Abstract
We have investigated the contribution of specific TATA-binding protein (TBP)–TATA interactions to the promoter activity of a constitutively expressed silkworm tRNACAla gene and have also asked whether the lack of similar interactions accounts for the low promoter activity of a silk gland-specific tRNASGAla gene. We compared TBP binding, TFIIIB-promoter complex stability (measured by heparin resistance), and in vitro transcriptional activity in a series of mutant tRNACAla promoters and found that specific TBP-TATA contacts are important for TFIIIB-promoter interaction and for transcriptional activity. Although the wild-type tRNACAla promoter contains two functional TBP binding sequences that overlap, the tRNASGAla promoter lacks any TBP binding site in the corresponding region. This feature appears to account for the inefficiency of the tRNASGAla promoter since provision of either of the wild-type TATA sequences derived from the tRNACAla promoter confers robust transcriptional activity. Transcriptional impairment of the wild-type tRNASGAla gene is not due to reduced incorporation of TBP into transcription complexes since both the tRNACAla and tRNASGAla promoters form transcription complexes that contain the same amount of TBP. Thus, the deleterious consequences of the lack of appropriate TBP-TATA contacts in the tRNASGAla promoter must come from failure to incorporate some other essential transcription factor(s) or to stabilize the complete complex in an active conformation.
The silkworm Bombyx mori provides a clear example of regulated tRNA gene expression. The demand for fibroin, the principal protein of silk, requires highly efficient transcription and translation of the fibroin gene in cells of the silk gland. Translational efficiency in these cells is maximized by the quantitative adaptation of the tRNA population to the composition of fibroin: 44% glycine, 29% alanine, and 12% serine (5, 29, 30, 42). In the case of tRNAAla, enrichment is achieved both by increasing the level of the constitutive type of tRNAAla (tRNACAla) and by synthesizing an additional, silk gland-specific, type (tRNASGAla) (31, 44). In vitro studies of representative tRNACAla and tRNASGAla genes have revealed transcription properties consistent with the patterns of tRNACAla and tRNASGAla accumulation in vivo. That is, in a variety of extracts from non-silk gland cells, the tRNACAla gene directs transcription much more efficiently than does the tRNASGAla gene, but in concentrated extracts from silk gland, the two genes are equally efficient (60). To understand how these two genes are differentially regulated, we have investigated the basis of the transcriptional impairment of the tRNASGAla gene that is observed under typical in vitro conditions.
Transcription of silkworm tRNAAla genes is driven by both internal and external promoter elements (26, 36, 56). The critical difference between the tRNACAla and tRNASGAla genes is in the interaction of their 5′ flanking promoter elements with the transcription factor complex, TFIIIB (47). Although the tRNASGAla gene can direct the addition of TFIIIB to a TFIIIC/D-promoter complex, as judged by band shift, upstream extension of the TFIIIC/D footprint is not observed when TFIIIB binds the tRNASGAla promoter as it is when TFIIIB binds the tRNACAla promoter (59). TFIIIB is a multiprotein complex containing a general transcription factor, TATA-binding protein (TBP), and its RNA polymerase III (Pol III)-specific associated factors (reviewed in references 10, 41, and 54). In yeast, TFIIIB is thought to consist of three components, TBP, BRF, and B" (19, 38). In higher eukaryotes, the human homologue of BRF has been cloned and shown to function with TBP in transcription of VAI, tRNA, and 5S RNA genes (32, 53), and a putative Drosophila BRF homologue has been identified by association with TBP (50). No homologue of yeast B" has yet been identified in another system.
TFIIIB is the initiation factor for Pol III (18) and is analogous to the Pol II-specific TBP-containing factor, TFIID. The means by which these two factors are recruited to promoters are not the same, however. TFIID is recruited to classical, TATA-containing, Pol II promoters by direct interaction between the TBP subunit and an upstream TATA element, and the promoter activity in such cases is strongly affected by mutation of the TATA element (57). The mechanism of TFIIIB association with Pol III promoters that drive 5S or tRNA transcription is not so well understood, largely because these promoters typically lack an obvious TBP binding site. It has been suggested, therefore, that TBP either contacts such Pol III templates through interactions that are not sequence specific or is incorporated into the transcription complex entirely through protein-protein interactions (46). This interpretation is supported by the fact that a mutation in the DNA binding domain of yeast TBP that renders it deficient for Pol II transcription does not impair either 5S or tRNA transcription (40).
On the other hand, silkworm Pol III promoters frequently contain AT-rich sequences that resemble TBP binding sites (36), although they vary in sequence and location with respect to the transcription initiation site. To determine whether the different abilities of tRNACAla and tRNASGAla promoters to interact with TFIIIB result from differences in their interaction with TBP, we analyzed the effects of promoter mutation on TBP binding, on the stability of TFIIIB-promoter complexes, and on transcriptional activity. For the tRNACAla promoter, we find that TBP binds within an AT stretch between −32 and −23, and optimal interaction increases both TFIIIB-promoter complex stability and transcriptional activity. In contrast, although the wild-type tRNASGAla promoter binds TBP with reasonable affinity, the TBP binding site is not optimally positioned and does not stabilize the TFIIIB-promoter complex or contribute to promoter activity. Our results indicate that specific TBP-TATA contact is a key determinant of tRNACAla gene promoter strength and that the lack of this contact at the required location explains the relative inefficiency of the tRNASGAla promoter.
MATERIALS AND METHODS
Cloned genes used in this work.
The wild-type tRNACAla and tRNASGAla promoter constructs were described previously (59), and mutants were created either by PCR with mutagenic primers or by recombinant PCR (11).
Recombinant silkworm TBP.
Recombinant silkworm TBP was expressed in Escherichia coli and purified as previously described (35).
Crude extracts and Pol III transcription fractions.
Transcription was catalyzed by extracts derived from B. mori ovaries (33). Fractionated transcription machinery was derived from silk gland extracts. Pol III was fractionated as described in reference 34, and TFIIIC/D was fractionated as described in reference 47. TFIIIB was isolated from the 300 mM KCl elution step from the heparin-Sepharose column used to isolate Pol III, and fractions containing TFIIIB activity were pooled and dialyzed against buffer H containing 50 mM KCl.
Assays. (i) TBP-TATA binding.
Binding reactions and analysis by gel retardation were performed as previously described (35). For competition assays, the DNA fragments used were PCR amplified products containing either the adenovirus major late promoter (Ad MLP) (145 bp) or derivatives of the tRNACAla (131 bp, extending from −91 to +40) or the tRNASGAla (138 bp, extending from −98 to +40) promoter. In each reaction mixture, radioactively labeled Ad MLP fragment at a concentration of 1 nM was incubated with increasing amounts of nonradioactive competitor fragments in the presence of an amount of recombinant TBP (8 nM total protein based on Bradford assay) that was found empirically to be limiting. From at least three experiments, the data points obtained in the presence of competitor were normalized to the points obtained in the absence of competitor, and the mean and standard deviation for each point were calculated. The most probable straight line (based on a least squares fit) was drawn through the data points plotted as the reciprocal of the fraction bound versus the molar ratio of competitor to labeled fragment. Relative affinity of different competitors was then obtained from the relative slopes of these lines.
(ii) Heparin-resistant TFIIIB-promoter complexes.
The interaction between TFIIIB and promoter DNA was examined by quantitating heparin-resistant TFIIIB-promoter complexes. A 262-bp tRNAAla gene-containing DNA fragment (extending from −91 to +171) was amplified by PCR, using radioactively labeled primers. A transcription factor complex was assembled on this fragment in a 20-μl reaction mixture that contained 5 fmol of labeled DNA, 4 μg of dG-dC, 5 μl of TFIIIC/D, and, if included, 7.5 μl of TFIIIB. The final concentrations of buffer components were 70 mM KCl, 30 mM Tris-HCl (pH 7.5), 4 mM MgCl2, 13% glycerol, and 3 mM dithiothreitol. After incubation for 1 h, the complex was separated from unbound components by native gel electrophoresis as described in reference 59. Components other than TFIIIB were removed by adding heparin to a final concentration of 100 ng/μl for 20 to 30 s before loading the gel.
(iii) Quantitative binding analysis.
Band shift gels were scanned on a Storm 860 PhosphorImager at 200-μm resolution and analyzed by ImageQuant v1.1 software (Molecular Dynamics). The number of heparin-resistant TFIIIB-promoter complexes formed on each template was first normalized to the number of unstripped complexes (TFIIIB/C/D-promoter complexes) formed on the same template and then expressed as a percentage of the number of heparin-resistant complexes on a wild-type tRNACAla promoter measured in the same experiment.
(iv) Footprinting.
Footprints of bound transcription factors were obtained by DNase I as described previously (59) or hydroxyl radical cleavage (4).
(v) In vitro transcription.
Transcription reactions were performed as previously described (36). Each 20-μl reaction mixture contained 5 μl of oocyte extract, 2 ng of gene-containing plasmid, and nonspecific DNA (pUC13M [36]) to bring the total amount of DNA to 200 ng. Transcripts were detected autoradiographically after resolution by gel electrophoresis as described elsewhere (33).
(vi) Transcription complex isolation and Western blotting.
Binding reaction mixtures assembled and incubated as described above for heparin-resistant TFIIIB-promoter complexes were loaded onto a 1.5% agarose gel in 50 mM Tris-borate (pH 8.0)–5 mM EDTA–5% glycerol. To optimize resolution, the agarose gel was prepared in a vertical apparatus containing a layer of 5% acrylamide at the bottom to retain the agarose. The gel was prerun at 150 V for 60 min, and the samples were fractionated at 150 V for 60 to 90 min. Band shift complexes were detected autoradiographically and excised from the gel. Agarose was removed by digestion with Gelase (Epicentre Technologies) at 42°C overnight according to the manufacturer's directions. The samples were concentrated by precipitation with 10% trichloroacetic acid (Sigma) and resuspended in the sodium dodecyl sulfate-containing sample buffer used for standard protein gels (39). The amount of complex was standardized by quantitating the labeled DNA fragment, and proteins in the complexes were resolved by sodium dodecyl sulfate–11% polyacrylamide gel electrophoresis for Western analysis. Generally 2 to 3 fmol of complex was obtained from a standard 20 μl band shift reaction. Western analysis was performed by using a Bio-Rad modular mini-protein II system following the standard procedure (39). TBP was detected by incubating with antibodies against silkworm TBP, followed by chemiluminescence detection of goat anti-rabbit immunoglobulin G (heavy plus light chain)–horseradish peroxidase conjugate (Bio-Rad) (51).
RESULTS
Recombinant silkworm TBP is able to bind both tRNACAla and tRNASGAla upstream promoters but at different positions.
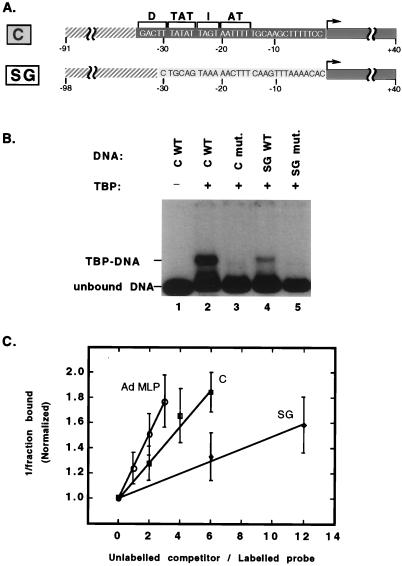
To understand the role of TBP-TATA contacts in tRNACAla and tRNASGAla upstream promoters, we first asked whether silkworm TBP by itself is able to bind these AT-rich, but different, sequences (Fig. 1A). Band shift assays revealed that recombinant silkworm TBP binds both wild-type promoters (Fig. 1B, lanes 2 and 4). To determine the relative affinities of tRNACAla and tRNASGAla promoters for TBP, we compared the ability of these promoters to compete with the Ad MLP TATA element for TBP binding. Silkworm TBP was previously determined to bind Ad MLP with the same affinity as does yeast TBP (2 × 109 M−1) (12, 35). As shown in Fig. 1C, silkworm TBP binds both tRNA promoters with high affinity, about twofold (tRNACAla) and sixfold (tRNASGAla) below its affinity for Ad MLP.
FIG. 1.
Cloned silkworm TBP binds both tRNACAla
and tRNASGAla promoters. (A) Diagram of the DNA
fragments used for TBP binding assays. The tRNACAla and
tRNASGAla promoters are designated as C and SG,
respectively, in all figures. Vector
(
) and
wild-type tRNACAla
( ) or
tRNASGAla
(
) or
tRNASGAla
( ) DNA
are shown. Upstream promoter sequences are in capital letters, and
positions relative to the transcription initiation site (arrow) are
numbered. The D (distal), TAT (TATAT), I (intervening), and AT (AATTTT)
regions of the tRNACAla promoter are delineated by
brackets at the top. (B) Cloned silkworm TBP binds specifically to both
tRNACAla and tRNASGAla promoters.
Silkworm TBP (5 nM) was incubated with radioactively labeled wild-type
(C WT; SG WT) or mutant tRNAAla (C mut.,
−29CGGC−26; SG mut.,
−10CGGC−7) promoter fragments (0.2 nM), and
the complexes were resolved on a nondenaturing gel. The locations of
the mutations on both promoters are within the protected sequences
bracketed in Fig. 2. The positions of TBP-bound fragments (TBP-DNA) and
unbound fragments (unbound DNA) are marked on the left. (C) Relative
affinities of TBP for the Ad MLP and the tRNACAla and
tRNASGAla promoters, determined by competition band
shift assays. Silkworm TBP and radioactively labeled Ad MLP fragments
were incubated with increasing amounts of unlabeled DNA fragments
corresponding to the Ad MLP or the tRNAAla promoters
diagrammed in panel A. Data points corresponding to the reciprocal of
the fraction of labeled DNA bound from seven experiments were
normalized to the same origin, and the mean and standard deviation for
each point were calculated. The relative affinities of
tRNACAla and tRNASGAla promoters to Ad
MLP were estimated by comparing the slopes of least-square-fitted
straight lines: tRNACAla/Ad MLP ≅ 1/2 and
tRNASGAla/Ad MLP ≅ 1/6.
) DNA
are shown. Upstream promoter sequences are in capital letters, and
positions relative to the transcription initiation site (arrow) are
numbered. The D (distal), TAT (TATAT), I (intervening), and AT (AATTTT)
regions of the tRNACAla promoter are delineated by
brackets at the top. (B) Cloned silkworm TBP binds specifically to both
tRNACAla and tRNASGAla promoters.
Silkworm TBP (5 nM) was incubated with radioactively labeled wild-type
(C WT; SG WT) or mutant tRNAAla (C mut.,
−29CGGC−26; SG mut.,
−10CGGC−7) promoter fragments (0.2 nM), and
the complexes were resolved on a nondenaturing gel. The locations of
the mutations on both promoters are within the protected sequences
bracketed in Fig. 2. The positions of TBP-bound fragments (TBP-DNA) and
unbound fragments (unbound DNA) are marked on the left. (C) Relative
affinities of TBP for the Ad MLP and the tRNACAla and
tRNASGAla promoters, determined by competition band
shift assays. Silkworm TBP and radioactively labeled Ad MLP fragments
were incubated with increasing amounts of unlabeled DNA fragments
corresponding to the Ad MLP or the tRNAAla promoters
diagrammed in panel A. Data points corresponding to the reciprocal of
the fraction of labeled DNA bound from seven experiments were
normalized to the same origin, and the mean and standard deviation for
each point were calculated. The relative affinities of
tRNACAla and tRNASGAla promoters to Ad
MLP were estimated by comparing the slopes of least-square-fitted
straight lines: tRNACAla/Ad MLP ≅ 1/2 and
tRNASGAla/Ad MLP ≅ 1/6.
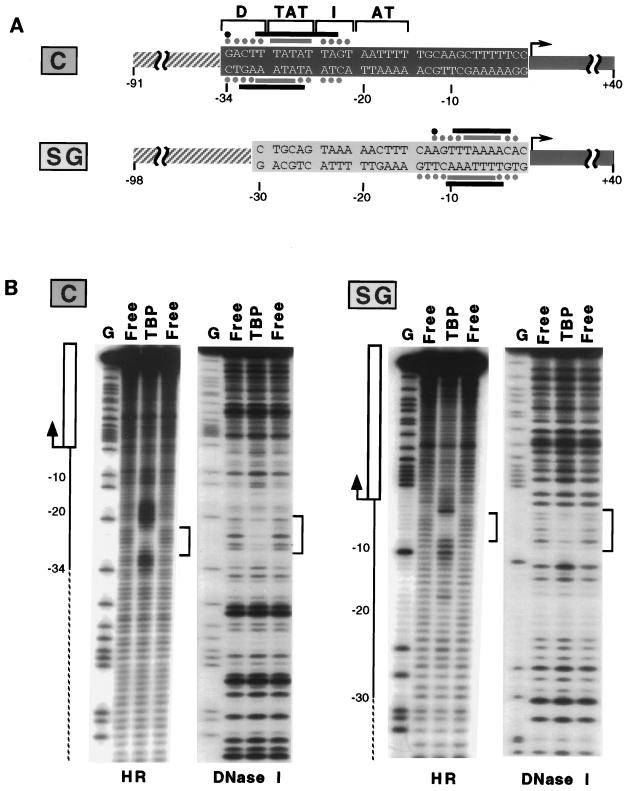
To determine the location of bound TBP, we used both hydroxyl radical and DNase I footprints. As shown in Fig. 2, TBP binds the two promoters in different places, ∼−32 to −23 in the tRNACAla promoter and −10 to −3 in the tRNASGAla promoter, and at each site generates both protection from and hypersensitivity to cleavage. Loss of TBP binding upon mutation of these sequences confirmed that they are specifically required for binding (Fig. 1B, lanes 3 and 5). The footprints in Fig. 2 are typical in showing ill-defined boundaries for the tRNACAla binding site. The inefficiency of DNase I cleavage of AT-rich sequences is partly responsible, but even hydroxyl radical cleavage, which is not base specific, does not display a sharp boundary between protected and unprotected sequences. In contrast, the tRNASGAla sequence protected from hydroxyl radical cleavage is sharply defined, and the boundaries are unambiguous.
FIG. 2.
TBP binds tRNACAla and tRNASGAla promoters in different positions. (A) Diagram of the DNA fragments used for TBP binding assays. Diagrammatical symbols are the same as Fig. 1A. Footprints obtained on both strands by hydroxyl radical (light gray) or DNase I (black) cleavage are summarized by bars (protection) and dots (hypersensitivity). (B) Gels showing the cleavage of the noncoding strand of the two promoters either by DNase I or by hydroxyl radicals (HR). Lanes: G, partial chemical cleavage at G residues; Free and TBP, cleavage of unbound and TBP-bound fragments, respectively. The extent of each promoter is shown by the solid line on the left with the initiation site labeled with an arrow, the extent of the primary transcript indicated by a rectangle, and vector sequences shown by dashed lines. Protected sequences are delineated by brackets on the right.
Silkworm TBP can bind alternative overlapping sites in the tRNACAla promoter.
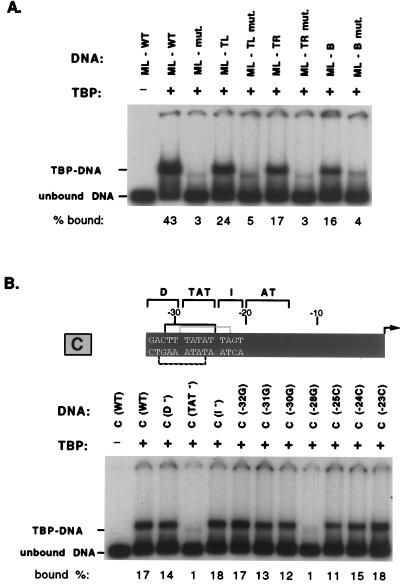
Since the 10-bp protected region in the tRNACAla promoter (Fig. 2) is longer than the sequence of 7 specific bp defined by mutational and crystallographic data (15), we wondered whether this region might contain more than one TBP binding site. Heterogeneity in the location of bound TBP would account both for the longer protected region and for the fuzziness of the footprint boundaries. Inspection of all of the 7-bp sequences within the TBP footprint revealed three candidates, based on TBP binding preferences inferred from Pol II transcription (57) or direct binding measurements (45, 58). Two candidates read on the top strand are −31TTTATAT−25 (TL [top left]) and −29TATATTA−23 (TR [top right]), and one read on the bottom strand is −26TATAAAG−32 (B [bottom]). We considered the other 7-bp segments unlikely because they contain either C or A in the first position or A in the third position, and these departures from a canonical TATA are known to reduce TBP binding (45). We tested the binding ability of the three strongest candidates by using each of them to replace the wild-type TATA sequence in Ad MLP. Band shift assays verified that TBP is able to bind each of these sequences with nearly equal affinity, although we consistently observed more binding to TL than to TR or B. Mutant sequences confirmed the specificity of the observed binding (Fig. 3A).
FIG. 3.
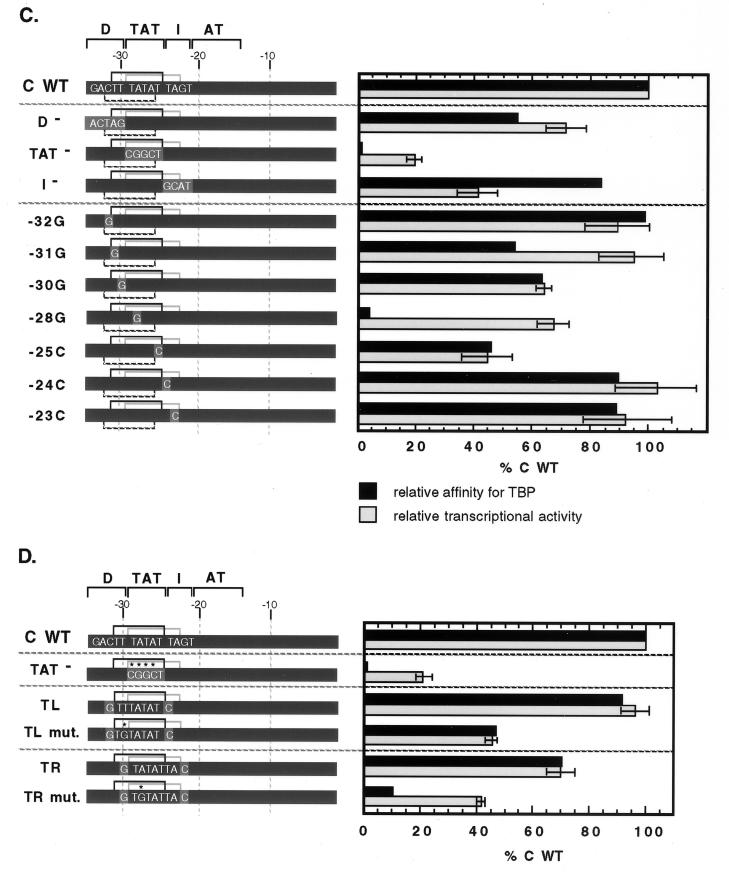
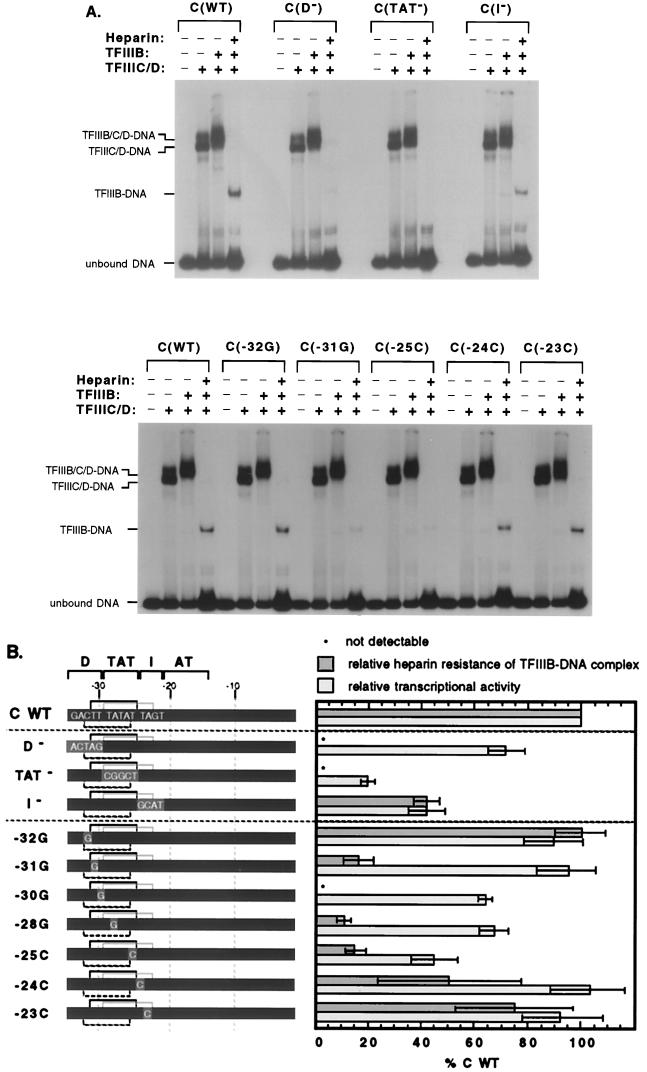
Specific TBP-TATA interaction contributes to wild-type tRNACAla promoter activity. (A) Cloned silkworm TBP is able to interact with at least three TATA sequences within the footprinted region of tRNACAla promoter. Three potential binding sites, two (−31TTTATAT−25 [TL] and −29TATATTA−23 [TR]) read on the top strand and one (−26TATAAAG−32 [B]) read on the bottom strand, were cloned into Ad MLP to replace its wild-type TATAAAA (ML-WT) sequence and tested for TBP binding. For each construct, patterns of TBP binding to wild-type and mutant versions (ML mut., TGTAAAA; ML-TL mut., TGTATAT; ML-TR mut., TGTATTA; ML-B mut., TATAAAC) were compared. Silkworm TBP (5 nM) was incubated with radioactively labeled fragments (0.2 nM), and the complexes were resolved on a nondenaturing gel. The kind of labeled fragment is marked at the top, and the positions of TBP-bound fragments (TBP-DNA) and unbound fragments (unbound DNA) are marked on the left. The percent labeled DNA bound, shown below each lane, is an average of multiple experiments. (B) Effects of promoter mutations on TBP binding. The positions of the TBP binding sequences within the tRNACAla promoter are diagrammed at the top, and a representative band shift gel illustrating the effect of all the promoter mutations on TBP binding is shown below. All symbols are the same as in previous figures. The percent labeled DNA bound is the average of multiple experiments. (C) Quantitative comparison of the effects of mutation on affinity for TBP and transcriptional activity. The wild-type (WT) promoter is represented by a dark gray bar, and mutant sequences are shown in capital letters on a lighter gray background. The two top-strand TATA sequences are delineated by black and gray brackets above each promoter. The relative affinity for TBP was determined by at least three competition assays as described in Fig. 1C. (D) Quantitative comparison of the effects of mutation on affinity for TBP and transcriptional activity for promoter constructs containing an isolated TATA sequence. In the constructs, sequences of the isolated wild-type or mutant TATA elements are shown in capital letters. The G-C base pairs used to isolate each TATA element are shown against a medium gray background, and asterisks indicate positions within the TATA that were mutated.
To determine whether TBP has a preference for one of these sequences in the context of the tRNACAla promoter, we examined the effects on binding of block and single base pair mutations in and around the TBP footprint. As shown in Fig. 3B, binding to all three potential binding sites should be affected by substitution of the TAT region, and the strong deleterious effect of the TAT− mutation supports this idea. In contrast, only two sites (TL and B) in D (distal region) mutants and only one (TR) in I (intermediate region) mutants should be affected. Figure 3B shows that neither a D nor an I mutation reduces affinity for TBP. These results are consistent with the possibility that TBP is capable of binding multiple sites within the tRNACAla promoter and that binding is not significantly impaired in mutants that retain at least one functional site. Since the block mutations vary in the extent to which they alter different binding sites, however, we could not exclude the possibility of a single strongly preferred site. Therefore, we dissected the region more systematically with single G-C or C-G base pair substitutions at positions −32, −31, −30, −28, −25, −24, and −23. As shown in Fig. 3B and C, mutations at −32, −24, and −23, which affect either site B (−26TATAAAG−32) or site TR (−29TATATTA−23) but not site TL (−31TTTATAT−25), have no effect on TBP binding. Mutations at −31 and −30, which affect sites TL and B but not site TR, reduce TBP binding, but only modestly (to ∼60% of the wild-type level). Similarly, a mutation at −25, which affects both TR and TL but not B, reduces TBP binding somewhat (to ∼50% of the wild-type level). In contrast, a mutation at position −28, which affects key residues in all three binding sites, reduces TBP binding dramatically (to 5% of the wild-type level). Taken together, the data argue that TBP is capable of binding alternative sites within the tRNACAla promoter and that binding is greatly impaired only by mutations that alter all of the sites. The modest reduction caused by mutations at −31, −30, and −25 is consistent with the idea, suggested in Fig. 3B, of inherent affinity differences among the three sites. Specifically, for these mutants, the remaining unimpaired site (TR or B) is expected to be slightly weaker than the TL site available in a wild-type promoter.
Specific TBP-TATA interaction contributes to wild-type tRNACAla promoter activity.
We next wanted to know whether TBP interaction with any or all of these sites matters for transcription. If so, mutations that reduce TBP binding should also reduce transcription. As shown in Fig. 3C, the overall patterns of mutant effects on TBP binding and transcription are similar, although in most cases, the effects on transcription are smaller. For instance, the TAT− mutant essentially eliminates TBP binding and reduces transcription to ∼20% of the wild-type level. Single base pair substitutions at −32, −24, or −23, which do not affect TBP binding, do not affect transcription, and three of the substitutions that reduce TBP binding (−30, −28, and −25) also reduce transcription. Of these, the mutation at −25 is most deleterious to transcription. Since position −25 is outside the bottom-strand TATA, this result suggests that transcription does not depend strongly on the bottom TATA. Consistent with this interpretation, there is no transcriptional phenotype associated with a mutation that is unique to the bottom TATA (−32). Since all of the mutations that do have transcriptional phenotypes affect one or both of the top-strand TATAs, it appears that the interaction of TBP with these sites is transcriptionally important.
To test the function of the top-strand TATAs directly, we designed constructs in which a tRNACAla promoter was provided with only one or the other of them. As controls, we constructed mutant versions of these isolated TATAs that were expected to have reduced affinity for TBP because of sequence changes at the second position (−31TGTATAT−25 and −29TGTATTA−23). Figure 3D shows that both of the wild-type isolated TATAs have affinities for TBP close to that of the wild-type tRNACAla promoter, although TBP binds slightly better to TL than to TR. Mutation of either site reduces binding. Figure 3D also shows that both of the isolated wild-type TATAs support efficient transcription, with a preference for TL (96% ± 5% of the wild-type level) over TR (69% ± 5% of the wild-type level), and that mutation of either TATA reduces transcriptional activity. These results suggest that, as part of a transcription complex, TBP can interact productively with either of the two top-strand TATAs.
Both specific TBP-TATA interaction and optimal geometry are required for full stability of TFIIIB-promoter complexes.
Since TBP functions in Pol III transcription as part of the TFIIIB complex, the role of specific TBP-TATA contacts may be to ensure proper association of TFIIIB with the template. If so, mutations that disrupt the sites contacted by TBP in the transcription complex should weaken the TFIIIB-template interaction. As a sensitive probe of this interaction, we wanted to analyze complexes consisting simply of TFIIIB and DNA, without the potentially stabilizing influence of the remainder of the transcription complex. We had to circumvent the problem, however, that silkworm TFIIIB does not bind to tRNA promoters by itself. As in all other systems tested, prior binding by TFIIIC (TFIIIC/D in the silkworm system) is required (reviewed in references 6, 10, and 54).
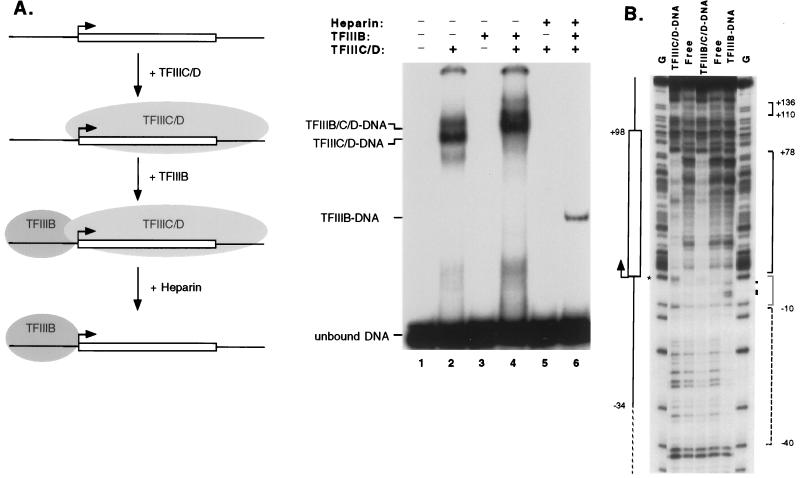
We therefore generated TFIIIB-promoter complexes by using heparin or KCl to strip complete complexes of the other transcription factors, an approach that was pioneered with yeast (18). Figure 4A shows the generation of heparin-stripped complexes containing silkworm transcription factors bound to the tRNACAla promoter. DNase I footprints of these complexes (Fig. 4B) demonstrate that the binding of TFIIIC/D alone protects sequences downstream from the tRNACAla transcription initiation site and causes hypersensitivity at the initiation site (+1), whereas the addition of TFIIIB extends protection to at least −35, as previously reported (59). The complexes that remain after stripping contain TFIIIB only, since upstream (−10 to −40) but not downstream sequences are protected. The stripped complexes are also hypersensitive to DNase I at three new sites, −2, −5, and −6, and their overall patterns of protection and hypersensitivity strongly resemble those of yeast TFIIIB-promoter complexes (18, 20).
FIG. 4.
Heparin treatment of tRNACAla transcription complexes yields TFIIIB-promoter complexes. (A) Diagram of the procedure and the heparin resistance of TFIIIB-promoter complexes. Radioactively labeled tRNACAla promoter fragments (5 fmol) were incubated with a fraction containing TFIIIC/D or with this fraction plus TFIIIB for 1 h. These mixtures were either loaded directly on a nondenaturing gel (lanes 2 and 4) or treated with heparin (100 ng/μl) for 20 to 30 s prior to loading (lanes 5 and 6). After electrophoresis, the complexes were visualized by autoradiography. The identities of the major protein-DNA complexes (indicated at the left) were determined by DNase I footprinting. (B) DNase I footprints of the transcription factor-promoter complexes shown in panel A. The identities of the protein-DNA complexes are shown at the top. Lanes marked G show partial chemical cleavage at G residues. The full extent of promoter sequences is shown by the solid line on the left, with the transcription initiation site labeled with an arrow and the extent of the primary transcript indicated by a rectangle. Sequences protected by TFIIIC/D alone (solid brackets) or by TFIIIB (dashed bracket) in the presence or absence of TFIIIC/D are indicated on the right. DNase I hypersensitivity induced by TFIIIC or TFIIIB is marked by an asterisk or dots, respectively. The light gray bracket shows the hypersensitive region of TFIIIB-promoter complexes.
In preliminary experiments, we had observed promoter-specific variation in the amounts of stripped TFIIIB-promoter complex obtained. Since we started with identical amounts of complete complex, the variation had to reflect differences in the ability of TFIIIB bound to certain promoters to withstand challenge by heparin or gel electrophoresis. We reasoned, therefore, that quantitation of the amounts of TFIIIB-promoter complex that survive stripping would measure the relative stability of the TFIIIB interaction with different promoters.
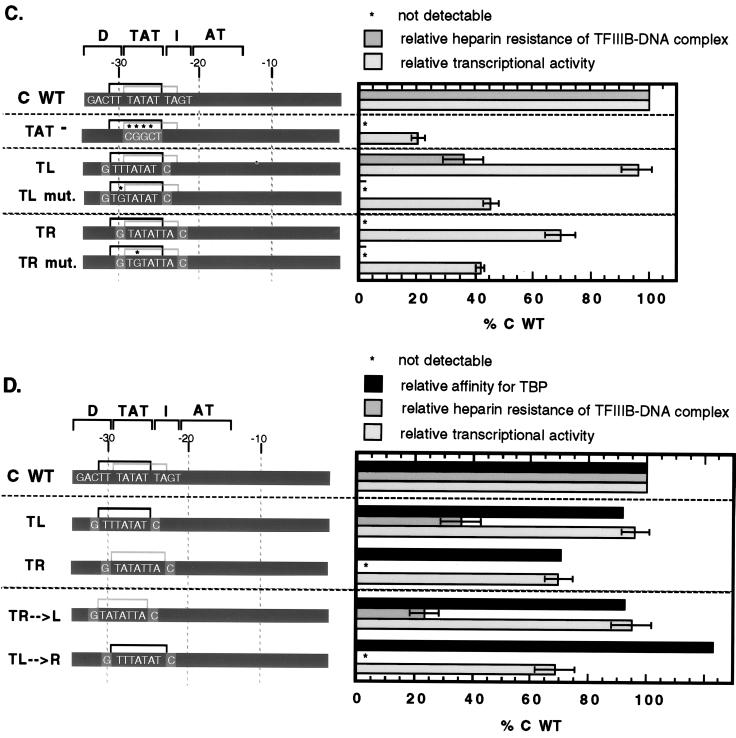
Using this assay, we carried out a systematic analysis of the effects of tRNACAla promoter mutations on the stability of TFIIIB-promoter complexes. Figure 5A shows a representative sample of the primary data, and Fig. 5B quantitates all of the data and compares them to the effects of the same mutations on transcriptional activity. Mutation of either the TAT or the D region eliminates detectable heparin-resistant TFIIIB-promoter complexes, whereas mutation of the I region reduces the amount only twofold. We estimate the lower limit of detection to be ∼10% of the amount of complex formed on the wild-type tRNACAla promoter. Single mutations from −31 to −25 all have strong deleterious effects on TFIIIB-promoter complex stability (<20% of wild-type heparin resistance remains), but mutations outside this region (−32, −24, and −23) have much smaller effects or none at all. These results suggest that the wild-type TL TATA (−31TTTATAT−25) makes a major contribution to the formation of TFIIIB-promoter complexes that are stable to heparin. This idea is reinforced by the results with constructs containing isolated TATAs, shown in Fig. 5C. The two constructs containing wild-type TATAs differ significantly in the way they bind TFIIIB. Heparin-resistant TFIIIB-promoter complexes are detectable on promoters containing the isolated TL sequence but not on those containing the isolated TR sequence.
FIG. 5.
Both specific TBP-TATA interaction and optimal geometry are required for full stability of TFIIIB-promoter complexes. (A) Effects of tRNACAla promoter block and point mutations on the heparin resistance of TFIIIB-promoter complexes. The formation and resolution of transcription factor-promoter complexes were as described for Fig. 4. The kind of promoter (wild type [WT] or mutant) is shown at the top, and the resolved complexes (TFIIIB-DNA, TFIIIC/D-DNA, and TFIIIB/C/D-DNA) and unbound DNA are indicated on the left. Two mutants (−30G and −28G) were not included in the experiment shown. (B) Quantitative comparison of the effects of mutation on affinity for TBP and on the heparin resistance of TFIIIB-promoter complexes. The promoter constructs are the same as in Fig. 3. The relative affinity for TBP was determined by at least three competition assays as described for Fig. 1C. The relative heparin resistance of TFIIIB-promoter complexes was determined by measuring the amount of protein-DNA complex after heparin treatment. The means and standard deviations based on at least three determinations are shown. (C) Quantitative comparison of the affinity for TBP and the heparin resistance of TFIIIB-promoter complexes for promoter constructs containing an isolated TATA sequence. TL and TR are the constructs containing the isolated sequence, −31TTTATAT−25 or −29TATATTA−23, respectively. TL mut. and TR mut. are mutants in which the second position of the wild-type TATA has been replaced by G. Mutants in which the seventh position was replaced by C also exhibited reduced TBP binding and TFIIIB-DNA complex stability (data not shown). (D) Quantitative comparison of the affinity for TBP, the heparin resistance of TFIIIB-promoter complexes, and transcriptional activity for tRNACAla promoter constructs containing interchanged isolated TATA sequences. TR→L (−31TATATTA−25) and TL→R (−29TTTATAT−23) are constructs containing isolated TATA sequence with switched positions.
Based on the preferences of Drosophila Pol III (52), we anticipated that the second-position T might give an advantage to the TL sequence over the TR sequence. To test the importance of sequence versus position directly, however, we interchanged the sequences of these isolated TATAs. The results (Fig. 5D) show that position is the most important determinant of both TFIIIB-promoter complex stability and transcriptional activity. With either the TTTATAT or the TATATTA sequence located at the left position (−31 to −25), heparin-resistant TFIIIB-promoter complexes are detectable, whereas with either sequence located at the right position (−29 to −23), they are not. Moreover, although the effects on transcriptional activity are not as large, the pattern is the same. Thus, there are at least two requirements for the formation of heparin-resistant TFIIIB-promoter complexes: specific TBP-TATA interactions and proper placement of the TATA relative to other promoter elements.
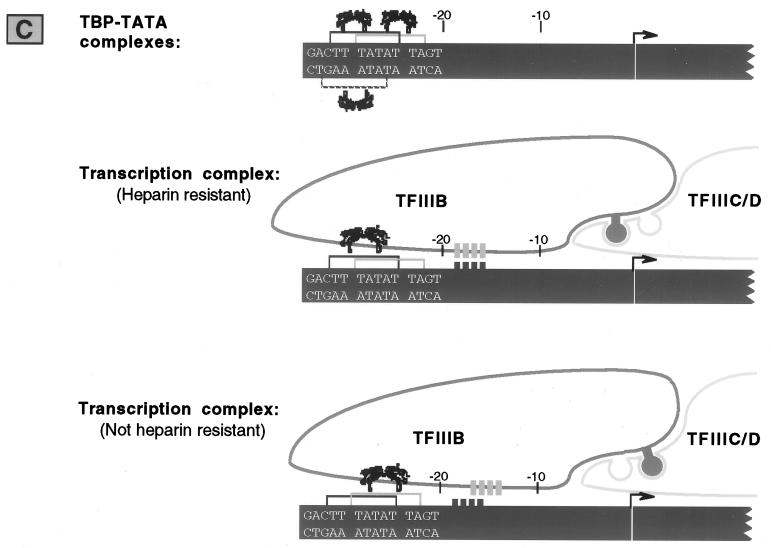
As summarized in Fig. 6, we find restriction in the use of alternative TBP-TATA interactions by different complexes formed on the tRNACAla promoter. Of the three sites that support binding by TBP alone, only two are used in transcription complexes and only one is able to stabilize the TFIIIB-promoter interaction to heparin challenge.
FIG. 6.
All TBP-TATA interactions are not equally effective for transcriptional activity and heparin resistance on the tRNACAla promoter. Cloned TBP is able to interact with three different sequences, but only two of them (TL and TR) are used in full transcription complexes, and only one (TL) can confer heparin resistance to TFIIIB-promoter complexes. The observation that differently spaced sites are effective for transcription but not TFIIIB-promoter stability suggests alternative linkages between TFIIIB and TFIIIC/D (diagrammed as knobs and holes) and stabilization of TFIIIB by sequences outside the TATA box (short vertical bars) (see Discussion).
The natural TBP binding sequence does not contribute positively to wild-type tRNASGAla promoter activity.
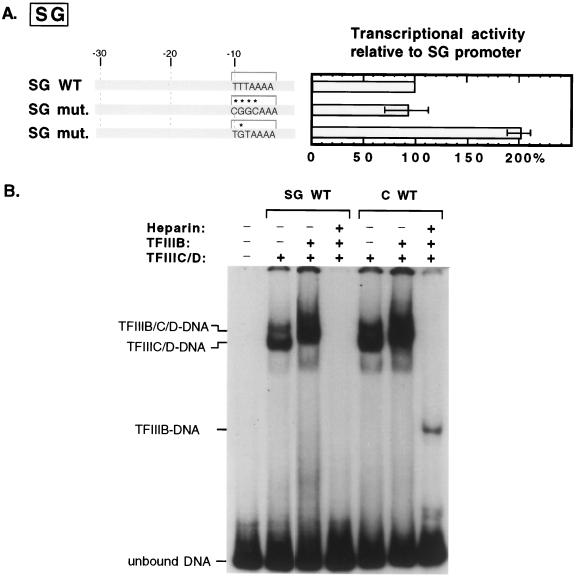
To determine whether the TBP-TATA interaction contributes to tRNASGAla promoter activity, we determined the transcriptional consequence of mutating the TBP binding site in the tRNASGAla promoter. The −10TTTAAAA−4 binding sequence was changed either to −10CGGCAAA−4 or, to minimize extraneous effects of altered base composition, to −10TGTAAAA−4. A band shift experiment confirmed that isolated silkworm TBP was no longer able to bind (Fig. 1B, lane 5, and data not shown). As shown in Fig. 7A, however, neither mutation had a deleterious effect on promoter activity. In fact, the activity of the −10TGTAAAA−4 mutant was slightly elevated. Thus, the TBP binding site in the tRNASGAla promoter does not play the same positive role as it does in the tRNACAla promoter. To determine whether this site stabilizes the TFIIIB-promoter interaction, we subjected transcription factor complexes on the tRNASGAla gene to heparin challenge. We anticipated that these complexes might not resist this challenge because we already knew that although they are stable to gel electrophoresis, the TFIIIB within them does not protect upstream sequences from DNase I cleavage (59). Indeed, as shown in Fig. 7B, the wild-type tRNASGAla promoter does not support a heparin-resistant complex.
FIG. 7.
Specific interaction of TBP with the natural TATA element at −10 does not contribute positively to tRNASGAla promoter activity. (A) Transcriptional activity of mutant tRNASGAla promoters containing substitutions in the TBP binding site (Fig. 2) is plotted relative to the activity of a wild-type tRNASGAla promoter. The wild-type (WT) tRNASGAla promoter is represented by a light gray bar, and its natural or mutant (mut.) TATA sequences are shown in capital letters delineated by a bracket, with asterisks indicating positions altered in the mutants. (B) The wild-type tRNASGAla promoter does not support a heparin-resistant complex. Transcription factor-promoter complexes were formed on the tRNASGAla (SG WT) and tRNACAla (C WT) genes and challenged with heparin as described for Fig. 4A. The identities of the major protein-DNA complexes are indicated at the left.
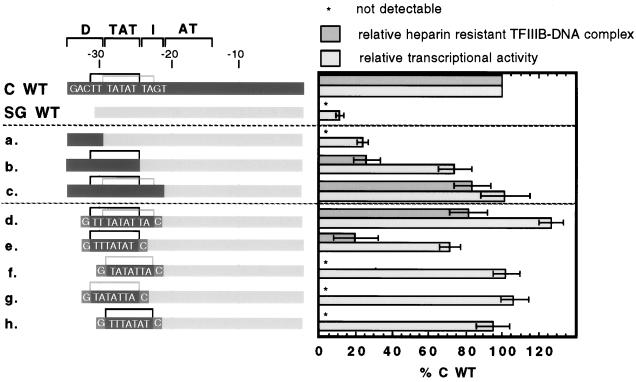
Lack of a properly positioned TBP binding site causes the low activity of the wild-type tRNASGAla promoter.
Since the natural TBP binding site does not contribute to tRNASGAla promoter activity and does not confer heparin resistance, we wondered whether an optimally positioned site would do so. To investigate this possibility, we designed a series of chimeric constructs in which segments of the tRNACAla gene upstream promoter replaced the corresponding parts of the tRNASGAla promoter. As shown in Fig. 8 (constructs a through c), replacement of the D region alone has no significant effect on either heparin resistance or transcription. However, replacement of both D and TAT elements increases the heparin resistance to ∼30% and transcriptional activity to ∼70% that of the wild-type tRNACAla promoter, and addition of the I element brings heparin resistance to ∼80% and transcriptional activity to 100% of wild-type levels. The construct that includes simply both TATAs (construct d) also displays ∼80% of the heparin resistance and 100% of the transcriptional activity of a wild-type tRNACAla promoter. In contrast, the four constructs that each contain a single isolated TBP binding site (constructs e through h) show that either TTTATAT or TATATTA, positioned as it is in the wild-type tRNACAla promoter (either −31 to −25 or −29 to −23), can dramatically stimulate transcription but confers little or no resistance to heparin. Transcriptionally, three of the four constructs are as active as the wild-type tRNACAla promoter, and the other one is ∼70% as active, but heparin resistant complexes are undetectable on three constructs and are only ∼20% of the wild-type level on the other. Overall, the results are similar to those for isolated TATAs within the context of the tRNACAla promoter. The difference is that the isolated TATAs within the tRNASGAla promoter confer less heparin resistance and, as judged by their transcriptional activities, exhibit less position preference than they do in the tRNACAla promoter.
FIG. 8.
Introduction of a TATA element within the −32 to −23 region increases tRNASGAla promoter activity. The heparin resistance of TFIIIB-promoter complexes and transcriptional activities of wild-type (WT) tRNACAla, tRNASGAla, and tRNAC/SGAla chimeric promoters are shown. In the diagrams of promoter constructs, wild-type tRNACAla and tRNASGAla promoters are shown by dark and light gray bars, respectively. The locations of the two functional TATA elements of the wild-type tRNACAla promoter are shown by dark or light gray top brackets. The sequences of these elements introduced into the tRNASGAla promoter (constructs d to h) are shown in capital letters, and the G-C base pairs used to isolate each TATA element are shown against a medium gray background. The heparin resistance of TFIIIB-promoter complexes was determined as described for Fig. 5.
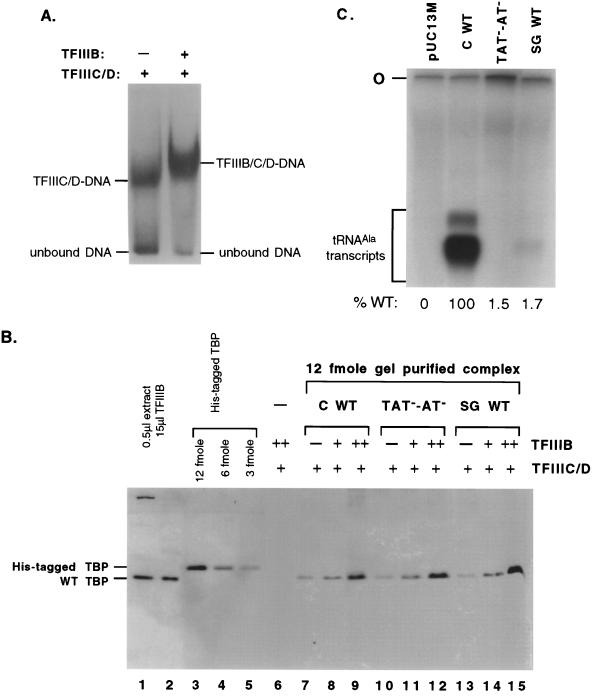
The same amount of TBP is present in transcription complexes formed on wild-type tRNACAla and tRNASGAla genes.
Why does the lack of a properly positioned TBP binding site reduce the efficiency of tRNASGAla transcription? There are at least two possibilities. The probability of incorporating TBP during transcription complex assembly might be reduced, generating a large proportion of incomplete, hence inactive, complexes. Alternatively, protein-protein interaction might suffice for TBP incorporation, but the absence of TBP-TATA contacts might preclude incorporation of another factor or allow a conformation that is incompatible with transcription. To distinguish between these two possibilities, we used antibodies to compare the TBP content of transcription complexes formed on tRNACAla and tRNASGAla genes. In each case, complexes were formed at subsaturating (1.5 μl) or saturating (7.5 μl) TFIIIB concentrations in the presence of 5 μl of TFIIIC/D. The resulting complexes were separated from unbound DNA and proteins by electrophoresis through agarose (Fig. 9A) and were then retrieved by enzymatic digestion of the gel. The amount of TBP in the complexes was determined by quantitative Western analysis. The amount of complex analyzed in each case was standardized by reference to the radioactively labeled template present in the complexes.
FIG. 9.
Amounts of TBP contained in tRNACAla and tRNASGAla transcription complexes are the same. (A) Transcription factor-promoter complexes were formed on the tRNACAla gene (as shown in Fig. 4A) and were resolved on a 1.5% agarose gel. The resolved complexes (TFIIIC/D-DNA and TFIIIB/C/D-DNA) and unbound DNA are indicated at the sides. (B) Western analysis of TBP in TFIIIC/D and TFIIIB/C/D complexes. The TFIIIC/D and TFIIIB/C/D complexes were formed on wild-type tRNACAla (C WT), mutant tRNACAla (TAT−-AT−) or wild-type tRNASGAla (SG WT) with either 0 (−), 1.5 (+), or 7.5 (++) μl of TFIIIB in the presence of 5 μl of TFIIIC/D. The complexes were resolved on an agarose gel as shown in panel A, detected autoradiographically, and isolated from the gel by digestion with an agarose-digesting enzyme. After concentration, the amount of complex was standardized by quantitating the labeled DNA fragment, and the proteins of the isolated complexes were examined by Western analysis using antibodies to silkworm TBP and a chemiluminescence detection method. The amount of TBP signal was compared to known amounts of cloned His-tagged TBP run in parallel (lane 3 to 5). The positions of wild-type (WT) TBP and His-tagged TBP are indicated on the left. (C) Transcription reactions were performed under the same buffer conditions as used for binding but with the addition of 5 μl of Pol III and nucleotides (33). Templates (200 ng of plasmid) were those used in panel B, and their parental plasmid (pUC13M) was used as a control. Transcription products were fractionated on an 8% polyacrylamide denaturing gel; autoradiography and quantitation of the transcripts were performed as previously described (33). The percentage of transcription rate relative to the wild-type tRNACAla gene is shown below each lane. “O” denotes the origin; transcript position is marked with a bracket.
We analyzed complexes formed on three different templates: the wild-type tRNACAla gene, a tRNACAla upstream promoter mutant, and a wild-type tRNASGAla gene. The result is shown in Fig. 9B. Known amounts of recombinant His-tagged silkworm TBP were used as quantitative standards (lanes 3 to 5). Each Western analysis was performed with 12 fmol of complex, and complexes formed with saturating input TFIIIB contain about 12 fmol of TBP, as judged by comparison with the His-tagged standards. The small amounts of TBP found in complexes formed without input TFIIIB are likely to be from the TFIIIB that contaminated the TFIIIC/D fraction, based on assays of complementing transcription activity (data not shown). Comparison of lanes 9, 12, and 15 shows that the same amounts of TBP are incorporated into transcription complexes formed on the three different templates. Thus, a quantitative difference in TBP incorporation into transcription complexes does not explain the difference between tRNACAla and tRNASGAla promoter activity, which, under these conditions, differed at least 50-fold. As shown in Fig. 9C, the transcription rate from tRNASGAla complexes was only 1.7% of the rate from the same number of tRNACAla complexes.
DISCUSSION
The TBP-TATA interaction stabilizes TFIIIB binding and enhances promoter function.
Our results show that specific interaction between TBP and a TATA element is key to tRNAAla promoter function. In the naturally robust tRNACAla promoter, TATA mutations that reduce TBP binding also reduce transcription, and in the naturally weak tRNASGAla promoter, provision of a properly positioned wild-type TATA element confers high-level promoter activity. In both cases, the transcriptional effects appear to be mediated through the TFIIIB complex since the stability of TFIIIB-promoter complexes is decreased by mutations that weaken the natural TATA region in the tRNACAla promoter and is enhanced by addition of that region to the tRNASGAla promoter.
Quantitatively, the effects on transcription and TFIIIB-promoter complex stability are quite different. Single base pair changes do not reduce transcription more than ∼2-fold, but they destabilize TFIIIB-promoter complexes at least 5- and as much as 50-fold. The different sizes of the functional units probed by these two assays probably accounts for the difference. Transcriptional activity is tested with the full transcription complex, which in the silkworm system is bound to a stretch of at least 150 bp of DNA (56, 59). In contrast, TFIIIB complexes contain only a subset of transcription factors (three in yeast [17, 19, 38]) and contact only ∼30 bp of template DNA. Thus, the loss of a few protein-DNA contacts within the TATA element is expected to have a greater impact on the stability of complexes that contain TFIIIB by itself than on the stability of the full transcription complex. Interestingly, the effects of mutations in the I region (I−, −24C, and −23C), as well as the behavior of constructs containing isolated TATAs, suggest that the TBP-TATA interaction, though necessary, is not sufficient for full stability of TFIIIB-promoter complexes. Additional ability to resist heparin challenge is provided by sequences 3′ to TL. This additional stabilization is represented schematically in Fig. 6, and current experiments are aimed at defining the limits of sequences responsible for it. Deformability may be an important characteristic of these sequences, given the increase in stability of yeast TFIIIB-DNA complexes that is caused by greater DNA flexure downstream of the TATA element (8). Both BRF and B" are candidates for proteins that provide stabilizing function. BRF is known to stabilize TBP binding to a wild-type Pol III promoter through sequences surrounding the TATA element (3, 28), and B" confers heparin resistance when added to a TBP-BRF-DNA complex (24).
Our finding that Bombyx transcription requires specific TBP-TATA interactions differs from the main idea that has emerged from earlier work with yeast. There, the absence of 5′ flanking mutations among tRNA gene mutants selected for reduced expression (25), as well as the observed insensitivity of promoter activity to deliberate alteration of upstream tRNA sequences (22), argued against sequence-specific upstream promoter elements in this organism. On the other hand, our results fit with the observation that in yeast, the presence or absence of an ectopic TATA element alters the efficiency of an artificial tRNA promoter (13). What accounts for the differences among these observations? One possibility is that specific TBP-TATA interactions occur in all systems but are more apparent in the Bombyx system because of its unusually strong dependence on upstream promoter elements. In yeast, transcription complex formation relies more heavily on interactions with the B box than with 5′ flanking sequences, whereas in the Bombyx system, the reverse is true (7, 43). Consequently, changes in the interaction of yeast transcription machinery with upstream promoter elements must be drastic to be manifest as transcription defects. Changes having the required impact in yeast may be more easily created in the context of an artificial promoter consisting of a single TATA embedded in GC-rich DNA than in natural tRNA promoters whose AT richness within the TFIIIB binding site could provide redundant TATA function (20).
Another apparent difference between the yeast and silkworm systems is in the sensitivity of TFIIIB binding to changes in promoter sequence. In the silkworm system, relatively subtle changes can eliminate the heparin resistance of TFIIIB-promoter complexes, but in yeast, even drastic changes do not do so (13, 23). This could indicate a fundamental difference in the nature of TFIIIB-promoter interaction in the two systems but could also simply reflect the higher threshold of detection in yeast, where the fraction of active transcription complexes that yields heparin-resistant TFIIIB-promoter complexes can be close to unity (13, 18, 20). Variation in the amount of heparin-resistant TFIIIB-promoter complex has been noted in yeast as a function of changes in DNA sequence or conformation (1), and in both systems, the proportion of complexes that is heparin resistant varies among different TFIIIB preparations (13; M. J. Martinez, unpublished data). Proteolysis during preparative manipulations may explain the nonuniformity of TFIIIB preparations since N-terminal truncation of BRF is known to generate TFIIIB that retains transcriptional activity but can no longer bind DNA in a heparin-resistant fashion (16). Thus, TFIIIB-promoter interactions in yeast and silkworms may be similar—an idea that is supported by a comparison of footprint data. In both cases, TFIIIB protects sequences between ∼−10 and −40 but leaves the transcription start site exposed, or even hypersensitive, to DNase (18, 24). TFIIIB-induced hypersensitivity just upstream from the start site is especially apparent in the silkworm system. It is worth noting that despite the contribution of optimal TBP-TATA interaction, silkworm TFIIIB does not bind detectably to tRNA promoters on its own (Fig. 4B, lane 3). Whether this represents a fundamental difference from yeast TFIIIB, which does bind TATA-containing U6 promoters directly (14, 55), or is the consequence of a higher limit of detection is not clear.
Multiple protein-DNA geometries are compatible with promoter function.
Our results argue that although specific TATA-TBP interaction is important for transcription, there is some leeway in the geometry that is allowed. For both the wild-type tRNACAla promoter and the TATA-containing derivatives of the tRNASGAla promoter, TATA elements 2 bp apart are nearly equivalent in the ability to direct transcription. Since these two TATAs differ markedly in the capacity to stabilize stripped transcription complexes containing only TFIIIB, their equivalence in the context of the full transcription complex suggests flexibility in the articulation between TFIIIB and other parts of the complex (illustrated in Fig. 6). Our results thus fit with indications in yeast of a flexible linkage between TFIIIB and TFIIIC (13). In that system, when natural 5′ flanking DNA is replaced by a GC stretch, promoter activity is strongly influenced by the presence or absence of a single TATA element within the stretch but is quite tolerant of variations in the position of the TATA. Since DNA between the TATA and the downstream TFIIIC binding region remains protected from DNase digestion even when it is 10 bp longer than normal, it has been suggested that the TFIIIB/C complex is capable of stretching to accommodate different spacing of promoter elements, probably via the nearly equivalent interactions that can be made between BRF and successive tetratricopeptide repeats in Tfc4, the 120-kDa subunit of TFIIIC (2, 13, 21).
Lack of a properly placed TATA enfeebles the tRNASGAla promoter.
In contrast to the tRNACAla promoter, the wild-type tRNASGAla promoter lacks a TATA element at the appropriate location and is thereby enfeebled under typical in vitro conditions. Why does the absence of a TATA have this effect? We have eliminated the most obvious possibility, namely, that the efficiency of TBP incorporation into tRNASGAla transcription complexes is reduced. Transcription complexes formed on tRNACAla and tRNASGAla genes contain the same amounts of TBP, but they direct transcription at very different rates. Under the conditions used for this comparison, the transcription rate from tRNASGAla complexes was at least 50-fold lower than that from the same number of tRNACAla complexes.
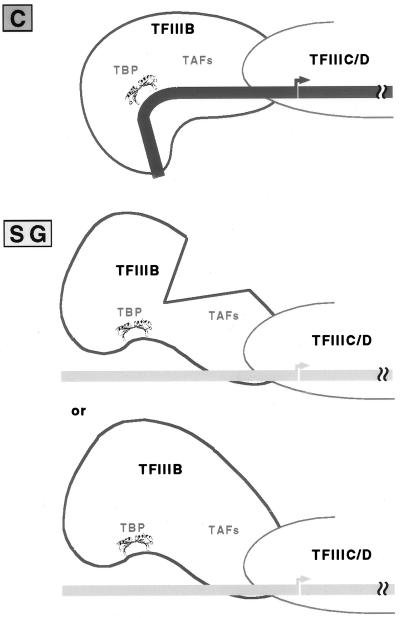
Thus, we are left with two possibilities. Either the lack of TBP-TATA interactions precludes incorporation of some other required factor, or it allows complete complexes to adopt an inactive conformation (Fig. 10). The DNA in transcriptionally active TFIIIB-promoter complexes in yeast is sharply bent at ∼−30, the middle of the TFIIIB binding site (1, 8, 27), and our preliminary results indicate that silkworm TFIIIB-promoter complexes formed on the wild-type tRNACAla promoter are similarly distorted (M. J. Martinez, unpublished data). Possibly, in the absence of specific TBP-TATA interactions, the template in a tRNASGAla transcription complex adopts a less distorted conformation that is an unsuitable substrate for polymerase.
FIG. 10.
Working model for the interaction of TFIIIB with tRNACAla and tRNASGAla promoters. Specific binding of TBP to the tRNACAla promoter stabilizes TFIIIB binding, possibly by inducing a bend in the upstream promoter. The resulting complex is transcriptionally active. In contrast, the lack of specific interaction between TBP and the tRNASGAla upstream promoter either prevents incorporation of another transcription factor(s) (top) or results in a complex with an alternative conformation that is not an effective substrate for RNA Pol III (bottom).
Our findings suggest a mechanism for the differential activity of tRNACAla and tRNASGAla promoters that is observed under typical in vitro conditions. This basal state, in which the tRNACAla promoter is on and the tRNASGAla promoter is off, presumably corresponds to the situation in cells other than the silk gland. If so, how is the tRNASGAla promoter activated in silk gland cells? Increases in TBP concentration have been seen to stimulate Pol III transcription (48), and, in fact, tRNACAla promoter output can be increased in Drosophila cells that overexpress TBP (49). It is unlikely, however, that increased TBP concentration is sufficient to rescue tRNASGAla transcription in the silk gland, since tRNASGAla transcription complexes contain normal amounts of TBP but are nonetheless functionally impaired. Indeed, the tRNASGAla promoter does not respond in vivo to elevated levels of TBP that are sufficient to stimulate the tRNACAla promoter (49). Moreover, the presence of a TBP binding site in the wrong part of the tRNASGAla promoter could potentially allow TBP to play a negative role, by interfering with the proper binding of TFIIIB. The increased promoter activity that results in vitro from elimination of the site is consistent with a negative role.
Alternatively, the actual in vivo role of the resident tRNASGAla TATA element could be positive. For instance, the site might be capable of binding a complex that is distinct from the traditional TBP-containing TFIIIB. In Drosophila, there are complexes containing TBP-related factors (TRF1 and TRF2) that are expressed in a cell-type-specific pattern and that associate with a subset of tRNA genes (9, 37). If comparable complexes exist in Bombyx, and if their distribution is tissue specific, the silk gland-specificity of the tRNASGAla promoter could be explained.
ACKNOWLEDGMENTS
This work was supported by NIH Public Health Service grants (GM25388 and GM32851) to K.U.S. M.J.M. is an NIH trainee supported by Graduate Training in Genetics grant GM07413.
We thank Diane Hawley, Heather Dunstan, and Fakhruddin Palida for critically reading the manuscript, and we thank Nan Ahnert and Gusti Zeiner for making some of the mutant promoter constructs.
REFERENCES
- 1.Braun B R, Kassavetis G A, Geiduschek E P. Bending of the Saccharomyces cerevisiae5S rRNA gene in transcription factor complexes. J Biol Chem. 1992;267:22562–22569. [PubMed] [Google Scholar]
- 2.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 3.Colbert T, Lee S, Schimmack G, Hahn S. Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol Cell Biol. 1998;18:1682–1691. doi: 10.1128/mcb.18.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon W J, Hayes J J, Levin J R, Weidner M F, Dombroski B A, Tullius T D. Hydroxyl radical footprinting. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- 5.Garel J P, Mandel P, Chavancy G, Daillie J. Functional adaptation of tRNAs to fibroin biosynthesis in the silkgland of Bombyx moriL. FEBS Lett. 1970;7:327–329. doi: 10.1016/0014-5793(70)80196-x. [DOI] [PubMed] [Google Scholar]
- 6.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 7.Geiduschek E P, Tocchini-Valentini G P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- 8.Grove A, Kassavetis G A, Johnson T E, Geiduscheck E P. The RNA polymerase III-recruiting factor TFIIIB induces a DNA bend between the TATA box and the transcriptional start site. J Mol Biol. 1999;29:1429–1440. doi: 10.1006/jmbi.1998.2347. [DOI] [PubMed] [Google Scholar]
- 9.Hansen S K, Takada S, Jacobson R H, Lis J T, Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–188. [Google Scholar]
- 12.Hoopes B C, LeBlanc J F, Hawley D K. Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multi-step pathway. J Biol Chem. 1992;267:11539–11547. [PubMed] [Google Scholar]
- 13.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Alternative outcomes in assembly of promoter complex: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 14.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juo Z S, Chiu T K, Leiberman P M, Baikalov I, Berk A J, Dickerson R E. How proteins recognize the TATA box. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 16.Kassavetis G A, Bardeleben C, Kumar A, Ramirez E, Geiduschek E P. Domain of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol Cell Biol. 1997;17:5299–5306. doi: 10.1128/mcb.17.9.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassavetis G A, Bartholomew B, Blanco J A, Johnson T E, Geiduschek E P. Two essential components of the Saccharomyces cerevisiaetranscription factor TFIIIB: transcription and DNA-binding properties. Proc Natl Acad Sci USA. 1991;88:7308–7312. doi: 10.1073/pnas.88.16.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiaeTFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 19.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Cloning, expression, and function of TFC5, the gene encoding the B" component of the Saccharomyces cerevisiaeRNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 22.Koski R A, Allison D S, Worthington M, Hall B D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4gene transcription. Nucleic Acids Res. 1982;10:8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A, Grove A, Kassavetis G A, Geiduschek E P. Transcription factor IIIB: the architecture of its DNA complex, and its roles in initiation of transcription by RNA polymerase III. Cold Spring Harbor Symp Quant Biol. 1998;63:121–129. doi: 10.1101/sqb.1998.63.121. [DOI] [PubMed] [Google Scholar]
- 24.Kumar A, Kassavetis G A, Geiduschek E P, Hambalko M, Brent C J. Functional dissection of the B" component of RNA polymerase III transcription factor IIIB: a scaffolding protein with multiple roles in assembly and initiation of transcription. Mol Cell Biol. 1997;17:1868–1880. doi: 10.1128/mcb.17.4.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurjan J, Hall B D, Gillam S, Smith M. Mutations at the yeast SUP4 tRNATyrlocus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980;20:701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- 26.Larson D, Bradford-Wilcox J, Young L S, Sprague K U. A short 5′ flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci USA. 1983;80:3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Léveillard T, Kassavetis G A, Geiduschek E P. Saccharomyces cerevisiae transcription factors IIIB and IIIC bend the DNA of a tRNAGlngene. J Biol Chem. 1991;266:5162–5168. [PubMed] [Google Scholar]
- 28.Librizzi M D, Brenowitz M, Willis I M. The TATA element and its context affect the cooperative interaction of TATA-binding protein with the TFIIB-related factor, TFIIIB70. J Biol Chem. 1998;273:4563–4568. doi: 10.1074/jbc.273.8.4563. [DOI] [PubMed] [Google Scholar]
- 29.Lucas F, Shaw J T B, Smith S G. The silk fibroins. Adv in Protein Chem. 1958;13:107–242. doi: 10.1016/s0065-3233(08)60599-9. [DOI] [PubMed] [Google Scholar]
- 30.Matsuzaki K. Fractionation of amino acid-specific s-RNA from silkgland by methylated albumin column chromatography. Biochem Biophys Acta. 1966;114:222–226. doi: 10.1016/0005-2787(66)90303-0. [DOI] [PubMed] [Google Scholar]
- 31.Meza L, Araya A, Leon G, Krauskopf M, Siddiqui M A Q, Garel J P. Specific alanine tRNA species associated with fibroin biosynthesis in the posterior silk-gland of Bombyx moriL. FEBS Lett. 1977;77:255–260. doi: 10.1016/0014-5793(77)80246-9. [DOI] [PubMed] [Google Scholar]
- 32.Mital R, Kobayashi R, Hernandez R. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton D G, Sprague K U. In vitrotranscription of a silkworm 5S RNA gene requires an upstream signal. Proc Natl Acad Sci USA. 1984;81:5519–5522. doi: 10.1073/pnas.81.17.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ottonello S, Rivier D H, Doolittle G M, Young L S, Sprague K U. The properties of a new polymerase III transcription factor reveal that transcription complexes can assemble by more than one pathway. EMBO J. 1987;6:1921–1927. doi: 10.1002/j.1460-2075.1987.tb02452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouyang C, Sprague K U. Cloning and characterization of the TATA-binding protein of the silkworm Bombyx mori. Gene. 1998;221:207–213. doi: 10.1016/s0378-1119(98)00460-0. [DOI] [PubMed] [Google Scholar]
- 36.Palida F A, Hale C, Sprague K U. Transcription of a silkworm tRNACAlagene is directed by two AT-rich upstream sequence elements. Nucleic Acids Res. 1993;21:5875–5881. doi: 10.1093/nar/21.25.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabenstein M D, Zhou S, Lis J T, Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci USA. 1999;96:4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts S, Miller S J, Lane W S, Lee S, Hahn S. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schultz M C, Reeder R H, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 41.Sharp P A. TATA-binding protein is a classless factor. Cell. 1992;68:819–821. doi: 10.1016/0092-8674(92)90023-6. [DOI] [PubMed] [Google Scholar]
- 42.Sprague K U. The Bombyx morisilk proteins: characterization of large polypeptides. Biochemistry. 1975;14:925–931. doi: 10.1021/bi00676a008. [DOI] [PubMed] [Google Scholar]
- 43.Sprague K U. Transcription of eukaryotic tRNA genes. In: Söll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C.: ASM Press; 1995. pp. 31–50. [Google Scholar]
- 44.Sprague K U, Hagenbüchle O, Zuniga M C. The nucleotide sequence of two silk gland alanine tRNAs: implications for fibroin synthesis and for initiator tRNA structure. Cell. 1977;11:561–570. doi: 10.1016/0092-8674(77)90074-5. [DOI] [PubMed] [Google Scholar]
- 45.Starr D B, Hoopes B C, Hawley D K. DNA bending is an important component of site-specific recognition by the TATA binding protein. J Mol Biol. 1995;250:434–446. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- 46.Struhl K. Duality of TBP, the universal transcription factor. Science. 1994;263:1103–1104. doi: 10.1126/science.8108728. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan H S, Young L S, White C N, Sprague K U. Silk gland-specific tRNAAla genes interact more weakly than constitutive tRNAAlagenes with silkworm TFIIIB and polymerase III fractions. Mol Cell Biol. 1994;14:1806–1814. doi: 10.1128/mcb.14.3.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trivedi A, Vilalta A, Gopalan S, Johnson D L. TATA-binding protein is limiting for both TATA-containing and TATA-lacking RNA polymerase III promoters in Drosophilacells. Mol Cell Biol. 1996;16:6909–6916. doi: 10.1128/mcb.16.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trivedi A, Young L S, Ouyang C, Johnson D L, Sprague K U. A TATA element is required for tRNA promoter activity and confers TATA-binding protein responsiveness in DrosophilaSchneider-2 cells. J Biol Chem. 1999;274:11369–11375. doi: 10.1074/jbc.274.16.11369. [DOI] [PubMed] [Google Scholar]
- 50.Vilalta A, Trivedi A, Wang Z, Roeder R G, Johnson D L. An RNA polymerase III-defective mutation in TATA-binding protein disrupts its interaction with a transcription factor IIIB subunit in Drosophilacells. J Biol Chem. 1997;272:18087–18092. doi: 10.1074/jbc.272.29.18087. [DOI] [PubMed] [Google Scholar]
- 51.Voelker R, Barkan A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 1995;14:3905–3914. doi: 10.1002/j.1460-2075.1995.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Jensen R C, Stumph W E. Role of TATA box sequence and orientation in determining RNA polymerase II/III transcription specificity. Nucleic Acids Res. 1996;24:3100–3106. doi: 10.1093/nar/24.15.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White R J. RNA polymerase III transcription. 2nd ed. Austin, Tex: R. G. Landes Co.; 1998. [Google Scholar]
- 55.Whitehall S K, Kassavetis G A, Geiduschek E P. The symmetry of the yeast U6 RNA gene's TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 56.Wilson E T, Larson D, Young L S, Sprague K U. A large region controls tRNA transcription. J Mol Biol. 1985;183:153–163. doi: 10.1016/0022-2836(85)90209-8. [DOI] [PubMed] [Google Scholar]
- 57.Wobbe C R, Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong J M, Bateman E. TBP-DNA interactions in the minor groove discriminate between A:T and T:A base pairs. Nucleic Acids Res. 1994;22:1890–1896. doi: 10.1093/nar/22.10.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young L S, Ahnert N, Sprague K U. Silkworm TFIIIB binds both constitutive and silk gland-specific tRNAAlapromoters but protects only the constitutive promoter from DNase I cleavage. Mol Cell Biol. 1996;16:1256–1266. doi: 10.1128/mcb.16.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young L S, Takahashi N, Sprague K U. Upstream sequences confer distinctive transcriptional properties on genes encoding silkgland-specific tRNAAla. Proc Natl Acad Sci USA. 1986;83:374–378. doi: 10.1073/pnas.83.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]