Abstract
The Xenopus embryo is a classical vertebrate model for molecular, cellular and developmental biology. Despite many advantages of this organism, such as large egg size and external development, imaging of early embryonic stages is challenging due to non-transparent cytoplasm. Staining and imaging of thin tissue sections is one way to overcome this limitation. Here we describe a step-by-step protocol that combines cryosectioning of gelatin-embedded embryos with immunostaining and imaging. The purpose of this protocol is to examine various cellular and tissue markers after the manipulation of protein function. This protocol can be carried out within a two-day period and allows to detect many antigens by immunofluorescence.
Keywords: Cryosectioning, immunostaining, gelatin embedding, immunofluorescence, Xenopus early development
Materials
Reagents
Antibodies for the protein of interest.
Secondary goat anti-mouse IgG antibodies conjugated to Alexa Fluor488, A11029 (1:200, Thermo Fisher).
Secondary donkey anti-rabbit IgG antibodies conjugated to Cy3, 711-165-152 (1:200, Jackson ImmunoResearch).
Bovine Serum Albumin (BSA), fraction V.
Blocking Buffer: 1x PBS + 1.2 % BSA + 6 % heat inactivated goat serum, filtered through 0.2 μm Nalgene filter.
Cold water fish gelatin (CWFG) (Sigma, G7041). Prepare 45 % (w/v) stock in water (incubate overnight at 37 °C) and 15 % CWFG solution with 15 % sucrose in water. Dissolve at 37°C until completely homogeneous with no air bubbles. For longer storage, add 10 % sodium azide to 0.02 % final concentration.
Dent’s fixative (80% methanol/20% dimethylsulfoxide, DMSO) (Dent et al. 1989).
Donkey or goat serum (Sigma). Heat inactivated for 30 min at 60°C.
Marc’s Modified Ringer’s solution (MMR): 100 mM NaCl, 2 mM KCl, 1 mM MgCl, 2 mM CaCl, 5 mM HEPES, pH7.5 (Newport and Kirschner 1982).
MMR Embryo buffer: 0.1x MMR in water.
Nail polish
Phosphate buffered saline (PBS) buffer, pH 7.4.
Trichloroacetic acid (TCA), 2 % in water (Sigma T-6399)
Vectashield antifade mounting media for fluorescence (Vector Laboratories), liquid or hard set.
Xenopus laevis embryos at the desired stage of development.
Equipment
Dissecting stereomicroscope
Dumont Biologie tip #55 fine forceps (Fine Science Tools, 11255–20).
Coverslip 24 × 60 mm (StatLab SL 102460).
Cryostat (Leica CM3050 S)
Cryostat sample holders
Cryostat sectioning blades, Polycut LP (StatLab, CUT8100).
Cryostat embedding molds, 10×10×5 mm (Tissue-Tek Cryomold, Sakura 4565).
Freezer −80°C
Glass vials, 5 ml.
Hydrophobic barrier pen (PAP pen)
Nutator (Adams) or horizontal shaker
Slide chamber for immunostaining
Paint brushes
Plastic slide holders
Superfrost Plus Slides (Thermo Fisher) or Millennia 2.0 Adhesion Slides StatLab, SL 318L).
Tissue-Plus O.C.T (optimal cutting temperature) compound 4586 (Scigen).
Zeiss AxioImager with the Apotome attachment or equivalent fluorescence or confocal microscope.
Method
This protocol is based on the following publications (Fagotto and Gumbiner 1994; Chalmers et al. 2003) with modifications introduced by our laboratory (Dollar et al. 2005; Ossipova et al. 2007). It is suitable for immunodetection of many embryonic antigens and favorably compares with paraffin sections (Fischer et al. 2008) or cryosections after the embedding into 30 % sucrose-OCT compound as recommended for frog oocytes (Neil and Mowry 2018), zebrafish and chick embryos (Westerfield 2007; Khudyakov and Bronner-Fraser 2009). One advantage of CWFG over the OCT compound is that tissue morphology is preserved better and that embryos can be easily oriented prior to sectioning. However, in situ hybridization works better with OCT-based sections.
Fixation
-
1
Remove the vitelline membrane from Xenopus embryos at the desired stage, using forceps in 1 % agarose dish filled with 0.1 × MMR. Detailed protocols for in vitro fertilization, Xenopus embryo culture and microinjections have been described elsewhere (Sive 1998).
-
2
Place embryos into 5 ml glass vials with the Dent’s fixative that has been chilled on ice. Wash with Dent’s twice to remove water completely. Fix overnight at −20°C. The fixed embryos can be stored at −20°C. Note that the optimal immunostaining may require alternative fixatives, such as 3.7 % formaldehyde (Kim et al. 2012; Chu et al. 2016) or 2 % trichloroacetic acid (Nandadasa et al. 2009; Ossipova et al. 2014; Ossipova et al. 2020).
-
3
Rinse embryos twice in 1x PBS for 5 to 10 min at room temperature (RT).
Embedding and cryosectioning
-
4
Add the embedding solution (15 % CWFG with 15 % sucrose), making sure that the embryos are completely submerged. Equilibrate the vials for 15–20 min at RT, then incubate for 24 hrs at 4°C. The embedded samples can be stored for up to 2 weeks at 4°C.
-
5
Place fresh 15 % CWFG with 15 % sucrose into the embedding chamber. Transfer 5–7 embryos into the center of the mold. Fill up the mold completely with CWFG. To keep track of multiple groups, label the sample.
-
6
Orient the embryos under a stereoscope with a gel-loading pipet tip. Sectioning will start from the bottom side of the gelatin block.
-
7
Place the mold with embryos in a dry ice bucket. Freeze the gelatin block containing embryos on dry ice for 10–20 min. Once frozen, the samples must be sectioned within the same day.
-
8
Release the frozen block from the mold. This is accomplished by making an incision with a razor blade and removing the plastic surrounding the sample.
-
9
Place a sample holder into the cryostat. The cryostat object and chamber temperature (OT and CT) should be set at −19°C and −25°C, respectively.
-
10
Attach the block with embryos to the sample holder. Place some liquid OCT on the holder, attach the frozen block to the holder in a way to start the sectioning with the bottom of the block that has smooth surface. Let it solidify completely for 5–10 min in the cryostat.
-
11
Equilibrate sample for at least 30 min in the cryostat (optional).
-
12
Cut 10–12 μm sections at −19°C OT, −25°C CT. The corner of the block is presented to the blade and as the sections are cut they are stabbed with a toothpick. This is done immediately as the first part of the section comes off the blade’s edge (Figure 1). First, trim the block at 20–30 μm thickness. Once the embryos become visible, set the sectioning thickness to 10–12 μm. Adjust the sample holder to produce properly oriented uniform sections.
-
13
Finish cutting the section and immediately transfer it to Fisherbrand Superfrost Plus or StatLab Millennia 2.0 slides with a wooden toothpick. The slide should be at RT and placed close to the knife, ready to receive the sections. Waiting too long to transfer the section may result in section melting and rolling. Label slides with a pencil to avoid label removal during subsequent acetone treatment.
-
14
Store slides at −80°C after sectioning. Since the immunostaining signal goes down after prolonged storage (more than 2 months), use freshly prepared sectioned embryos for best results.
Figure 1.
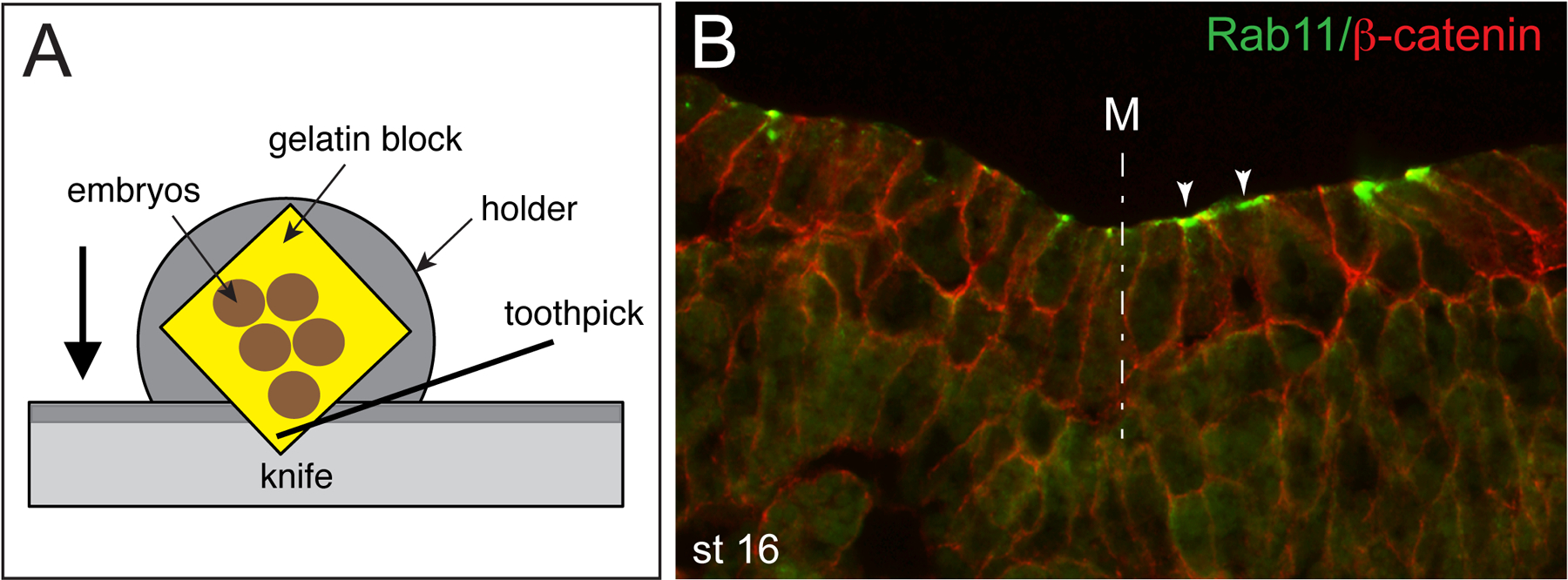
A. Cryosectioning of gelatin-embedded Xenopus embryos. Section is picked with a toothpick from the sectioning blade to be transferred to a microscope slide. Arrow indicates the direction of movement of the specimen attached to the holder. B. Double staining of the neural plate with mouse monoclonal antibodies specific for Rab11 (1:100, Invitrogen) and rabbit polyclonal antibody against β-catenin (1:200, Sigma). Goat secondary antibodies (1:100, Invitrogen) were against mouse or rabbit IgG conjugated to Alexa Fluor 488 or Alexa Fluor 555, respectively. Rab11 is apically localized (arrowheads) in neuroepithelial cells. Midline, M, is indicated (dashed line).
Indirect immunofluorescence
-
15
Dry slides at RT for 1 hr in the fume hood after transferring them from −80°C freezer.
-
16
Place the slides in a histology glass slide holder and remove gelatin by dipping it in a glass reservoir filled with acetone for 5–7 min.
-
17
Dry slides in the fume hood for 10 min to prevent the sections float off the slides during subsequent steps. To reduce the amount of primary antibody, use PAP pen to draw a ring around sections and let it dry completely.
-
18
Place slides horizontally in a humidity chamber with wet Kimwipe tissue at the bottom. Make sure there is enough liquid in the humidity chamber for overnight incubation. Rehydrate slides in PBS for 5 min at RT.
-
19
Add 300–400 μl of the blocking solution (1x PBS + 1.2 % BSA + 6 % heat-inactivated goat or donkey serum) on top of each slide with a 1 ml pipette tip. Make sure slides are not touching each other or the Kimwipe tissue. Block for 60 min at RT in the humidity chamber. Note: For optimal blocking, use serum from the species, in which the secondary antibodies have been raised.
-
20
Replace the blocking solution with 300 to 350 μl of the primary antibody diluted in fresh blocking solution. The optimal dilution of the primary antibody is determined in prior experiments or recommended by the manufacturer. Incubate in the humidity chamber overnight at 4°C (or for 3–4 hrs at RT). Note: Do not forget to include a control sample with no primary antibody.
-
21
Wash 3–4 times with PBS (20 min each, 1 hr total time) at RT. Dry slides in a vertical position on a paper towel. Reduce washing time to 30 min total in case of 3–4 hrs incubation with primary antibody. If the staining produces high background, add 0.1% Triton X-100 or 1–5% DMSO to both the blocking and the washing solutions.
-
22
Place the slides in the humidity chamber and cover each slide with 350 μl of properly diluted fluophore-conjugated secondary antibody in the blocking solution. Incubate for 2 hrs at RT. This and following steps are carried out in the dark by covering the humidity chamber with aluminium foil. If required, add DAPI (0.5 μg/ml final concentration) to the secondary antibody mix.
-
23
For double immunostaining, incubate the samples simultaneously with two primary antibodies raised in different animals (for example, mouse monoclonal anti-GFP and rabbit polyclonal anti-Myc). Use Alexa Fluor488-conjugated goat (or donkey) anti-mouse IgG and Cy3-conjugated donkey anti-rabbit IgG secondary antibodies. Blocking and washing steps remain the same.
-
24
Wash 3 times (20 min each) with PBS at RT.
-
25
Mount the samples with 2–3 drops of the Vectashield mounting medium. Care must be taken when placing the coverslip, as the frozen sections are very fragile. Avoid making air bubbles. If using the liquid set mounting medium, seal your slides with a nail polish.
-
26
The mounted sections are now ready for imaging using regular fluorescence or confocal microscope. The immunostained samples can be stored for several weeks in the dark at 4°C.
Troubleshooting
Problem: Sections are not easily coming off the knife or are rolling.
Solution. Cool down the cryostat (it may not be cold enough) or replace the knife. Rolling of sections may indicate that the block is too large. In that case trimming the block would help. Use correct gelatin powder to prepare gelatin solution. Clean the blade surface and the stage from unused sections using a paint brush, otherwise freshly cut sections would stick to the blade.
Problem: The tissue is detaching from the slides during immunostaining.
Solution: Replace the slides with a different brand of slides containing positively charged surface or purchase a new batch.
Problem: High nonspecific background from antibody incubations.
Solution: To reduce background, it is recommended to add 0.1 % Triton X-100 or 1–5 % DMSO to the washing and/or blocking solutions. When using a commercial antibody, always check manufacturer-suggested fixation and blocking conditions. Some commercial antibodies that are useful for Xenopus embryo staining, their dilutions and fixation conditions are listed in Table 1.
Table 1. Selected antibodies that are suitable for staining of cryosectioned Xenopus tissues.
(antigen, dilution, source, species of origin, special fixation conditions if any).
| β-catenin (1:200, Sigma, rabbit) |
| Centrin-2 (1:200, home-made, rabbit) |
| GFP (1:200, B-2, Santa Cruz, mouse) |
| Protein kinase Cζ (1:200, C-20, Santa Cruz, rabbit) |
| Rab11 (1:100, Zymed, rabbit and BD biosciences, mouse; TCA fixative) |
| Sox3 (1:200, 5H6, Alfandari lab/DSHB, mouse) |
| Acetylated-tubulin (1:200, 6-11B-1, Sigma, mouse, Dent’s fixative) |
| α-tubulin (1:100, B-5-1-2, Sigma, mouse, Dent’s fixative) |
| γ-tubulin (1:150, GTU-88, Sigma, mouse, Dent’s fixative) |
| Vangl2 (1:200, home-made, rabbit, TCA fixative) |
| ZO-1 (1:200, Invitrogen, mouse; Zymed, rabbit) |
ACKNOWLEDGEMENTS
We thank Nancy Papalopulu and Chris Wylie for sharing their laboratory immunostaining protocols. This work in the Sokol laboratory has been supported by the grants from the National Institutes of Health GM122492, HD092990, DE027665 and NS100759.
References
- Chalmers AD, Strauss B, Papalopulu N. 2003. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development 130: 2657–2668. [DOI] [PubMed] [Google Scholar]
- Chu CW, Ossipova O, Ioannou A, Sokol SY. 2016. Prickle3 synergizes with Wtip to regulate basal body organization and cilia growth. Scientific reports 6: 24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent JA, Polson AG, Klymkowsky MW. 1989. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development 105: 61–74. [DOI] [PubMed] [Google Scholar]
- Dollar GL, Weber U, Mlodzik M, Sokol SY. 2005. Regulation of Lethal giant larvae by Dishevelled. Nature 437: 1376–1380. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Gumbiner BM. 1994. Beta-catenin localization during Xenopus embryogenesis: accumulation at tissue and somite boundaries. Development 120: 3667–3679. [DOI] [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, Zeller R. 2008. Paraffin embedding tissue samples for sectioning. CSH Protoc 2008: pdb prot4989. [DOI] [PubMed] [Google Scholar]
- Khudyakov J, Bronner-Fraser M. 2009. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev Dyn 238: 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lake BB, Haremaki T, Weinstein DC, Sokol SY. 2012. Rab11 regulates planar polarity and migratory behavior of multiciliated cells in Xenopus embryonic epidermis. Dev Dyn 241: 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandadasa S, Tao Q, Menon NR, Heasman J, Wylie C. 2009. N- and E-cadherins in Xenopus are specifically required in the neural and non-neural ectoderm, respectively, for F-actin assembly and morphogenetic movements. Development 136: 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil CR, Mowry K. 2018. Fluorescence In Situ Hybridization of Cryosectioned Xenopus Oocytes. Cold Spring Harb Protoc 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. 1982. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30: 675–686. [DOI] [PubMed] [Google Scholar]
- Ossipova O, Kim K, Lake BB, Itoh K, Ioannou A, Sokol SY. 2014. Role of Rab11 in planar cell polarity and apical constriction during vertebrate neural tube closure. Nature communications 5: 3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O, Mancini P, Sokol S. 2020. Direct imaging of core PCP proteins in Xenopus embryos. Methods in Molecular Biology (in press). [DOI] [PubMed] [Google Scholar]
- Ossipova O, Tabler J, Green JB, Sokol SY. 2007. PAR1 specifies ciliated cells in vertebrate ectoderm downstream of aPKC. Development 134: 4297–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H, Grainger RM, Harland RM. 1998. The early development of Xenopus laevis: A laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Westerfield M. 2007. The Zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene. [Google Scholar]


