Abstract
Here, we present a karyotype, a chromosome-scale genome assembly, and a genome annotation from the ctenophore Hormiphora californensis (Ctenophora: Cydippida: Pleurobrachiidae). The assembly spans 110 Mb in 44 scaffolds and 99.47% of the bases are contained in 13 scaffolds. Chromosome micrographs and Hi-C heatmaps support a karyotype of 13 diploid chromosomes. Hi-C data reveal three large heterozygous inversions on chromosome 1, and one heterozygous inversion shares the same gene order found in the genome of the ctenophore Pleurobrachia bachei. We find evidence that H. californensis and P. bachei share thirteen homologous chromosomes, and the same karyotype of 1n = 13. The manually curated PacBio Iso-Seq-based genome annotation reveals complex gene structures, including nested genes and trans-spliced leader sequences. This chromosome-scale assembly is a useful resource for ctenophore biology and will aid future studies of metazoan evolution and phylogenetics.
Keywords: ctenophore, Ctenophora, genomics, comb jelly, chromosome-scale, heterozygosity, Iso-Seq, Hi-C, PacBio, inversion
Introduction
Ctenophore genome assemblies have been key to understanding the early evolution of animals. The draft genomes of the ctenophores Mnemiopsis leidyi and Pleurobrachia bachei showed that many important animal developmental and neuron-specific genes did not evolve until the common ancestor of the bilateria (Ryan et al. 2013; Moroz et al. 2014). Years after publication, these two ctenophore genomes remain crucial for studying the evolution of gene families and developmental pathways in the ancestor to all animals (Sebé-Pedrós et al. 2018; Fernández and Gabaldón 2020; Tikhonenkov et al. 2020; Wang et al. 2020), and for studying the evolution of genome regulation within animals (Gaiti et al. 2017; Bråte et al. 2018).
It remains controversial whether ctenophores or sponges are sister to the rest of animals (Simion et al. 2017; Shen et al. 2017; Whelan et al. 2017; Laumer et al. 2019). Therefore, it is unclear on what ancestral evolutionary branch some metazoan characters evolved, such as neurons and the mesoderm. One method that could possibly resolve the phylogenetic position of ctenophores and sponges is comparing whole-chromosomes (Sacerdot et al. 2018). However, the M. leidyi and P. bachei assemblies are not chromosome-scale. Furthermore, karyotypes are not known for M. leidyi, P. bachei, or any other ctenophore.
In contrast to the hundreds of published chromosome-scale genome assemblies from vertebrates and other bilterians, there are currently only three from nonbilaterian animals: the freshwater sponge Ephydatia (Kenny et al. 2020), the cnidarian Rhopilema (Li et al. 2020; Nong et al. 2020), and the cnidarian Nematostella (Zimmermann et al. 2020). The disparity in the number of bilaterian versus nonbilaterian chromosome-scale assemblies can be partly explained by the difficulties of isolating nucleic acids from nonbilaterians (Dawson et al. 1998; Simister et al. 2011). Also, nonbilaterians tend to have highly heterozygous genomes (Leffler et al. 2012), which complicates standard approaches to genome assembly (Kajitani et al. 2014). The assemblies from Ephydatia, Rhopilema, and Nematostella were possible only due to recent advances in long-read sequencing and the advent of Hi-C data for whole-genome scaffolding (Burton et al. 2013; Rice and Green 2019).
Hormiphora californensis is a globular 2 cm ctenophore abundant in the temperate Pacific Ocean with several attractive features for experimental work (Matthews and Vosshall 2020). This species is readily cultured in aquaria (Patry et al. 2019), has a life cycle as short as 2 weeks, produces hundreds to thousands of eggs per spawning event, and has easily observed embryonic development (Freeman 1977). In addition, there are established CRISPR-Cas9 genome-editing methods for other ctenophore species that may be adaptable to Hormiphora (Presnell and Browne 2021). The genus Hormiphora is in the same family, the Pleurobrachiidae, as the ctenophore P. bachei. Given these useful traits, and the availability of the P. bachei genome, we selected H. californensis for chromosome-scale genome assembly.
Here, we report a karyotype, chromosome-scale genome assembly, and a manually curated genome annotation of H. californensis individual Hc1. Using Hi-C data from Hc1 and Hc3, we present evidence for three heterozygous inversions that span 73% of one Hormiphora chromosome. We find that there are several inversion breakpoints in common between Hormiphora and Pleurobrachia. We estimate the indel and SNP heterozygosity of H. californensis. We use Iso-Seq based annotation to resolve hundreds of complex nested intronic (NI) genes, and find that trans-spliced leaders are common in ctenophore mRNAs. The H. californensis genome assembly, annotation, and sequencing data will be a valuable resource for comparative genomics and evolutionary studies.
Materials and methods
Sample collection
We sampled two H. californensis individuals, Hc1 and Hc2, wild-caught from the Monterey Bay (Table 1). We also sampled a third individual, called Hc3, from the seventh generation of a lab-reared culture at the Monterey Bay Aquarium. The aquarium culture’s provenance was a single broadcast spawning event from ten individuals wild-caught in the Monterey Bay. Hc1 and Hc2 samples were collected with the ROV Ventana aboard the Monterey Bay Aquarium Research Institute’s R/V Rachel Carson, and from a Tucker trawl aboard MBARI’s R/V Western Flyer. Samples were flash frozen in liquid nitrogen after allowing the gut to clear. Samples were collected under the State of California Department of Fish and Wildlife collecting permit SC-4029. Additional details are included in Table 1 and Supplementary Figure S1. An individual collected by Tucker trawl, Hc1, was selected as the sole source for DNA and RNA sequencing for the genome assembly and annotation.
Table 1.
Hormiphora californensis sample collection details
| Biosample accession | Name | Depth | Collec. temp. | Collection method | Collec. date | Latitude, longitude DMS | mtDNA accession |
|---|---|---|---|---|---|---|---|
| SAMN12924379 | Hc1 | 0-520m | 6°C–13°C | Tucker Trawl | 2016-12-13 |
36° 41' 42'' N, 122° 5' 22'' W |
MN544300 |
| SAMN12924380 | Hc2 | 230m | 8.9°C | ROV Ventana | 2013-11-11 |
36° 41' 48'' N, 122° 2' 36'' W |
MN544301 |
| SAMN16124402 | Hc3 | 0m | 12°C | F7 from Monterey Bay Aquarium | 2020-06-05 |
36° 41' 48'' N, 122° 2' 36'' W |
NA |
Karyotyping
We prepared H. californensis chromosomes from embryos to produce a karyotype. To collect embryos we placed Tucker trawl-collected H. californensis individuals in 200 mL of 12°C filtered seawater, adapted the animals to darkness for 4 h, then induced spawning with light (Patry et al. 2019). The embryos were concentrated into 10 mL of seawater using a 40 µm mesh Fisherbrand Sterile Cell Strainer, then were incubated at 12°C for 6 h to allow development to approximately the 64-cell stage. The embryos were fixed using a protocol for chromosome spread preparation for Nematostella vectensis (Guo et al. 2018). The slides of chromosome preparations were stained using DAPI, mounted with Fluoromount-G, then stored at 4°C until imaging. Micrographs of chromosome spreads were collected with a 100x objective and 1.5x diopter on a Leica DM5500 B microscope with a DAPI excitation light and filter at the UC Santa Cruz Life Sciences Microscopy Center.
Data preparation
In total, we constructed 13 Illumina and PacBio DNA and RNA sequencing libraries. Eleven of these libraries were from the individual used for genome assembly and annotation, Hc1. The remaining two libraries were one Illumina WGS of individual Hc2, and a Hi-C library of individual Hc3. The preparation protocols used for the Chicago (Putnam et al. 2016) and Hi-C (Adams et al. 2020) libraries were from published methods. Briefly, from Hc1 we collected 247x coverage of PacBio WGS CLR reads, 573x coverage of Illumina WGS reads, 1956x coverage of Chicago and Hi-C reads, 28 Gbp of Illumina RNA-seq reads, and 2.5 million Iso-Seq transcripts. The mean read length for both the PacBio Sequel I CLR and the PacBio Sequel II Iso-Seq data was 2.7 kb (Supplementary Figure S2). The Iso-Seq read length distribution roughly matched the size distribution of the input RNA (Supplementary Figure S3). Sequencing was performed at the University of California Davis (UCD) DNA Technologies Core, Fulgent Genetics, MedGenome Inc., or at the University of Utah. The raw data are available on NCBI under BioProject PRJNA576068. Details for each library are available in Table 2. Reads were trimmed and prepared for genome assembly and genome annotation. For details, see the Supplementary section Sequencing data preparation.
Table 2.
A summary of the H. californensis SRAs sequenced for this study
| Individual | SRR | Data type | Total GB | Number of reads (or pairs)—Millions | Physical coverage |
|---|---|---|---|---|---|
| Hc1 | SRR10237148, SRR10237149, SRR10237137 | PacBio WGS CLR | 27.4 | 9.7 | 247.7 |
|
| |||||
| SRR10237134 | 10X Chromium | 22.3 | 74.3 | 201.4 | |
|
| |||||
| SRR10237129, SRR10237130, SRR10237131, SRR10237132, SRR10237133 | Illumina WGS—NEBNext UII | 36.1 | 120.2 | 325.8 | |
|
| |||||
| SRR10237128 | Chicago—DpnII | 10.7 | 35.6 | 96.6 | |
|
| |||||
| SRR10237146, SRR10237147 | Chicago—MluCI | 20.9 | 69.8 | 189.2 | |
|
| |||||
| SRR10237145, SRR13784183 | HiC—DpnII—rep 1 | 50.0 | 166.5 | 451.3 | |
|
|
|||||
| SRR10237144, SRR13784182 | HiC—DpnII—rep 2 | 68.1 | 226.8 | 614.8 | |
|
|
|||||
| SRR10237139, SRR13784181 | HiC—MluCI—rep 1 | 38.1 | 127.1 | 344.5 | |
|
|
|||||
| SRR10237138, SRR13784180 | HiC—MluCI—rep 2 | 28.8 | 95.9 | 260.0 | |
|
|
|||||
| SRR10237136 | TruSeq RNA Library Prep Kit v2 (stranded) | 28.8 | 96.0 | NA | |
|
|
|||||
| SRR10403849, SRR10403581 | Iso-Seq Express | 6.0 | 2.5 | NA | |
|
| |||||
| Hc2 | SRR10237135 | Illumina TruSeq Nano DNA | 12.8 | 64.0 | 115.7 |
|
| |||||
| Hc3 | SRR12632403, SRR13784179 | HiC—DpnII | 70.2 | 233.9 | 633.8 |
Each row is a single library. For individual Hc1, the total physical coverage of DNA WGS reads is 573.5x, and the coverage for Hi-C and Chicago data is 1956.4x. More detailed information can be found in Supplementary data S1.
De novo transcriptome assembly
The trimmed Hc1 Illumina RNA-seq data were assembled using Trinity v2.5.1 (Grabherr et al. 2011) with the parameter –SS_lib_type RF. Transcripts that contained adapters or vector contamination in the NCBI contamination database were removed. The assembly is available on the NCBI Transcriptome Shotgun Assembly archive, accession GHXS00000000.
Mitochondrial genome and phylogeny
We assembled the mitochondrial genomes of H. californensis individuals Hc1 and Hc2 using PacBio and Illumina reads, using canu v2.1.1 (Koren et al. 2017) and pilon v1.22 (Walker et al. 2014). To determine the phylogenetic position of H. californensis individuals Hc1 and Hc2 we constructed an 18S tree, a COX1 nucleotide tree, and a multi-locus mitochondrial protein tree. See the Supplementary materials sections Mitochondrial Genome Assembly and Annotation, and Phylogeny construction.
Genome assembly
Genome size estimation
K-mers were counted from trimmed Illumina WGS H. californensis reads and from publicly available P. bachei WGS libraries (SRR116669 and SRR116670) (Moroz et al. 2014) using jellyfish v2.2.10 (Marçais and Kingsford 2011) with options -C-s1000000000-k21. Genome sizes of both species were estimated using the k-mer count histograms on the GenomeScope2 server (Ranallo-Benavidez et al. 2020).
Genome assembly
The genome was assembled using wtdbg2 v2.4 (Ruan and Li 2019). The assembly was then polished with arrow v2.2 (github.com/PacificBiosciences/gcpp), then with pilon v1.22 (Walker et al. 2014). Haplotigs were removed with Purge Haplotigs v1.0.4 (Roach et al. 2018). Dovetail Genomics HiRise vAug2019 was used to scaffold the haplotig-purged assembly with the trimmed Chicago and Hi-C reads. Scaffolds with a mean coverage of less than 100, or having greater than 50% GC, were removed from the assembly using BlobTools v1.1.1 (Laetsch and Blaxter 2017). Assembly gaps were closed with LRGapcloser (Xu et al. 2019). The assembly was polished with pilon. See the Genome Assembly section of the Supplementary methods for additional details.
Genome quality assessment
We calculated the final assembly statistics such as the number of scaffolds, contigs, and the N50, using the program fasta_stats included with the Meraculous assembler (Chapman et al. 2011). We also assessed the completeness of the assembly by calculating the percent of PacBio Sequel subreads and full-length nonchimeric (FLNC) transcripts that mapped to the assembly. We also used a custom python script to calculate the percent of bases of each read type that mapped to the assembly. We performed a self-to-self genome alignment using LASTZ v1.04.03 (Harris 2007) to check for erroneously duplicated regions. To check for uncollapsed haplotypes or regions with many indels we used samtools mpileup v1.7 (Li et al. 2009) and chep commit 60c4312 (github.com/conchoecia/chep).
Characterizing chromosomal inversions
We generated a Hi-C heatmap to check for genome misassemblies. For details, see the Hi-C heatmap generation Supplementary section. We noticed three strong off-diagonal bowtie-shaped Hi-C hotspots on Chromosome 1. If this type of signal arises from a misassembly, then the misassembly can be corrected by inverting the bowtie-shaped region of the Hi-C matrix. Heterozygous inversions are not correctable by the same process (Corbett-Detig et al. 2019; Chida et al. 2020). We used PretextView to combinatorially invert sections of the Hi-C matrix to attempt to remove the off-diagonal signal.
Genome variant calling and phasing
To find diploid variants in the genome, we mapped PacBio CLR and Illumina WGS reads to the genome with minimap2 v2.17 (Li 2017) and BWA-MEM v0.7.17 (Li 2013), called variants using freebayes v1.3.2-38 (Garrison and Marth 2012) and gnu parallel v20161222 (Tange 2011), then filtered the VCF to only include diploid calls. We then phased the variants using Picard v2.25.1 (“Picard Toolkit” 2016) and HapCUT2 v1.3.1 (Edge et al. 2017). See the section Genome Variant Calling and Phasing in the Supplementary methods for parameters.
Genome annotation
Manual genome annotation
We annotated the genome by manually curating transcript models generated from several datasets. The transcript sets were generated with PacBio Iso-Seq and Illumina RNA-seq reads as input for BRAKER v2.14 (Hoff et al. 2019), AUGUSTUS v3.3.3 (Stanke et al. 2004), and GeneMark-ES/ET v4.65 (Hoff et al. 2016). The PacBio Iso-Seq data were used as input for PacBio Cupcake tools v8.0 (github.com/Magdoll/cDNA_Cupcake), StringTie v2.0.4 (Pertea et al. 2015), and Pinfish commit b6f3c06 (github.com/nanoporetech/pinfish). See the Genome Annotation and Transcript phasing sections of the Supplementary materials for details on how each program was run.
Genome annotation consisted of three rounds. In annotation round 1, we reviewed the genome in IGV or JBrowse and manually verified the StringTie transcripts that were generated with the PacBio Iso-Seq data. Specifically, we identified if the StringTie transcripts were fused, correct, or fragmented by comparing them to the FLNC PacBio Iso-Seq reads. If the mapping pattern of PacBio Iso-seq reads suggested that a StringTie transcript was a fusion between two or more adjacent transcripts, then we replaced the fused StringTie transcript with Pinfish, AUGUSTUS, or GeneMark-ES/ET transcripts. The replacement transcripts were selected if they matched the gene structure of the PacBio Iso-Seq reads that mapped to the same locus. If a StringTie transcript was only a partial gene, also evidenced by the PacBio Iso-Seq reads, then the partial StringTie transcript was replaced with a correct Pinfish, AUGUSTUS, or GeneMark-ES/ET transcript. StringTie transcripts that did not match a transcript observed in the PacBio Iso-Seq data were removed. If Iso-Seq reads were mapped to a locus, but the locus had no representative StringTie transcript, then a matching Pinfish, AUGUSTUS, or GeneMark-ES/ET transcript was added to the annotation. StringTie transcripts that were grouped together by StringTie, but actually represented multiple genes with mutually exclusive exons, were split into multiple genes. At this stage the annotation contained genes and transcripts representing the complement of PacBio Iso-Seq data derived from the adult Hc1.
In annotation round 2, AUGUSTUS gene models generated from hints that included Illumina RNA-seq reads were added to the annotation if they did not overlap with the transcripts from round 1.
In annotation round 3, we sought to find life-stage-specific and tissue-specific transcripts in the H. californensis genome that may not have been present in the RNA sample from the adult Hc1. Gene models were generated by mapping P. bachei transcripts to the H. californensis genome. The resulting gene models were removed if they did not contain an ORF in the H. californensis genome, or if they overlapped with H. californensis annotation round 1 or round 2 genes. Gene models were only included in the annotation if their ORF’s protein product had a blastp v2.10.0+ (Altschul et al. 1997) hit to publicly available ctenophore transcriptomes with an e-value of less than 1e-10.
For each transcript in the annotation, we generated haplotype-resolved protein sequences. See the Genome Annotation and Transcript phasing sections of the Supplementary materials for more details.
Annotation completeness assessment
We used gVolante and BUSCO Eukaryota v3 to calculate the BUSCO score of the protein models from our annotation, the de novo transcriptome, and the genome assembly (Simão et al. 2015; Nishimura et al. 2017).
TAD calling and boundary analysis
We called topologically associating domains (TADs) using the HOMER Hi-C analysis pipeline (Heinz et al. 2018). The TADs were called with 1/4 kb bin resolutions and 10/40 kb windows. We masked regions around Hi-C heatmap irregularities such as off-diagonal signal that appeared to be due to inversions. This signal confounds the discovery of TADs in well-assembled genome regions. We calculated TAD separation scores with HiCExplorer v3.6 (Ramírez et al. 2018) using Cooler v0.8.10 (Abdennur and Mirny 2020).
We applied HOMER’s de novo motif discovery pipeline to 1.5 kb regions on either side of each TAD boundary (Heinz et al. 2018). For motif discovery, we selected background regions that exhibited minimal local change in TAD separation score, as these regions least resemble TAD boundaries.
We noticed that TADs tended to occur near gene boundaries. To test the significance of this observation we performed a permutation test. We first measured the median distance between TAD boundaries and the nearest gene. The background distribution was calculated by 1000 permutations of randomly placing TADs across the genome using the same size distribution as our observed TADs, then measuring the distance to the nearest gene.
Identification of nested intronic genes
Nested intronic genes were identified using chep gff_to_intron_bed.py (github.com/conchoecia/chep, commit 60c4312), allowing for a 15% overlap with the host exons at both the 5ʹ and 3ʹ ends of the nested transcript. We excluded the longest 0.5% of introns from the analysis to avoid counting the introns from trans-spliced splice leaders.
Repeats and centromeres
We used Tandem Repeats Finder v March 13, 2006 (Benson 1999) and EDTA v1.8.3 (Ou et al. 2019) to identify repeats and transposable elements.
Comparative analyses
Whole-genome heterozygosity estimation
The single nucleotide heterozygosity of Hc1 was estimated by only using sites that had exactly 178x Illumina WGS read mapping depth. This depth, 178x, was the mode of the mapping depth for the whole genome, and thus represents sites at which reads from both haplotypes mapped. We implemented this method, first described in Saremi et al. (2019), in a purpose-built software package called chep (github.com/conchoecia/chep). We also measured the heterozygosity of the Hc1 exonic, intronic, and intergenic regions on individual chromosomes using chep.
We measured the heterozygosity of Hc1, Hc2, and P. bachei individual SAMN00216730 by counting 21-mers with jellyfish v2.2.10 (Marçais and Kingsford 2011) then using the resulting spectrum in GenomeScope 2 (Ranallo-Benavidez et al. 2020), by using vcftools’ –het option (v0.1.17), and by using angsd realSFS (v0.921) (Danecek et al. 2011; Korneliussen et al. 2014). We were not able to measure the heterozygosity of M. leidyi because the sequencing libraries were derived from multiple individuals.
Analysis of the HiC-scaffolded Pleurobrachia genome
The P. bachei genome assembly was recently scaffolded using Hi-C data (Hoencamp et al. 2021). The new, scaffolded assembly did not include a genome annotation. We identified the protein positions in the P. bachei genome assembly using tblastn and the previously published P. bachei protein sequences (Moroz et al. 2014). We also looked for orthologous scaffolds between the H. californensis and P. bachei genomes by plotting the protein coordinates of reciprocal best blastp hits between the proteins of the two genomes. For comparative analysis, we generated a P. bachei Hi-C heatmap by mapping the P. bachei Hi-C reads, SRR13364273 (Hoencamp et al. 2021), to the new assembly using the same protocol as for H. californensis. See the Hi-C heatmap generation Supplementary section for details.
Microsynteny between ctenophores
The H. californensis proteins were queried against the M. leidyi and P. bachei proteins using diamond blastp v0.9.24 (Buchfink et al. 2015). The gene positions and the diamond blastp table were then used to identify collinear blocks of genes using the purpose-built Python script microsynteny.py (github.com/wrf/genomeGTFtools). We required a minimum of 3 consecutive genes, allowed for up to 5 intervening genes, and allowed a maximum distance of 30 kb to the next gene.
Results and discussion
Genome sequencing and assembly
To determine the ploidy of H. californensis and to estimate its genome size, we computed k-mer spectra from H. californensis and P. bachei WGS libraries. Libraries from both species had two major k-mer peaks. The lower-coverage peak was larger than the higher-coverage peak in both species. This pattern is consistent with the k-mer spectra of other highly heterozygous diploid organisms (Ranallo-Benavidez et al. 2020). From the k-mer spectrum, the predicted 1C genome size of H. californensis was 96–98 Mb (Supplementary Figure S4), which is close to the predicted P. bachei genome size, 97.5 Mb (Supplementary Figure S5).
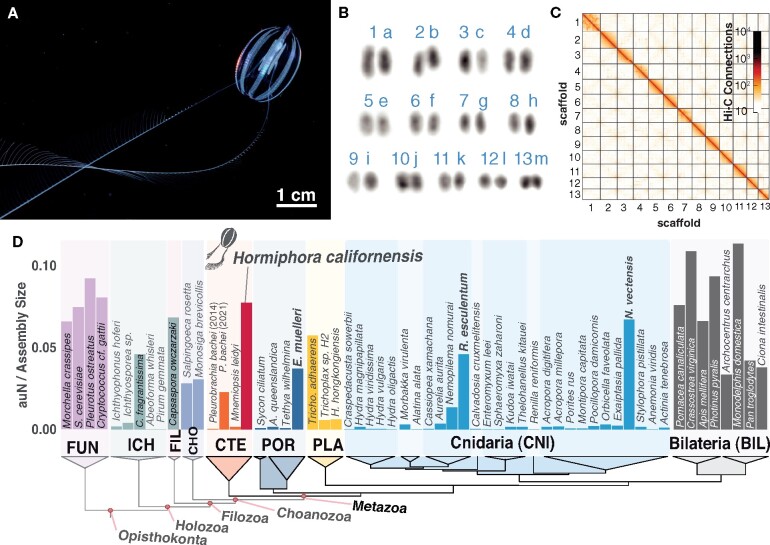
We aimed to generate a chromosome-scale reference genome for H. californensis in which each chromosome is represented by a composite sequence obtained by combining both haplotypes. This genome sequence was assembled from PacBio long reads, polished with Illumina short paired-end reads, and scaffolded with in vitro and in vivo chromatin conformation capture reads from a single individual, Hc1 (Methods). The H. californensis genome assembly totaled 110.6 Mb in 44 scaffolds and 351 contigs. Half of the sequence is present in scaffolds longer than 8.5 Mb (N50), with 2.76 gaps per Mb within scaffolds. The 13 longest scaffolds comprise 99.47% of the assembly, ranging in size from 10.3 to 6.4 Mb. These 13 long scaffolds match the microscopy-based karyotype of n = 13, detailed below. The remaining 31 scaffolds were each shorter than 50 kb and represent short unplaced sequences. We found no detectable contamination from marine bacteria or gut contents based on the blobtools results (Supplementary Figure S6). The genome dotplot made with D-Genies (Cabanettes and Klopp 2018) did not reveal erroneously duplicated assembly regions (Supplementary Figure S7). 95.32% of the PacBio Sequel subreads (Supplementary Table S1), and 99.02% of PacBio Sequel II Iso-Seq FLNC transcripts mapped to the 13 largest scaffolds.
We note that our 110 Mb H. californensis assembly is substantially shorter than the published P. bachei assembly (156 Mb) despite similar size estimates based on k-mer spectra. Our analysis of the P. bachei assembly, included in the Supplemental text, suggests that over half of the reported P. bachei scaffolds are unmerged haplotypes.
Variant calling and phasing
We called variants using freebayes after mapping Hc1 PacBio CLR and Illumina WGS reads to the H. californensis reference genome. After filtering we identified 2.24 million heterozygous single nucleotide or indel variants. These variants were phased using PacBio CLR, Chicago, and Hi-C reads, resulting in phased blocks of variants that spanned more than 99% of the length of each chromosome-length scaffold. Of the 2.24 million diploid variants, 1.75 million (77.9%) were in the chromosome-scale phased variant blocks. The high density of phased variants, one for every 63 bp of genome assembly, suggests that the H. californensis data may be a useful benchmarking candidate for phased, or diploid, genome assemblers.
Mitochondrial genome and phylogeny
We assembled and annotated the mitochondrial genomes of Hc1 and Hc2, two individuals from the same Monterey Bay population, and collected 3 years apart. The mitochondrial genomes (mtDNA) from Hc1 and Hc2 were 99.6% identical. The H. californensis mtDNA is 71.5% identical to the mtDNA of the closely related P. bachei, and 80% identical to P. bachei when only considering coding regions. The H. californensis mitochondrial genome has a 1.8 kb insertion relative to P. bachei, between COX2 and 16S (Supplementary Figure S8). The percent identity between the H. californensis and P. bachei mtDNA confirms they are distinct species, despite their similar morphology. Phylogenetic analysis of P. bachei and H. californensis mtDNA is also consistent with these being distinct but closely related species (Supplementary Figure S9).
Despite the distinct mtDNA of H. californensis and P. bachei, phylogenetic trees based on 18S rRNA and COX1 show that H. californensis falls within the Pleurobrachia clade. Furthermore, other Hormiphora species are sister to Pleurobrachia. These results suggest that the genus Hormiphora as currently defined may be polyphyletic. Future taxonomy work should consider reassigning H. californensis to the genus Pleurobrachia.
Karyotype
The karyotype has not been previously described for any ctenophore species. We used microscopy of DAPI-stained chromosome spreads to determine that the H. californensis genome is composed of n = 13 chromosome pairs (Figure 1B, Supplementary Figure S10). Four of the images correspond to a 2n of 26, and the remaining images have counts within 3 chromosomes of 26. This count, 13 pairs, is consistent with the 13 multi-megabase scaffolds in the de novo genome assembly presented here.
Figure 1.
Karyotype and genome assembly quality. (A) H. californensis with its tentacles extended during feeding. (B) One karyotype image (rearranged and color-inverted) of a H. californensis chromosome preparation that contained 13 chromosome pairs. (C) The H. californensis Hi-C map, showing thirteen chromosome-scale scaffolds. (D) Genome contiguity normalized by genome size [auN/Assembly Size, per (Li 2020)] from all available nonbilaterian holozoan genomes and select fungal and bilaterian genomes, with tree topology based on (Whelan et al. 2017). Bolded species names are nonbilaterians with chromosome-scale genome assemblies. BIL, bilaterians; CHO, choanoflagellates; CNI, cnidarians; CTE, ctenophores; FIL, Filasteria; FUN, Fungi; ICH, Ichthyosporea; PLA, placozoans; POR, Porifera.
Genome annotation
We manually annotated the genome using gene models generated with Hc1 Iso-Seq reads, Illumina RNA-seq data, and P. bachei transcripts. The long Iso-Seq reads capture, in many cases, complete cDNA sequences and represent transcripts from a single haplotype. These features allowed us to produce haplotype-specific transcripts and proteins.
Our approach to annotating the H. californensis genome identified 14,265 protein-coding genes, of which 13,235 are supported by Iso-Seq reads (Supplementary Table S2). The BUSCO complete plus fragmented score was 96% (303 Eukaryotic genes— Supplementary Table S3). We found that 96% of the Pleurobrachia proteins with orthologs in other ctenophores also have an ortholog in the H. californensis annotation. We note that due to the haplotype redundancy of the P. bachei assembly, many annotated P. bachei genes are reported in allelic copies, which therefore overestimates the gene count of this species (Supplementary materials).
In addition, the H. californensis genome contains 619 protein-coding genes that have orthologs in other ctenophore transcriptomes, but do not appear in either the M. leidyi or P. bachei genomes (Supplementary Table S2). Of those 619 genes, 122 had blastp hits to nr, and included genes with a wide variety of functions such as DNA-binding proteins, calmodulins, histones, proteases, and more. Among these 122 genes we did not find any evidence for the presence of the neural and mesoderm-component genes reported to be missing from ctenophores (Ryan et al. 2013).
We found 1729 cases where two or more neighboring P. bachei gene models, and 1200 cases where M. leidyi gene models, appear to be fragments of a larger gene based on orthology with H. californensis. For example, the pecanex gene (2096 amino acids in H. californensis) appears to be split into four proteins in the M. leidyi annotation (Supplementary Figure S11).
97.7% of the eukaryotic BUSCOs were complete or partial in the translated Iso-Seq FLNC data, and 99.0% were complete or partial in the Illumina RNA-seq de novo transcriptome. Because these values are higher than the 96% complete or partial BUSCOs from the genome annotation, it is possible that the genome annotation does not capture the full complement of H. californensis genes. Future annotation iterations will benefit from Iso-Seq sequencing of different tissues and developmental stages.
Tandem Repeats Finder (Benson 1999) identified 14 Mb (13%) of the genome as repeats, none of which were identifiable as centromeric. Thus, we are unable to annotate or further describe centromeres in these genomes.
Topologically associating domains and 3D genome structure
Genome analyses using Hi-C data have shown that in many species, chromatin is organized in segments of close proximity that are known as TADs (Lieberman-Aiden et al. 2009).
We used proximity ligation data to identify and characterize TADs in H. californensis and found evidence that the H. californensis genome contains small TADs with a median length of 60 kb. The H. californensis TADs are significantly smaller than human TADs (median length 1.15 Mb) (McArthur and Capra 2021). Despite the fact that the Ephydatia and Drosophila genomes are comparable in size to the H. californensis genome, the mean H. californensis TAD length is half the length of the TADs in those two species (Hou et al. 2012; Kenny et al. 2020). We found that TAD boundaries tend to occur in the noncoding DNA bordering genes (P = 0.001, permutation). The mean distance from a TAD boundary to a gene is 7.8 kb.
We used HOMER to search, de novo, for DNA motifs in the sequences flanking the outside of TADs. In these sequences flanking TADs we found enriched motifs, most of which resembled the RNA polymerase II-binding motifs of known transcription factors. The six most-enriched motifs were homeodomain and MYB-related transcription factor binding sites, which are conserved in eukaryotes. Homeodomain and MYB-related transcription factor genes, as well as RNA polymerase II, were present in the H. californensis genome annotation.
Analysis of the HiC-scaffolded Pleurobrachia genome
We assessed the quality of the P. bachei genome assembly that was recently scaffolded using Hi-C data (Hoencamp et al. 2021). This assembly was generated by linking together contigs and scaffolds from the original P. bachei assembly (Moroz et al. 2014). The authors reported that they found 13 or more putative chromosomal scaffolds, but did not provide further description or analysis of the assembly.
The scaffolded P. bachei genome contains 157.1 Mbp in 20,121 scaffolds and 39,072 contigs. The 13 largest scaffolds contain 81.5 Mb of sequence, and appear to be chromosome-scale in the Hi-C map. Those scaffolds contain 69.4 Mb of contigs, and 12.2 Mb of stretches of Ns. Given that the predicted 1C size of the P. bachei genome is 96.6 Mbp (see results above), the assembly size of the contigs in the 13 largest scaffolds relative to the predicted size is 72%. The 81.5 Mbp in the 13 largest scaffolds is 51.9% of the total assembly size (Supplementary Figure S12).
To further investigate the completeness and correctness of the 13 chromosome-scale P. bachei scaffolds, we performed synteny comparisons with the H. californensis genome assembly. The Moroz et al. (2014) and Hoencamp et al. (2021)P. bachei genomes do not include genome annotations, although Moroz et al. (2014) included 19,002 P. bachei protein models from the genome sequence. Using those 19,002 proteins we identified 9714 genes on the 13 largest P. bachei scaffolds. This is 68.2% of the total number of genes found on the 13 largest Hormiphora scaffolds. We performed a reciprocal-best blastp search between the P. bachei and H. californensis proteins and plotted their coordinates in 2-dimensions to visualize regions of macrosynteny, also called an Oxford dot plot. This plot revealed that each of the 13 largest P. bachei and H. californensis scaffolds predominantly had reciprocal best blastp hits on only one scaffold of the other species (Supplementary Figure S13). We did not find evidence for chromosomal fusions or fissions between the karyotype of the two animals.
In summary, we find that the scaffolded P. bachei assembly contains 13 scaffolds that correspond to the 13 chromosomes of H. californensis. However, as a large fraction of the P. bachei genome is not represented in these 13 scaffolds as measured by overall assembled genome sequence or gene content, the P. bachei genome would likely be greatly improved using a contemporary long-read contig assembly approach, followed by chromosome-scale scaffolding.
Heterozygous chromosome inversions and microsynteny
Chromosome 1 of H. californensis individual Hc1 contains three large heterozygous chromosomal inversions (Figure 2A, Supplementary Figure S14). Each inversion is approximately 2 Mbp, or 20% of chromosome 1. Together, these putative inversions span 73% of the length of chromosome 1. These are unlikely to be assembly errors, since inverting the Hi-C heatmap around the errors does not remove the off-diagonal signal (Supplementary Figure S14). All six breaks of between-haplotype synteny appear to occur between genes, and outside of TAD boundaries. The Hi-C matrix from individual Hc3 does not have off-diagonal hotspots (Figure 2C), suggesting that both haplotypes of Hc3 chromosome 1 match the genome assembly sequence. Large heterozygous inversions can prevent recombination over large chromosomal regions (Kirkpatrick 2010; McBroome et al. 2020), therefore these two haplotypes of H. californensis chromosome 1 may be segregating independently. Large heterozygous inversions between the haplotypes in one individual are not prevalent in vertebrate species, but have been observed before in the genomes of other invertebrates, such as in the mosquito Anopheles gambiae (Corbett-Detig et al. 2019).
Figure 2.
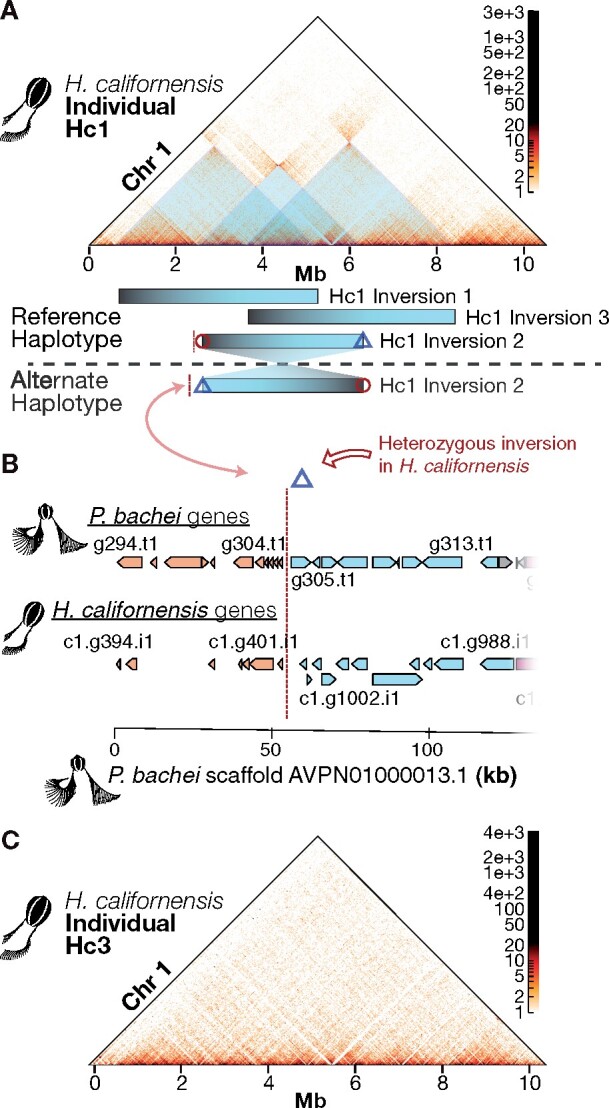
Heterozygous inversions on H. californensis chromosome 1. (A) Three heterozygous overlapping inversions are present on chromosome 1 in the Hc1 genome. Black/blue bars show the spans of each heterozygous inversion. (B) The alternative haplotype of Hc1 inversion 2, indicated by the close proximity of the blue diamond and red line, has the same gene order as the P. bachei genome. The x-axis is genome coordinates of P. bachei scaffold AVPN01000013.1. Orange H. californensis and P. bachei genes to the left of the vertical red dotted line are orthologous and have microsynteny. Blue genes to the right of the vertical red dotted line are orthologous and have microsynteny. (C) Hi-C map of H. californensis individual Hc3 shows concordance with Hc1 chromosome 1, but no off-diagonal Hi-C evidence for heterozygous inversions.
We examined whether the inversion breakpoints in H. californensis chromosome 1 also occurred in the genome of the closely related P. bachei. Three of these breakpoints, including an exact match to Hc1 heterozygous inversion 2, were found to occur in the P. bachei genome. Hc1 inversion 2, in which H. californensis gene 355 on chromosome 1 lies next to H. californensis gene 864 (sequentially numbered) on the alternate haplotype, reflects the gene order in the P. bachei genome on scaffold AVPN01000013.1 (Figure 2B). The H. californensis inversion breakpoint at position 5.20 Mb is also a point of synteny mismatch in P. bachei, in which the gene on one side of the P. bachei synteny break matches a synteny breakpoint in H. californensis (H. californensis gene c1.g741). However, the gene on the opposite side of the synteny break (H. californensis gene c1.g424) does not match any of the gene intervals from inversions 1, 2, or 3 from H. californensis. These results suggest that chromosomal inversions may not only exist between different ctenophore species, but also may be prevalent within a single population of one species.
Gene colinearity analyses suggest that H. californensis and M. leidyi only share limited gene microsynteny. The largest identifiable blocks of gene colinearity only contained four genes in common. Given the extensive gene rearrangements seen between the closely related species H. californensis and P. bachei, it is not surprising to find the lack of gene colinearity between the distantly related H. californensis and M. leidyi.
The largest colinear block was over 5.8 Mbp of H.californensis-P.bachei chromosome 5, encompassing 964 genes in H. californensis. However, most other chromosomes were significantly rearranged between the two species.
Comparative analyses
Heterozygosity
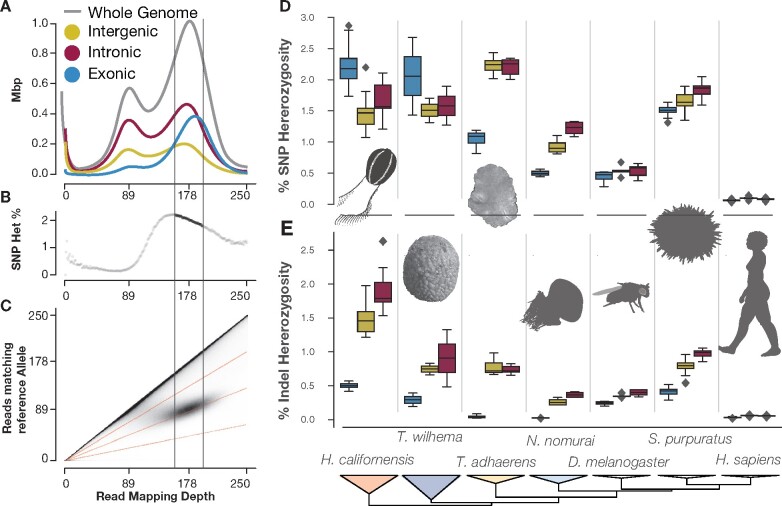
We measured the heterozygosity of the intronic, exonic, and intergenic regions of H. californensis and six other metazoan species (Figure 3, Supplementary Figure S15 and Tables S4 and S5) using a method that avoids mis-estimation due to genome assembly errors or inaccurate heterozygous site calls in a VCF file (Saremi et al. 2019). H. californensis had a high combined single nucleotide and indel heterozygosity rate—approximately 3.2% overall, and a per-chromosome rate of between 2.4% and 4.7%. The overall single-nucleotide heterozygosity was 2%. These analyses also revealed that both H. californensis and the sponge Tethya wilhelma (Mills et al. 2018) had high SNP heterozygosity in exons, but depressed SNP heterozygosity in both intergenic and intronic regions (Figure 3, D and E). This pattern is contrary to other species, where heterozygosity of the introns and intergenic regions is higher than in the exons. This pattern in our data is likely due to short Illumina reads from one allele not mapping to regions with high combined SNP and indel heterozygosity (Figure 3A), therefore placing an artificial ceiling on the measurable heterozygosity. Using long, accurate reads such as PacBio HiFi data, or measuring heterozygosity with a diploid genome assembly, should overcome these shortcomings.
Figure 3.
The Hormiphora genome is highly heterozygous and contains many indels in noncoding regions. (A) The histogram of the number of bases in a genome assembly (y-axis) with a specific mapping depth (x-axis) using Illumina WGS reads. The mode of the mapping depth of the Illumina WGS reads was 178x, so bases with close to 178x mapping coverage have reads from both haplotypes. Therefore, bases with a mapping coverage around 89x have reads mapped from a single haplotype. (B) The whole-genome heterozygosity calculated at each read mapping depth. The heterozygosity value for each mapping depth (x-axis) is the number of sites where only one half of the mapped reads match the reference allele, divided by the total number of sites at that depth. (C) The 2D histogram showing that bi-allelic sites are centered around 178x read mapping depth. Also, the 2D histogram is one way to visualize the data input for panel (B). (D–E) Box-and-whiskers plots show the distribution of SNP (D) or indel (E) heterozygosity in each chromosome-size scaffold. In H. californensis, the intergenic and intronic regions have a reduced SNP heterozygosity, but an elevated rate of indels. This pattern differs from the correlation between SNP and indel heterozygosity found in species with lower overall heterozygosity.
Ubiquity of trans-spliced leader sequences
A 2010 study of ctenophore and cnidarian ESTs showed that these phyla have extensive 5′-capping trans-splicing (Derelle et al. 2010). However, this study lacked genomes to examine the origins of the leader sequence (Derelle et al. 2010). One prevalent feature of H. californensis genome organization was gene clusters sharing a 5′ exon, but otherwise having different exons, and seemingly nonoverlapping transcripts. The shared first exons were between 35-48bp long, and were all >90% identical to 5ʹGAGTTTCAAACTTTTCAACACTACTTTAAACAAATTAATTTGAG 3ʹ. We identified 718 of these leader sequences in the H. californensis genome. The leader sequence was found on 56% of our Iso-Seq reads. This appears to be the result of trans-splicing of a leader sequence (Boroni et al. 2018). The Iso-Seq reads lacking the leader sequence may be sheared at the 5′ end, as is common in full-length cDNA library preparation. Thus, 56% represents a lower bound for the true percentage of H. californensis mRNAs with trans-spliced leaders. The shared exons we identified in the genome may be a result of the leader sequences on the Iso-Seq reads being artifactually mapped to the nearest spliced-leader locus 5ʹ of the transcript in the genome assembly.
In M. leidyi, although it was not reported previously, we found several examples of gene clusters with shared first exons using a de novo M. leidyi transcriptome (SRX993241) mapped to the M. leidyi genome. The leader motif from Derelle et al. (2010) was also identified in the M. leidyi genome 491 times. Both the M. leidyi and H. californensis leader sequences end in a TGAG motif, part of a mostly conserved 5ʹAATTTGAG 3ʹ motif. Over half of the annotated transcripts in H. californensis begin with AG.
Nested intronic genes in the Metazoa
Using 1058 eukaryotic genomes, including all genomes available on NCBI RefSeq, we quantified the percent of exonic basepairs that are from NI genes—genes whose transcripts are within the boundaries of a single intron of another gene (Figure 4). In the H. californensis genome, we found 1654 genes hosting one or more NI genes. There were 2357 NI genes inside the 1654 host genes (Supplementary data). We estimated that H. californensis has 12.24% of exonic bases in NI genes, similar to the rate found in primate and some arthropod genomes. From the 2357 NI genes we identified 484 doubly-nested genes, which are NI genes within another NI gene. We found that 1109 NI genes are flanked by transposable elements (TEs) on at least one side of the gene, and 176 NI genes are flanked on both sides by TEs. Many NI genes are also parallel with the host gene, necessitating a complex transcription or splicing system to separately process the two genes. Parallel NI genes have been observed before in other taxa, such as human (Yu et al. 2005) and fly (Henikoff et al. 1986).
Figure 4.
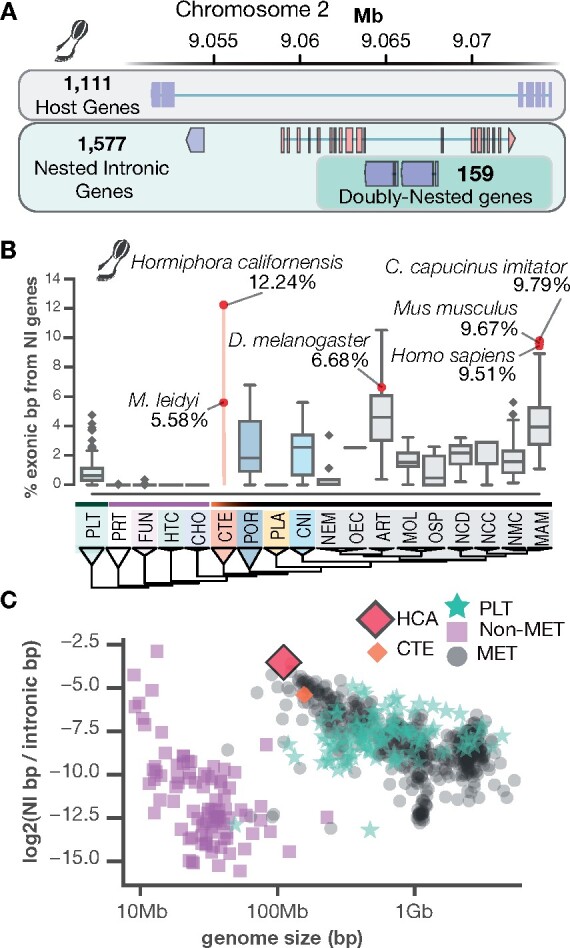
An analysis of NI genes in 1058 eukaryote genomes. (A) NI genes are genes that are contained within the introns of other genes. In this example from the H. californensis genome, four genes are nested within a single host gene. Two of the genes are doubly nested—they are within the intron of another NI gene. Blue genes are coded on the antisense strand relative to the reference sequence and red genes are coded on the sense strand. (B) The percent of all exonic bp that are in NI genes. Protists and nonmetazoans have a negligible proportion compared to animals. Abbreviations: ART, panarthropoda; CHO, choanoflagellates; CNI, cnidarians; CTE, ctenophores; FUN, fungi; HCA, H. californensis; HTC, holozoa through choanoflagellates; MAM, mammals; MET, Metazoa; MOL, molluscs; NCD, nonchordate deuterostomes; NCC, noncraniata chordates; NEM, nematodes; NMC, nonmammalian chordates; OEC, other ecdysozoa; OSP, other spiralians; PLA, placozoans; PLT, plants; POR, Porifera; PRT, other protists. (C) As animal and protist genomes become smaller, a higher proportion of the exonic bases are in NI genes.
We observed that NI genes are largely absent from the genomes of protists, fungi, and other nonmetazoan opisthokonts such as the choanoflagellates (Figure 4C). This observation could be due to genome annotation errors in those clades, or NI genes may have undergone genomic expansions in the metazoan last common ancestor (LCA), and in the plant LCA. We also found that smaller animal genomes tend to have a higher percent of exonic bases in NI genes (Figure 4C). Given that NI genes introduce additional ways to control transcription, such as antisense transcription competition (Yu et al. 2005), the punctuated appearance of these genes in the metazoa is possibly one of the complex transcriptional control mechanisms that evolved in the ancestor to all animals. High-quality genome assemblies and annotations of outgroup species to the metazoa will be necessary to determine the extent to which nested genes are a feature of metazoan genomes.
Data availability
All sequencing reads, transcriptomes, and the genome assembly are available for download via NCBI BioProject PRJNA576068. Hc1 and Hc2 mitochondrial sequences are available through NCBI accessions MN544300 and MN544301. Additional data are available through G3 figshare: https://doi.org/10.25387/g3.15170382. Custom programs used in this publication, and annotation guidelines, are available at github.com/conchoecia/hormiphora and Zenodo DOI: 10.5281/zenodo.4074309. No tissue from these samples is available, as it was consumed during library preparation.
Author Contributions
D.T.S., W.R.F., R.E.G., and S.H.D.H. conceived and designed the study. D.T.S. and L.M.C. collected sequencing data. D.T.S assembled the genome and transcriptomes. D.T.S and W.R.F. annotated the genome. D.T.S., W.R.F., and J.D.M. performed analyses on the data. D.T.S, W.R.F., J.D.M., and R.E.G. wrote the manuscript. All authors contributed to the revision and review of the manuscript.
Funding
This work was supported by the David and Lucile Packard Foundation, the Monterey Bay Aquarium Research Institute, the University of California Biomolecular Engineering and Bioinformatics department; United States National Science Foundation DEB-1542679 to S.H.D.H.; the United States National Science Foundation GRFP DGE 1339067 to D.T.S.
Conflicts of interest
R.E.G. is a cofounder and paid consultant of Dovetail Genomics. All other authors declare no competing interests.
Acknowledgments
The authors thank the crew, ROV pilots, and ship captains of the R/V Rachel Carson and R/V Western Flyer for assistance in collecting animals. They would also like to thank Daniel Rokhsar, Karolin Luger, Joseph Ryan, Manabu Bessho-Uehara, and May Roberts for critical feedback on the manuscript. We also thank Wyatt Patry at the Monterey Bay Aquarium for providing Hormiphora tentacles. They also thank Ben Abrams of the UCSC Life Sciences Microscopy Center for his suggestions for producing chromosome images.
Literature cited
- Abdennur N, Mirny LA.. 2020. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 36:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M, McBroome J, Maurer N, Pepper-Tunick E, Saremi NF, et al. 2020. One fly-one genome: chromosome-scale genome assembly of a single outbred Drosophila melanogaster. Nucleic Acids Res. 48:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 27:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroni M, Sammeth M, Gava SG, Jorge NAN, Macedo AM, et al. 2018. Landscape of the spliced leader trans-splicing mechanism in Schistosoma mansoni. Sci Rep. 8:3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bråte J, Neumann RS, Fromm B, Haraldsen AAB, Tarver JE, et al. 2018. Unicellular origin of the animal MicroRNA machinery. Curr Biol. 28:3288–3295.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12:59–60. [DOI] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, et al. 2013. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 31:1119–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanettes F, Klopp C.. 2018. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ. 6:e4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, Ho I, Sunkara S, Luo S, Schroth GP, et al. 2011. Meraculous: de novo genome assembly with short paired-end reads. PLoS One. 6:e23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chida AR, Ravi S, Jayaprasad S, Paul K, Saha J, et al. 2020. A near-chromosome level genome assembly of Anopheles stephensi. Front Genet. 11:565626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Said I, Calzetta M, Genetti M, McBroome J, et al. 2019. Fine-mapping complex inversion breakpoints and investigating somatic pairing in the Anopheles gambiae species complex using proximity-ligation sequencing. Genetics. 213:1495–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, et al. ; 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics. 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MN, Raskoff KA, Jacobs DK.. 1998. Field preservation of marine invertebrate tissue for DNA analyses. Mol Mar Biol Biotechnol. 7:145–152. [PubMed] [Google Scholar]
- Derelle R, Momose T, Manuel M, Da Silva C, Wincker P, et al. 2010. Convergent origins and rapid evolution of spliced leader trans-splicing in Metazoa: insights from the Ctenophora and Hydrozoa. RNA. 16:696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge P, Bafna V, Bansal V.. 2017. HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 27:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández R, Gabaldón T.. 2020. Gene gain and loss across the metazoan tree of life. Nat Ecol Evol. 4:524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman G. 1977. The establishment of the oral-aboral axis in the ctenophore embryo. Development. 42:237–260. [Google Scholar]
- Gaiti F, Calcino AD, Tanurdžić M, Degnan BM.. 2017. Origin and evolution of the metazoan non-coding regulatory genome. Dev Biol. 427:193–202. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv. [q-bio.GN]. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Accorsi A, He S, Guerrero-Hernández C, Sivagnanam S, et al. 2018. An adaptable chromosome preparation methodology for use in invertebrate research organisms. BMC Biol. 16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved Pairwise Alignment of Genomic DNA. The Pennsylvania State University.
- Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, et al. 2018. Transcription elongation can affect genome 3D Structure. Cell. 174:1522–1536.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Keene MA, Fechtel K, Fristrom JW.. 1986. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell. 44:33–42. [DOI] [PubMed] [Google Scholar]
- Hoencamp C, Dudchenko O, Elbatsh AMO, Brahmachari S, Raaijmakers JA, et al. 2021. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science. 372:984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M.. 2016. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 32:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lomsadze A, Borodovsky M, Stanke M.. 2019. Whole-genome annotation with BRAKER. Methods Mol Biol. 1962:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG.. 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 48:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, Francis WR, Rivera-Vicéns RE, Juravel K, de Mendoza A, et al. 2020. Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat Commun. 11:3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve? PLoS Biol. 8:e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML.. 2017. BlobTools: Interrogation of genome assemblies. F1000Res. 6:1287. [Google Scholar]
- Laumer CE, Fernández R, Lemer S, Combosch D, Kocot KM, et al. 2019. Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc Biol Sci. 286:20190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, Meyer WK, Ségurel L, et al. 2012. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 10:e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. [Google Scholar]
- Li H. 2017. Minimap2: pairwise alignment for nucleotide sequences. arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2020. auN: a new metric to measure assembly contiguity. https://lh3.github.io/2020/04/08/a-new-metric-on-assembly-contiguity
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. ; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Gao L, Pan Y, Tian M, Li Y, et al. 2020. Chromosome-level reference genome of the jellyfish Rhopilema esculentum. Gigascience. 9:giaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 27:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Vosshall LB.. 2020. How to turn an organism into a model organism in 10 “easy” steps. J Exp Biol. 223:jeb218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur E, Capra JA.. 2021. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am J Hum Genet. 108:269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroome J, Liang D, Corbett-Detig R.. 2020. Fine-scale position effects shape the distribution of inversion breakpoints in Drosophila melanogaster. Genome Biol Evol. 12:1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills DB, Francis WR, Vargas S, Larsen M, Elemans CP, et al. 2018. The last common ancestor of animals lacked the HIF pathway and respired in low-oxygen environments. eLife. 7:e31176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature. 510:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura O, Hara Y, Kuraku S.. 2017. gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics. 33:3635–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nong W, Cao J, Li Y, Qu Z, Sun J, et al. 2020. Jellyfish genomes reveal distinct homeobox gene clusters and conservation of small RNA processing. Nat Commun. 11:3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patry WL, Bubel MK, Hansen C, Knowles T.. 2019. Diffusion tubes: a method for the mass culture of ctenophores and other pelagic marine invertebrates. bioRxiv. 751099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, et al. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toolkit Picard, 2016. Broad institute, GitHub repository. https://github.com/broadinstitute/picard
- Presnell JS, Browne WE.. 2021. Krüppel-like factor gene function in the ctenophore Mnemiopsis leidyi assessed by CRISPR/Cas9-mediated genome editing. Development. dev.199771. [DOI] [PubMed] [Google Scholar]
- Putnam NH, O'Connell BL, Stites JC, Rice BJ, Blanchette M, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, et al. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC.. 2020. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 11:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice ES, Green RE.. 2019. New approaches for genome assembly and scaffolding. Annu Rev Anim Biosci. 7:17–40. [DOI] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR.. 2018. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 19:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H.. 2019. Fast and accurate long-read assembly with wtdbg2. bioRxiv. 530972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen A-D, Moreland RT, et al. ; NISC Comparative Sequencing Program. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 342:1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C, Louis A, Bon C, Berthelot C, Crollius HR.. 2018. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 19:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saremi NF, Supple MA, Byrne A, Cahill JA, Coutinho LL, et al. 2019. Puma genomes from North and South America provide insights into the genomic consequences of inbreeding. Nat Commun. 10:4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Chomsky E, Pang K, Lara-Astiaso D, Gaiti F, et al. 2018. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat Ecol Evol. 2:1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X-X, Hittinger CT, Rokas A, Minh BQ, Braun EL.. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol. 1:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simion P, Philippe H, Baurain D, Jager M, Richter DJ, et al. 2017. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr Biol. 27:958–967. [DOI] [PubMed] [Google Scholar]
- Simister RL, Schmitt S, Taylor MW.. 2011. Evaluating methods for the preservation and extraction of DNA and RNA for analysis of microbial communities in marine sponges. J Exp Mar Bio Ecol. 397:38–43. [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B.. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 32:W309–W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange O. 2011. Gnu parallel-the command-line power tool. USENIX Magazine. 36:42–47. [Google Scholar]
- Tikhonenkov DV, Hehenberger E, Esaulov AS, Belyakova OI, Mazei YA, et al. 2020. Insights into the origin of metazoan multicellularity from predatory unicellular relatives of animals. BMC Biol. 18:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang L, Lian S, Qin Z, Zhu X, et al. 2020. Evolutionary transcriptomics of metazoan biphasic life cycle supports a single intercalation origin of metazoan larvae. Nat Ecol Evol. 4:725–736. [DOI] [PubMed] [Google Scholar]
- Whelan NV, Kocot KM, Moroz TP, Mukherjee K, Williams P, et al. 2017. Ctenophore relationships and their placement as the sister group to all other animals. Nat Ecol Evol. 1:1737–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G-C, Xu T-J, Zhu R, Zhang Y, Li S-Q, et al. 2019. LR_Gapcloser: a tiling path-based gap closer that uses long reads to complete genome assembly. Gigascience. 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Ma D, Xu M.. 2005. Nested genes in the human genome. Genomics. 86:414–422. [DOI] [PubMed] [Google Scholar]
- Zimmermann B, Robb SMC, Genikhovich G, Fropf WJ, Weilguny L, et al. 2020. Sea anemone genomes reveal ancestral metazoan chromosomal macrosynteny. bioRxiv. 10.30.359448. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequencing reads, transcriptomes, and the genome assembly are available for download via NCBI BioProject PRJNA576068. Hc1 and Hc2 mitochondrial sequences are available through NCBI accessions MN544300 and MN544301. Additional data are available through G3 figshare: https://doi.org/10.25387/g3.15170382. Custom programs used in this publication, and annotation guidelines, are available at github.com/conchoecia/hormiphora and Zenodo DOI: 10.5281/zenodo.4074309. No tissue from these samples is available, as it was consumed during library preparation.