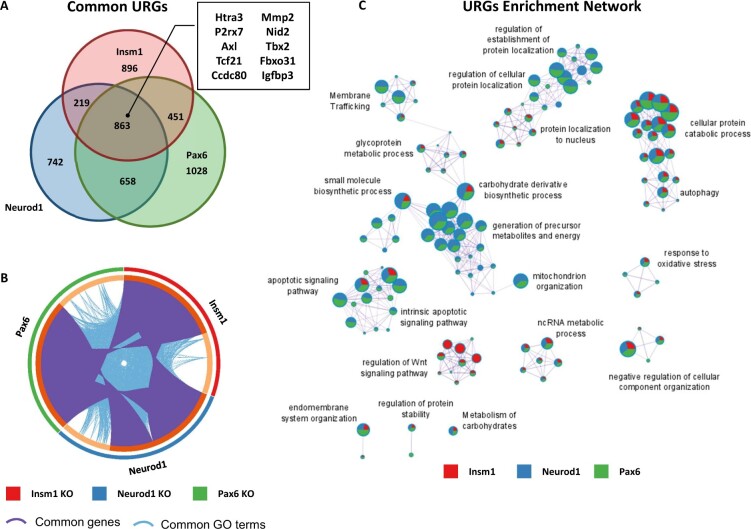
Enriched GO terms and pathways common for genes downregulated in Insm1, Neurod1, and Pax6 KO pancreatic endocrine cells. (A) Venn diagram shows overlap between downregulated genes (DRGs) (P-value < 0.05 cutoff) in each of the pairwise comparisons: Insm1 KO, Neurod1 KO, and Pax6-KO versus Insm1+/− datasets. A subset of commonly downregulated genes is highlighted within a boxed insert. (B) Circos plot representing genes (purple curves) and GO terms/pathways (blue curves) that are shared between DRGs from the three comparisons. (C) Network depiction of the enriched gene ontology terms shared between DRGs from the three comparisons. Node size is proportional to the number of genes in GO or pathway category, with pie charts indicating a proportion of genes from each comparison in that GO term. Metascape analyses were run using default parameters.
Figure 5.
Enriched GO terms and pathways common for genes upregulated in Insm1, Neurod1, and Pax6 KO pancreatic endocrine cells. (A) Venn diagram shows overlap between upregulated genes (URGs) (P-value < 0.05 cutoff) in each of the pairwise comparisons: Insm1 KO, Neurod1 KO, and Pax6 KO versus Insm1+/− datasets. A subset of commonly downregulated genes is highlighted within a boxed insert. (B) Circos plot representing genes (purple curves) and GO terms/pathways (blue curves) that are shared between URGs from the three comparisons. (C) Network depiction of the enriched gene ontology terms shared between URGs from the three comparisons. Node size is proportional to the number of genes in GO or pathway category, with pie charts indicating a proportion of genes from each comparison in that GO term. Metascape analyses were run using default parameters.
Next, to identify commonalities in the functional annotations of genes dysregulated in each gene set, we performed Metascape analyses for both the DRGs and URGs. These analyses revealed many genes and functional GO terms and pathways shared between dysregulated genes from each gene set (Figures 4B and 5B). Of the 620 commonly regulated DRGs (14.07% shared), the enriched GO term and signaling pathway network includes such categories as regulation of mRNA metabolic processes and translation (Akap17b, Clk1, Srsf3, Srsf6), cell cycle regulation (Bbx, Cdk6, Cdc14b), and chromatin organization (Kmt2a, Setd5, Kdm7a) (Figure 4C and Supplementary Table S2). Other common DRGs included regulators of insulin secretion and signaling (Ins1, Nnat, Ucn3, Insr), transcriptional regulators (St18, Neurod1, Rfx3, Myt1l, Runx1t1, Onecut2), kinases (Akt3, Mapk8, Taok1, Prkcb), and anti-apoptotic genes (Xiap, Bcl-W) (Supplementary Table S1). While it is not surprising that these TFs regulate other TFs that are known to be involved in pancreas development and function, it is important to note their role in affecting the expression of cell cycle regulators and chromatin organization. Tight regulation of these processes has long been linked to cellular development, expansion, and differentiation, and their dysregulation likely contributes to the observed developmental delays and perturbed ratios of islet cell types (Dalton 2015; Kim et al. 2015; Boward et al. 2016; Soufi and Dalton 2016; Krentz et al. 2017).
The shared functional network for 863 URGs (17.7% shared) in all three KOs contains GO terms and pathways for ncRNA processing (Aars, Ctu1, Lsm6, Tsen2), apoptotic signaling pathway (Casp9, Tradd, Tnfrsf21, Ddx47), protein localization (Ipo4, Ipo9, Timm29), and response to oxidative stress (Pink1, Selenon, Fancc) (Figure 5C and Supplementary Table S2). Genes involved in Wnt-signaling pathways (Frzb, Lzts2) and other TFs (Sox10, Tbx2, Foxa2) were also upregulated (Supplementary Table S1). The upregulation of genes associated with apoptosis and oxidative stress response, in combination with the downregulation of anti-apoptotic genes, coincides with the increases in apoptosis in Neurod1 and Pax6 KO pancreata. Likewise, increases in the expression of members of the Wnt signaling pathway and early endocrine TFs are reminiscent of morphological developmental delays (Sharon et al. 2019). This is particularly true of those observed in Insm1 KO embryos which are able to generate endocrine cells, but many of those are unable to properly differentiate and mature into hormone expressing endocrine cells (Mellitzer et al. 2006; Osipovich et al. 2014).
Metascape analysis also identified protein–protein interaction (PPI) enrichments common to all three gene sets. Enriched subnetworks of DRGs known to form PPIs included members of the ATF2- (Atf2, Pou2f1, Nf1, Brca1), cohesin- (Smca1, Smc3, Atrx, Mecp2), and histone acetylation-complexes (Kat2b, Crebbp, Clock). (Supplementary Figure S7, A, C, and E). The PPI subnetworks of URGs involved in NF-κB signaling (Ikbkb, Ikbkg, Rnf31, Rbck1) and histone modification (Tuba1a, Hdac6, Kat5b) (Supplementary Figure S8, A and C). Interestingly, NF-κB signaling has been recently shown to regulate β-cell proliferation and apoptosis, and in turn β-cell mass, during development (Sever et al. 2021). Together, these analyses indicate that while there are differences in the genes dysregulated by Insm1, Neurod1, and Pax6, there are many shared functionalities, both from a GO and PPI subnetwork perspective.
Prediction of direct targets of Insm1 and Neurod1 critical for endocrine cell development
To predict direct targets of Insm1 and Neurod1 in developing endocrine cells, we utilized previously reported ChIP-Seq datasets (Jia et al. 2015). Consistent with published data, our analysis showed that Insm1 and Neurod1 share a large proportion of binding sites (11,409 sites across the genome, 81.9% of total binding sites) (Supplementary Figure S9A). Integration of ChIP-Seq data with our RNA-Seq data on dysregulated genes suggests that 4180 and 3789 genes, respectively, may be direct targets of Insm1 and Neurod1 in endocrine progenitor cells (Supplementary Figure S9, B and C and Table S3). We further compared the lists of direct target genes and found 1,719 (27.5% shared) genes bound and dysregulated by both Insm1 and Neurod1. These included genes involved in peptide hormone response and secretion (G6pc2, Glp1r, Sytl4, Stxbp5l), zinc finger proteins (Zfp326, Zbtb33, Zbdf2), rhythmic processes (Clock, Dhx9, Kcnd2), and mRNA processing (Celf4, Tra2a, Akap17b, Zfp871, Luc7l2, Ddx17) (Supplementary Figure S9D). These findings indicate that of the genes dysregulated in our Insm1 and Neurod1 KO endocrine progenitor cells, 89% and 82% are predicted to be direct targets, respectively, based on the presence of binding sites within 5 kb of the transcriptional start site.
Insm1, Neurod1, and Pax6 KO endocrine cells exhibit dysregulated RNA splicing
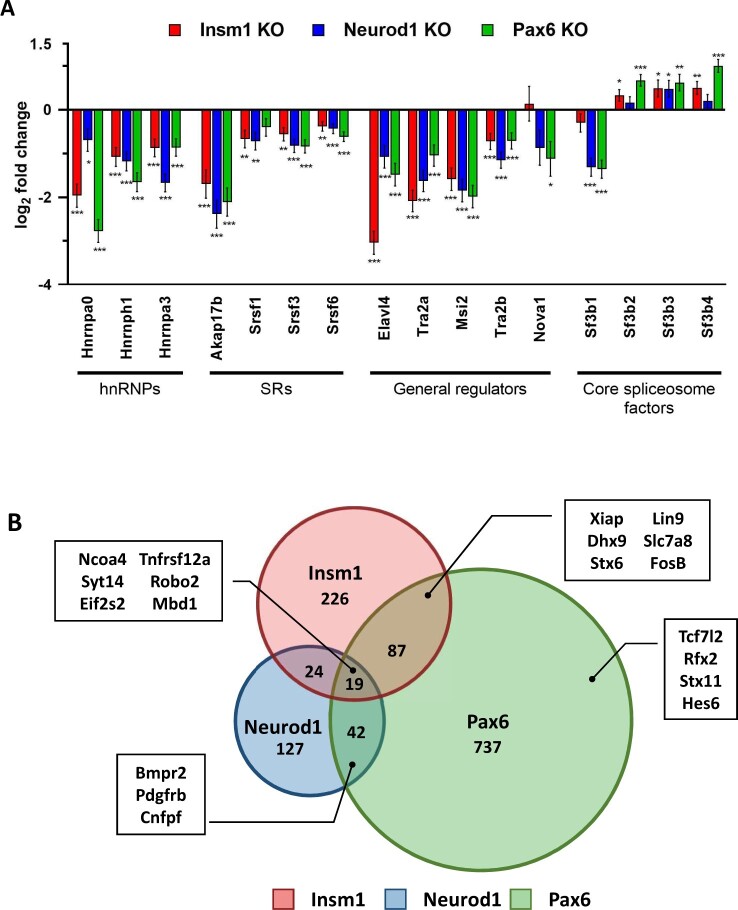
Among the enriched GO terms and signaling pathways of dysregulated genes found in all three of the KO gene sets was the regulation of RNA metabolic processes, which includes the regulation of alternative splicing. To determine whether mRNA splicing was affected in the Insm1, Neurod1, and Pax6 KO cells, we first examined the expression of several RBPs known to regulate mRNA splicing. We found several RBPs with similarly dysregulated expression patterns, including Elavl3, Elavl4, Tra2A, and Tra2B (Figure 6A). The expression of other differential RNA splicing factors such as serine/arginine-rich proteins that generally act as activators of splicing (Srsf1, Srsf3), heterogeneous nuclear ribonucleoproteins (hnRNPs) with repressive functions (Hnrnpa0, Hnrnph1, Hnrnpha3), and members of the spliceosome itself (Sfb1, Sfb2, Sfb3, Sfb4) are also dysregulated. Importantly, ChIP-Seq data indicate that Insm1 and Neurod1 bind at the promoter regions of many of the same RBPs whose expression levels are dysregulated in our KO data. For example, Elavl3/4, Nova1, Tra2a/b, and Srsf1, in addition to being downregulated, are all bound by both Insm1 and Neurod1 at their promoter regions (Supplementary Figure S10). To determine if the dysregulation of RBP expression in the absence of Insm1, Neurod1, or Pax6 was correlated with differential splicing events, we used our RNA-Seq datasets to perform a computational alternative splicing analysis using JunctionSeq. The results indicated hundreds of differential splicing events in Insm1 KO (356), Neurod1 KO (212), and Pax6 KO (885) datasets (adj. P-value ≤ 0.01, log2 FC > |1|) (Figure 6B and Supplementary Table S4). Of the 1,262 total differentially spliced genes, 172 were shared between pairs of TFs, with Pax6 itself being alternatively spliced in Insm1 and Pax6 KO datasets (Supplementary Table S4). Additionally, Tcf7l2, which has strong polymorphism associations with T2D and is known to have isoform-specific effects on β-cell function, is also differentially spliced in the Pax6 KO samples (Prokunina-Olsson, 2009; Osmark, 2009; Le Bacquer, 2011; Zhang, 2021). However, only 19 (1.5%) differential splicing events were common to all three KOs (Ncoa4, Syt14, Tnfrsf12a, Mov10) (Supplementary Table S4). Since some genes have several possible transcriptional isoforms, interpreting alternative splicing data is challenging. Therefore, we chose to take a closer look at a few genes that were differentially spliced in our KO datasets, and which have only a few RNA splicing variants.
Figure 6.
Insm1, Neurod1, and Pax6 regulate alternative splicing in pancreatic endocrine cells. (A) Many important RNA binding proteins (RBPs) are dysregulated in in Insm1, Neurod1, and Pax6 KOs. Bar graph of the log2 fold change of a subset of dysregulated RBPs. Asterisks indicate P-values of *<0.05, **<0.01, ***<0.001. (B) Venn diagram demonstrates overlaps between differentially spliced genes in Insm1, Neurod1, and Pax6 KO RNA-Seq datasets. Examples of alternatively spliced genes are highlighted with a box insert. Differential splicing events between the knock-out and control groups were identified by performing JunctionSeq analysis on the RNA-Seq datasets. A cutoff of P-adj. value ≤0.01 and log2 fold change ≥|1| was used.
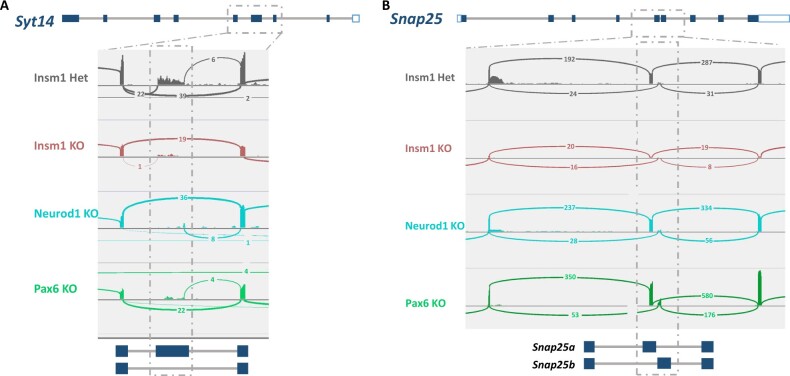
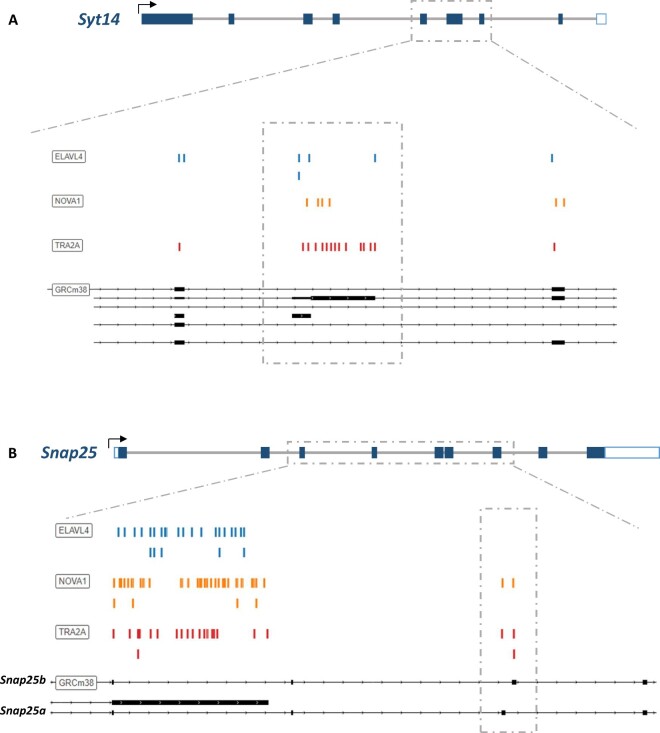
Syt14 is a Ca2+-independent synaptotagmin required for the exocytosis and trafficking of secretory vesicles. Interestingly differential splicing patterns were revealed for Syt14 across all three KO datasets, specifically a decrease in expression of the Ensemble splice variant 205. To quantitatively visualize these differences, we generated splice junction plots (Sashimi plots), that tally the exon junction read counts. Sashimi plots of Syt14 reveal a marked decrease of junction reads at exon 4 of the Sty14-205 splice variant (Figure 7A). To determine if dysregulated RBPs show binding preferences near the differentially spliced loci might cause these splicing differences, we used the online oRNAment database to identify local binding motifs (Benoit Bouvrette et al. 2020) and found that TRA2A, NOVA1, and ELAVL4 all show binding patterns at exon 4, as well as adjacent exons 3 and 5 (Figure 8A). Importantly, expression of Tra2A and Elavl4 are downregulated in all three KO gene sets, and Nova1 downregulated in Pax6 KO suggesting their potential involvement in differential splicing of Syt14 in developing endocrine cells (Figure 6A).
Figure 7.
Identification of differential splicing events in Syt14 and Snap25 genes. The gene structures of (A) Syt14 and (B) Snap25 are illustrated above corresponding Sashimi plots of differentially spliced regions. Sashimi plots show the representative splice junctions as arcs that connect the exons in each of the datasets. The numbers found on each of the corresponding arcs indicate the junction depth, or reads spanning the exon junctions. Dashed boxes highlight the regions of alternative splicing, which are again depicted below the Sashimi plot to provide a visual reference for read alignments. Sashimi plots were generated using the Integrated Genomic Viewer (IGV).
Figure 8.
Differentially spliced regions of Syt14 and Snap25 genes are bound by dysregulated RBPs. (A) Schematic of Syt14 gene structure and corresponding binding sites of the RBPs ELAVL4, NOVA1, and TRA2A near the differentially spliced regions. (B) Schematic of Snap25 and corresponding binding sites of ELAVL4, NOVA1, and TRA2A at the differentially spliced site. Colored dashes above the indicated gene region represent the putative binding site of the given RPB. Binding sites were identified via the online oRNAment database.
Similarly, Snap 25 is a critical SNARE protein involved in regulating exocytosis of synaptic vesicles in the brain and hormones in pancreatic islet cells (Kadkova et al. 2019; Liang et al. 2020). Two isoforms, Snap25a and Snap25b, have been identified that differ based on the alternative usage of two different exon 5 sequences (5a and 5b), with corresponding proteins differing in only 9 amino acids (Kadkova et al. 2019). We observe that the Snap25b isoform is increased in the Pax6 KO (P-value < 0.001) and decreased in Insm1 KO (P-value < 0.01) datasets, as indicated in the associated Sashimi plots (Figure 7B). RBP binding site analysis of this locus indicated TRA2A and NOVA1 binding sites at both 5a and 5 b exons, and ELAVL4 sites at nearby upstream exons (Figure 8B). Together, these findings indicate that Insm1, Neurod1, and Pax6 are required for the expression of RBPs that bind in the vicinity of exons whose proper splicing may be essential for the function of pancreatic endocrine cells.
Discussion
Insm1, Neurod1, and Pax6 both independently and coordinately govern pre-endocrine cell gene expression
Insm1, Neurod1, and Pax6 are individually essential for the formation of functional pancreatic endocrine cells (Naya et al. 1997; St-Onge et al. 1997; Gierl et al. 2006; Gu et al. 2010; Osipovich et al. 2014; Jia et al. 2015; Mitchell et al. 2017). Here, we systematically compared the effects of deleting Insm1, Neurod1, and Pax6 in a Insm1GFPCre heterozygous background based on the morphology, gene expression, and differential RNA splicing of nascent pancreatic endocrine cells. By doing so, we obtained new insights into the vital roles that each of these factors has in the development of pancreatic endocrine cells.
Role of Insm1
Consistent with previously published results we showed that Insm1 KOs have defects in endocrine cell differentiation that were manifested in decreased numbers of Ins- and Gcg-positive and increased number of Ppy-positive endocrine cells. This is accompanied by decreased endocrine cell proliferation, as well as changes in expression of genes that affect these processes (Gierl et al. 2006; Osipovich et al. 2014). Our comparative transcriptional analysis revealed both similarities and differences in the genes regulated by Insm1 and Neurod1 and Pax6. For example, Insm1, but not Neurod1 or Pax6, activates the expression of Fev and Mnx1 in endocrine cells, TFs that are known to influence the numbers and types of endocrine cells, as well as islet morphology. Insm1 is specifically important for expression of many genes involved in inducible secretory function of endocrine cells, including protein secretion and Ca2+ regulated vesicle exocytosis. This is consistent with the role of Insm1 in the proliferative and secretory functions of mature endocrine cells (Tao et al., 2018). Our new data, together with re-analysis of previously reported ChIP-Seq data, suggests that 89% of genes dysregulated in endocrine progenitor cells of Insm1 KO mice contain binding sites for Insm1 with 5 kb of the transcriptional start site.
Role of Neurod1
Mice that lack Neurod1 exhibit an increased number of Ppy-expressing endocrine cells without significantly altering the proportion of other differentiated endocrine cell types. However, Neurod1 KO mice exhibit both a profound decrease in proliferation and an increase in apoptosis of endocrine cells, thereby causing a marked reduction in overall number of endocrine cells. These findings are consistent with previous studies, supporting the hypothesis that a lack of proliferative cells and increase in apoptosis are primary causes for the reduced total number of endocrine cells observed (Naya et al. 1997; Romer et al. 2019). Likewise, Romer et al. (2019) recently reported a decrease in α- and β-cell numbers only after the time ∼P0 stage. Their quantifications at E17 show no differences in the cell numbers or ratios of Ins- or Gcg-expressing cells, indicating that the specific reduction in α- and β-cells occurs during late embryonic to early postnatal stages after islet differentiation has largely concluded, which is consistent with our findings.
Our transcriptional profiling results also suggest that Neurod1 promotes proliferation and maturation through the stimulation of genes important for cell cycle regulation and chromatin modifications, processes that are important for cell differentiation and function (Pauklin and Vallier 2013; Chen and Dent 2014; Dowen et al. 2014; Krentz et al. 2017). Simultaneously, the observed upregulation of apoptotic and autophagy genes likely contributes to an increase in cell death, further decreasing the total number of endocrine cells.
The regulation of endocrine specific TFs (Fev, Mnx1) and other genes important for cellular function further support a role for Neurod1 in mature cells as is suggested both by the expression of Neurod1 in adult β- and α-cells and that the conditional ablation of Neurod1 in β-cells impairs their maturation (Naya et al. 1997; Gu et al. 2010; Mastracci et al. 2013). Additionally, ChIP-Seq data indicates that Neurod1 binds within 5 kb of 82% of the genes dysregulated in this analysis, strongly suggesting that it is the primary effector of the observed changes. Together these findings indicate the critical role of Neurod1 in propagating enough fully functional islet cells during development via the promotion of cellular proliferation and activation of key TFs.
Role of Pax6
It is well established that Pax6 is necessary for the specification of pancreatic α-cells, so the marked reduction of Gcg-positive cells (3.3% vs. 15% in controls) in the Pax6 KO animals is not a surprise (Gosmain et al. 2010). However, our data have revealed that endocrine cells lacking Pax6 not only have a decrease in the expression of Gcg, but also a marked dysregulation of other genes important for α-cell development and function, such as Pou3f4 (or Brn4), Pcsk2, Slc30a8, Irx2, and Smarca1. Besides its function in α-cells, Pax6 is important for development and function of adult β-cells, which is supported by our findings of decreased expression of Ins1, Ins2, and Insr in Pax6 KOs (Gosmain et al. 2012; Mitchell et al. 2017; Swisa et al. 2017). While the morphological phenotypes of the islets we observed are in line with previously published results, due to the design of our study we cannot exclude a combinatorial effect caused by the haplosufficiency of Insm1 in mice containing the Insm1GFPCre allele (Sander et al. 1997; St-Onge et al. 1997; Gosmain et al. 2010). Indeed, and as discussed below, the difficulty we had in obtaining Pax6 KO animals at E18.5, which contained only one Insm1GFPCre allele, could be due defects in the developing central nervous system where both Pax6 and Insm1 are expressed (Farkas et al. 2008; Lan and Breslin 2009; Osipovich et al. 2014). Furthermore, it was previously shown that Pax6 RNA is alternatively spliced, and that β- and α-cells preferably express different splicing variants (Singer et al. 2019). Thus, our finding that Insm1 affects the processing of Pax6 mRNA also affects the interpretation of our results. Despite this, the downregulation of genes involved in chromatin organization, cell cycle regulation, and cell growth in the Pax6 KOs, similar to Neurod1 KOs, likely contributes to the reduction in endocrine cell expansion, while an increase in apoptotic genes correlates to the increase in cell death.
Regulatory overlap of Insm1, Neurod1, and Pax6
Recently, we have shown that Insm1 and Neurod1 exhibit a high degree of co-expression during endocrine differentiation, and that Pax6 is similar except that it continues to be expressed in adult β-cells (Osipovich et al. 2021). These co-expression relationships are consistent with Insm1, Neurod1, and Pax6 being important components of the GRN in nascent pancreatic endocrine cells. The genes regulated by these three factors also share many functional assignments, such as mRNA processing and alternative splicing and cell cycle regulation. These similarities likely underpin the broadly similar morphological phenotypes when each is knocked-out, which includes a reduced overall number of pro-endocrine cells, reduced endocrine cell proliferation and increased apoptosis, as well as defects in differentiation toward different kinds of hormone-expressing cells.
Our findings strongly point to a negative epistasis between Insm1 and Pax6, as already noted, due to the fewer than expected numbers of Insm1GFPCre/+; Pax6fl/fl animals at E18.5. Consistent with this, a recent report showed that mice with only a single functional Insm1 allele (Insm1+/−) have impaired β-cell proliferation that results in impaired glucose tolerance in adult animals (Tao et al. 2018). However, the authors observed no impairments in β-cell mass at birth and argued that Insm1 haploinsufficiency has little to no effect on embryonic endocrine development (Tao et al. 2018). Previous Pax6 KO studies have also not reported any unexpected difficulties in obtaining Pax6 null embryos, and animals died just after birth (Sander et al. 1997; St-Onge et al. 1997; Ashery-Padan et al. 2004; Heller et al. 2004). Nonetheless, the reduced number of Pax6 KO embryos at E18.5 appears to be due to a detrimental synergistic effect, or negative epistasis, between Insm1 and Pax6 that impairs embryo survival.
In addition to the greater lethality of Pax6 KO embryos, our data show an increase in both apoptotic events at E18.5, an increase in expression of pro-apoptotic genes and a decrease in expression of anti-apoptotic genes at E15.5. Previous studies of global Pax6 KO mice have all observed a marked decrease in hormone-positive endocrine cells, but none quantified apoptosis events (Sander et al. 1997; St-Onge et al. 1997; Ashery-Padan et al. 2004; Heller et al. 2004). Moreover, Ashery-Padan et al. (2004), who utilized a Cre-Lox-based lineage tracing strategy to quantify the proportion of Pax6 expressing cells relative to total pancreatic tissue at E18.5, reported the loss of hormone expression but no difference in total cell number. While apoptosis was not specifically quantified, their study suggests that pro-endocrine cells form and expand but then fail only to express islet hormones (Ashery-Padan et al. 2004). If so, then our observations are consistent with Insm1 and Pax6 having redundant functions, perhaps in the suppression pro-apoptotic genes. In the future, alternate strategies that do not rely on the Insm1GFPCre the allele and that maintain full expression of Insm1 will need to be utilized to explore how these two genes interact with each other.
Dysregulation of RBP gene expression and identification of alternative splicing events in Insm1, Neurod1, and Pax6 KO animals
Alternative RNA splicing enables a single gene to give rise to multiple mRNA products that encode different protein products. The function of these different protein products can vary greatly and impact cellular function (Alvelos et al. 2018). Recent studies have suggested the importance of the unique splicing program innate to pro-endocrine cells, and how the functional outputs help steer the developmental process, yet it remains incompletely understood (Juan-Mateu et al. 2018; Singer et al. 2019; Colli et al. 2020; Alvelos et al. 2021).
Our analyses indicate Insm1, Neurod1, and Pax6 each regulate the expression of RBPs that mediate different RNA splicing events, including expression of Elavl4, Tra2A, Msi1 and Msi2, and Nova1. Consistent with this, we identified multiple global differential slicing events within KO datasets and, specifically, in the Syt14 and Snap25 genes. Synaptotagmin 14 (Syt14) is a member of the SYT family that is necessary for exocytosis of secretory vesicles, and mutations in this gene have been associated with neurodegenerative disorders in humans (Doi et al. 2011). Our data revealed that Syt14 is differentially spliced in all three of the KO mice studied. Similarly, Synaptosome Associated Protein 25, or Snap25, is a member of the SNARE complex. It is necessary for insulin secretion, is known to be downregulated in T2D islet cells, and is differentially spliced in both Insm1 and Pax6 KO endocrine cells (Sadoul et al. 1995; Gonelle-Gispert et al. 1999; Ostenson et al. 2006; Daraio et al. 2017). It is well established that Snap25 has two primary mRNA isoforms that have slightly different localization patterns and functional traits (Nagy et al. 2005; Daraio et al. 2017; Irfan et al. 2019). Considering this information, it is likely that the differential splicing of these both Syt14 and Snap25 contributes to the impaired function of KO islets. Moreover, it also highlights the importance of alternative splicing in endocrine cell development and indicate that Insm1, Neurod1, and Pax6, besides having large effects on the transcriptome, also control differential splicing events through their regulation of RBPs and thereby affect endocrine cell function in this manner. In the future, it would be beneficial to determine the effects of disrupting the TF binding sites at the promoter regions of RBPs on gene expression levels. Likewise, more work needs to be done to determine the role of specific binding activity of RBPs at their target sites within alternatively spliced genes and if those RBPs maintain endocrine-specific functions.
In conclusion, the present study provides additional insights into the roles for Insm1, Neurod1, and Pax6 in driving endocrine development and differentiation. While each of these TFs has its own unique regulatory functions, there is also functional overlap between these factors based on the large number of commonly dysregulated genes represented in our data. Importantly, we show that the differential expression of RBPs correlates with differential splicing events that can either be shared or common to each gene KO, demonstrating a novel role shared by each factor. Finally, our data demonstrate the broad impact of eliminating each TF, further illustrating their individual roles and importance in establishing the GRN within pancreatic endocrine cells.
Data availability
RNA sequencing data are available at ArrayExpress with the accession number E-MTAB-10262. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables provided.
Supplementary material is available at G3 online.
Supplementary Material
Acknowledgments
We thank the staff of both the Vanderbilt Cell Imaging Shared Resource and the Vanderbilt Technologies for Advanced Genomics (VANTAGE) for their expert assistance.
Funding
This study was supported by funding from the National Institutes of Health to MAM (DK72473 and DK89523), to VANTAGE (CA68485, EY08126, and RR030956), and to the Vanderbilt Cell Imaging Shared Resource (CA68485, DK20593, DK58404, DK59637, and EY08126). K.D.D. received support from the Molecular Endocrinology Training Program (T32 DK07563).
Conflicts of interest
The authors declare that there is no conflict of interest.
Author contribution
K.D.D. and A.B.O. performed experiments and analyzed data. J.P.C. performed RNA-Seq data processing, alignment, and differential expression analyses. K.D.D., A.B.O., and M.A.M. wrote and edited the manuscript. M.A.M. supervised the experiments.
Literature cited
- Alvelos MI, Bruggemann M, Sutandy FR, Juan-Mateu J, Colli ML, et al. 2021. The RNA-binding profile of the splicing factor SRSF6 in immortalized human pancreatic β-cells. Life Sci Alliance. 4:e202000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvelos MI, Juan-Mateu J, Colli ML, Turatsinze JV, Eizirik DL.. 2018. When one becomes many-alternative splicing in beta-cell function and failure. Diabetes Obes Metab. 20 (Suppl 2):77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Benitez CM, Kim SK.. 2013. Gene regulatory networks governing pancreas development. Dev Cell. 25:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P.. 2000. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 14:2701–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R, Zhou X, Marquardt T, Herrera P, Toube L, et al. 2004. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev Biol. 269:479–488. [DOI] [PubMed] [Google Scholar]
- Bechard ME, Bankaitis ED, Hipkens SB, Ustione A, Piston DW, et al. 2016. Precommitment low-level Neurog3 expression defines a long-lived mitotic endocrine-biased progenitor pool that drives production of endocrine-committed cells. Genes Dev. 30:1852–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit Bouvrette LP, Bovaird S, Blanchette M, Lecuyer E.. 2020. Ornament: a database of putative RNA binding protein target sites in the transcriptomes of model species. Nucleic Acids Res. 48:D166–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boward B, Wu T, Dalton S.. 2016. Concise review: control of cell fate through cell cycle and pluripotency networks. Stem Cells. 34:1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA.. 2008. Pdx1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol. 316:74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo F, Romero JP, Rubio A.. 2019. Upstream analysis of alternative splicing: a review of computational approaches to predict context-dependent splicing factors. Brief Bioinform. 20:1358–1375. [DOI] [PubMed] [Google Scholar]
- Chen T, Dent SY.. 2014. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wu H, Wang S, Koito H, Li J, et al. 2009. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat Neurosci. 12:1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Kraus MR, Lemaire LA, Yoshimoto M, Vemula S, et al. 2012. Dual lineage-specific expression of Sox17 during mouse embryogenesis. Stem Cells. 30:2297–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colli ML, Ramos-Rodriguez M, Nakayasu ES, Alvelos MI, Lopes M, et al. 2020. An integrated multi-omics approach identifies the landscape of interferon-alpha-mediated responses of human pancreatic beta cells. Nat Commun. 11:2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S. 2015. Linking the cell cycle to cell fate decisions. Trends Cell Biol. 25:592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daraio T, Bombek LK, Gosak M, Valladolid-Acebes I, Klemen MS, et al. 2017. SNAP-25b-deficiency increases insulin secretion and changes spatiotemporal profile of Ca2+ oscillations in β cell networks. Sci Rep. 7:7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, et al. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, Yoshida K, Yasuda T, Fukuda M, Fukuda Y, et al. 2011. Exome sequencing reveals a homozygous SYT14 mutation in adult-onset, autosomal-recessive spinocerebellar ataxia with psychomotor retardation. Am J Hum Genet. 89:320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, et al. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejarque M, Cervantes S, Pujadas G, Tutusaus A, Sanchez L, et al. 2013. Neurogenin3 cooperates with Foxa2 to autoactivate its own expression. J Biol Chem. 288:11705–11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, et al. 2008. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron. 60:40–55. [DOI] [PubMed] [Google Scholar]
- Fiszbein A, Kornblihtt AR.. 2017. Alternative splicing switches: important players in cell differentiation. Bioessays. 39:1600157. [DOI] [PubMed] [Google Scholar]
- Gierl MS, Karoulias N, Wende H, Strehle M, Birchmeier C.. 2006. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 20:2465–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonelle-Gispert C, Halban PA, Niemann H, Palmer M, Catsicas S, et al. 1999. SNAP-25a and -25b isoforms are both expressed in insulin-secreting cells and can function in insulin secretion. Biochem J. 339 (Pt 1):159–165. [PMC free article] [PubMed] [Google Scholar]
- Gopel SO, Kanno T, Barg S, Rorsman P.. 2000. Patch-clamp characterisation of somatostatin-secreting cells in intact mouse pancreatic islets. J Physiol. 528:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmain Y, Katz LS, Masson MH, Cheyssac C, Poisson C, et al. 2012. Pax6 is crucial for beta-cell function, insulin biosynthesis, and glucose-induced insulin secretion. Mol Endocrinol. 26:696–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmain Y, Marthinet E, Cheyssac C, Guerardel A, Mamin A, et al. 2010. Pax6 controls the expression of critical genes involved in pancreatic {alpha} cell differentiation and function. J Biol Chem. 285:33381–33393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F.. 2000. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA. 97:1607–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Stein GH, Pan N, Goebbels S, Hornberg H, et al. 2010. Pancreatic beta cells require Neurod to achieve and maintain functional maturity. Cell Metab. 11:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, Melton DA.. 2002. Direct evidence for the pancreatic lineage: Ngn3+ cells are islet progenitors and are distinct from duct progenitors. Development. 129:2447–2457. [DOI] [PubMed] [Google Scholar]
- Heller RS, Stoffers DA, Liu A, Schedl A, Crenshaw EB 3rd, et al. 2004. The role of Brn4/Pou3f4 and Pax6 in forming the pancreatic glucagon cell identity. Dev Biol. 268:123–134. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huang HP, Liu M, El-Hodiri HM, Chu K, Jamrich M, et al. 2000. Regulation of the pancreatic islet-specific gene Beta2 (Neurod) by neurogenin 3. Mol Cell Biol. 20:3292–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfan M, Gopaul KR, Miry O, Hokfelt T, Stanton PK, et al. 2019. SNAP-25 isoforms differentially regulate synaptic transmission and long-term synaptic plasticity at central synapses. Sci Rep. 9:6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson DA, Philipson LH.. 2007. Action potentials and insulin secretion: new insights into the role of Kv channels. Diabetes Obes Metab. 9 (Suppl 2):89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery N, Richardson S, Chambers D, Morgan NG, Harries LW.. 2019. Cellular stressors may alter islet hormone cell proportions by moderation of alternative splicing patterns. Hum Mol Genet. 28:2763–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Ivanov A, Blasevic D, Muller T, Purfurst B, et al. 2015. Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic beta-cell function. EMBO J. 34:1417–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan-Mateu J, Alvelos MI, Turatsinze JV, Villate O, Lizarraga-Mollinedo E, et al. 2018. Srp55 regulates a splicing network that controls human pancreatic beta-cell function and survival. Diabetes. 67:423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadkova A, Radecke J, Sorensen JB.. 2019. The SNAP-25 protein family. Neuroscience. 420:50–71. [DOI] [PubMed] [Google Scholar]
- Kim YH, Larsen HL, Rue P, Lemaire LA, Ferrer J, Grapin-Botton A.. 2015. Cell cycle-dependent differentiation dynamics balances growth and endocrine differentiation in the pancreas. PLoS Biol. 13:e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krentz NAJ, van Hoof D, Li Z, Watanabe A, Tang M, et al. 2017. Phosphorylation of Neurog3 links endocrine differentiation to the cell cycle in pancreatic progenitors. Dev Cell. 41:129–142.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan MS, Breslin MB.. 2009. Structure, expression, and biological function of Insm1 transcription factor in neuroendocrine differentiation. FASEB J. 23:2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bacquer O, Shu L, Marchand M, Neve B, Paroni F, et al. 2011. TCF7L2 splice variants have distinct effects on beta-cell turnover and function. Hum Mol Genet. 20:1906–1915. doi: 10.1093/hmg/ddr072. Epub 2011 Feb 28. PMID: 21357677. [DOI] [PubMed] [Google Scholar]
- Liang T, Qin T, Kang F, Kang Y, Xie L, et al. 2020. SNAP23 depletion enables more SNAP25/calcium channel excitosome formation to increase insulin exocytosis in type 2 diabetes. JCI Insight. 5:e129694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB.. 2010. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 11:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Ishizu Y, Itoh M, Lassmann T, Hasegawa A, et al. ; FANTOM consortium. 2015. Mapping mammalian cell-type-specific transcriptional regulatory networks using KD-CAGE and ChIP-seq Data in the TC-YIK cell line. Front Genet. 6:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning KS, Cooper TA.. 2017. The roles of RNA processing in translating genotype to phenotype. Nat Rev Mol Cell Biol. 18:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci TL, Anderson KR, Papizan JB, Sussel L.. 2013. Regulation of Neurod1 contributes to the lineage potential of Neurogenin3+ endocrine precursor cells in the pancreas. PLoS Genet. 9:e1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald E, Li J, Krishnamurthy M, Fellows GF, Goodyer CG, et al. 2012. Sox9 regulates endocrine cell differentiation during human fetal pancreas development. Int J Biochem Cell Biol. 44:72–83. [DOI] [PubMed] [Google Scholar]
- Mellitzer G, Bonne S, Luco RF, Van De Casteele M, Lenne-Samuel N, et al. 2006. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 25:1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RK, Nguyen-Tu MS, Chabosseau P, Callingham RM, Pullen TJ, et al. 2017. The transcription factor Pax6 is required for pancreatic beta cell identity, glucose-regulated ATP synthesis, and Ca dynamics in adult mice. J Biol Chem. 292:8892–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE.. 1999. Neurod is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 13:1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Milosevic I, Fasshauer D, Muller EM, de Groot BL, et al. 2005. Alternative splicing of SNAP-25 regulates secretion through nonconservative substitutions in the snare domain. Mol Biol Cell. 16:5675–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, et al. 1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in Beta2/Neurod-deficient mice. Genes Dev. 11:2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipovich AB, Dudek KD, Greenfest-Allen E, Cartailler J, Manduchi E, et al. 2021. A developmental lineage-based gene co-expression network for mouse pancreatic β-cells reveals a role for Zfp800 in pancreas development. Development. 148:dev196964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmark P, Hansson O, Jonsson A, Rönn T, Groop L, et al. 2009. Unique splicing pattern of the TCF7L2 gene in human pancreatic islets. Diabetologia. 52:850–854. doi: 10.1007/s00125-009-1293-z. Epub 2009 Feb 27. PMID: 19247628. [DOI] [PubMed] [Google Scholar]
- Osipovich AB, Long Q, Manduchi E, Gangula R, Hipkens SB, et al. 2014. Insm1 promotes endocrine cell differentiation by modulating the expression of a network of genes that includes Neurog3 and Ripply3. Development. 141:2939–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostenson CG, Gaisano H, Sheu L, Tibell A, Bartfai T.. 2006. Impaired gene and protein expression of exocytotic soluble n-ethylmaleimide attachment protein receptor complex proteins in pancreatic islets of type 2 diabetic patients. Diabetes. 55:435–440. [DOI] [PubMed] [Google Scholar]
- Pan FC, Brissova M, Powers AC, Pfaff S, Wright CV.. 2015. Inactivating the permanent neonatal diabetes gene Mnx1 switches insulin-producing beta-cells to a delta-like fate and reveals a facultative proliferative capacity in aged beta-cells. Development. 142:3637–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S, Vallier L.. 2013. The cell-cycle state of stem cells determines cell fate propensity. Cell. 155:135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Welch C, Hansson O, Adhikari N, Scott LJ, et al. 2009. Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet. 18:3795–3804. doi: 10.1093/hmg/ddp321. Epub 2009 Jul 14. PMID: 19602480; PMCID: PMC2748888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer AI, Singer RA, Sui L, Egli D, Sussel L.. 2019. Murine perinatal beta-cell proliferation and the differentiation of human stem cell-derived insulin-expressing cells require Neurod1. Diabetes. 68:2259–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul K, Lang J, Montecucco C, Weller U, Regazzi R, et al. 1995. SNAP-25 is expressed in islets of Langerhans and is involved in insulin release. J Cell Biol. 128:1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, et al. 1997. Genetic analysis reveals that Pax6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 11:1662–1673. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzgebel VM, Scheel DW, Conners JR, Kalamaras J, Lee JE, Anderson DJ, et al. 2000. Expression of Neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 127:3533–3542. [DOI] [PubMed] [Google Scholar]
- Sever D, Hershko-Moshe A, Srivastava R, Eldor R, Hibsher D, et al. 2021. NF-kappaB activity during pancreas development regulates adult beta-cell mass by modulating neonatal beta-cell proliferation and apoptosis. Cell Death Discov. 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N, Vanderhooft J, Straubhaar J, Mueller J, Chawla R, et al. 2019. Wnt signaling separates the progenitor and endocrine compartments during pancreas development. Cell Rep. 27:2281–2291.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer RA, Arnes L, Cui Y, Wang J, Gao Y, et al. 2019. The long noncoding RNA paupar modulates Pax6 regulatory activities to promote alpha cell development and function. Cell Metab. 30:1091–1106.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Dalton S.. 2016. Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Development. 143:4301–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P.. 1997. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 387:406–409. [DOI] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, et al. 2008. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 11:1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sund NJ, Vatamaniuk MZ, Casey M, Ang SL, Magnuson MA, Stoffers DA, et al. 2001. Tissue-specific deletion of Foxa2 in pancreatic beta cells results in hyperinsulinemic hypoglycemia. Genes Dev. 15:1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisa A, Avrahami D, Eden N, Zhang J, Feleke E, et al. 2017. Pax6 maintains beta cell identity by repressing genes of alternative Islet cell types. J Clin Invest. 127:230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Zhang Y, Ma L, Deng C, Duan H, et al. 2018. Haploinsufficiency of Insm1 impairs postnatal baseline beta-cell mass. Diabetes. 67:2615–2625. [DOI] [PubMed] [Google Scholar]
- Wang H, Gauthier BR, Hagenfeldt-Johansson KA, Iezzi M, Wollheim CB.. 2002. Foxa2 (HNF3beta) controls multiple genes implicated in metabolism-secretion coupling of glucose-induced insulin release. J Biol Chem. 277:17564–17570. [DOI] [PubMed] [Google Scholar]
- Wang S, Yan J, Anderson DA, Xu Y, Kanal MC, et al. 2010. Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Dev Biol. 339:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe VO, Venkitaraman AR.. 2016. RNA processing and genome stability: Cause and consequence. Mol Cell. 61:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu L, Xu X. 2021. The role of transcription factor 7-like 2 in metabolic disorders. Obes Rev. 22:e13166. doi: 10.1111/obr.13166. Epub 2020 Dec 4. PMID: 33615650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are available at ArrayExpress with the accession number E-MTAB-10262. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables provided.
Supplementary material is available at G3 online.