Abstract
The prognosis of pancreatic cancer is poor because patients are usually asymptomatic in the early stage and the early diagnostic rate is low. Therefore, in this study, we aimed to identify potential prognosis-related genes in pancreatic cancer to improve diagnosis and the outcome of patients. The mRNA expression profile data from The Cancer Genome Atlas database and GSE79668, GSE62452, and GSE28735 datasets from Gene Expression Omnibus were downloaded. The prognosis-relevant genes and clinical factors were analyzed using Cox regression analysis and the optimal gene sets were screened using the Cox proportional model. Next, the Kaplan-Meier survival analysis was used to evaluate the relationship between risk grouping and patient prognosis. Finally, an optimal gene-based prognosis prediction model was constructed and validated using a test dataset to discriminate the model accuracy and reliability. The results showed that 325 expression variable genes were identified, and 48 prognosis-relevant genes and three clinical factors, including lymph node stage (pathologic N), new tumor, and targeted molecular therapy were preliminarily obtained. In addition, a gene set containing 16 optimal genes was identified and included FABP6, MAL, KIF19, and REG4, which were significantly associated with the prognosis of pancreatic cancer. Moreover, a prognosis prediction model was constructed and validated to be relatively accurate and reliable. In conclusion, a gene set consisting of 16 prognosis-related genes was identified and a prognosis prediction model was constructed, which is expected to be applicable in the clinical diagnosis and treatment guidance of pancreatic cancer in the future.
Keywords: pancreatic cancer, expression variable genes, prognosis, prognosis prediction model
Introduction
Pancreatic cancer, which is the most fatal malignancy of the digestive system, is highly aggressive with a poor prognosis, leading to considerable morbidity and mortality worldwide and it continues to be a major health challenge (Forster et al. 2020; Nevala-Plagemann et al. 2020). The 5-year survival rate of pancreatic cancer is <5%, and 85% of patients die within 12 months of diagnosis because the asymptomatic early stages generally lead to late detection (Qian et al. 2019). Conventional risk factors such as advanced age, alcohol consumption, tobacco use, obesity, and history of diabetes mellitus and chronic pancreatitis are associated with the etiology of pancreatic cancer (Rawla et al. 2019). In addition, molecular biomarkers play an important role in the development and prognosis of pancreatic cancer. The discovery of effective biomarkers is essential for the detection of pancreatic cancer at an early stage, which may contribute to improving the prognosis and developing new therapeutic strategies.
Recently, numerous abnormally expressed genes have been identified in pancreatic cancer based on a microarray technology, which can detect subsets of genes that could be potential biomarkers for cancer diagnosis (Nonogaki et al. 2010). For instance, AKT serine/threonine kinase 1 (AKT 1) regulates growth factor-induced cell survival and its phosphorylated form (p-Akt1) is involved in the carcinogenesis of pancreatic cancer (Liu et al. 2010). This molecule has been detected at high levels in patients and is correlated with a lower primary tumor size and extent tumor (T) stage, which may be a favorable prognostic factor for pancreatic cancer (Liu et al. 2010). Song et al. (2020) found that protein arginine methyltransferase 1 (PRMT1) promotes pancreatic cancer growth and is predictive of poor prognosis. Saif et al. (2007) reported that P16 deletion may potentially inhibit cyclin D, CDK4, and CDK6 function and regulate cell-cycle progression. Furthermore, P16 deletion is significantly associated with shorter average survival times and acts as a predictive marker for poor prognosis in patients with pancreatic ductal adenocarcinoma (Luo et al. 2013). In addition, many studies suggest that the abnormal expression of growth factors or their receptors may affect cellular functions and the tumorigenicity of pancreatic cancer (Ebert et al. 1995; Gnatenko et al. 2018). It has been reported that high plasma levels of the transforming growth factor (TGF)- β1 in pancreatic cancer patients is associated with advanced stages of the disease and significantly shorter survival times (Javle et al. 2014).
Genes with variable expressions that are related to the prognosis of pancreatic cancer have been identified in many studies. However, screening for new indicators and the construction of prognostic models based on additional prognosis-related genes, appears to be a more sensitive strategy than using signal genes for the early detection of pancreatic cancer. Moreover, the predictive accuracy of these potential prognostic biomarkers has been largely limited by the sample size in previous studies. Therefore, in the present investigation, the mRNA expression profiles extracted from The Cancer Genome Atlas (TCGA) database were used to identify significant prognosis-related genes and clinical factors. Then, a risk prediction model for prognosis was constructed based on optimal genes and clinical factors, and three datasets (GSE79668, GSE62452, and GSE28735) from the Gene Expression Omnibus (GEO) database were downloaded to validate the correctness and reliability of the prognostic prediction models.
Methods
Data sources and data preprocessing
The mRNA-seq expression profile data of pancreatic cancer were downloaded from the TCGA database (https://gdc-portal.nci.nih.gov/), which is based on the Illumina HiSeq 2000 RNA sequencing platform and used as a training dataset in this analysis. A total of 168 samples, including 164 pancreatic cancer tumors and four normal samples, were included in this dataset. Gene expression profiles of GSE79668 (Kirby et al. 2016), GSE62452 (Yang et al. 2016), and GSE28735 (Zhang et al. 2012) were obtained from the National Center for Biotechnology Information (NCBI) GEO database (http://www.ncbi.nlm.nih.gov/geo/) (Barrett et al. 2005). The GSE79668, GSE62452, and GSE28735 profiles were used as three independent validation datasets and included information on 51, 69, and 45 pancreatic cancer tumors, respectively. The distributions of the clinical information for the training and validation datasets are listed in Table 1. The CEL files of the GSE62452 and GSE28375 datasets produced from Affymetrix platform were downloaded and the raw data were processed using the oligo package (version 1.44.0) in R3.4.1 (http://www.bioconductor.org/packages/release/bioc/html/oligo.html) (Parrish and Spencer 2004). This process included background correction using the MAS method, supplementation of missing values using the median method, and quantile normalization (Irizarry et al. 2003). Furthermore, the gene count data of GSE79668 were acquired from the GEO database based on the Illumina HiSeq 2000 RNA sequencing platform (Supplementary File S1, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79668), and the gene transcript sequence counts for all genes across all tissues using GENCODE v. 9.
Table 1.
Distributions of clinical information for training set and testing sets
| TCGA (N = 164) | GSE79668 (N = 51) | GSE62452 (N = 69) | GSE28735 (N = 45) | |
|---|---|---|---|---|
| Age (mean ± SD) | 64.28 ± 11.31 | 64.04 ± 11.57 | — | — |
| Gender (Male/Female/-) | 86/65/13 | 32/19 | — | — |
| Chronic pancreatitis history (Yes/No/-) | 13/110/41 | — | — | — |
| Diabetes history (Yes/No/-) | 32/95/37 | 22/29 | — | — |
| Alcohol (Yes/No/-) | 91/52/21 | — | — | — |
| Tobacco (Never/Reform/Current/-) | 54/54/17/39 | — | — | — |
| New tumor (patients had new tumor event after initial treatment) (Yes/No/-) | 54/89/21 | — | — | — |
| Pathologic_metastasis (M, the defined absence or presence of distant spread or metastases to locations via vascular channels or lymphatics) (M0/M1/-) | 68/2/94 | 48/1 | — | — |
| Pathologic_node (N, the stage of cancer based on the nodes present) (N0/N1/-) | 40/107/17 | 14/37 | — | — |
| Pathologic_tumor (T, the size or contiguous extension of the primary tumor) (T1/T2/T3/T4/-) | 8/16/124/2/14 | 3/12/31/5 | — | — |
| Pathologic_stage (the extent of a cancer, especially whether the disease has spread from the original site to other parts of the body) (I/II/III/IV/-) | 17/128/2/2/14 | — | 4/46/13/6 | — |
| Radiation therapy (Yes/No/-) | 37/102/25 | — | — | — |
| Targeted molecular therapy (Yes/No/-) | 98/45/21 | — | —- | — |
| Dead (Death/Alive/-) | 83/66/15 | 45/6 | 49/16 | 29/13 |
| Overall survival months (mean ± SD/-) | 18.56 ± 15.27 | 26.78 ± 26.12 | 20.21 ± 16.69 | 17.41 ± 12.07 |
“-” indicates relevant information is missing.
Preliminary screening of expression variable genes
The expression variable genes were preliminarily identified based on the coefficient of variation (CV) value, which is a random variable defined as a ratio obtained by comparing the full range, mean deviation, standard deviation, and mean values (Koopmans et al. 1964). Both the dispersion and average value of the variable can affect the CV, which reflects the fluctuations in data size (Koopmans et al. 1964). The mRNAs were obtained after filtering genes with expression median values <1 in the samples retrieved from the TCGA database. The CV value of the gene expression of samples in the TCGA training dataset was calculated using the genefilter package (version 1.58.1) (Gentleman et al. 2009) in R3.4.1 (https://bioconductor.org/packages/release/bioc/html/genefilter. html), based on a threshold CV value >0.7.
Prognosis-related gene and clinical factor identification
After screening for expression variable genes and their corresponding samples, the clinical information was extracted for follow-up analysis. The univariate and multivariate Cox regression analyses were performed to identify significant prognosis-related genes and clinical factors using the R3.4.1 language survival package (version 2.41.3, http://bioconductor.org/packages/survival/) (Wang et al. 2016). Statistical significance was set at P < 0.05.
Following the extraction of the expression values of the variable genes from the TCGA database, a coupled two-way clustering analysis based on centered Pearson’s analysis (Eisen et al. 1998) was conducted to identify similarities among these genes using the pheatmap package (version 1.0.8) in R3.4.1 (Wang et al. 2014) (https://bioconductor.org/packages/release/Bioc/html/pheatmap.html).
Optimal prognostic gene identification
After inputting the matrix of the gene expression values obtained from previous screening steps, the associations between optimal genes and the prognosis of pancreatic cancer were analyzed using the Cox proportional hazards (PHs) model based on L1–penalized regularization in the penalized package (version 0.9.50, http://bioconductor.org/packages/penalized/) in the R3.4.1 language (Tibshirani 1997; Goeman 2010). The optimal parameter “lambda” in the screening model was obtained using 1000 cross-validation likelihood (cvl) cycle calculations.
Risk prediction model construction and validation
Risk prediction model construction based on optimal genes
The risk prediction model of the optimal genes was constructed based on the prognostic coefficient of the Cox-PH generated using L1–penalized regularization and the gene expression levels (clinical factors) in the TCGA training set. The prognosis index (PI) of each sample was calculated using the following formula: PI score = ∑Coefgene/clinical × Expgene/clinical, where Coefgene/clinical represents the coefficient of the identified genes (clinical factors), Expgene/clinical represents the expression level of the genes (clinical factors) in the TCGA training set, and the median PI score was used as the boundary to divide the samples into high- and low-risk groups in the training dataset.
The Kaplan-Meier (KM) survival analysis in the R3.4.1 language survival package (version 2.41.3, http://bioconductor.org/packages/survival/) was used to evaluate the relationship between risk grouping and the overall survival (OS) time of pancreatic cancer patients (Goeman 2010). Finally, three datasets were used to validate the correctness and reliability of the prognostic prediction models using survival and receiver operating characteristic (ROC) curve analyses (Hajian-Tilaki 2013).
Risk prediction model construction based on optimal genes and clinical factors
To identify the significant clinical factors, the Cox-PH model was also constructed again using the clinical information of the samples provided by the TCGA training dataset. We used the same method to construct a risk prediction model for optimal genes and clinical factors and calculated the PI score of each sample. Then, we divided the samples into high- and low-risk groups by setting the median PI score as the dividing point. Similarly, the association between risk grouping and the survival ratio of patients was analyzed again using the survival analysis, and the accuracy of prognosis of the training dataset was evaluated using an ROC curve analysis.
Results
Preliminary screening of variable mRNA
Expression values for 13,238 mRNAs were retained after filtering out genes with median expression values <1 in the TCGA database. Then, 325 expression variable genes from 13,238 mRNAs were obtained using a CV > 0.7 as the threshold.
Screening for prognosis-related genes and clinical factors
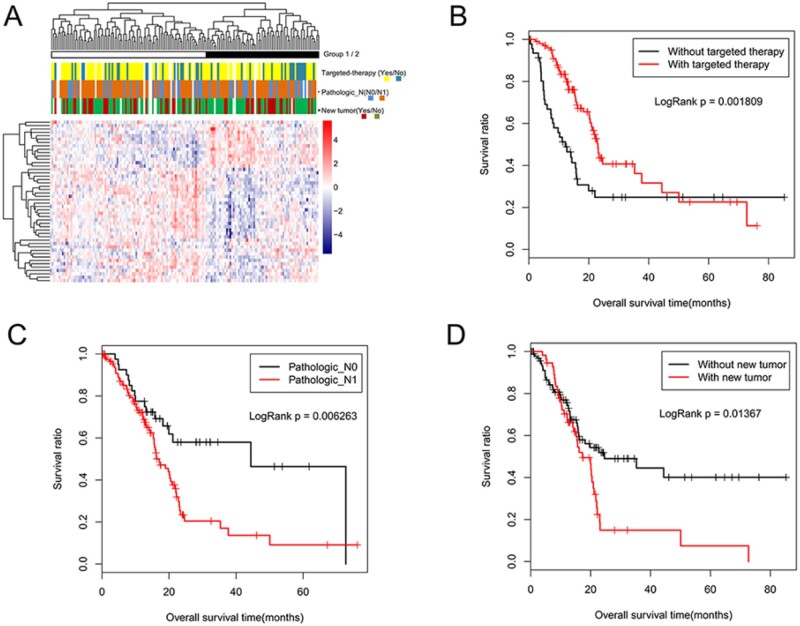
The Cox univariate regression analysis identified 129 prognosis-related genes from 325 expression variable genes. A set of 48 genes significantly associated with the prognosis of pancreatic cancer were identified using the Cox multivariate analysis (Table 2). Similarly, the following three independent prognostic clinical factors were identified: pathologic N (P = 0.05), new tumor (P = 0.01), and targeted molecular therapy (P = 0.00) (Table 3). The survival curve analysis showed that there were statistically significant differences in OS time between patients with and without targeted molecular therapy (logRank P = 0.00, Figure 1B), pathologic N0 compared with pathologic N1 (logRank P = 0.01, Figure 1C), and between patients with and without new tumors (log-rank P = 0.01, Figure 1D).
Table 2.
Prognosis relevant genes obtained via cox univariate and multivariate analysis
| Gene | P-value | Hazard ratio | Lower 0.95 | Upper 0.95 | Gene | P-value | Hazard ratio | Lower 0.95 | Upper 0.95 |
|---|---|---|---|---|---|---|---|---|---|
| HTR3A | 1.24E-06 | 0.426 | 0.30176 | 0.6015 | SFTPA2 | 0.004024 | 1.8497 | 1.21644 | 2.8125 |
| LUZP2 | 2.24E-06 | 2.0719 | 1.5321 | 2.802 | GSTM1 | 0.004456 | 1.1257 | 1.0375 | 1.2215 |
| TRHDE | 1.29E-05 | 0.5313 | 0.39987 | 0.7059 | CRYBA2 | 0.005296 | 1.7281 | 1.1765 | 2.5384 |
| TF | 1.61E-05 | 1.9314 | 1.432 | 2.6049 | MUC17 | 0.006349 | 0.8364 | 0.7358 | 0.9509 |
| KCNMB2 | 2.40E-05 | 7.603 | 2.96625 | 19.4879 | UPK1B | 0.007586 | 1.2045 | 1.0507 | 1.3808 |
| FABP6 | 4.55E-05 | 0.6701 | 0.55282 | 0.8123 | CALY | 0.009702 | 0.5868 | 0.3918 | 0.8789 |
| FAM150A | 0.000116 | 0.4829 | 0.33349 | 0.6993 | NKX2 | 0.010313 | 1.9606 | 1.1721 | 3.2795 |
| DNAH3 | 0.000116 | 0.4031 | 0.25397 | 0.6398 | NXF3 | 0.010715 | 1.3481 | 1.07175 | 1.6958 |
| TMEM63C | 0.000214 | 2.7768 | 1.617 | 4.7685 | MMP3 | 0.012372 | 1.5115 | 1.09353 | 2.0892 |
| CDH19 | 0.000253 | 0.4374 | 0.28086 | 0.6812 | HBA1 | 0.013372 | 0.7897 | 0.65504 | 0.9522 |
| ST18 | 0.000329 | 0.2055 | 0.08663 | 0.4873 | SVOP | 0.015743 | 0.3359 | 0.13858 | 0.8143 |
| REG4 | 0.000523 | 1.3042 | 1.1224 | 1.5153 | HEPACAM2 | 0.016981 | 0.766 | 0.6154 | 0.9534 |
| EGF | 0.000677 | 1.9088 | 1.31479 | 2.7712 | TNNT2 | 0.017516 | 1.5525 | 1.08001 | 2.2316 |
| LRRC24 | 0.001067 | 0.4726 | 0.30162 | 0.7404 | NLRP2 | 0.017973 | 0.8446 | 0.7344 | 0.9714 |
| MAL | 0.001079 | 2.2394 | 1.38108 | 3.6311 | FPR2 | 0.018503 | 0.5301 | 0.31264 | 0.8989 |
| KIF19 | 0.001551 | 1.787 | 1.24737 | 2.5602 | KCNJ3 | 0.022995 | 1.7697 | 1.08191 | 2.8948 |
| CTSG | 0.001583 | 0.465 | 0.28918 | 0.7478 | CCDC141 | 0.030918 | 0.5583 | 0.32888 | 0.9479 |
| RFX6 | 0.001695 | 0.3283 | 0.16376 | 0.6581 | G6PC2 | 0.033274 | 0.6703 | 0.4638 | 0.9688 |
| FCRL5 | 0.001762 | 0.537 | 0.36366 | 0.7928 | BIRC7 | 0.034584 | 1.3956 | 1.02446 | 1.9011 |
| FCGR3B | 0.001964 | 2.2492 | 1.34631 | 3.7576 | KLK8 | 0.03853 | 1.2119 | 1.0102 | 1.4539 |
| CD177 | 0.002024 | 1.4554 | 1.14682 | 1.8471 | PI16 | 0.039332 | 1.2375 | 1.0105 | 1.5155 |
| CCK | 0.002994 | 0.6613 | 0.50323 | 0.8689 | CUZD1 | 0.040752 | 0.691 | 0.4849 | 0.9846 |
| DMRTC1B | 0.003835 | 2.9336 | 1.41439 | 6.0847 | NEUROD1 | 0.045108 | 1.8698 | 1.0137 | 3.4486 |
| FER1L6 | 0.003862 | 0.7896 | 0.6728 | 0.9269 | PSAPL1 | 0.045527 | 1.2431 | 1.00434 | 1.5386 |
Table 3.
Independent prognosis relevant clinical factors obtained via cox univariate and multivariate analysis
| Clinic characteristics | Univariable cox regression |
Multivariable cox regression |
||
|---|---|---|---|---|
| P-value | HR (95% CI) | P-value | HR (95%CI) | |
| Age (Above/Below 60) | 0.1013 | 1.483(0.923–2.385) | — | — |
| Gender (Male/Female) | 0.6876 | 0.915 (0.594–1.409) | — | — |
| Pathologic_M (M0/M1) | 0.3855 | 1.872 (0.444–7.895) | — | — |
| Chronic pancreatitis history (Yes/No) | 0.8403 | 1.079 (0.513–2.269) | — | — |
| Diabetes history (Yes/No) | 0.7064 | 0.896 (0.504–1.591) | — | — |
| Alcohol (Yes/No) | 0.6921 | 1.099 (0.688–1.757) | — | — |
| Tobacco (Never/Reform/Current) | 0.3998 | 1.114 (0.867–1.431) | — | — |
| Pathologic_T (T1/T2/T3/T4) | 0.02309* | 1.696 (1.07–2.689) | 0.6734 | 0.859(0.425–1.738) |
| Pathologic_Stage (I/II/III/IV) | 0.03476* | 1.569 (1.04–2.367) | 0.135 | 1.863(0.824–4.215) |
| Radiation therapy (Yes/No) | 0.01989* | 0.508 (0.284–0.908) | 0.4666 | 0.778(0.398–1.526) |
| Pathologic_N (N0/N1) | 0.006263* | 2.09 (1.218–3.585) | 0.04824* | 1.805(0.927–3.516) |
| New tumor (Yes/No) | 0.01367* | 1.753 (1.116–2.755) | 0.01218* | 1.921(1.153–3.199) |
| Targeted molecular therapy (Yes/No) | 0.001809* | 0.493 (0.313–0.776) | 0.000356* | 0.362(0.207–0.633) |
P < 0.05 indicates statistical significance.
Figure 1.
Screening for prognosis-related genes and clinical factors. (A) Two-way hierarchical clustering analysis of 48 prognosis relevant genes. First row represents cluster 1 (black bars) and 2 (white bars), and rows two to four represent patients treated with targeted molecular therapy (yellow and blue represent those with and without targeted therapy, respectively), pathologic N stage (blue and orange represent N0 and N1, respectively), and new tumor (red and green represent patients with and without new tumors, respectively). (B) OS time according to targeted molecular therapy groups: red and black represent patients with and without targeted therapy, respectively. (C) OS time according to pathologic groups: red and black represent patients with pathologic N1 and N0, respectively. (D) OS time according to new tumor groups: red and black represent patients with and without new tumors.
Hierarchy cluster analysis was conducted according to the similarity of the expression values of 48 independent prognosis-related genes. The results showed that samples in the TCGA database were, clearly, divided into clusters 1 and 2, which contained 97 and 67 pancreatic cancer samples, respectively (Figure 1A). In addition, the distribution of clinical information of the two clusters was determined using the chi-square test, and the results are shown in Table 4. A statistically significant association between new tumors and the formation of clusters was observed (P = 0.02). However, there was no significant correlation between the formation of clusters and any other clinical factors (pathologic N and targeted molecular therapy).
Table 4.
Statistics of clinical factors in clusters 1 and 2 based on clustering analysis of 48 independent prognosis-related genes
| Clinic characteristics | Cluster1 | Cluster2 | X-squared | P-value | ||
|---|---|---|---|---|---|---|
| New tumor (Yes/No) | 22 | 56 | 29 | 31 | 5.0649 | 0.02441* |
| Pathologic_N (N0/N1) | 29 | 54 | 12 | 49 | 3.3096 | 0.06888 |
| Targeted molecular therapy (Yes/No) | 54 | 23 | 39 | 18 | 5.00E-04 | 0.9819 |
P < 0.05 indicates statistical significance.
Screening of optimal prognosis genes
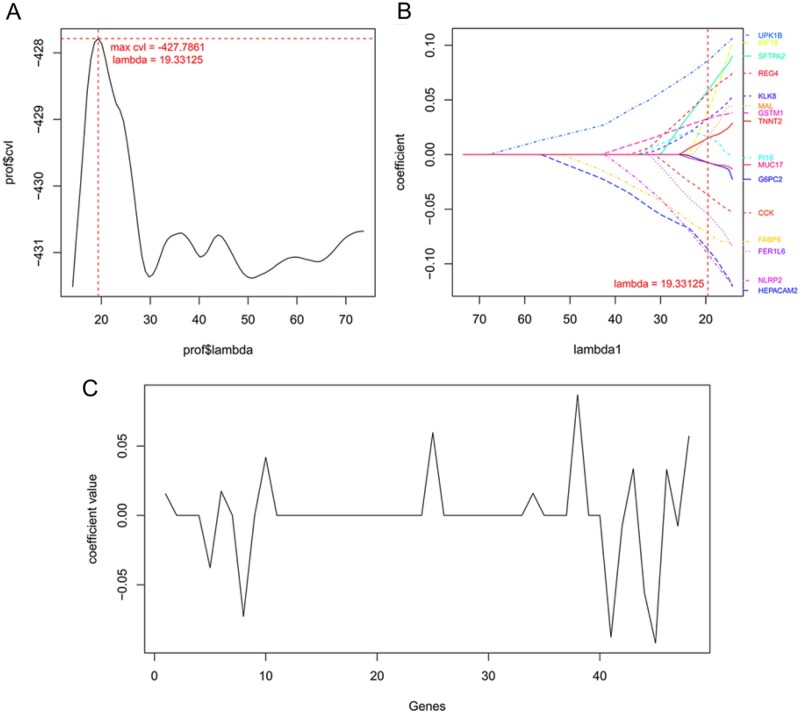
A Cox-PH model was constructed to identify the optimal genes related to the prognosis of pancreatic cancer, and the lambda score (19.33) was obtained from 1000 cvl cycle calculations (Figure 2A). As shown in Table 5, of these 48 genes, 16 optimal genes (CCK, FABP6, FER1L6, G6PC2, GSTM1, HEPACAM2, KIF19, KLK8, MAL, MUC17, NLRP2, PI16, REG4, SFTPA2, TNNT2, and UPK1B) associated with the prognosis of pancreatic cancer were identified (Table 5). Furthermore, the distribution of the gene coefficients is shown in Figure 2, B and C.
Figure 2.
Screening of optimal prognosis genes. (A) Curve for lambda parameter based on cross-validation likelihood screening. Horizontal and vertical axes represent different values for lambda and cross-validation likelihood (cvl), respectively, and the red dotted line indicates lambda = 19.33 when cvl = −427.79. (B) Coefficient of optimal prognosis-related genes based on Cox-PH model and each color represents a different gene. (C) Distribution of gene coefficient. Horizontal and vertical axes represent 48 prognosis-related genes and coefficient of each gene, respectively.
Table 5.
The prognosis of pancreatic cancer relevant genes and clinical factors
| Features | Coef in coxPH | Hazard ratio | P-values |
|---|---|---|---|
| Gene features | |||
| FABP6 | −0.0728 | 0.6701 | 4.55E-05 |
| REG4 | 0.0570 | 1.3042 | 0.000523 |
| MAL | 0.0175 | 2.2394 | 0.001079 |
| KIF19 | 0.0419 | 1.787 | 0.001551 |
| CCK | −0.0377 | 0.6613 | 0.002994 |
| FER1L6 | −0.0557 | 0.7896 | 0.003862 |
| SFTPA2 | 0.0597 | 1.8497 | 0.004024 |
| GSTM1 | 0.0331 | 1.1257 | 0.004456 |
| MUC17 | −0.0076 | 0.8364 | 0.006349 |
| UPK1B | 0.0868 | 1.2045 | 0.007586 |
| HEPACAM2 | −0.0877 | 0.766 | 0.016981 |
| TNNT2 | 0.0155 | 1.5525 | 0.017516 |
| NLRP2 | −0.0920 | 0.8446 | 0.017973 |
| G6PC2 | −0.0075 | 0.6703 | 0.033274 |
| KLK8 | 0.0336 | 1.2119 | 0.03853 |
| FABP6 | −0.0728 | 0.6701 | 4.55E-05 |
| Clinic features | |||
| Pathologic_N | 0.6794 | 1.805 | 0.04824 |
| New tumor | 0.4940 | 1.921 | 0.01218 |
| Targeted molecular therapy | −0.9077 | 0.362 | 0.000356 |
Construction and validation of risk prediction models
Risk prediction model construction based on optimal genes
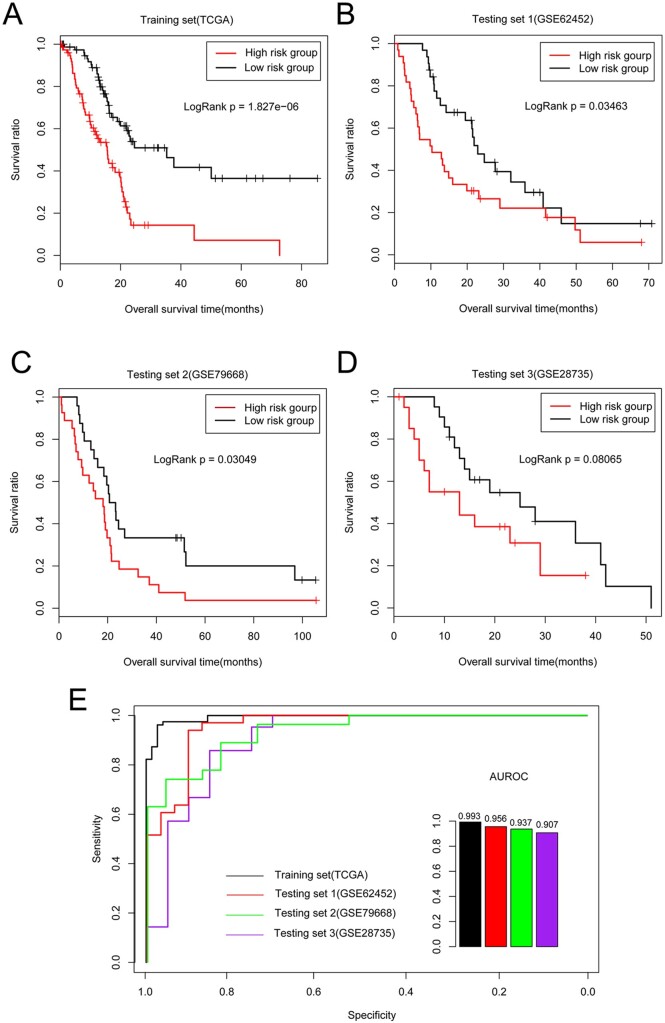
The training dataset included 164 patients who were divided into the high- and low-risk groups according to the median PI score (0.12). The mean OS time was 21.3 ± 17.26 months and 12.42 ± 10.73 months for the low- (82 of 164 samples) and high-risk (82 of 164 samples) groups, respectively according to clinical information. The survival curve analysis revealed a significant difference in OS time between the high- and low-risk groups (P < 0.01, Figure 3A). The area under the ROC (AUROC) curve analysis value was 0.99, indicating that our risk prediction model for prognosis might be relatively accurate and in agreement with real-life conditions in estimating the prognosis based on the samples (Figure 3E).
Figure 3.
Risk prediction model construction based on optimal genes. OS time of patients in (A) TCGA training, (B) GSE62452, (C) GSE79668, and (D) GSE28735 validation datasets assigned to low-risk group (black) compared with high-risk group (red). (E) ROC curve for all datasets based on optimal gene combination. Black, red, green, and purple curves represent TCGA training, GSE62452, GSE79668, and GSE28735 validation datasets, respectively.
Then, the risk model of the optimal genes was further validated using three validation datasets. The patient samples in the GSE62452 dataset were divided into high- and low-risk groups, and 32 patients in the low-risk group showed a longer mean OS time (23.98 ± 16.02 months) than that of the 33 in the high-risk group (16.55 ± 16.73 months). The patients in the GSE79668 dataset were also divided into high- and low-risk groups according to the median PI score (0.81), and the mean OS time of the 25 low-risk patients was higher (34.04 ± 29.63 months) than that of the 26 high-risk patients (20.33 ± 21.06 months). There were significant differences in survival rates between the high- and low-risk pancreatic cancer groups represented in the GSE62452 (Figure 3B) and GSE79668 datasets (Figure 3C, both P = 0.03). In addition, the AUROC curve analysis values for accurate discrimination of the prognosis of pancreatic cancer patients in the GSE62452 and GSE79668 datasets were 0.96 and 0.94, respectively.
The patients in the GSE28735 dataset were also divided into high- and low-risk groups (21 samples each) according to the median PI score (0.44), and they showed mean OS times of 13.14 ± 10.44 and 21.6 ± 12.31 months, respectively. However, no significant correlation between risk grouping and the OS time of the pancreatic cancer patients was observed according to the survival curve analysis of this dataset (P = 0.08, Figure 3D). The AUROC curve value for accurate analysis of the GSE28735 dataset was 0.91.
Risk model construction based on optimal genes and clinical factors
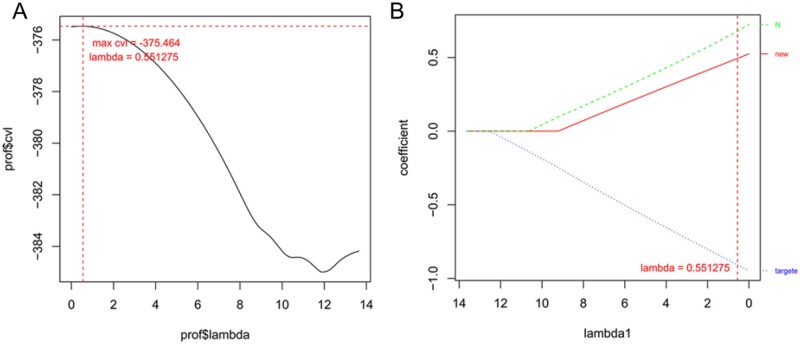
Overall, the following three clinical factors related to the prognosis of pancreatic cancer were identified from the TCGA training dataset: pathologic N, new tumor, and targeted molecular therapy based on the Cox-PH model (Figure 4B), and the results were consistent with those of the Cox univariate and multivariate analyses. The lambda score was 0.55 after 1000 cvl cycle calculations (Figure 4A). The result of the discriminant analysis of the two risk groups (82 samples each) using the clinical information showed that the mean OS time was longer for the low-risk group (19.02 ± 15.02 months) than it was for the high-risk group, which showed a poor prognosis (14.93 ± 11.68 months). The results of the survival curve analysis suggested that risk grouping was significantly associated with the prognosis of pancreatic cancer (P < 0.01, Figure 5A).
Figure 4.
Risk model construction based on optimal genes and clinical factors. (A) Curve for lambda parameter based on cross-validation likelihood (cvl) screening. Horizontal and vertical axes represent different values for lambda and cvl, respectively, and red dotted line indicates lambda = 0.55 when the cvl = −375.46. (B) Distribution of coefficients for prognosis-related clinical factors based on Cox-PH model of the L1–penalized regularized algorithm.
Figure 5.
Validation of correctness and reliability of prognostic prediction models. OS time of patients in TCGA training dataset assigned to high-risk group (black) compared with low-risk group (red) based on (A) clinical factors and (B) combination of optimal genes and clinical factors. (C) ROC curve of prognosis model based on optimal genes and clinical factors.
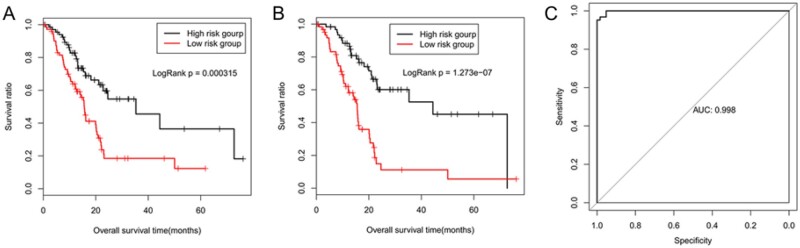
To further estimate the prognosis of pancreatic cancer, a risk model was constructed using the optimal genes and three clinical factors. The mean OS time of the low-risk group was higher (21.81 ± 14.97 months) than that of the high-risk group (13.78 ± 11.5 months, 82 samples each) and the KM curve analysis showed that there was significant difference in survival rate between low and high-risk groups (P < 0.01; Figure 5B), indicating that degrees of risk for these genes and factors were significantly related to the prognosis of pancreatic cancer. In addition, the AUROC value was 0.99 (Figure 5C), reflecting the accuracy and reliability of the prognostic prediction system.
Discussion
A total of 325 genes with high expression variability were preliminarily identified between tumor and normal tissue samples. Furthermore, 48 genes and the following three clinical factors, pathologic N, new tumor, and targeted molecular therapy relevant to prognosis, were screened using data from the TCGA database. In addition, a risk prediction model containing 16 optimal genes related to prognosis was constructed and validated to be relatively accurate and reliable according to the results of the KM curve analysis of the GSE79668 and GSE62452 validation datasets. Among these genes, FABP6, MAL, KIF19, and REG4 were significantly correlated with the prognosis of pancreatic cancer and are expected to be further applied in the clinical diagnosis of pancreatic cancer and treatment guidance.
The most significant prognosis-related gene in our data was the fatty acid binding protein 6 (FABP6), which is a highly conserved cytoplasmic protein that binds bile acids and participates in the metabolism of enterohepatic bile acid (Fisher et al. 2009). Bile acids have been reported to regulate cell growth and proliferation, and abnormal levels are implicated in hepatic inflammation and tumorigenesis (Li and Apte 2015). Thus, genes associated with bile acids are also likely to be involved in cancer pathogenesis. Ohmachi et al. (2006) demonstrated higher expression levels of FABP6 mRNA in colorectal cancer tissues than in adenomas and metastatic lymph nodes, and the tumor size was smaller with high expression levels of FABP6. In addition, colorectal cancer cells transfected with FABP6 showed weaker invasiveness and lower levels of apoptosis than mock cells did (Ohmachi et al. 2006). Fang et al. (2007) reported that a novel transcript of FABP6 may protect colon cancer cells from apoptosis through activation of the nuclear factor (NF)-κB pathway. In this study, we found that FABP6 may act as a favorable biomarker for the prognosis of pancreatic cancer according to the multivariate Cox PH regression analysis. Further comprehensive investigations of the mechanism mediating the involvement of FABP6 in pancreatic cancer development are needed and will be included in our future work.
The Mal, T cell differentiation protein (MAL) is mainly localized in the endoplasmic reticulum and plasma membrane of T cells, and regulates T cell differentiation (Rancaño et al. 1994). MAL has been reported as a tumor suppressor gene, and its increased expression may be implicated in the reducing the pathogenesis of head and neck squamous cell carcinoma (HNSCC) via suppression of cell proliferation, invasiveness, and tumor growth (Cao et al. 2010). In addition, MAL expression is correlated with prolonged disease-free survival in breast and gastric cancers (Buffart et al. 2008; Horne et al. 2009). However, the predictive accuracy of MAL in cancer prognosis remains controversial. For instance, upregulated MAL mRNA levels were significantly associated with shorter OS and progression-free survival in a cohort of high-grade serious epithelial ovarian carcinoma patients (Zanotti et al. 2017). This previous finding is consistent with our observation that MAL, which has high expression variability, was identified as one of the optimal genes related to the poor prognosis of pancreatic cancer. Our observation suggests that MAL might play an important role in the etiology of pancreatic cancer.
Kinesin superfamily proteins (KIFs) facilitate the transport of mRNAs, protein complexes, and organelles in an ATP- and microtubule-dependent manner (Brendza et al. 2000). KIFs are essential for mitosis and meiosis (Vicente and Wordeman 2015) and any abnormalities in mitosis cause cell death, gene deletion, and even induce carcinogenesis (Yu and Feng 2010; Zhu et al. 2014). Thus, the detection of abnormal kinesin protein expression could be utilized as a biomarker for early tumor diagnosis or to predict the survival of patients with tumors. Numerous studies have reported that altered expression of kinesins is related to the development and progression of various human tumors (Corson et al. 2007; Taniwaki et al. 2007;Lukong and Richard 2008). For instance, altered KIF14 mRNA expression has been shown to be a prognostic indicator for patients with breast and lung cancers (Corson and Gallie 2006). KIFC1 is essential for the viability of certain extra-centrosome-containing cancer cells (Cai et al. 2009). Chen et al. (2017) reported that KIF19 is significantly associated with the prognosis of hepatocellular carcinoma. The results showed that KIF19 expression was significantly correlated with the prognosis of pancreatic cancer, which implies it has a role in carcinogenesis and may have clinical value as a biomarker.
Another significantly poor prognosis-related gene identified in our data was regenerating family member 4 (REG4), a member of the regenerating (REG) islet-derived family of proteins. REG4 has been demonstrated to be highly expressed in ovarian (Lehtinen et al. 2016), colorectal (Zhu et al. 2015), rectal (He et al. 2014), and gastric (Miyagawa et al. 2008) cancers, and acts as a predictive biomarker for early diagnosis and prognosis. High serum REG4 levels in pancreatic ductal adenocarcinoma at tumor stages IA–IIA are associated with a worse survival rate than that of patients with grade I tumors (Saukkonen et al. 2018). The REG4-induced epidermal growth factor receptor (EGFR)/AKT/cAMP-response element binding protein (CREB) signaling pathway is involved in macrophage polarization to the M2 phenotype, which may promote pancreatic cancer cell proliferation and invasion, and regulate the invasion of extra-pancreatic and lymph vessels (Ma et al. 2016). In addition, a study reported that REG4 promotes the proliferation and invasiveness of pancreatic cancer cells by upregulating the invasion-related genes matrix metalloproteinase 7 (MMP-7) and MMP-9 (He et al. 2012). These findings suggest that REG4 may serve as a useful biomarker for the prognosis of pancreatic cancer.
In addition, the three clinical factors, pathologic N, new tumor, and targeted molecular therapy relevant to the prognosis of pancreatic cancer were screened in this study. In addition, a prognosis prediction model based on optimal genes was constructed, which was verified in the independent validation datasets GSE79668 and GSE62452. However, the model was not verified in the GSE28735 validation dataset (P = 0.08), which might have been due to the limited dataset sample size. Furthermore, a prediction model based on optimal genes and clinical factors was also constructed, and a significant difference was observed in the survival ratio between the high- and low-risk groups, and the AUROC value was 0.99, indicating the correctness and reliability of our prognostic prediction models.
This study used three datasets to verify the prediction model. However, this study had some limitations that are worth mentioning. First, although the trend of the result is consistent, no significant correlation between risk grouping and the OS time of pancreatic cancer was observed according to KM survival curve analysis in GSE28735 dataset. In addition, the comprehensive model was not validated because the clinical information on the pathologic N, new tumor, and targeted molecular therapy were not included in the three validation datasets at the same time. Therefore, more comprehensive data validation needs to be conducted in future studies. In addition, relevant experiments need to be performed to verify the multiple candidate targets identified in this study.
Conclusions
In summary, in this study, 325 expression variable genes and a gene set consisting of 16 prognosis-related genes were identified from data retrieved from the TCGA database. The risk prediction model for prognosis based on optimal genes was validated to be relatively accurate and reliable in the GSE79668 and GSE62452 validation datasets according to the results of the KM curve analysis. Among these genes, FABP6, MAL, KIF19, and REG4 were significantly associated with the prognosis of pancreatic cancer and can expected to be further applied in the clinical diagnosis of pancreatic cancer and in the guidance of its treatment.
Data availability
The data used to support the findings of this study are available from the public databases, including TCGA database (https://gdc-portal.nci.nih.gov/), GEO database (http://www.ncbi.nlm.nih.gov/geo/, containing datasets of GSE79668, GSE62452, GSE28735). Supplementary material is available at figshare: https://doi.org/10.25387/g3.15104160.
Acknowledgments
Conception and design of the research: Y.G., M.Q., and Z.C.; acquisition of data: H.S. and X.Z.; analysis and interpretation of data: H.S. and X.Z.; statistical analysis: H.S. and X.Z.; obtaining funding: Y.G. and M.Q.; drafting the manuscript: Z.C.; revision of the manuscript for important intellectual content: Y.G. and M.Q. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81972280 and 81972290), the “Rising Stars of Medical Talent” Youth Development Program (Grant No. yyxx201901), and the Outstanding Clinical Discipline Project of Shanghai Pudong (Grant No. PWYgy2018-02).
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, et al. 2005. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 33:D562–D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Duffy JB, Saxton WM.. 2000. A function for kinesin I in the posterior transport of Oskar mRNA and Staufen protein. Science. 289:2120–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffart TE, Overmeer RM, Steenbergen RD, Tijssen M, van Grieken NC, et al. 2008. MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. Br J Cancer. 99:1802–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Weaver LN, Ems-McClung SC, Walczak CE.. 2009. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell. 20:1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Zhang ZY, Xu Q, Sun Q, Yan M, et al. 2010. Epigenetic silencing of MAL, a putative tumor suppressor gene, can contribute to human epithelium cell carcinoma. Mol Cancer. 9:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li S, Zhou S, Cao S, Lou Y, et al. 2017. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther. 13:651–659. [DOI] [PubMed] [Google Scholar]
- Corson TW, Gallie BL.. 2006. KIF14 mRNA expression is a predictor of grade and outcome in breast cancer. Int J Cancer. 119:1088–1094. [DOI] [PubMed] [Google Scholar]
- Corson TW, Zhu CQ, Lau SK, Shepherd FA, Tsao MS, et al. 2007. KIF14 messenger RNA expression is independently prognostic for outcome in lung cancer. Clin Cancer Res. 13:3229–3234. [DOI] [PubMed] [Google Scholar]
- Ebert M, Yokoyama M, Friess H, Kobrin MS, Büchler MW, et al. 1995. Induction of platelet-derived growth factor A and B chains and over-expression of their receptors in human pancreatic cancer. Int J Cancer. 62:529–535. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D.. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Dean J, Smith JW.. 2007. A novel variant of ileal bile acid binding protein is up-regulated through nuclear factor-kappaB activation in colorectal adenocarcinoma. Cancer Res. 67:9039–9046. [DOI] [PubMed] [Google Scholar]
- Fisher E, Grallert H, Klapper M, Pfäfflin A, Schrezenmeir J, et al. 2009. Evidence for the Thr79Met polymorphism of the ileal fatty acid binding protein (FABP6) to be associated with type 2 diabetes in obese individuals. Mol Genet Metab. 98:400–405. [DOI] [PubMed] [Google Scholar]
- Forster T, Huettner FJ, Springfeld C, Loehr M, Kalkum E, et al. 2020. Cetuximab in pancreatic cancer therapy: a systematic review and meta-analysis. Oncology. 98:53–60. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Huber W, Hahne F.. 2009. Genefilter: Methods for Filtering Genes from Microarray Experiments. R Package Version 1.24.2., Bioconductor 2.6. http://bioc.ism.ac.jp/2.6/bioc/html/genefilter.html. [Google Scholar]
- Gnatenko DA, Kopantzev EP, Sverdlov ED.. 2018. Variable effects of growth factors on developmental gene expression in pancreatic cancer cells. Dokl Biochem Biophys. 481:217–218. [DOI] [PubMed] [Google Scholar]
- Goeman JJ. 2010. L1 penalized estimation in the Cox proportional hazards model. Biom J. 52:70–84. [DOI] [PubMed] [Google Scholar]
- Hajian-Tilaki K. 2013. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 4:627–635. [PMC free article] [PubMed] [Google Scholar]
- He HL, Lee YE, Shiue YL, Lee SW, Lin LC, et al. 2014. Overexpression of REG4 confers an independent negative prognosticator in rectal cancers receiving concurrent chemoradiotherapy. J Surg Oncol. 110:1002–1010. [DOI] [PubMed] [Google Scholar]
- He XJ, Jiang XT, Ma YY, Xia YJ, Wang HJ, et al. 2012. REG4 contributes to the invasiveness of pancreatic cancer by upregulating MMP-7 and MMP-9. Cancer Sci. 103:2082–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne HN, Lee PS, Murphy SK, Alonso MA, Olson JA Jr, et al. 2009. Inactivation of the MAL gene in breast cancer is a common event that predicts benefit from adjuvant chemotherapy. Mol Cancer Res. 7:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4:249–264. [DOI] [PubMed] [Google Scholar]
- Javle M, Li Y, Tan D, Dong X, Chang P, et al. 2014. Biomarkers of TGF-β signaling pathway and prognosis of pancreatic cancer. PLoS One. 9:e85942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby MK, Ramaker RC, Gertz J, Davis NS, Johnston BE, et al. 2016. RNA sequencing of pancreatic adenocarcinoma tumors yields novel expression patterns associated with long-term survival and reveals a role for ANGPTL4. Mol Oncol. 10:1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans LH, Owen DB, Rosenblatt JI.. 1964. Confidence intervals for the coefficient of variation for the normal and log normal distributions. Biometrika. 51:25–32. [Google Scholar]
- Lehtinen L, Vesterkvist P, Roering P, Korpela T, Hattara L, et al. 2016. REG4 is highly expressed in mucinous ovarian cancer: a potential novel serum biomarker. PLoS One. 11:e0151590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Apte U.. 2015. Bile acid metabolism and signaling in cholestasis, inflammation, and cancer. Adv Pharmacol. 74:263–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng Sun SH, Sun SJ, Huang C, Hu HH, et al. 2010. Phosph-Akt1 expression is associated with a favourable prognosis in pancreatic cancer. Ann Acad Med Singap. 39:548–547. [PubMed] [Google Scholar]
- Lukong KE, Richard S.. 2008. Breast tumor kinase BRK requires kinesin-2 subunit KAP3A in modulation of cell migration. Cell Signal. 20:432–442. [DOI] [PubMed] [Google Scholar]
- Luo Y, Tian L, Feng Y, Yi M, Chen X, et al. 2013. The predictive role of p16 deletion, p53 deletion, and polysomy 9 and 17 in pancreatic ductal adenocarcinoma. Pathol Oncol Res. 19:35–40. [DOI] [PubMed] [Google Scholar]
- Ma X, Wu D, Zhou S, Wan F, Liu H, et al. 2016. The pancreatic cancer secreted REG4 promotes macrophage polarization to M2 through EGFR/AKT/CREB pathway. Oncol Rep. 35:189–196. [DOI] [PubMed] [Google Scholar]
- Miyagawa K, Sakakura C, Nakashima S, Yoshikawa T, Fukuda K, et al. 2008. Overexpression of RegIV in peritoneal dissemination of gastric cancer and its potential as A novel marker for the detection of peritoneal micrometastasis. Anticancer Res. 28:1169–1179. [PubMed] [Google Scholar]
- Nevala-Plagemann C, Hidalgo M, Garrido-Laguna I.. 2020. From state-of-the-art treatments to novel therapies for advanced-stage pancreatic cancer. Nat Rev Clin Oncol. 17:108–123. [DOI] [PubMed] [Google Scholar]
- Nonogaki K, Itoh A, Kawashima H, Ohno E, Ishikawa T, et al. 2010. A preliminary result of three-dimensional microarray technology to gene analysis with endoscopic ultrasound-guided fine-needle aspiration specimens and pancreatic juices. J Exp Clin Cancer Res. 29:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmachi T, Inoue H, Mimori K, Tanaka F, Sasaki A, et al. 2006. Fatty acid binding protein 6 is overexpressed in colorectal cancer. Clin Cancer Res. 12:5090–5095. [DOI] [PubMed] [Google Scholar]
- Parrish RS, Spencer HJ III. 2004. Effect of normalization on significance testing for oligonucleotide microarrays. J Biopharm Stat. 14:575–589. [DOI] [PubMed] [Google Scholar]
- Qian L, Li Q, Baryeh K, Qiu W, Li K, et al. 2019. Biosensors for early diagnosis of pancreatic cancer: a review. Transl Res. 213:67–89. [DOI] [PubMed] [Google Scholar]
- Rancaño C, Rubio T, Correas I, Alonso MA.. 1994. Genomic structure and subcellular localization of MAL, a human T-cell-specific proteolipid protein. J Biol Chem. 269:8159–8164. [PubMed] [Google Scholar]
- Rawla P, Sunkara T, Gaduputi V.. 2019. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 10:10–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif MW, Karapanagiotou L, Syrigos K.. 2007. Genetic alterations in pancreatic cancer. World J Gastroenterol. 13:4423–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saukkonen K, Hagström J, Mustonen H, Lehtinen L, Carpen O, et al. 2018. Prognostic and diagnostic value of REG4 serum and tissue expression in pancreatic ductal adenocarcinoma. Tumour Biol. 40:1010428318761494. [DOI] [PubMed] [Google Scholar]
- Song C, Chen T, He L, Ma N, Li JA, et al. 2020. PRMT1 promotes pancreatic cancer growth and predicts poor prognosis. Cell Oncol (Dordr). 43:51–62. [DOI] [PubMed] [Google Scholar]
- Taniwaki M, Takano A, Ishikawa N, Yasui W, Inai K, et al. 2007. Activation of KIF4A as a prognostic biomarker and therapeutic target for lung cancer. Clin Cancer Res. 13:6624–6631. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. 1997. The lasso method for variable selection in the Cox model. Statist Med. 16:385–395. [DOI] [PubMed] [Google Scholar]
- Vicente JJ, Wordeman L.. 2015. Mitosis, microtubule dynamics and the evolution of kinesins. Exp Cell Res. 334:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Cao C, Ma Q, Zeng Q, Wang H, et al. 2014. RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 14:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Wang Y, Hang B, Zou X, Mao JH.. 2016. A novel gene expression-based prognostic scoring system to predict survival in gastric cancer. Oncotarget. 7:55343–55351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, He P, Wang J, Schetter A, Tang W, et al. 2016. A novel MIF signaling pathway drives the malignant character of pancreatic cancer by targeting NR3C2. Cancer Res. 76:3838–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Feng YM.. 2010. The role of kinesin family proteins in tumorigenesis and progression: potential biomarkers and molecular targets for cancer therapy. Cancer. 116:5150–5160. [DOI] [PubMed] [Google Scholar]
- Zanotti L, Romani C, Tassone L, Todeschini P, Tassi RA, et al. 2017. MAL gene overexpression as a marker of high-grade serous ovarian carcinoma stem-like cells that predicts chemoresistance and poor prognosis. BMC Cancer. 17:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, et al. 2012. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 7:e31507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Han Y, Yuan C, Tu W, Qiu G, et al. 2015. Overexpression of Reg4, alone or combined with MMP-7 overexpression, is predictive of poor prognosis in colorectal cancer. Oncol Rep. 33:320–328. [DOI] [PubMed] [Google Scholar]
- Zhu X, Mei J, Wang Z.. 2014. Aurora-A kinase: potential tumor marker of osteosarcoma. J Cancer Res Ther. 10 (Suppl.):C102–C107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the public databases, including TCGA database (https://gdc-portal.nci.nih.gov/), GEO database (http://www.ncbi.nlm.nih.gov/geo/, containing datasets of GSE79668, GSE62452, GSE28735). Supplementary material is available at figshare: https://doi.org/10.25387/g3.15104160.