Abstract
Aberration in nuclear morphology is one of the hallmarks of cellular transformation. However, the processes that, when mis-regulated, result aberrant nuclear morphology are poorly understood. In this study, we carried out a systematic, high-throughput RNAi screen for genes that affect nuclear morphology in Caenorhabditis elegans embryos. The screen employed over 1700 RNAi constructs against genes required for embryonic viability. Nuclei of early embryos are typically spherical, and their NPCs are evenly distributed. The screen was performed on early embryos expressing a fluorescently tagged component of the nuclear pore complex (NPC), allowing visualization of nuclear shape as well as the distribution of NPCs around the nuclear envelope. Our screen uncovered 182 genes whose downregulation resulted in one or more abnormal nuclear phenotypes, including multiple nuclei, micronuclei, abnormal nuclear shape, anaphase bridges, and abnormal NPC distribution. Many of these genes fall into common functional groups, including some that were not previously known to affect nuclear morphology, such as genes involved in mitochondrial function, the vacuolar ATPase, and the CCT chaperonin complex. The results of this screen add to our growing knowledge of processes that affect nuclear morphology and that may be altered in cancer cells that exhibit abnormal nuclear shape.
Keywords: nuclear envelope, paired nuclei, micronuclei, anaphase bridges, cytokinesis, C. elegans, vacuolar ATPase, CCT chaperonin, ER morphology
Introduction
The nucleus is bound by a nuclear envelope (NE) composed of two nuclear membranes, an inner nuclear membrane (INM) and an outer nuclear membrane (ONM), that are connected at sites of nuclear pore complexes (NPCs). The ONM is continuous with the endoplasmic reticulum (ER), while the INM interacts with the underlying lamina and chromatin-associated proteins (Cohen-Fix and Askjaer 2017; Ungricht and Kutay 2017; De Magistris and Antonin 2018). The lamina provides rigidity to the nucleus and helps in various nuclear processes including chromatin organization, DNA damage repair, and transcriptional regulation (Gruenbaum and Foisner 2015; De Leeuw et al. 2018; dos Santos and Toseland 2021). The nucleus undergoes dramatic changes during the cell cycle. Most eukaryotic organisms follow an “open” mitosis scheme where the lamina and NPCs disassemble and the nuclear membranes become highly fenestrated, abolishing the permeability barrier that existed between the cytoplasm and nucleoplasm (Cohen-Fix and Askjaer 2017; Ungricht and Kutay 2017; Luckner and Wanner 2018; Rahman et al. 2020). This process of nuclear envelope breakdown (NEBD) is regulated by CDK1, PLK1, and other mitotic kinases, leading to the disassembly of the NPCs and nuclear lamina (Heald and McKeon 1990; Laurell et al. 2011; Rahman et al. 2015; Linder et al. 2017; Martino et al. 2017; Velez-Aguilera et al. 2020). Once mitosis is over, the NE reappears around the chromosomes to establish a single nucleus in each daughter cell, and it then expands during interphase (Lee et al. 2000; Cohen-Fix and Askjaer 2017; Ungricht and Kutay 2017).
Defects in nuclear morphology are associated with abnormal nuclear function (Mukherjee et al. 2016; Romero-Bueno et al. 2019). Mutations in components of the nuclear lamina lead to several diseases (a.k.a. laminopathies) such as Emery-Dreifuss muscular dystrophy, limb girdle muscular dystrophy, dilated cardiomyopathy, Dunnigan-type familial partial lipodystrophy, and Hutchinson-Gilford progeria syndrome (HGPS) (Worman et al. 2010; Dobrzynska et al. 2016; Donnaloja et al. 2020; Pathak et al. 2021), all of which are associated with abnormal nuclear shape. Alterations in nuclear morphology have also been associated with cancer (Mukherjee et al. 2016), and irregularity in nuclear shape and size is used in cancer diagnosis (Zink et al. 2004). Changes in nuclear shape and size have also been observed in normal aging (Haithcock et al. 2005) and conditions associated with premature aging (Goldman et al. 2004). Importantly, however, whether defects in nuclear structure are the cause or consequence of the above-mentioned disease states and conditions remains to be elucidated. What is missing is an understanding of what regulates nuclear size and shape so that nuclear morphology can be manipulated and the consequences explored.
For these reasons, there is interest in identifying genes that when down-regulated or inactivated lead to changes in nuclear morphology. We chose to do an RNAi screen for genes whose downregulation affects nuclear morphology using C. elegans early embryos: C. elegans is amenable to high throughput RNAi screens by feeding, and nuclei of the early embryo are large and nearly spherical, facilitating detection of morphological perturbations. Furthermore, the C. elegans strain we used expressed a fluorescently tagged subunit of the NPC, allowing sensitive detection of nuclear shape alterations, as well as alteration in NPC distribution. Finally, unlike siRNA screens done in tissue culture cells, RNAi screens in C. elegans are in a physiological developmental context. The high degree of conservation of NE and NPC proteins between nematodes and vertebrates makes it likely that the information garnered from a C. elegans screen will help better understand processes that affect mammalian nuclear morphology.
Based on the assumption that defects in nuclear morphology will likely affect embryonic viability, our screen employed RNAi against genes that were reported at the time to be essential for embryonic viability. This screen differed from earlier C. elegans screens in two ways: first, some of the earlier screens used either differential interference contrast (DIC) microscopy, which provides less information on nuclear shape abnormalities (Gonczy et al. 2000; Piano et al. 2000; Zipperlen et al. 2001; Sonnichsen et al. 2005), or used chromosome detection methods [e.g., DAPI staining, fluorescently-tagged histone proteins; (Colaiacovo et al. 2002; Green et al. 2011)], that only indirectly inform on nuclear morphology. Second, earlier screens that surveyed nuclei encompassed a different set of genes [e.g., genes whose downregulation leads to sterility (Green et al. 2011), genes on a particular chromosome (Fraser et al. 2000; Gonczy et al. 2000), or examined a different developmental stage (Colaiacovo et al. 2002; Green et al. 2011)]. Indeed, our screen uncovered many essential genes and pathways not previously known to affect nuclear morphology. Overall, we have identified 182 genes to be required for normal nuclear morphology in C. elegans. This screen has already led to two studies describing the involvement of spliceosome components in NPC distribution and the requirement of the TCA cycle in progression past the first embryonic G2 phase (Joseph-Strauss et al. 2012; Rahman et al. 2014). The present study describes the rest of the results from this screen, including the involvement of unexpected processes, such as the vacuolar ATPase and components of the CCT chaperonin complex, in nuclear morphology.
Materials and methods
Screen design
At the time of primary screening, 1713 genes essential for embryonic development were listed on WormBase and were present in our IPTG-inducible RNAi feeding library (Open Biosystems, Huntsville, AL) (https://wormbase.org/species/all/phenotype/WBPhenotype:0000050#042315--10). The vector backbone was pL4440-DEST and the RNAi sequences included in each plasmid are indicated in Supplementary Table S1. To monitor the changes in nuclear morphology, the screen was performed on strain OCF3 {unc-119(ed3); jjIs1092[pNUT1::npp-1::gfp + unc-119(+)]; ltIs37 [pAA64: pie-1p::mCherry::his-58 + unc-119 (+)]} expressing GFP tagged NPP-1 (NPP-1::GFP), a subunit of the NPC, to visualize the NE, and histone H2B fused to the monomeric red fluorescent protein mCherry (H2B::mCherry) to visualize chromatin (Golden et al. 2009). The screen was performed by feeding L4 stage worms with bacteria expressing dsRNA from the RNAi feeding library for 48–52 h at 20°C. For the control RNAi, worms were fed with dsRNA against smd-1 (Golden et al. 2009). For making RNAi plates, a single colony of dsRNA-expressing bacteria was inoculated in 360 µl Luria-Bertani (LB) liquid medium with carbenicillin (50 µg/ml) and tetracycline (20 µg/ml) in 96 deep-well plates. This culture was spread on RNAi plates containing IPTG (6 mM) and carbenicillin (50 µg/ml). For the primary screen, 3 L4s were fed on each RNAi plate and 2 animals were scored for any abnormality in nuclear morphology after 48 h of treatment. The remaining worm was allowed to lay embryos to determine embryonic viability. This resulted in 325 RNAi clones with a putative effect on nuclear morphology.
For the secondary screen, the identity of all 325 clones was verified by sequencing. A single colony of dsRNA-expressing bacteria was inoculated in 1–2 ml LB liquid medium with 50 µg/ml of ampicillin and grown overnight at 37°C. On the next day, 0.1% starter inoculum from the overnight culture was used for inoculating 5 ml of LB liquid medium with 50 µg/ml ampicillin, and was grown at 37°C. After reaching OD600 of around 0.5 (∼4–5 h), 10 µl of 0.5 M IPTG were added and the induced culture was grown for ∼4 h at 37 °C. The culture was then spun at 5000 g for 5 min at room temperature and the pellet was resuspended in 1 ml LB liquid medium with 50 µg/ml ampicillin. Around 200 µl of this culture were spread on each RNAi plate and allowed to grow for 1–2 days at 25°C. The RNAi condition used in secondary screen was found to be more effective than the one in the primary screen. Approximately 15–20 L4-stage larvae were transferred to RNAi plates, and embryos were examined after 48–52 h of RNAi. To determine embryonic lethality, 4–5 adult hermaphrodites were transferred to new RNAi plates following 48–52 h treatment and removed 3–6 h thereafter. Hatching was scored 24 h later. The screen was repeated at least 3 times. This narrowed the list to 182 genes whose downregulation consistently led to one or more nuclear phenotypes (Supplementary Table S1). For each treatment, embryos from at least 10 hermaphrodites were scored.
Scoring nuclear phenotypes
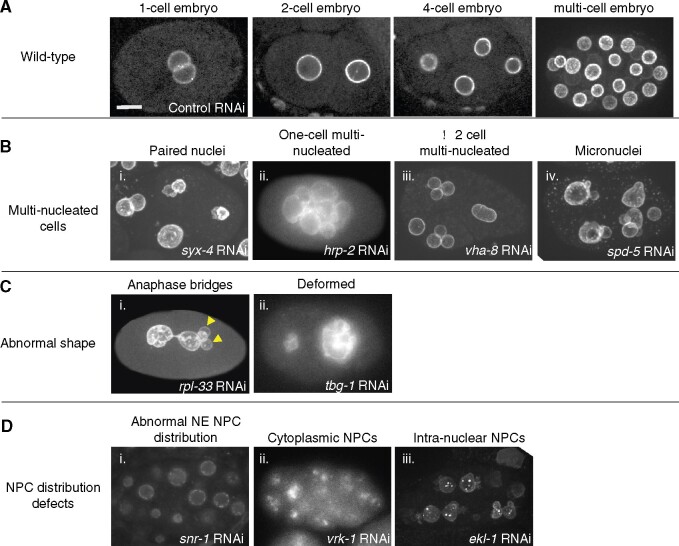
Nuclear phenotypes were scored manually. The vast majority of abnormal nuclei could be classified into 9 different phenotypes which fell into 3 broad categories (Figure 1). Category 1: multi-nucleate cells, including (i) paired nuclei, (ii) one-cell multi-nucleated embryos, (iii) ≥2 cell multi-nucleated embryos, and (iv) micronuclei; Category 2: abnormally shaped nuclei, including (i) anaphase bridges, and (ii) deformed nuclei; Category 3: NPC distribution defects, including (i) abnormal NE distribution of NPCs, (ii) cytoplasmic distribution of NPCs, and (iii) intra-nuclear NPCs (see Results and Discussion for more detail). To qualify as a positive hit, at least 7% of the embryos analyzed per RNAi treatment had to exhibit one or more of these phenotypes.
Figure 1.
Classes of abnormal nuclear phenotypes. Shown are representative images of C. elegans embryos expressing NPP-1 fused to GFP (NPP-1::GFP) from worms treated with RNAi against the indicated genes. (A) Wild type phenotype on control RNAi: examples of 1-, 2-, 4-, and multi-cell embryos (some nuclei are on an out-of-focus plane).(B) The multi-nucleated cell phenotype: (i) paired nuclei (some nuclei in this particular case are also deformed), (ii) multiple nuclei in a 1-cell embryo, (iii) Multi-nucleated cells in a 2 cell embryo, and (iv) micronuclei (along with nuclei). (C) The abnormal nuclear shape phenotype: (i) anaphase bridges (accompanied here by micronuclei, arrowheads) and (ii) deformed nuclei. (D) NPC distribution defect phenotypes: abnormal distribution of NPP-1::GFP (i) on the nuclear envelope, (ii) in the cytoplasm, or (iii) inside the nucleus. Scale bar (for all images) = 10 μm.
Examining ER phenotype
As a tertiary screen, we looked at the ER morphology following RNAi of select genes. This was performed on strain OCF5 {unc-119(ed3); ojIs23 [SP12::GFP + unc-119(+)]; ltIs37[pAA64: pie-1p::mCherry::his-58 + unc-119(+)]} expressing an ER signal peptide, SP12, tagged with GFP and histone H2B tagged with mCherry marking the chromatin (Golden et al. 2009). The results of RNAi treatments against dhc-1, dyci-1, rpl-21, rps-22, cct-2, cct-6, vha-8, and vha-14 are shown in Figures 6 and 7 and the raw data are provided in Supplementary Image Collections S1-S9. The RNAi treatment conditions were similar to the ones used for the secondary screen.
Figure 6.
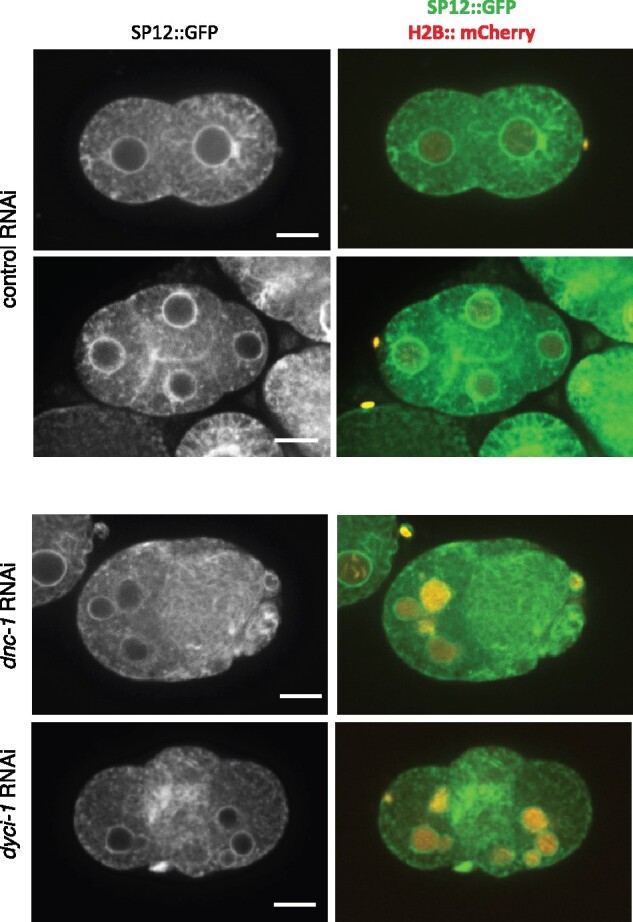
Inactivation of dynein/dynacin proteins has minimal effect on ER structure. Examples of C. elegans embryos expressing SP12::GFP (an ER marker) and histone H2B:: mCherry (chromatin), treated with control RNAi against smd-1 (top two rows) or RNAi against dynacin (dnc-1) or dynein (dyci-1) subunits (bottom two rows). Scale bar = 10 µm. See Supplementary Image Collections S1–S3 for additional examples and confocal microscopy data.
Figure 7.
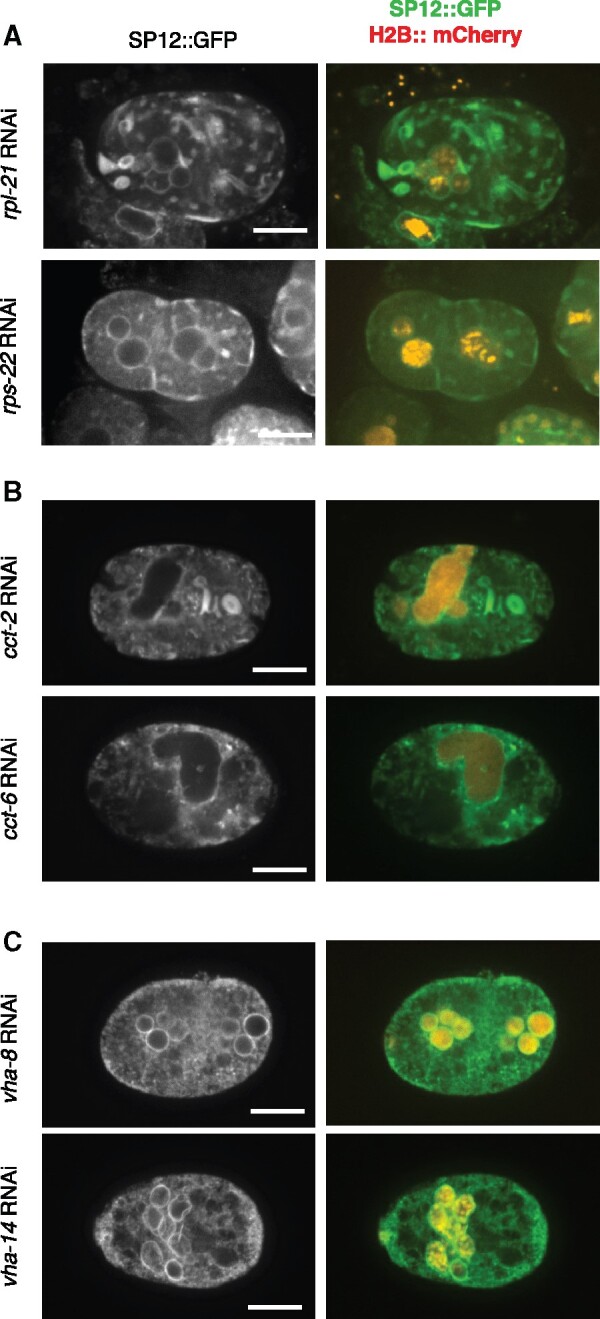
The effects of down-regulating translation machinery, chaperonin, and the vacuolar ATPase on ER morphology. Representative images of C. elegans embryos as in Figure 6 following RNAi treatment against genes involved in translation (A), chaperonin (B), or vacuolar ATPase (C). Scale bar = 10 µm. Supplementary Image Collections S4–S9 for additional examples and confocal microscopy data.
Microscopy and image analysis
Images of the primary screen were obtained using a Nikon E800 microscope, equipped with a 60x 1.4 NA Plan Apo objective, using a charge-coupled device camera (CCD; C4742-95; Hamamatsu Photonics) and operated by IPLab 3.9.5 software (BD Biosciences). Images of the secondary screen were obtained using a Nikon Eclipse TE2000U microscope equipped with a 60x 1.4 NA Plan Apo objective. This system was outfitted with a Spectral Applied Research LMM5 laser merge module to control the output of four diode lasers (excitation at 405, 491, 561, and 655 nm), a Yokogawa CSU10 spinning-disk unit, and a Hamamatsu C9100-13 EM-CCD camera. Images were acquired using IPLab 4.0.8 software (BD Biosciences). Images of ER in Figures 6 and 7 were taken using a Nikon confocal Ti2 fitted with a Nikon water 60x 1.2-NA Apo Plan objective, a Yokagawa CSU-X1 spinning disk and a photometrix Prime 95B camera. Image acquisition was done using Elements software (Nikon Instruments, Inc.). Image processing was done with FIJI (ImageJ) (Schindelin et al. 2012).
Gene ontology overrepresentation analysis
Statistical overrepresentation analysis of gene ontology (GO) terms was analyzed using PANTHER Classification System v16.0 (http://pantherdb.org; Mi et al. 2019). WormBase IDs were entered for input and the Fisher’s Exact test with the default false discovery rate (FDR) calculation settings was used to determine enriched GO terms.
Results and discussion
Downregulation of 182 C. elegans genes led to abnormal nuclear morphology or abnormal NPC distribution
To identify genes whose downregulation affects nuclear morphology, we carried out a large-scale RNAi screen in C. elegans, visualizing nuclei in embryos. We targeted genes essential for embryonic development reasoning that severe alterations in nuclear morphology would likely result in embryonic lethality. At the time this screen was initiated, nearly 2000 genes, when down-regulated, were known to cause embryonic lethality. Of those, 1713 were present in our RNAi feeding library. To monitor the changes in nuclear morphology, the screen was performed using a transgenic line expressing C. elegans homolog of Nup54 (NPP-1), a subunit of the NPC, fused to GFP (NPP-1::GFP) to visualize the NE, and histone H2B fused to the monomeric red fluorescent protein mCherry (H2B::mCherry) to visualize chromatin (Golden et al. 2009; Joseph-Strauss et al. 2012). NPP-1::GFP also allowed us to identify the genes that affect NPC distribution.
The screen was performed by feeding L4 stage worms with bacteria expressing dsRNA from the RNAi feeding library, as described under Materials and Methods. All 1713 RNAi treatments were subjected to visual inspection of nuclear morphology as well as embryonic lethality. RNAi against a large set of genes, about two-thirds of the entire collection, did not lead to a robust embryonic lethal phenotype. There could be multiple reasons for this: first, the efficiency of feeding RNAi is likely less than that of other forms of gene inactivation (e.g., dsRNA injection, mutations) on which the reported embryonic lethal phenotype could have been based. Second, the time of RNAi treatment was not optimized, and it is possible that longer exposures may have resulted in a higher faction of RNAi constructs causing robust lethality. Finally, our cutoff for an embryonic lethal phenotype was ≥15% of dead embryos, and in theory some of the reported embryonic lethal phenotypes could have been below this rate. In total, downregulation of 529 genes by RNAi caused embryonic lethality of at least 15%. Of those, in the primary screen, downregulation of 325 genes by RNAi led to abnormal nuclear morphology. Of note, the abnormal nuclear morphology was not a nonspecific result of embryonic lethality because (a) we observed a variety of abnormal nuclear phenotypes (see below) and (b) not all RNAi treatments that led to embryonic lethality also resulted in abnormal nuclear morphology. None of the RNAi treatments that did not cause embryonic lethality resulted in an abnormal nuclear morphology. In addition, 25 genes, when down-regulated, led to phenotypes that precluded assessing the nuclear morphology of an early embryo, such as meiotic arrest, unfertilized embryos or lack of NPP-1::GFP expression. The initial 325 genes were rescreened to verify the phenotype(s) and ensure reproducibility, as described under Materials and Methods. This narrowed down the list to 182 genes that, when down-regulated, consistently exhibited a robust nuclear morphology phenotype (Supplementary Table S1), as discussed below.
Nuclear morphology defects could be divided into 3 major categories
In wild-type C. elegans early embryos, nuclei are spherical and the NPCs are distributed evenly around the NE, as shown by NPP-1::GFP localization (Figure 1A) (Golden et al. 2009). In our screen, deviation from the normal nuclear phenotype resulted in one or more defects that could be divided into three major phenotypic categories. It should be noted that many RNAi treatments resulted in multiple nuclear phenotypes (see Figure 1 for examples and also below), which is not uncommon (Fraser et al. 2000).
Category 1: multi-nucleate cells:
This category included: (i) Paired nuclei, as seen by the presence of two attached nuclei in each cell, likely due to a defect in pronuclear fusion after fertilization (Audhya et al. 2007; Galy et al. 2008; Nishi et al. 2008; Rivers et al. 2008; Golden et al. 2009; Gorjánácz and Mattaj 2009; Noatynska et al. 2010; Rahman et al. 2015; 2020); (ii) one-cell multi-nucleated embryos, likely caused by multiple nuclear divisions without intervening cytokinesis (Swan et al. 1998; Gonczy et al. 1999; Echard et al. 2004); (iii) ≥2 cell multi-nucleated embryos, which could be due to cytokinesis defects at later stage embryos (Green et al. 2011); and (iv) micronuclei, one or more smaller sized nuclei along with a relatively regular-sized nucleus, which could have resulted from chromosome fragmentation or defective chromosome segregation (Strome et al. 2001) (Figure 1B).
Category 2: abnormally shaped nuclei:
This category included: (i) anaphase bridges, likely a result of defects in chromosome segregation, such as improper attachment of chromosomes to the spindle or chromosome catenation that precludes full segregation (chromatin image data not shown) (Moore et al. 1999; Hagstrom et al. 2002; Bembenek et al. 2013); and (ii) deformed nuclei, which could have resulted from defective lamina, alteration in chromatin architecture or defects in the cytoskeleton, among other reasons (Figure 1C).
Category 3: NPC distribution defects:
This category included: (i) abnormal NE distribution of NPCs, shown by the mislocalization of NPP-1::GFP at the NE, as described previously (Joseph-Strauss et al. 2012); (ii) cytoplasmic distribution of NPCs, as seen by cytoplasmic localization of NPP-1::GFP, possibly due to defects in NE reassembly or proper NPC subunit targeting, and (iii) intra-nuclear NPCs, as seen by the mislocalization of NPP-1::GFP inside the nucleus and that could have also resulted from an improper NE reassembly and perhaps internal nuclear membrane structures (Figure 1D).
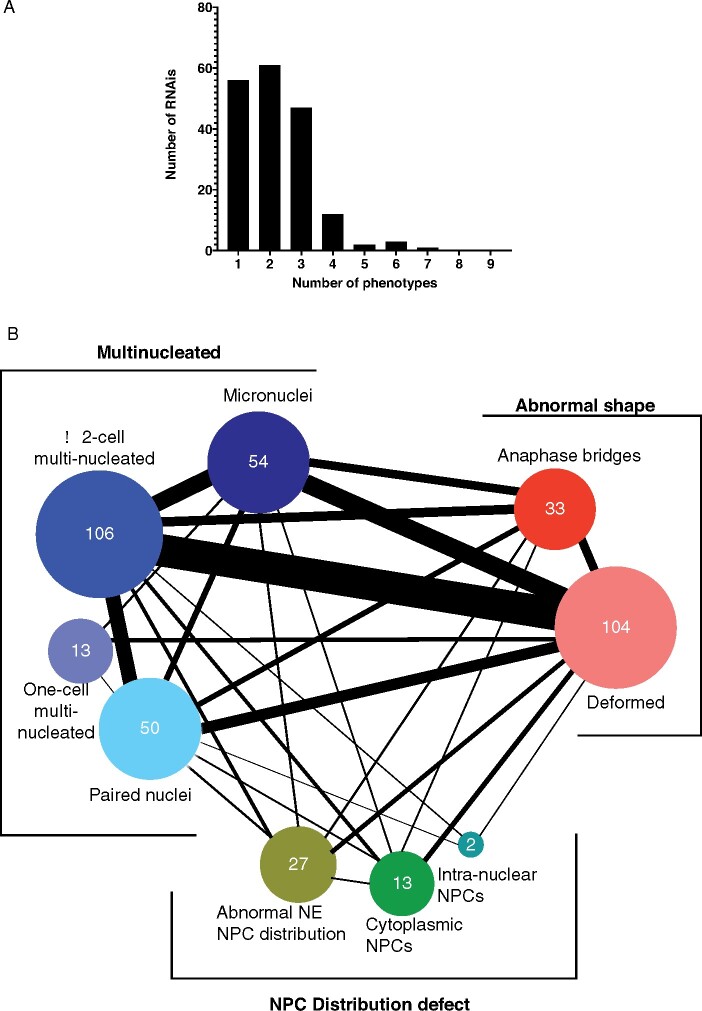
Embryos from each RNAi treatment were scored for these 9 phenotypic categories (Supplementary Table S1). Out of the 182 genes whose downregulation resulted in a consistent abnormal nuclear morphology, the most common defect was the multi-nucleated cell phenotype (RNAi against 147 genes). Downregulation of 119 genes caused an abnormal nuclear shape, while downregulation of 39 genes caused NPC distribution defects (Supplementary Table S1). As noted above, in most cases downregulation of one gene resulted in multiple nuclear phenotypes, and only about a third of the RNAi treatments resulted in a single abnormal nuclear phenotype (Figure 2A). For example, RNAi against cks-1, a cyclin-dependent kinase (Polinko and Strome 2000), led to 7 different nuclear phenotypes (Supplementary Table S1). Depletion of ∼30% of genes led to at least 2 nuclear phenotypes, and ∼25% resulted in 3 different nuclear phenotypes (Figure 2A). Some of the phenotypes tended to occur together (Figure 2B and Supplementary Table S2). For example, deformed nuclei were concurrent in 50% of embryos with paired nuclei (26/50), ∼70% of ≥2-cell multi-nucleated embryos (71/106), ∼75% of one-cell multi-nucleated embryos (9/13), and ∼75% of embryos with micronuclei (40/54). Micronuclei were most common in embryos also exhibiting anaphase bridges (18/33, 55%), followed by embryos with deformed nuclei (40/104, 38%), ≥ 2-cell multi-nucleated embryos (36/106, 33%), 1-cell multi-nucleated embryos (4/13, 31%) and embryos with paired nuclei (12/50, 24%). RNAi treatments that led to uneven NPC distribution exhibited few other nuclear phenotypes, but those that led to cytoplasmic or intra-nuclear NPCs were often accompanied by other nuclear defects, and in particular multi-nuclei in ≥2 cell embryos [54% for embryos with cytoplasmic NPCs (7/13) and 100% for embryos with intra-nuclear NPCs (2/2)].
Figure 2.
Distribution of abnormal nuclear phenotypes among the 182 RNAi treatments. (A) Bar graph showing the number of RNAi treatments that resulted in the indicated number of phenotypes. (B) Connectivity of phenotypic categories. The thickness of each connecting line corresponds to the number of genes that, when down-regulated, result in both phenotypes. See Supplementary Table S2 for the exact number of shared phenotypes.
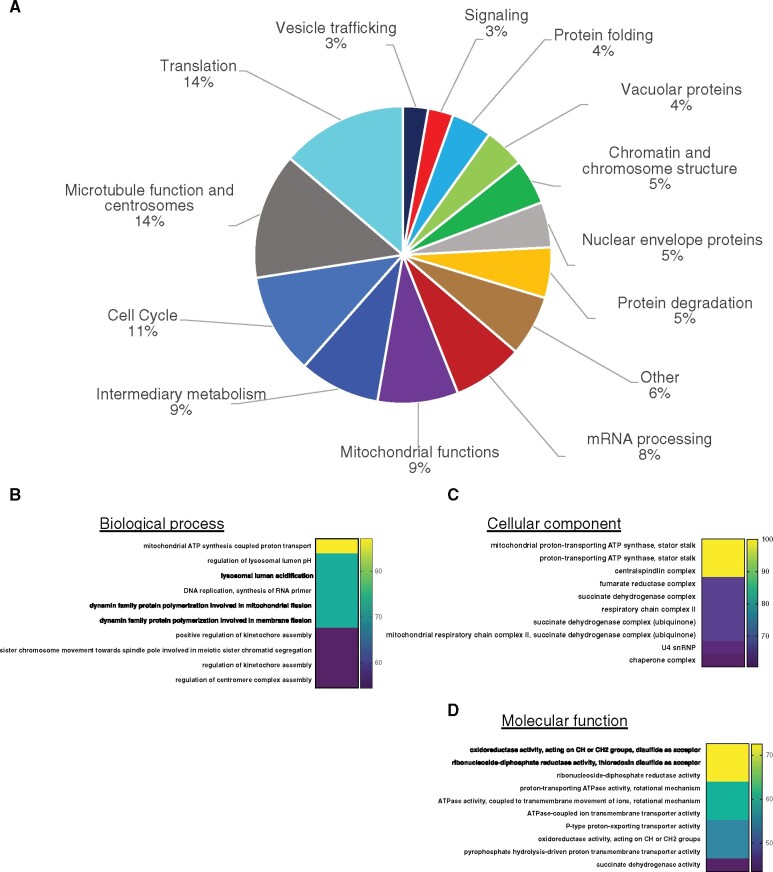
The relationship between phenotype and gene function
Our screen uncovered multiple genes in 13 distinct functional categories, plus a number of genes that could not be placed in a larger functional category (Figure 3A and Supplementary Table S1). Moreover, the screen uncovered clusters of functionally related genes, either within the same complex same or in the same pathway. To determine the significance of these data, we analyzed the biological functions of our gene list by manual curation based on primarily C. elegans literature and homology to genes in other organisms with known or predicted function (Figure 3A and Supplementary Table S1). Our functional assignments were in good agreement with the ascribed functions on WormBase. To determine the overrepresentation of a functional class in our dataset compared to the C. elegans genome, we performed a statistical overrepresentation test of GO terms associated with those 182 genes using PANTHER Classification System v16.0. PANTHER recognized 181 genes out of 182 (Mi et al. 2019). Figure 3, B–D shows some of the most highly enriched biological process, cellular component, and molecular function classes (the complete lists, including P-values, are shown in Supplementary Tables S3–S5). Mitochondrial ATP synthesis coupled transport (GO: 0042776) is the most enriched GO biological process (>87-fold enrichment) relative to the C. elegans reference genome among the biological classes (Figure 3B and Supplementary Table S3). Consistently, the mitochondrial functions gene class formed one of the largest in our manual classification (Figure 3A and Supplementary Table S1). Lysosomal acidification was highly enriched among the biological process, cellular component, and molecular function classifications (Figure 3, B–D and Supplementary Table S3–S5). There is no doubt that our screen has missed many genes that could affect nuclear morphology, either because they were not present in our RNAi collection or because the feeding RNAi treatment did not sufficiently down-regulate their activity. Nonetheless, the genes that were uncovered serve as a basis for further investigation into the involvement of specific processes in affecting nuclear morphology.
Figure 3.
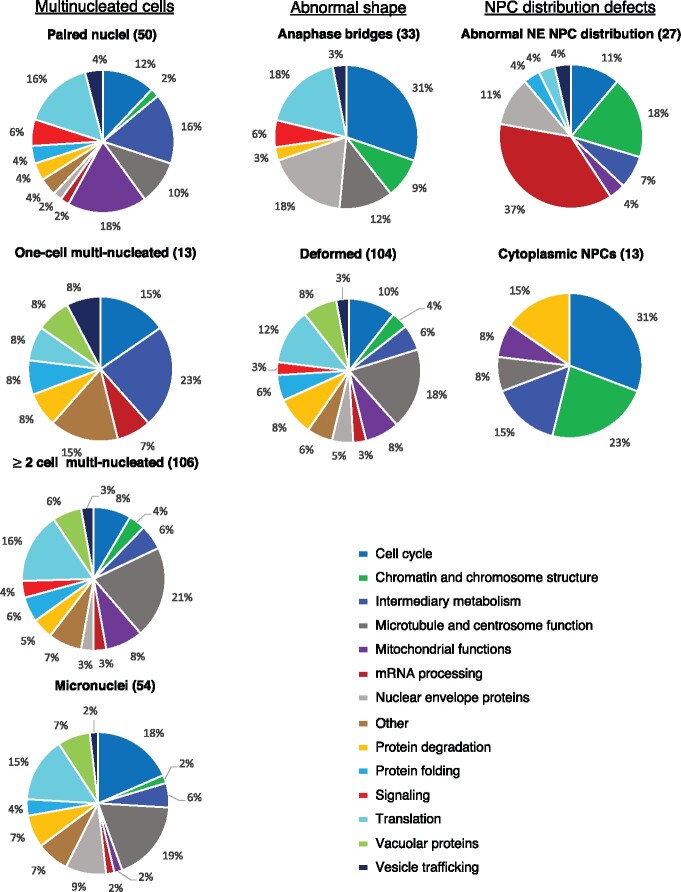
Functional categories of genes whose downregulation results in one or more abnormal nuclear or NPC phenotype. (A) The relative abundance (in percentage) of each functional category within the 182 hits is indicated. (B–D) Statistical overrepresentation analysis of GO terms using the PANTHER Classification System. The top statistically significant overrepresented subclasses based on Biological Process (B), Cellular Component (C), and Molecular Function classes (D) are shown. The number on the right represents fold enrichment. See Supplementary Tables S3–S5 for the complete list and P-values.
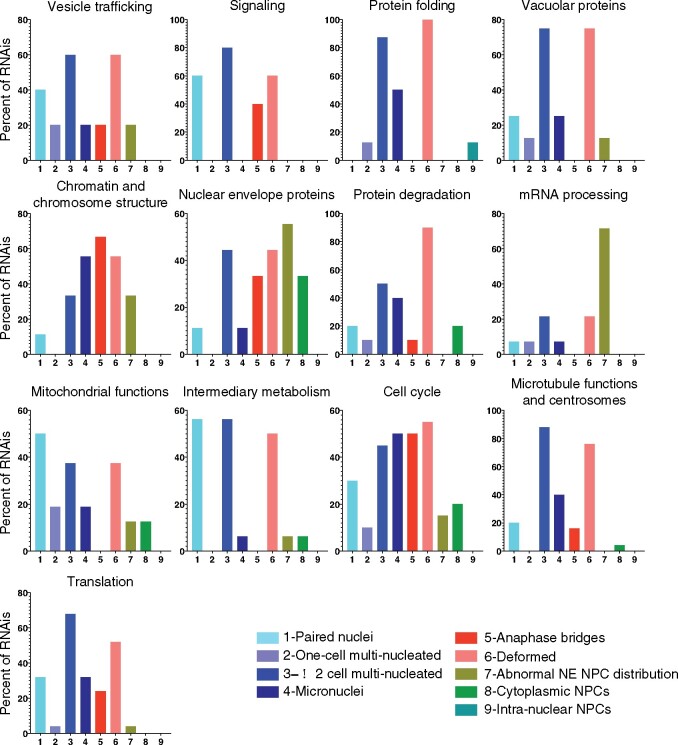
As alluded to above, our screen uncovered genes in processes whose involvement in nuclear morphology was unexpected. We previously reported the surprising link between downregulation of splicing factors and a defect in NPC distribution (Joseph-Strauss et al. 2012; Supplementary Table S1), and between TCA cycle genes and the accumulation of embryos in the 1-cell stage with paired nuclei due to a block in mitotic progression (Rahman et al. 2014; Supplementary Table S1). In addition, we found that downregulation of translation, chaperonin, vacuolar function, intermediary metabolism, and mitochondrial function also affected nuclear morphology (Figures 3A and 4; Supplementary Table S1). This revealed that a defect in a variety of core cellular processes can cause alteration in nuclear morphology, a finding that could be relevant to the abnormal nuclear morphology often seen in cancer cells. Below we discuss the relationship between nuclear morphology and several of the functionally related genes. Importantly, different functional categories displayed distinct patterns of abnormal nuclear phenotypes: for example, the most prevalent phenotype of the protein degradation functional group was deformed nuclei, whereas the most common phenotypes of the mRNA processing and the chromatin and chromosome functions groups were abnormal NE NPC distribution and anaphase bridges, respectively (Figure 4). This supports the conclusion that the abnormal nuclear phenotype is not merely a consequence of dying cells, and suggests that distinct processes impact the nucleus differently. Interestingly, we observed similar phenotypic signatures in disparate functional categories, such as the ones resulting from the inactivation for protein folding genes and vacuolar genes (Figure 4), suggesting that the underlying cause for abnormal nuclear morphology may be similar. In contrast, the NPC distribution defect phenotypes were mostly seen when mRNA processing and NE genes were down-regulated, but the rest of the phenotypes of these two functional groups are quite dissimilar.
Figure 4.
Functional categories of genes whose downregulation results in one or more abnormal nuclear or NPC phenotype. The prevalence of each of the 9 phenotypes observed within each functional category: Phenotypic categories are on the x-axis, and the percent of RNAis within a functional category displaying a particular phenotype is on the y-axis.
Cell cycle and chromosome segregation-related functions:
Cell cycle genes were expected to affect nuclear morphology, and downregulation of genes in this category sometimes led to pleotropic effects due to their involvement in a wide range of processes (Figure 4 and Supplementary Table S1). Within the cell cycle functional category, DNA replication genes and kinetochore genes, when down-regulated, led predominantly to anaphase bridges (7/7 genes, Supplementary Table S1), likely because of defects in segregating incompletely replicated of mis-aligned chromosomes. Not surprisingly, nuclear morphology was affected by the downregulation of numerous genes associated with spindle/microtubule function, such as genes coding for of α and β tubulin (tba-2, tba-4, and tbb-2), centrosome proteins (tbg-1, gip-2, spd-5, sas-5, sas-6, and syz-4) and dynein and dynactin subunits (dlc-1, dyci-1, dli-1, nud-1, and dnc-1), as well as other motor proteins (zen-4, klp1-18, and klp-19) (Supplementary Table S1). These genes are involved in the precise positioning and orientation of the mitotic spindle necessary for both chromosome segregation and cytokinesis (Carminati and Stearns 1997; Shaw et al. 1997; Skop and White 1998; Gonczy et al. 1999; Karki and Holzbaur 1999; Dujardin and Vallee 2002; Nguyen-Ngoc et al. 2007; Moore et al. 2008; Markus and Lee 2011; Collins et al. 2012; Laan et al. 2012; Kotak and Gonczy 2013; Pintard and Bowerman 2019). Indeed, downregulation of 88% of genes (22 out of 25) in the microtubule and centrosome function category exhibited multi-nucleated cells in embryos with 2 or more cells (Figure 4 and Supplementary Table S1). Likewise, the most common phenotypes observed when genes within the “chromatin and chromosome structure” category were down-regulated were micronuclei, anaphase bridges and deformed nuclei, consistent with the effect of chromosome mis-segregation on nuclear morphology (Figure 4 and Supplementary Table S1).
Protein translation:
The translation machinery is comprised of ribosomal subunits, tRNA aminoacylation factors, translation factors, and more. This machinery also depends on genes that regulate the biogenesis of these factors, such as ribosome biogenesis genes. Downregulation of genes in this class has also appeared in other C. elegans genome-wide screens, displaying defects such as multi-nucleated embryos, aberrant cytoplasmic texture, and osmotic sensitivity (Gonczy et al. 2000; Sonnichsen et al. 2005; Green et al. 2011). We also observed a multi-nucleated phenotype: Downregulation of 21 of 25 genes involved in translation resulted in at least one of the multi-nucleated category phenotypes (Figure 4 and Supplementary Table S1). Moreover, Downregulation of 7 of 10 ribosomal genes and 5 of 7 tRNA synthetase genes (Supplementary Table S1) resulted in ≥ 2-cell multi-nucleated embryos. The range of phenotypes within the “translation” functional category was not uniform (Supplementary Table S1). For example, downregulation of over half of the ribosomal subunits picked up in our screen resulted in anaphase bridges (5/10, Supplementary Table S1). In contrast, none of seven different tRNA synthetases, and only 1 of 6 translation factors, exhibited anaphase bridges when down-regulated (Supplementary Table S1). Thus, while all these factors participate in translation, depletion of ribosomal proteins appear to affect the nucleus in ways that tRNA synthetases and translation factors do not (Supplementary Table S1). How this group of genes affects nuclear morphology is not known. A defect in translation could have resulted in a reduced level of one or more proteins required for nuclear integrity, making the involvement of translation in nuclear morphology indirect. Conversely, the association of ribosomes with the ER may have a direct effect on membrane conformation, such that a defect in the translation machinery could deform the ER, resulting, conceivably, in difficulties to undergo cytokinesis and leading to a multi-nuclear phenotype. The effect of downregulation of the translation machinery and genes from other functional categories on ER structure is examined below.
Chaperonin:
The eukaryotic cytosolic chaperonin containing TCP-1 (CCT) has a role in the folding of cytoskeletal components (Horwich et al. 2007). There are 8 CCT subunits (CCT-1 to CCT-8) encoded by the C. elegans genome (Leroux and Candido 1995, 1997; Matus et al. 2010). They are involved in tubulin folding and microtubule growth in the early embryo (Srayko et al. 2005; Lundin et al. 2008). Our screen picked up 5 cct genes: cct-1, cct-2, cct-5, cct-6, and cct-7. Downregulation of each of the 5 resulted in multi-nucleated embryos with deformed nuclei (Supplementary Table S1). This pattern of phenotypes is similar to that observed for defects in microtubule function and centrosomes (Figure 4), consistent with the nuclear morphology defect due to microtubule malfunction when cct genes are down-regulated.
Vacuolar ATPase:
Several subunits of the vacuolar-type H+ ATPase (V-ATPase) were also picked up in our screen. The V-ATPase is a large protein complex that acidifies intracellular compartments (Nelson 1992). These proteins comprise of two functional domains V1 (8 subunits A-H) and V0 (6 subunits a, c, c', c”, d, and e). The worm genome encodes all the subunits except c′. Our screen has picked up 8 of these subunits: vha-1 (V0 subunit c), vha-2 (V0 subunit c), vha-4 (V0 subunit b), vha-8 (V1 subunit E1), vha-9 (V1 subunit F), vha-11 (V1 subunit C1), vha-14 (V1 subunit D), and unc-32 (V0 subunit a1) (reviewed in Lee et al. 2010). Interestingly, when down-regulated, all but one resulted in a multi-nucleated phenotype (Supplementary Table S1). Previously, Choi et al. (2003) reported a clustered nuclei phenotype equivalent to our multi-nucleated phenotype upon VHA-8 depletion (Choi et al 2003). Furthermore, downregulation of 6 of the 8 vha genes that we isolated led to ≥2-cell multi-nucleated embryos that were often accompanied by deformed nuclei (Figure 4 and Supplementary Table S1). This was reminiscent of the range of phenotypes obtained when down-regulating microtubule and centrosome function genes (Figure 4), suggesting that defects in vacuolar function may result in chromosome segregation and/or cytokinesis defects.
Overall, different phenotypes were predominantly caused by defects in distinct cellular functions (Figure 5). For example, genes related to microtubule and centrosome functions made up 19 and 21% of genes that, when down-regulated, resulted in micronuclei and ≥2-cell multi-nucleated embryos, respectively, whereas they only made up 10% of paired nuclei embryo, and none of the 1-cell multi-nucleated embryos (Figure 5, left column). Not surprisingly, the relative weight of the functional categories that affected NPC distribution was distinct from those that affected nuclear morphology (Figure 5, compare right column with the middle and left columns): mRNA processing genes were prominent in the “abnormal NE NPC distribution” category (37%), but only a few percent or none at all in all other categories. Mitochondrial function genes also appeared in a subset of phenotypes. Thus, while most functional groups, when down-regulated, could lead to many different phenotypes, each phenotype had its own unique combination of functional groups.
Figure 5.
Different phenotypes are a result of the downregulation of different sets of genes. The contribution of each functional group to a given phenotype, as determined by the number of genes from a particular functional category that, when down-regulated, resulted in the indicated phenotypes. Functional groups are arranged clockwise, starting with cell cycle genes. For example, 37% of genes that lead to abnormal NPC NE distribution (top right) are involved in mRNA processing (red sector). The breakdown for the “Nuclear NPC” category is not shown as it encompasses only 2 genes.
Downregulation of some, but not all, genes that affect nuclear morphology also affects ER morphology
To gain further insight into how some of the unexpected functional groups may affect nuclear morphology, we examined the effect of their downregulation on ER organization, as the ER is continuous with the NE (Watson 1955). The underlying hypothesis was that if down-regulating genes of a particular functional category also affected ER morphology, then the effect on nuclear morphology could by affecting ER/nuclear membrane properties. An example of such a case is the downregulation of lipin (lpin-1 in C. elegans), a phosphatidic acid phosphatase that converts phosphatidic acid to diacylglycerol (Siniossoglou et al. 1998; Tange et al. 2002; Santos‐Rosa et al. 2005; Campbell et al. 2006; Han et al. 2006). In the absence of lipin activity the total amount of cellular phospholipids increases, resulting the ER expansion (Santos‐Rosa et al. 2005). Downregulation of C. elegans lpin-1 results in abnormal ER morphology and deformation of the nucleus (Golden et al. 2009; Gorjánácz and Mattaj 2009).
To this end, we repeated select RNAi treatments on worms expressing the ER signal peptide SP12 tagged with GFP (SP12::GFP) (Poteryaev et al. 2005). The ER in the early embryo forms a thin reticulate pattern with only rare patches of denser/sheet ER (Figure 6, control RNAi and Poteryaev et al. 2005). Under our RNAi conditions, downregulation of dynein and dynactin, a microtubule-dependent motor, resulted in largely normal ER with a mostly reticular structure, despite the abnormal number of nuclei (Figure 6 and see Supplementary Image Collections S1–S3 for raw images; the percent of embryos exhibiting a normal/mostly reticular ER structure were 100% (n = 15), 100% (n = 23), and 85% (n = 47) for control, dnc-1 and dyci-1 RNAi, respectively). This is perhaps not surprising given that the underlying defect is likely a failure in spindle positioning and cytokinesis. In contrast, downregulation of two different ribosome subunits, rpl-21 and rps-22, which also resulted in multi-nucleated cells, did affect ER structure, as seen by the increased abundance of ER patches and whorls (Figure 7A and Supplementary Image Collections S4 and S5; 100% of embryos affected and n = 17 in both cases). This suggests that the nuclear defect seen when translation is down-regulated could be a consequence of an ER defect. We speculate that a reduced abundance of ribosomes may affect ER morphology indirectly, due to a decrease in translation of one or more ER shaping factors or the induction of stress than may affect ER morphology (Mateus et al. 2018). Alternatively, a reduction in the number of ribosomes may affect ER morphology directly, as ribosomes associate with the ER and may contribute to its organization (Shibata et al. 2010).
Chaperonin is a chaperon required for actin and tubulin folding (Horwich et al. 2007). Downregulation of chaperonin subunits results in deformed nuclei that are often enlarged (Figure 7B and Supplementary Image Collections S6 and S7), a phenotype that is often seen when tubulin is down-regulated (Supplementary Table S1) and may be due to failure in chromosome segregation coupled with additional rounds of DNA replication. Chaperonin downregulation also results in abnormal ER morphology (Figure 7B and Supplementary Image Collections S6 and S7; 100% of embryos affected in both cases, n = 19 and 8 for RNAi against cct-2 and cct-6, respectively). Since actin is involved in maintaining ER morphology (Poteryaev et al. 2005) downregulation of chaperonin may affect nuclear and ER morphologies independently.
Finally, we examined the ER following downregulation of the vacuolar ATPase. Unlike downregulation of ribosome subunits of chaperonin, the ER in embryos down-regulated for VHA subunits largely retained its reticular structure and did not exhibit extensive areas of ER patches or whorls [Figure 7C and Supplementary Image Collections S8 and S9; 100% of embryos affected in both vha-8 (n = 8) and vha-14 (n = 21) RNAi]. The void regions within the ER may reflect an abnormal ER organization, or they could be regions occupied by large vesicles, such as lysosomes, not visible with the markers that were used. The overall reticular structure of the ER combined with the preponderance of embryos with multi-nucleated cells was reminiscent of the phenotype seen when spindle function and/or cytokinesis was defective (Figure 6 and Supplementary Table S1). This suggests that defects in the worm vacuolar ATPase may affect spindle function, positioning, and/or the machinery responsible for cytokinesis. Further studies are required to elucidate the connection between translation, chaperonin, and vacuolar ATPase and nuclear morphology.
Taken together, we have shown that RNAi against 182 genes, known for their requirement during embryogenesis, results in 3 major phenotypic categories related to nuclear morphology or NPC distribution. Most of these genes can be subdivided into 13 functional categories, and in several cases our screen uncovered genes for components of the same complex or pathway, all of which affect nuclear morphology or NPC distribution in a similar way. Some of these functional categories were not previously known to affect nuclear morphology. This is significant because one of the hallmarks of cancer is abnormal nuclear size and/or shape, and our data suggest that the underlying defects may originate in a wide range of biological processes. The screen has clearly missed some genes, but the list provided in this study can serve as a solid foundation for the discovery of additional proteins and processes that affect nuclear morphology, especially when partially or fully depleted.
Acknowledgments
The authors thank the members of the Cohen-Fix lab for advice and support.
Funding
The authors were supported by an intramural grant from National Institute of Diabetes & Digestive & Kidney Diseases, NIH#DK069012.
Conflicts of interest
The authors declare that there is no conflict of interest.
Data availability
All strains, plasmids, and reagents are available upon request.
Supplementary Table S1, containing the list of genes whose down regulation resulted in a nuclear morphology defect, and Supplementary Image Collections S1–S9, containing the raw data of confocal ER images described in Figures 6 and 7, are available via GSA figshare: https://doi.org/10.25387/g3.14998272.
Literature cited
- Audhya A, Desai A, Oegema K.. 2007. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 178:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembenek JN, Verbrugghe KJ, Khanikar J, Csankovszki G, Chan RC.. 2013. Condensin and the spindle midzone prevent cytokinesis failure induced by chromatin bridges in C. elegans embryos. Curr Biol. 23:937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JL, Lorenz A, Witkin KL, Hays T, Loidl J, et alet al. 2006. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol Biol Cell. 17:1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T.. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 138:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KY, Ji YJ, Dhakal BK, Yu JR, Cho C, et alet al. 2003. Vacuolar-type H+-ATPase E subunit is required for embryogenesis and yolk transfer in Caenorhabditis elegans. Gene. 311:13–23. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Askjaer P.. 2017. Cell biology of the Caenorhabditis elegans Nucleus. Genetics. 205:25–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo MP, Stanfield GM, Reddy KC, Reinke V, Kim SK, et alet al. 2002. A targeted RNAi screen for genes involved in chromosome morphogenesis and nuclear organization in the Caenorhabditis elegans germline. Genetics. 162:113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL.. 2012. Cell cycle–regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell. 23:3380–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw R, Gruenbaum Y, Medalia O.. 2018. Nuclear Lamins: thin filaments with major functions. Trends Cell Biol. 28:34–45. [DOI] [PubMed] [Google Scholar]
- De Magistris P, Antonin W.. 2018. The dynamic nature of the nuclear envelope. Curr Biol. 28:R487–R497. [DOI] [PubMed] [Google Scholar]
- Dobrzynska A, Gonzalo S, Shanahan C, Askjaer P.. 2016. The nuclear lamina in health and disease. Nucleus. 7:233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnaloja F, Carnevali F, Jacchetti E, Raimondi MT.. 2020. Lamin A/C mechanotransduction in laminopathies. Cells. 9:1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos Á, Toseland CP.. 2021. Regulation of nuclear mechanics and the Impact on DNA damage. Int J Mol Sci. 22:3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB.. 2002. Dynein at the cortex. Curr Opin Cell Biol. 14:44–49. [DOI] [PubMed] [Google Scholar]
- Echard A, Hickson GR, Foley E, O'Farrell PH.. 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 14:1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, et alet al. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 408:325–330. [DOI] [PubMed] [Google Scholar]
- Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, et alet al. 2008. A role for gp210 in mitotic nuclear-envelope breakdown. J Cell Sci. 121:317–328. [DOI] [PubMed] [Google Scholar]
- Golden A, Liu J, Cohen-Fix O.. 2009. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci. 122:1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, et al. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 101:8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, et alet al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 408:331–336. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, et al. 1999. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J Cell Biol. 144:927–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjánácz M, Mattaj IW.. 2009. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J Cell Sci. 122:1963–1969. [DOI] [PubMed] [Google Scholar]
- Green RA, Kao HL, Audhya A, Arur S, Mayers JR, et alet al. 2011. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 145:470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R.. 2015. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 84:131–164. [DOI] [PubMed] [Google Scholar]
- Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ.. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, et alet al. 2005. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA. 102:16690–16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Wu WI, Carman GM.. 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 281:9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, McKeon F.. 1990. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 61:579–589. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Fenton WA, Chapman E, Farr GW.. 2007. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 23:115–145. [DOI] [PubMed] [Google Scholar]
- Joseph-Strauss D, Gorjanacz M, Santarella-Mellwig R, Voronina E, Audhya A, et alet al. 2012. Sm protein down-regulation leads to defects in nuclear pore complex disassembly and distribution in C. elegans embryos. Dev Biol. 365:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL.. 1999. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol. 11:45–53. [DOI] [PubMed] [Google Scholar]
- Kotak S, Gonczy P.. 2013. Mechanisms of spindle positioning: cortical force generators in the limelight. Curr Opin Cell Biol. 25:741–748. [DOI] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, Van Duijn M, et alet al. 2012. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 148:502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, et alet al. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 144:539–550. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL.. 2000. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 11:3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Li W, Ryu SE, Rhim T, Ahnn J.. 2010. Vacuolar (H+)-ATPases in Caenorhabditis elegans: what can we learn about giant H+ pumps from tiny worms? Biochim Biophys Acta. 1797:1687–1695. [DOI] [PubMed] [Google Scholar]
- Leroux MR, Candido EPM.. 1995. Characterization of four new tcp-1-related cct genes from the nematode Caenorhabditis elegans. DNA Cell Biol. 14:951–960. [DOI] [PubMed] [Google Scholar]
- Leroux MR, Candido EPM.. 1997. Subunit characterization of the Caenorhabditis elegans chaperonin containing TCP-1 and expression pattern of the gene encoding CCT-1. Biochem Biophys Res Commun. 241:687–692. [DOI] [PubMed] [Google Scholar]
- Linder MI, Kohler M, Boersema P, Weberruss M, Wandke C, et alet al. 2017. Mitotic disassembly of nuclear pore complexes involves CDK1- and PLK1-mediated phosphorylation of key interconnecting nucleoporins. Dev Cell. 43:141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckner M, Wanner G.. 2018. Precise and economic FIB/SEM for CLEM: with 2 nm voxels through mitosis. Histochem Cell Biol. 150:149–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin VF, Srayko M, Hyman AA, Leroux MR.. 2008. Efficient chaperone-mediated tubulin biogenesis is essential for cell division and cell migration in C. elegans. Dev Biol. 313:320–334. [DOI] [PubMed] [Google Scholar]
- Markus SM, Lee WL.. 2011. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev Cell. 20:639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino L, Morchoisne-Bolhy S, Cheerambathur DK, Van Hove L, Dumont J, et alet al. 2017. Channel nucleoporins recruit PLK-1 to nuclear pore complexes to direct nuclear envelope breakdown in C. elegans. Dev Cell. 43:157–171.e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus D, Marini ES, Progida C, Bakke O.. 2018. Rab7a modulates ER stress and ER morphology. Biochim Biophys Acta Mol Cell Res. 1865:781–793. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Li X-Y, Durbin S, Agarwal D, Chi Q, et alet al. 2010. In vivo identification of regulators of cell invasion across basement membranes. Sci Signal. 3:ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Huang X, Ebert D, Mills C, et alet al. 2019. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc. 14:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Li J, Cooper JA.. 2008. Dynactin function in mitotic spindle positioning. Traffic. 9:510–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LL, Morrison M, Roth MB.. 1999. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J Cell Biol. 147:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee RN, Chen P, Levy DL.. 2016. Recent advances in understanding nuclear size and shape. Nucleus. 7:167–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. 1992. Structural conservation and functional diversity of V-ATPases. J Bioenerg Biomembr. 24:407–414. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gonczy P.. 2007. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 9:1294–1302. [DOI] [PubMed] [Google Scholar]
- Nishi Y, Rogers E, Robertson SM, Lin R.. 2008. Polo kinases regulate C. elegans embryonic polarity via binding to DYRK2-primed MEX-5 and MEX-6. Development. 135:687–697. [DOI] [PubMed] [Google Scholar]
- Noatynska A, Panbianco C, Gotta M.. 2010. SPAT-1/Bora acts with Polo-like kinase 1 to regulate PAR polarity and cell cycle progression. Development. 137:3315–3325. [DOI] [PubMed] [Google Scholar]
- Pathak RU, Soujanya M, Mishra RK.. 2021. Deterioration of nuclear morphology and architecture: A hallmark of senescence and aging. Ageing Res Rev. 67:101264. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ.. 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol. 10:1619–1622. [DOI] [PubMed] [Google Scholar]
- Pintard L, Bowerman B.. 2019. Mitotic Cell Division in Caenorhabditis elegans. Genetics. 211:35–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polinko ES, Strome S.. 2000. Depletion of a Cks homolog in C. elegans embryos uncovers a post-metaphase role in both meiosis and mitosis. Curr Biol. 10:1471–1474. [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A.. 2005. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell. 16:2139–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Chang IY, Harned A, Maheshwari R, Amoateng K, et alet al. 2020. C. elegans pronuclei fuse after fertilization through a novel membrane structure. J Cell Biol. 219:e201909137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Munzig M, Kaneshiro K, Lee B, Strome S, et alet al. 2015. Caenorhabditis elegans polo-like kinase PLK-1 is required for merging parental genomes into a single nucleus. Mol Biol Cell. 26:4718–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Rosu S, Joseph-Strauss D, Cohen-Fix O.. 2014. Down-regulation of tricarboxylic acid (TCA) cycle genes blocks progression through the first mitotic division in Caenorhabditis elegans embryos. Proc Natl Acad Sci USA. 111:2602–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers DM, Moreno S, Abraham M, Ahringer J.. 2008. PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. J Cell Biol. 180:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Bueno R, de la Cruz Ruiz P, Artal-Sanz M, Askjaer P, Dobrzynska A.. 2019. Nuclear organization in stress and aging. Cells. 8:664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, Rosa H, Leung J, Grimsey N, Peak‐Chew S, Siniossoglou S.. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24:1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et alet al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K.. 1997. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 139:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, et alet al. 2010. Mechanisms determining the morphology of the peripheral ER. Cell. 143:774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Santos, Rosa H, Rappsilber J, Mann M, Hurt E.. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17:6449–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, White JG.. 1998. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr Biol. 8:1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, et alet al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 434:462–469. [DOI] [PubMed] [Google Scholar]
- Srayko M, Kaya A, Stamford J, Hyman AA.. 2005. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell. 9:223–236. [DOI] [PubMed] [Google Scholar]
- Strome S, Powers J, Dunn M, Reese K, Malone CJ, et alet al. 2001. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 12:1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan KA, Severson AF, Carter JC, Martin PR, Schnabel H, et alet al. 1998. cyk-1: a C. elegans FH gene required for a late step in embryonic cytokinesis. J Cell Sci. 111:2017–2027. [DOI] [PubMed] [Google Scholar]
- Tange Y, Hirata A, Niwa O.. 2002. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J Cell Sci. 115:4375–4385. [DOI] [PubMed] [Google Scholar]
- Ungricht R, Kutay U.. 2017. Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol. 18:229–245. [DOI] [PubMed] [Google Scholar]
- Velez-Aguilera G, Nkombo Nkoula S, Ossareh-Nazari B, Link J, Paouneskou D, et al. 2020. PLK-1 promotes the merger of the parental genome into a single nucleus by triggering lamina disassembly. eLife. 9:e59510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML. 1955. The nuclear envelope; its structure and relation to cytoplasmic membranes. J Biophys Biochem Cytol. 1:257–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Ostlund C, Wang Y.. 2010. Diseases of the nuclear envelope. Cold Spring Harb Perspect Biol. 2:a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA.. 2004. Nuclear structure in cancer cells. Nat Rev Cancer. 4:677–687. [DOI] [PubMed] [Google Scholar]
- Zipperlen P, Fraser AG, Kamath RS, Martinez-Campos M, Ahringer J.. 2001. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20:3984–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains, plasmids, and reagents are available upon request.
Supplementary Table S1, containing the list of genes whose down regulation resulted in a nuclear morphology defect, and Supplementary Image Collections S1–S9, containing the raw data of confocal ER images described in Figures 6 and 7, are available via GSA figshare: https://doi.org/10.25387/g3.14998272.