Abstract
The performance of a silica chip-based resequencing method, the Affymetrix HIV PRT 440 assay (hereafter referred to as the Affymetrix assay), was evaluated on a panel of well-characterized nonclade B viral isolates and on isolates exhibiting length polymorphisms. Sequencing of human immunodeficiency virus type 1 (HIV-1) pol cDNAs from clades A, C, D, E, and F resulted in clade-specific regions of base-calling ambiguities in regions not known to be associated with resistance polymorphisms, as well as a small number of spurious resistance polymorphisms. The Affymetrix assay failed to detect the presence of additional serine codons distal to reverse transcriptase (RT) codon 68 that are associated with multinucleoside RT inhibitor resistance. The increasing prevalence of non-clade B HIV-1 strains in the United States and Europe and the identification of clinically relevant pol gene length polymorphisms will impact the generalizability of the Affymetrix assay, emphasizing the need to accommodate this expanding pool of pol genotypes in future assay versions.
Until recently, determination of human immunodeficiency virus type 1 (HIV-1) genotypes has relied on either dye-terminator cycle sequencing of cloned viral genes or direct (consensus) sequencing of PCR products. However, the recent commercial introduction of chip-based resequencing techniques (5, 7, 8, 11) may offer certain advantages to high-throughput genotyping applications.
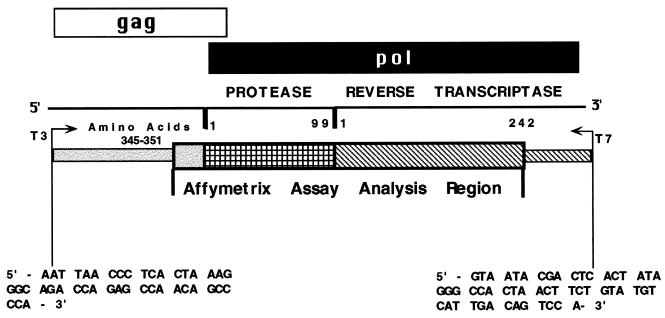
Key features of the Affymetrix GeneChip HIV PRT 440 assay (hereafter termed the Affymetrix assay) include high-resolution single-base microresequencing with an 18- to 20-fold redundancy of interrogation. The assay rationale (Fig. 1) employs targeted viral protease (PR) and reverse transcriptase (RT) gene sequences (together denoted as PRT sequences) which are reverse transcribed into cDNA by using a specific 3′ external primer. The resulting amplicon is transcribed in two separate reactions, using T3 and T7 RNA polymerases in the presence of fluorescently labeled UTP nucleotides, to yield a labeled cRNA of ∼1,200 bases. The cRNA is fragmented by hydrolysis and hybridized to the HIV PRT 440 GeneChip.
FIG. 1.
Schematic representation of the regions in the PR and RT genes of HIV-1 that are assessed by the Affymetrix assay. The cDNA amplicon contains codons 345 to 351 of gag, all 99 codons of the pol PR gene, and the first 242 codons of the RT region of pol. The location and orientation of the bacteriophage T3 and T7 RNA polymerase promoters within the amplicon are shown.
In contrast to traditional methods, which derive sequence de novo, the chip-based methods are designed only to confirm previously defined sequence information. It is not known how the performance of resequencing platforms, optimized for clade B virus, will be impacted by the increasing occurrence of length polymorphisms and of non-clade B templates in clinical populations being assessed by resistance genotyping. Since there are an increasing number of observations supporting the incidence of non-clade B viral strains in major metropolitan areas worldwide that hitherto virtually exclusively had only clade B strains, the importance of developing assays to detect and monitor such infections is growing (1–3). As the incidence of non-clade B viral subtypes increases, the need to have reliable tools to assess the impact of antiretroviral drugs on the treatment of patients will likewise become important. Already the awareness of this requirement has resulted in the modification of viral load assays, such as the Food and Drug Administration-approved Roche Amplicor Monitor assay, whose accuracy was initially limited to clade B templates only, to encompass the assessment of all strains (3, 10).
While it is established that the Affymetrix assay performs well on clade B templates (6, 7), it is imperative to evaluate its performance on a diverse panel of viral isolates in order to assess the universal utility of the method. To address this issue, we chose to evaluate the performance of the Affymetrix assay on a well-characterized panel of non-clade B viral isolates and on several isolates with confirmed length polymorphisms.
MATERIALS AND METHODS
Viral isolates.
Viral isolates derived from HIV-1 clades A to F were obtained from peripheral blood mononuclear cell cultures, with the amount of viral isolate RNA input for quantitation being normalized to the electron microscopy-based particle count (10). Amplicons were recovered from subjects shown by conventional cycle sequencing analysis (ABI; Applied Biosystems, Foster City, Calif.) of cDNA amplicons to have either the S69SS or S69SSS insertion, associated with multiple antiretroviral drug resistance (9), in the RT region of the pol gene. These cDNAs were reamplified with HIV PRT T3 and T7 primers as described by the manufacturer (Affymetrix, Inc.) except that the number of cycles was reduced to 25. The resulting cDNA was then subjected to the Affymetrix assay.
Viral RNA extraction.
Plasma samples were rapidly thawed in a 37°C water bath, and 0.5-ml aliquots of the samples were added separately to 1-ml volumes of 1× phosphate-buffered saline (Gibco-BRL, Bethesda, Md.) containing 5 μg of heat-inactivated bovine serum albumin (Boehringer Mannheim, Indianapolis, Ind.)/ml. Samples were centrifuged at 14,000 × g for 60 min at 4°C. The viral pellets were lysed by resuspending them in 800 μl of Tri-reagent (Molecular Research Center, Woodlands, Tex.), and the nucleic acids were purified in accordance with the manufacturer’s instructions. This method differs from the Qiagen method of extraction described in the Affymetrix assay manual and was determined to be optimal in terms of reproducible yield of high-quality RNA (data not shown). Purified RNA was stored at −80°C.
Amplification of viral template by RT PCR.
Twenty-three microliters of viral RNA was annealed with HIV PRT RT primer (5′-TTT CCC CAC TAA CTT CTG TAT GTC ATT GAC A-3′ [HIV-1MN sequence positions 3353 to 3323; GenBank accession no. M17449]) (Affymetrix, Inc., Santa Clara, Calif.) at 70°C for 3 min in a total reaction volume of 25 μl containing 0.2 μM HIV PRT RT primer. The RNA-DNA template was then subjected to reverse transcription in a 50-μl volume containing 10 mM MgCl2, 50 mM KCl, 10 mM dithiothreitol 100 mM Tris-HCl (pH 8.3), 0.5 mM concentrations of each deoxynucleoside triphosphate (dNTP; Pharmacia Biotech, Piscataway, N.J.), 30 U of RNAguard (Pharmacia Biotech), and 7 U of avian myeloblastosis virus RT (Gibco-BRL) at 45°C for 1 h. All incubations were carried out in a Perkin-Elmer model 9600 thermocycler (PE Applied Biosystems, Inc., Foster City, Calif.) with thin-walled microcentrifuge tubes. A 10-μl aliquot of the resultant cDNA was subjected to PCR with primers containing T3 and T7 promoter sequences. The sequence for the HIV PRT T3 (sense orientation) primer (Affymetrix, Inc.) is 5′-AAT TAA CCC TCA CTA AAG GGC AGA CCA GAG CCA ACA GCC CCA-3′ (HIV-1MN sequence positions 2145 to 2166; the sequence for the HIV PRT T7 (antisense orientation) primer (Affymetrix, Inc.) is 5′-GTA ATA CGA CTC ACT ATA GGG CCA CTA ACT TCT GTA TGT CAT TGA CAG TCC A-3′ (HIV-1MN sequence positions 3348 to 3318). HIV-1 sequences in both primers are underlined. PCR was performed in a 50-μl volume containing 10 μl of cDNA, 10 mM Tris-HCl (pH 8.0), 1.25 mM magnesium acetate, 0.5 mM dNTPs (Pharmacia Biotech), 0.25 μM HIV PRT T7 primer (Affymetrix, Inc.), 0.25 μM HIV PRT T3 primer (Affymetrix, Inc.), and 2 U of recombinant Tth DNA polymerase XL (Applied Biosystems, Inc., Foster City, Calif.). The cycling scheme was 1 min at 95°C followed by 50 cycles of 95°C for 15 s, 65°C for 40 s, and 72°C for 45 s, with a final extension of 10 min at 72°C. The resulting 1,245-bp amplicon includes sequences from codon −18 relative to the PR region of the pol gene, all 99 codons of the PR region, and codons 1 to 242 of the RT region of pol. This method differs from the one given in the Affymetrix assay package insert in having higher Mg ion and avian myeloblastosis virus RT enzyme concentrations. In addition, amplicon yield was determined by ethidium bromide staining of PCR products resolved on agarose gels and was assessed by using an AlphaImager digital imaging system and AlphaEase image analysis software (Alpha Innotech Corporation, San Leandro, Calif.) to give semiquantitative measurements of amplicon concentration. This was imperative for the optimal performance of subsequent assay steps.
In vitro transcription and RNA fragmentation.
Approximately 100 ng of amplified DNA was bidirectionally transcribed in vitro in two separate 20-μl reaction mixtures containing 100 ng of amplicon, 20 U of either T3 (sense) or T7 (antisense) RNA polymerase, and buffer, dithiothreitol, RNase inhibitors, and rNTPs and fluorescein-12-UTP at the concentrations recommended by the manufacturer of the RNA polymerase (Promega Scientific, Madison, Wis.). Reaction mixtures were incubated at 37°C for 90 min prior to analysis of the resulting cRNA by electrophoresis through 1% native agarose gels. For fragmentation, 19-μl volumes of sense and antisense cRNAs were separately incubated in 30 mM MgCl2 at 94°C for 30 min in a final reaction volume of 21.5 μl. Typically, 10 μl of fragmented sense and antisense cRNA was used in the hybridization reaction.
Hybridization of cRNA with the DNA microarray.
The fragmented, fluorescently labeled sense and antisense cRNA transcripts were hybridized to the HIV PRT 440s (sense) and HIV PRT 440a (antisense) probe arrays (Affymetrix, Inc.) in a hybridization mixture containing 500 μl of 5× SSPE buffer (20× SSPE is 2.98 M NaCl, 0.02 M EDTA, and 0.2 M NaPO4, pH 7.4 [Quality Biological Inc., Gaithersburg, Md.]), 0.05% Triton X-100 (Sigma Chemical Co., St. Louis, Mo.), and 5 μl (1.0 nM) of Control Oligo F1 (proprietary sequence; Affymetrix, Inc.). Hybridization on individual probe arrays was facilitated by using the Affymetrix GeneChip Fluidics 400 Station to execute the automated hybridization protocol at the desired temperatures (30°C for sense arrays and 35°C for antisense arrays) for 30 min. Following hybridization, the GeneChip arrays underwent stringent washings with appropriate buffers (6× SSPE–0.005% Triton X-100 for sense arrays and 7.5× SSPE–0.005% Triton X-100 for antisense arrays) that were executed automatically by the fluidics station.
Data analysis and interpretation.
Probe arrays were scanned with the HP GeneArray Scanner (Hewlett-Packard, Santa Clara, Calif.). The scanner, operated by the GeneChip software (Affymetrix, Inc.), interrogates each array of 90 μm by 90 μm probe cells and generates a comparative fluorescence value for each cell which is subsequently analyzed and reported. The data from the scanned array are analyzed by using the GeneChip software, version 3.0 (Affymetrix, Inc.). The nucleotide sequence is assessed for the presence of drug-associated resistance mutations by comparison to an HIV-1 pol gene reference library defined as wild type by the manufacturer and derived from the analysis of 200 drug-naive seropositive subjects infected with clade B virus.
Conventional cycle sequencing.
pol cDNAs were amplified from viral supernatants by PCR using the same primer locations as for the Affymetrix assay, ligated into the TA cloning vector pCR2.1 (Invitrogen Corporation, Carlsbad, Calif.), and introduced into Escherichia coli DH5α cells by electroporation. Transformants were selected either at random or by colony hybridization with a 32P-labeled pol probe. Plasmids containing cDNAs of the correct sizes were characterized by DNA sequence analysis using dye-terminator chemistry, affording complete double-strand coverage, and a model 373A automated DNA sequencer (Applied Biosystems, Inc.). Consensus sequences were developed by using Sequencher version 3.1 software (Gene Codes Corporation, Ann Arbor, Mich.), and multiple sequence alignments were constructed by using the MegAlign software package (DNAStar, Inc., Madison, Wis.). cDNAs encoding nonsense mutations were excluded from alignment.
GenBank accession numbers.
The sequences derived from cycle sequencing and described here have been deposited at GenBank under accession no. AF107368 to AF107402.
RESULTS
Performance of the Affymetrix assay on non-clade B HIV-1 cDNA.
A panel of highly characterized isolates representing HIV-1 clades A through F (10) was employed to evaluate the performance of the Affymetrix assay on non-clade B pol cDNA templates. pol cDNA was obtained from culture supernatants, and the resulting templates were sequenced by both the Affymetrix assay and ABI cycle sequencing technologies. pol cDNA was subjected to prior molecular cloning, and ABI sequence analysis was performed on five individual clones. Molecular-clone genotypes clustered uniquely with their cognate Affymetrix assay-derived data, and each subtype cluster formed a unique node on a phylogenetic tree (data not shown), eliminating the possibility of sequence contamination. Affymetrix assay sequencing of all non-clade B cDNAs resulted in clade-specific signature patterns of regions of ambiguous base calling (Fig. 2). The majority of these ambiguities fell outside regions of currently identified drug resistance polymorphisms as defined by clade B data.
FIG. 2.
Schematic representation of the discrete regions of nucleotide sequence ambiguity detected by the Affymetrix assay on non-clade B virus. Ambiguity is defined as either an inability to resolve the nucleotide or the report of a nucleotide that varies from the wild-type clade B pol gene sequence at that location. The HIV-1MN amino acid sequence numbering scheme for PR and RT is given. The boxes indicating regions of chip failure are drawn to scale and encompass a minimum of 10 contiguous probe cells or tiles.
All of these non-clade B isolates were obtained from subjects prior to the era of modern antiretroviral drug therapy. Surprisingly, a number of mutations known to be associated with drug resistance of clade B isolates were detected by the Affymetrix assay and are described in Table 1 as putative resistance mutations. Also listed in Table 1 are the codon locations associated with drug resistance of clade B virus but which the Affymetrix assay either could not resolve or reported a polymorphism not known to be associated with drug resistance (13). Except for the mutation involving the substitution of isoleucine for the methionine at position 36 (M36I) in the PR, none of the putative mutations reported by the Affymetrix assay were confirmed by conventional ABI sequence analysis, and hence they were interpreted as artifacts of the Affymetrix assay for non-clade B templates.
TABLE 1.
Drug resistance-associated mutations misidentified by the Affymetrix assay in 21 drug-naive non-clade B viral isolates
| Clade | Putative mutation(s)
|
Base pair no. resolved | Amino acid(s) incorrectly identified | |
|---|---|---|---|---|
| In protease | In RT | |||
| A | M36Ia | PR 20, 77 | RT L100Mb | |
| M36Ia | L210W | |||
| M36Ia | RT 210 | RT L100T | ||
| C | M36Ia | RT A98P | ||
| M36Ia | K65R | RT 210 | PR V77G | |
| M36Ia | K65R, K70R | RT 210 | ||
| M36Ia | L210W | RT K65E, M184R, R211G | ||
| PR K20E, M36R, V77G | ||||
| D | M36Ia | RT L100M | ||
| M36Ia | ||||
| E | K70R | PR 36 | ||
| K70R, L210W | RT 106 | PR M36R | ||
| M36Ia | K70R, L210W | |||
| M46I | K70R | PR 36 | PRT K20E, V776; RT D67G | |
| M36Ia | K70R | |||
| N88D | K70R | PR 36 | RT D67G | |
| K70R | PR 20, 36 | RT M41R, D67G | ||
| N88D | K70R | PR 36 | RT D67G | |
| F | K20R | PR 36, RT 210 | PR V77G | |
| PR 77 | ||||
Mutation independently confirmed by ABI sequencing.
The amino acid to the right of the codon location is not wild type, or the expected amino acid correlated with known drug resistance.
Detection of length polymorphisms by the Affymetrix assay.
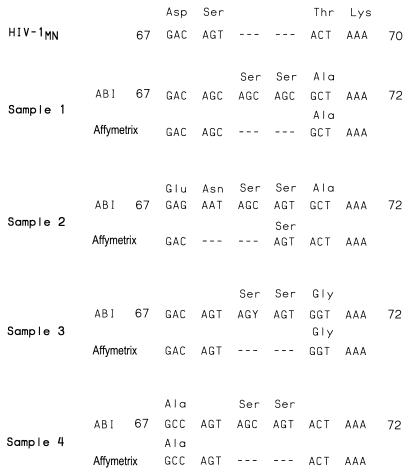
A length polymorphism consisting of the addition of sequence at codon 69 of RT has been reported which, when combined with zidovudine resistance mutations, confers high-level multinucleoside resistance (9). This polymorphism is associated with the addition of 1 or 2 serine codons, resulting in an S68→SS or S68→SSS genotypic change. The capability of the Affymetrix assay to detect the presence of this insertion was assessed on four pol cDNAs obtained from four individuals confirmed to have this length polymorphism by conventional cycle sequencing. As shown in Fig. 3, the Affymetrix assay failed to detect this polymorphism in all four samples tested. Amplicons containing this length polymorphism were easily detected after being annealed with wild-type amplicons, using the heteroduplex mobility assay (4) (data not shown).
FIG. 3.
Detection of length polymorphisms. pol cDNAs obtained from four individuals with multiple antiretroviral drug resistance and known serine codon insertions after RT codon 68 were sequenced by both the conventional cycle sequencing and Affymetrix sequencing technologies. All sequences are aligned with HIV-1MN beginning with RT codon 67. Codons divergent with HIV-1MN are translated. Gaps in the alignment are indicated by dashes.
DISCUSSION
In agreement with reports by other investigators (6, 7), we found little difference in the cDNA sequences obtained from the same plasma samples processed for either the Affymetrix assay or for conventional cycle sequencing by the ABI technology when the HIV-1 quasispecies was of clade B and lacked the single known pol length polymorphism. The Affymetrix assay was designed to sequence HIV-1 clade B pol cDNAs that lack length polymorphisms (7). This method is based on the confirmation of sequence by hybridization of the sample to the clade B-defined oligonucleotide array on the chip. Sequencing of pol cDNAs from clades A, C, D, E, and F HIV-1 strains resulted in clade-specific regions of base-calling ambiguities and a small number of incorrectly called resistance polymorphisms. These ambiguities mapped largely to regions not known to be associated with resistance polymorphisms, suggesting the failure of these sequences in the sample to hybridize to any probe cells on the chip. This explanation is supported by the observation that the areas of chip failure are in contiguous regions on the oligonucleotide array.
Since the Affymetrix assay is a hybridization technique, it could be postulated that manipulation of the hybridization conditions and subsequent washes as is done to optimize Southern and Northern blotting and other hybridization-based techniques might help resolve some of these ambiguities. However, the additional fact that the chip contains only a defined array of oligonucleotides and is capable of resequencing but not of de novo sequence determination minimizes the impact that manipulation of experimental conditions might have on rectification of the ambiguities. Since the oligonucleotide sequences for non-clade B virus are not represented on the chip, the only approach that could widen the applicability of the chip to non-clade B isolates would be to engineer a chip that contained non-clade B oligonucleotides.
The insertion of serine codons distal to RT codon 68 was not detected by this version of the Affymetrix assay. The ability to detect and sequence these regions may be important for thorough resistance genotyping. The fact that the Affymetrix assay cannot detect such sequences in clade B virus has significant implications with regard to its utility with the template for which the platform was designed and optimized. The failure of the chip to detect the insertion of additional sequence is due to the fact that the resequencing method is limited to the detection of previously defined sequence. While our observations confirm that the addition of sequence is not detected by the Affymetrix assay, the absence of sequence would likely result in ambiguities as well.
In summary, the increasing entry of non-clade B HIV-1 strains into geographic regions where health care systems will support the clinical use of antiretroviral drug resistance genotyping (12, 13) and the recognition of pol gene length polymorphisms that correlate with drug resistance will impact the generalizability of this assay in its current format. In its current configuration, the Affymetrix assay is not applicable for the accurate assessment of non-clade B virus or virus exhibiting length polymorphisms. Future-generation DNA microarrays should be developed to accommodate this expanding pool of pol genotypes.
ACKNOWLEDGMENTS
We thank Thomas Gingeras and Mark Hurt of Affymetrix, Inc., for helpful discussions, Linda Jagodzinski for manuscript review, and Deborah L. Birx for support.
This work was supported in part by Cooperative Agreement no. DAMD17-93-V-3004 between the U.S. Army Medical Research and Materiel Command and the Henry M. Jackson Foundation for the Advancement of Military Medicine.
REFERENCES
- 1.Alaeus A, Leitner T, Lidman K, Albert J. Most HIV-1 genetic subtypes have entered Sweden. AIDS. 1997;11:199–202. doi: 10.1097/00002030-199702000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Alaeus A, Lidman K, Sonnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Debyser Z, Van Wijngaerden E, Van Laethem K, Beuselinck K, Reynders M, De Clercq E, Desmyter J, Vandamme A M. Failure to quantify viral load with two of the three commercial methods in a pregnant woman harboring an HIV type 1 subtype G strain. AIDS Res Hum Retroviruses. 1998;14:453–459. doi: 10.1089/aid.1998.14.453. [DOI] [PubMed] [Google Scholar]
- 4.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 5.Fodor S P. Massively parallel genomics. Science. 1997;277:393–395. [Google Scholar]
- 6.Gunthard H F, Wong J K, Ignacio C C, Havlir D V, Richman D D. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res Hum Retroviruses. 1998;14:869–876. doi: 10.1089/aid.1998.14.869. [DOI] [PubMed] [Google Scholar]
- 7.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 8.Lipshutz R J. Using oligonucleotide probe arrays to assess genetic diversity. BioTechniques. 1995;19:442–447. [PubMed] [Google Scholar]
- 9.Mellors J W. 12th World AIDS Conference, Geneva, Switzerland. 1998. Review of Lago Maggiore resistance meeting. [Google Scholar]
- 10.Michael N L, Herman S A, Kwok S, Dreyer K, Wang J, Chistopherson C, Spadoro J P, Young K K Y, Polonis V, McCutchan F E, Carr J, Mascola J R, Jagodzinski L L, Robb M L. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J Clin Microbiol. 1999;37:2557–2563. doi: 10.1128/jcm.37.8.2557-2563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pease A C, Solas D, Sullivan E J, Cronin M T, Holmes C P, Fodor S P. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wainberg M A, Gu Z, Gao Q, Arts E, Geleziunas R, Bour S, Beaulieu R, Tsoukas C, Singer J, Montaner J. Clinical correlates and molecular basis of HIV drug resistance. J Acquired Immune Defic Syndr. 1993;6(Suppl. 1):S36–S46. [PubMed] [Google Scholar]
- 13.Wainberg M A, Salomon H, Spira B, Mercure L, Wainberg J, Nagai K, Bentwich Z, Montaner J. HIV resistance to anti-viral drugs. Braz J Med Biol Res. 1993;26:299–308. [PubMed] [Google Scholar]