Abstract
In consecutive serum samples from 25 tourists with acute dengue fever, virus-specific RNA was detected by using fully automated TaqMan reverse transcriptase PCR. For this amplification technique new primers and special fluorochrome-labeled probes had to be synthesized. During amplification the increasing amount of viral DNA could simultaneously be measured in the tightly sealed tubes. Dengue virus RNA was found in almost all patients (17 of 18), if the samples had been taken soon after the onset of symptoms and before anti-dengue virus antibody had been produced. RNA was detectable in only one of five persons who had anti-dengue virus immunoglobulin M (IgM) antibodies but not yet IgG antibodies. In 30 late samples with both IgG and IgM antibodies viral RNA was no longer demonstrable. In two early samples from two frequent travelers obtained 1 and 2 days after the onset of symptoms significant IgG antibody titers were present but there were no anti-dengue virus IgM antibodies. In these samples a viral load of >5 × 106 dengue virus RNA copies (dengue types 1 and 2) was detectable. These findings of a high viral load in the presence of anti-dengue virus IgG antibody are suggestive of a secondary dengue virus infection. In the 20 tourists (17 plus 1 plus 2) in whom viral RNA was found, the dengue virus serotype could be related to the area where the infection had taken place. Most of our patients came from southeast Asia and most frequently had dengue virus type 1 infections (8 of 20).
Dengue fever is endemic in most tropical and subtropical areas worldwide (9, 10, 26), and several hundred thousand dengue hemorrhagic fever cases are reported to occur annually. The increase in dengue fever in humans is paralleled by an increase in the prevalence of Aedes aegypti or Aedes albopictus (9, 10, 14, 24, 26). Due to the vast expansion of air travelling new dengue virus strains may be introduced into a susceptible population in the tropics (20, 21). Also tourists with dengue fever are now frequently seen in areas where dengue fever is not endemic and where physicians are not familiar with the disease (29). As symptoms of dengue fever are usually nonspecific, a reliable diagnosis is difficult to obtain unless virological techniques are included. Both dengue virus-specific immunoglobulin G (IgG) and IgM antibodies are usually found in the sera from patients with acute primary infections, while the IgM response may be low or sometimes even absent in secondary dengue fever (27). However, a strong antibody cross-reactivity exists among the flavivirus family. Therefore, the antibody response may be difficult to interpret with regard to an acute dengue fever, if other flavivirus infections cannot be excluded by clinical, laboratory, or epidemiological means. In contrast, the detection of dengue virus RNA by reverse transcriptase PCR (RT-PCR) in human serum or plasma samples is highly indicative of acute dengue fever (4, 5, 7, 16, 22, 30). Moreover, the latter method is able to identify the dengue virus serotype by demonstrating defined sequence homologies in the viral genomic RNA. Thus, information on the distribution of the four dengue virus serotypes and even of strains or quasispecies in tropical areas can be obtained (15, 17).
Unfortunately, the technique of RT-PCR is handicapped both by time-consuming nested amplification protocols and by false positive reactions which may in part be due to the contamination of dengue virus DNA in the laboratory. We have, therefore, applied a fully automated amplification protocol which sensitively detects all four serotypes but at the same time avoids DNA contamination. By using the TaqMan principle (8, 11, 13) the increase in dengue virus-specific DNA during amplification can be measured by simultaneously monitoring a fluorescence signal in the tightly sealed test tubes. Since the test tubes no longer need to be opened to quantitate the PCR product, a rather simple but highly specific and sensitive test procedure could be obtained which allowed us to operate with numerous serum samples.
MATERIALS AND METHODS
Serum samples.
From 61 tourists with dengue fever included in this study two to three consecutive serum samples could be obtained. Clinical data and the travel history of the patients were obtained by a questionnaire. Upon visiting a region in the tropics where dengue fever is endemic the patients had developed an acute fever with usually slightly elevated levels of aminotransferases and decreased thrombocyte counts. Dengue virus-specific IgM and IgG antibodies and/or fourfold anti-dengue virus IgG titer rises could be demonstrated in the consecutive serum samples of all patients.
Indirect IF antibody test.
The immunofluorescence (IF) test was performed by using cell smears of Vero-E6 cells infected with dengue virus type 1 for 5 days at 37°C. Infected cells were spread on multispot slides, thoroughly air dried, and then fixed in cold acetone at −20°C for 10 min. The slides were sealed in vacuum bags and stored at room temperature. Twofold serum dilutions starting with 1:10 were applied for 1 h. Fluorescein isothiocyanate-labeled anti-human conjugate was used for staining.
μ-Capture enzyme-linked immunosorbent assay (ELISA).
Anti-IgM-coated microtiter plates were prepared as described previously (28). Twofold serum dilutions beginning with 1:10 were incubated for 2 h at room temperature followed by incubating the antigen overnight. The antigen consisted of an undiluted supernatant of Vero cells infected with dengue virus type 1 for 5 days at 37°C. The antigen was stored frozen at −20°C. Then biotinylated anti-West Nile monoclonal antibody (our monoclonal WNT 15R4, cross-reacting with other flaviviruses) was applied (diluted 1:10,000 for 2 h, and finally streptavidin-peroxidase conjugate was used (diluted 1:5,000) before adding the substrate (chloronaphthol).
Selection of optimized primers and probes for the TaqMan PCR.
Since the conditions of the TaqMan amplification differ from ordinary RT-PCRs, it was not possible to use established amplification systems in our assay. Primers and probes were selected by scanning all known dengue virus genomes for a suitable target of amplification. This search was done by using the Primer Express software (PE Applied Biosystems, Foster City, Calif.). The possible targets of amplification were aligned to all known dengue virus sequences (DNA Star software; Perkin Elmer, Norwalk, Conn.), and the most conserved targets for each serotype were chosen for TaqMan RT-PCR (type 1 strain West Pac, accession no. U88535; type 2 strain 16681, accession no. U874; type 3 strain H87, accession no. M93130; type 4 strain H241, accession no. M14931).
In the 3′ nontranslated region of the dengue virus genome the highest level of conservation among the various dengue virus types can be observed (23, 30). However, the short sequences, which were almost identical for all four serotypes, could not be chosen for our primer design, because these regions contained high concentrations of AT nucleotides and inverted repeats. Therefore, suitable universal primer pairs for all four types could not be applied, and individual primers had to be chosen for the amplification of each serotype. For primers, probes, and test-specific parameters see Table 1.
TABLE 1.
Primers, fluorescence-labeled probes, and MgSO4 concentrations for TaqMan amplification
| Dengue virus subtype | Forward primer (concn) | Reverse primer (concn) |
|---|---|---|
| 1 | 5′ ATC CAT GCC CAT CAC CAA TG 3′ (30 pmol) | 5′ CAG GGA TCC ACA CCA CTG ATC 3′ (10 pmol) |
| 2 | 5′ ACA AGT CGA ACA ACC TGG TCC AT 3′ (40 pmol) | 5′ GCC GCA CCA TTG GTC TTC TC 3′ (10 pmol) |
| 3 | 5′ TGG CAA CAG GTC CCT TTC TG 3′ (30 pmol) | 5′ TGG CGT TGG ATG CTA GTC TAA GA 3′ (5 pmol) |
| 4 | 5′ GCG TGG TGA AGC CCC TAG AT 3′ (30 pmol) | 5′ GAA CAA CTA GTG AGC GGC CAT C 3′ (10 pmol) |
| TaqMan probe (concn) | Length of amplified DNA (bp) | MgSO4 concn (mM) |
|---|---|---|
| 5′ FAM AAC ATC TTC CCA ACT GGA TAC ATG GGT TTT GTC 3′TAMRAa (5 pmol) | 162 | 2.5 |
| 5′ FAM TGG GAT TTC CTC CCA TGA TTC CAC TGG 3′TAMRAa (5 pmol) | 177 | 3.5 |
| 5′ FAM AAG AAA GTT GGT TAG TTC CCT GCA GAC CCC A 3′TAMRA | 229 | 4 |
| 5′ FAM TGG CAC TTC CCT CCT CTT CTT GAA CGA C 3′TAMRA | 186 | 2.5 |
Probe binds to complementary strand.
Preparation of positive dengue virus RNA controls.
Standard positive RNA controls were prepared for all four dengue virus serotypes. For this reason the following virus preparations were used: dengue virus types 1 (West Pac) and 3 (NIH H87) had been propagated in baby mouse brain. Approximately 20 mg of mouse brain was dissolved in an RNA lysis buffer (buffer RTL in RNeasy kit; Qiagen, Hilden, Germany), transferred to a 2-ml matrix tube (BIO 101; Dianova, Hamburg, Germany), and twice processed for 20 s in a FastPrep 120 processor (Bio 101). The mixture was then applied to a spin column as described by the manufacturer (Qiagen). Dengue type 2 (16681) and 4 (H241) RNAs were purified from tissue culture supernatants with an activated silica membrane-based kit supplied by Qiagen (QIAamp viral RNA kit). RNA was eluted in diethylpyrocarbonate (DEPC)-treated water and used immediately. The latter procedure was also applied for the purification of human serum samples.
For the construction of the positive controls an RT-PCR was used. Five microliters of the extracted RNA was reverse transcribed for 1 h at 50°C (Superscript II; Life Technologies, Glasgow, United Kingdom) by using the 3′ primers as indicated in Table 1. After reverse transcription the cDNA was subjected to a modified “hot-start-touch-down” PCR. For this PCR 10 μl of the primer mix at the bottom of a thin-walled PCR tube (PE Applied Biosystems) was overlaid with a thin film of Hot Start wax (Biozym, Hamburg, Germany), on top of which the reaction mixture was placed. The PCR was started in a thermocycler (GeneAmp PCR system 9600; Perkin Elmer) at an annealing temperature exceeding the calculated temperature by 10°C. During the first ten cycles the annealing temperature was continuously decreased until the calculated final temperature was reached. During the remaining 30 cycles the constant calculated annealing temperature was applied.
The RT-PCR products were cloned into a PCR cloning vector (TOPO TA cloning kit; Invitrogen BV, Leek, The Netherlands). Positive clones were checked for the correct sequence and orientation by sequencing, with the ABI Prism 310 sequence detection system (PE Applied Biosystems). The resulting plasmids were designated pTADen1 to pTADen4. These plasmids were purified, linearized, and transcribed in vitro with T7 RNA polymerase (T7 transcription kit; MBI Fermentas, St. Leon-Rot, Germany). The RNA transcripts were treated with RNase-free DNase (Boehringer, Mannheim, Germany), extracted with the RNeasy minikit (Qiagen), and ethanol precipitated. The RNA was resuspended in DEPC-treated water, and the concentration was determined by spectrophotometric reading. The RNA was diluted in DEPC-treated water containing 1 μg of carrier RNA (poly[rA]; Boehringer) and stored frozen at −70°C. A defined number of RNA molecules was used for spiking negative human sera.
TaqMan procedure.
During TaqMan amplification an internal probe hybridizes within the region of specific amplification. This internal probe is labeled with two different dyes. When the two dyes are in close proximity, as is the case in an intact oligonucleotide probe, one of the dyes (TAMRA [N,N,N′,N′-tetramethyl-6-carboxyrhodamine]) acts as a quencher for the second fluorescent dye (FAM [5-carboxyfluorescein) by absorbing at the FAM emission spectra. The 5′ exonuclease activity of Taq polymerase will degrade an internally hybridizing probe during the course of PCR (13). The degradation of the probe leads to the separation of these two dyes in solution, with a subsequent increase in the level of fluorescence in the reaction mixture. The amount of fluorescence measured in a sample is proportional to the amount of specific PCR product generated (11). The amplified material is usually discarded without opening the test tubes. Thus, the contamination of the samples by amplified DNA can be completely avoided. Alternatively, for sequencing positive PCR mixtures from serum samples were opened at a strictly separated location and then analyzed by gel electrophoresis and cleaned (QIAex gel extraction kit; Qiagen).
The purified RNA of the serum samples was amplified in thin-walled MicroAmp optical tubes (PE Applied Biosystems). The one-step RT-PCR system (Life Technology Systems) in combination with the ABI Prism 7700 sequence detector (PE Applied Biosystems) was used for uninterrupted thermal cycling. A mastermix reaction solution was prepared and dispensed in 45-μl aliquots into the thin-walled MicroAmp optical tubes, allowing a continuous monitoring of the amount of amplified RNA. Five microliters of RNA extracted from the serum samples or from the positive controls was added to each tube. The final reaction mixture contained the sample RNA, the primers, the respective probe at picomole concentrations as indicated in Table 1; 150 μM concentrations of dATP, dCTP, dGTP, and dTTP (Life Technologies), 2.5 to 4 mM MgSO4 (Table 1), 15% glycerol, 10 mM Tris HCl (pH 8.3), and 1 U of Superscript II RT/Taq Mix (Life Technologies) in a final volume of 50 μl were used. Prior to amplification the RNA was reverse transcribed at 50°C for 30 min. This was followed by one cycle of denaturation at 94°C for 5 min. Next, PCR amplification proceeded with 40 cycles at 94°C for 15 s, 55°C for 1 min, and 72°C for 20 s. The complete procedure, including extraction of RNA, reverse transcription, and amplification, lasts about 5 h.
Monitoring during amplification.
The ABI Prism 7700 sequence detector is capable of analyzing the emitted fluorescence during amplification (on-line monitoring). A positive RT-PCR is measured by the cycle number required to reach the cycle threshold (Ct). The Ct is defined as 10 times the standard deviation of the mean baseline emission calculated for PCR cycles 3 to 15.
RESULTS
Adaptation of the dengue RT-PCR to the TaqMan conditions.
In contrast to other PCR techniques the TaqMan system (11, 13) makes use of a fluorescence-labeled probe that has to be digested by the nuclease activity of the polymerase to monitor the amplification process. For the digestion an almost complete hybridization of the probe to the target DNA is essential. Therefore, a highly conserved region of the dengue virus genome had to be chosen to allow optimum annealing not only of the primers but also of the labeled probe. Therefore, by using the TaqMan technique new primers and suitable probes had to be selected. Since even slight mismatches interfere with efficient TaqMan probing, published consensus primers (12, 19, 23, 30, 31) enclosing rather variable sequences could not be used for the detection of the dengue virus RNA. New primers and probes were found in the 3′ NS3 coding region of the dengue virus genome by using DNA Star software (Perkin Elmer). For an efficient amplification and probing four different RT-PCR protocols had to be applied. The individual primers and probes for each serotype amplification are listed in Table 1. Fortunately, the four TaqMan serotype RT-PCRs could be run in parallel by using identical time and temperature profiles. The concentration of MgSO4 and the amount of primers had to be adapted to the TaqMan conditions, and the optimum concentrations are also included in Table 1.
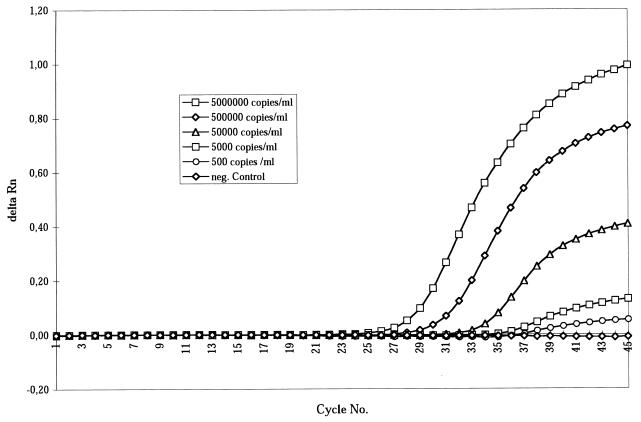
The sensitivity and specificity of the dengue virus RNA detection were evaluated by using fixed amounts of in vitro-transcribed dengue virus RNA of all four serotypes. When the RNA standards containing 5 × 106 molecules/ml were applied in 10-fold dilutions it turned out that about 500 RNA molecules/ml (corresponding to two to three molecules in 5 μl in the test tube) could be detected (Fig. 1). Only one of 120 assays gave a false negative result (confidence intervals, 95.4 to 99.9%). The specificity of the TaqMan amplification was controlled by using 20 serum samples from German patients in whom hepatitis C virus RNA had been detected with the quantitative Amplicor RT-PCR (Roche, Bâle, Switzerland). The samples were negative for dengue virus antibody, and besides a recent visit to tropical areas could be excluded. Upon TaqMan amplification dengue virus serotype RNA was not falsely detectable in any of the 20 sets of four test tubes.
FIG. 1.
Standardization of dengue virus type 2 TaqMan RT-PCR. Totals of 500 to 5 × 106 RNA molecules/ml were diluted in human sera from healthy subjects without anti-dengue virus antibodies. A minimum of 500 RNA molecules/ml, corresponding to two to three molecules in 5 μl of RNA extract in the amplification mix, could be detected after 38 cycles. On the y axis is shown the intensity of fluorescence (delta Rn) (FAM-TAMRA), and on the x axis is shown the number of cycles.
Detection of dengue virus RNA in human serum samples.
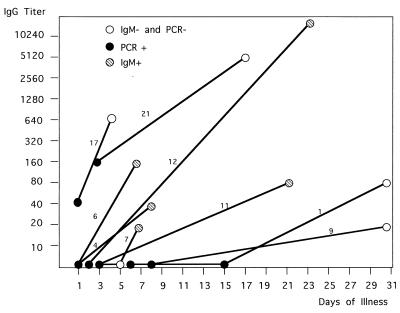
During the second half of 1998, serum samples from 1,332 European tourists returning from tropical areas were sent to our institute for anti-dengue virus antibody testing. Dengue fever was diagnosed in 172 of the tourists by detecting specific IgM antibody and/or a significant rise in anti-dengue virus IgG antibody titer. Consecutive serum samples were available from 61 patients. In 36 of the 61 patients both IgG and IgM antibodies were already present in the first serum sample. As it is unlikely to detect dengue virus in the presence of high neutralizing antibody titers (3) these 36 samples were not tested for dengue virus RNA. The serum samples from 18 patients who did not have any anti-dengue virus antibody at all yet in their early samples were subjected to TaqMan RT-PCR. We also included five patients with IgM antibody only and two with IgG antibody only. The first samples of the 25 (18 plus 5 plus 2) patients were obtained 1 to 8 days after the onset of symptoms. Dengue virus serotype RNA could be demonstrated in 20 of the 25 early serum samples, while in 30 consecutive late samples with high anti-dengue virus IgG titers collected between days 5 and 31 no viral RNA could be detected. In more detail, 17 of 18 early serum samples without any anti-dengue virus antibody contained dengue virus RNA as detected by TaqMan RT-PCR (94%). In early samples viral RNA was demonstrated in one of five patients in whom IgM was already detectable, and in another two early samples viral RNA was found in the presence of relatively high IgG titers but without specific IgM (patients 17 and 21 [Fig. 2]).
FIG. 2.
Kinetics of IgG antibody response in consecutive serum samples from nine representative patients. Each sample is represented by a circle. In addition to the IgG titers (y axis) a positive PCR is indicated by a black circle and a positive anti-dengue virus IgM response is indicated by a striped circle. Absence of IgM or RNA is indicated by a white circle. Patients 4, 12, and 21 had dengue virus type 1 infections; patients 1, 6, 9, 11, and 17 had type 2 infections; and patient 7 had a type 3 infection.
In Table 2 the viral serotypes and the immune response in the first and also in later (second or third) samples of the 20 RNA-positive patients are listed. Dengue virus type 1 RNA was found most frequently (nine sera), while dengue virus type 4 was not detectable in any of the samples. In late serum samples obtained 5 to 31 days after the onset of symptoms anti-dengue virus IgM antibodies could be detected in 17 of 20 subjects by μ-capture ELISA (Table 2), with titers from 1:20 up to 1:2,560, and IgG antibodies were found in all 20 patients. From the RT-PCR positive samples we tried to isolate dengue virus in tissue culture. However, retrospectively after storage at 4°C for at least 7 days dengue virus (types 1 and 2) could be cultivated in only two of them.
TABLE 2.
Distribution of dengue virus subtype RNA in serum samples from 20 patients with acute dengue fever
| Dengue virus subtype | No. of patients with dengue virus RNA in the first sample | No. of first samples with:
|
No. of second or third samples with:
|
Geographic distribution (no. of cases) | ||
|---|---|---|---|---|---|---|
| IgM | IgG | IgM | IgG | |||
| 1 | 12 | 0 | 1 | 11 | 12 | Southeast Asia (8) |
| Caribbean Islands (1) | ||||||
| Central Africa (1) | ||||||
| Samoa (1) | ||||||
| 2 | 6 | 1 | 1 | 4 | 6 | Southeast Asia (5) |
| Caribbean Islands (1) | ||||||
| 3 | 2 | 0 | 0 | 2 | 2 | Southeast Asia (2) |
| 4 | 0 | |||||
Due to the fact that the initial amount of target RNA molecules is directly correlated to the respective Ct, a rough estimation of viral load in the serum sample can be made (Fig. 1). A very high load of >106 RNA molecules/ml was found in both patients with suspected secondary infection (patient 17, dengue virus type 2; patient 21, dengue virus type 1), while a very low load of 103 molecules/ml was seen in the early sample of patient 3, in which a low concentration of specific IgM antibody had been detected.
In Fig. 2 the kinetics of the IgG antibody response of nine representative patients is shown. Positive results obtained by TaqMan PCR and by IgM antibody testing are also indicated. Viral RNA was detected only in the first samples collected during days 1 to 8 after the onset of symptoms. In some patients (1, 9, and 11 [Fig. 2]) the production of IgG antibody was delayed and the final titer did not exceed 1:80. In such patients a dengue virus type 2 infection was usually identified. In seven of the nine patients a seroconversion could be documented both by ELISA and IF. No IgG or IgM antibodies were present in the first serum samples from which the viral RNA could be amplified. In contrast, the sera of two patients (17 and 21 [Fig. 2]) had high IgG antibody levels, but no anti-dengue virus IgM antibodies were present on the 1st or 2nd day after the onset of symptoms. Both patients had been in southeast Asia at least twice and had acquired a dengue virus type 2 and type 1 infection, respectively. They did not recollect a previous dengue-like disease, and they did not report any flavivirus vaccinations. The initial anti-dengue virus IgG titer together with the missing anti-dengue virus IgM response and the high viral load is highly suggestive of a secondary infection in the two patients, although a preceding nondengue flavivirus infection could not be completely excluded.
In the patients in whom serotype-specific dengue virus RNA had been found, the travel history during the previous 3 weeks could be documented. As can be seen from Table 2, most of these patients had been visiting southeast Asia (usually Thailand). Here dengue virus types 1 and 2 were most prevalent. No dengue virus type 4 case was identified, although 2 of the 20 early serum samples were taken from patients after they had visited the Caribbean islands where dengue virus type 4 seems to be prevailing (1). Besides, we did not obtain any amplification data consistent with double infections or unspecific results, i.e., the other three serotype-specific reactions were always negative when a positive amplification reaction was found.
DISCUSSION
Dengue fever is a severe health problem in tropical areas, with two-thirds of the world population at risk of infection (24). Four virus serotypes and even more quasispecies can be differentiated both in the transmitting insect (A. aegypti) and in the blood of patients with acute dengue fever (26). During recent decades Europe has been free of dengue infections, but as a result of increasing tourism every year several thousand tourists return to European countries with acute dengue fever.
In the second half of 1998 we diagnosed 172 dengue patients. All patients were European tourists, most of whom had experienced a disease with high fever and headache for several days. Usually early serum samples were taken 4 to 8 days after the onset of symptoms. Therefore, early serum samples without dengue virus antibodies at all or with either IgG or IgM antibodies were obtained from only 25 patients (15%). In 20 early samples viral RNA could be demonstrated by TaqMan PCR, in one case even in the presence of low IgM antibody concentrations. Thus, the high sensitivity of the TaqMan procedure as documented by a detection limit of about three RNA molecules per assay is also reflected by the 94% positive PCR results in early samples without any anti-dengue virus antibodies.
Also false positive results were not observed when IgG antibody-negative samples from hepatitis C subjects or late samples from patients with dengue fever containing high IgG and IgM antibody titers were analyzed. Using the earlier nested RT-PCR protocols, including agarose gel electrophoresis (7, 29), contamination with dengue virus DNA was always difficult to avoid. In contrast, the TaqMan PCR procedure allows the monitoring of amplification in sealed test tubes. This constitutes the most important progress toward the elimination of false positive results due to cross-contamination. Besides, the dengue virus subtype can be identified directly during amplification, while the three remaining test tubes serve as internal negative controls. On the other hand, the method described here is not especially useful in tracing dengue viral quasispecies (17, 18, 25, 32) because highly conserved regions in the 3′ coding region of the dengue virus genome were the targets of our amplification procedure. When we sequenced our amplified material, it turned out that compared to the published sequences of the four serotypes only 2 to 12 base exchanges were observed (data not shown). For the construction of parsimony trees the E glycoprotein gene would be more suitable. For this reason a specially designed nested RT-PCR procedure (6, 25, 32) might be applied to our RNA samples.
Our results with the detection of viral RNA by using the TaqMan procedure confirm earlier findings showing that dengue virus can be readily detected in early serum samples taken before IgG antibodies are present (2, 3). Even in the presence of low IgM titers viral RNA may be detectable at least at an early point during IgM production.
In the European tourists living in areas where dengue fever is not endemic, primary infections will usually be seen. Accordingly, the typical immune response with both anti-dengue virus IgM and IgG antibody production was observed in almost all patients presented here. Only in a late sample (31 days after onset from patient 9) and during secondary infections (patients 17 and 21) specific IgM antibodies were not detectable either with our ELISA or with a commercial rapid dengue virus IgM test (PanBio, Brisbane, Australia). From a statistical point of view tourists are usually exposed to dengue virus only once during life, but due to repeated visits to tropical areas in the future secondary infections with new serotypes will be seen more often, even in tourists living in countries where dengue is not endemic. We report here two secondary infections (tourists 17 and 21 [Fig. 2]) for whom viral RNA could be detected in early serum samples in the presence of high dengue virus IgG antibody concentrations but without the production of specific IgM antibody. In both patients hemorrhagic signs were not present. Also symptoms consistent with primary dengue fever were not recollected by the patients. We speculate that cross-reacting anti-dengue virus antibodies present in the blood were unable to clear the virus but enhanced the production of dengue virus which was detected at a high concentration in both patients.
As we have demonstrated here, the TaqMan RT-PCR is a suitable tool to collect data on the viral load in the blood of dengue fever patients. A correlation of the viral load with clinical data may provide important information on the pathogenesis of the different dengue-associated syndromes such as dengue hemorrhagic fever and dengue shock syndrome.
ACKNOWLEDGMENTS
We thank P. C. Grauballe, Copenhagen, Denmark, and T. Jänisch, Heidelberg, Germany, for supplying serum samples and clinical data. We also appreciate the technical assistance of G. Rietdorf and C. Thomé-Bolduan.
This work was supported by the Bundesamt für Wehrtechnik und Beschaffung (grant M5916).
REFERENCES
- 1.Anonymous. Dengue outbreak associated with multiple serotypes—Puerto Rico, 1998. Morbid Mortal Weekly Rep. 1998;47:952–956. [PubMed] [Google Scholar]
- 2.Brown J L, Wilkinson R, Davidson R N, Wall R, Lloyd G, Howells J, Pasvol G. Rapid diagnosis and determination of duration of viraemia in dengue fever using a reverse transcriptase polymerase chain reaction. Trans R Soc Trop Med Hyg. 1996;90:140–143. doi: 10.1016/s0035-9203(96)90115-7. [DOI] [PubMed] [Google Scholar]
- 3.Chan S Y, Kautner I M, Lam S K. The influence of antibody levels in dengue diagnosis by polymerase chain reaction. J Virol Methods. 1994;49:315–322. doi: 10.1016/0166-0934(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 4.Chang G-J J, Trent D W, Vorndam A V, Vergne E, Kinney R M, Mitchell C J. An integrated target sequence and signal amplification assay, reverse transcriptase–PCR–enzyme-linked immunosorbent assay, to detect and characterize flaviviruses. J Clin Microbiol. 1994;32:477–483. doi: 10.1128/jcm.32.2.477-483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow V T. Molecular diagnosis and epidemiology of dengue virus infection. Ann Acad Med Singap. 1997;26:820–826. [PubMed] [Google Scholar]
- 6.Deubel V. Recent advances and prospective researchers on molecular epidemiology of dengue viruses. Mem Inst Oswaldo Cruz. 1992;87(Suppl. 5):133–136. doi: 10.1590/s0074-02761992000900021. [DOI] [PubMed] [Google Scholar]
- 7.Deubel V, Laille M, Hugnot J P, Chungue E, Guesdon J L, Drouet M T, Bassot S, Chevrier D. Identification of dengue sequences by genomic amplification: rapid diagnosis of dengue virus serotypes in peripheral blood. J Virol Methods. 1990;30:41–54. doi: 10.1016/0166-0934(90)90042-e. [DOI] [PubMed] [Google Scholar]
- 8.Förster V T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann Phys. 1948;2:55–75. [Google Scholar]
- 9.Gubler D J. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubler D J. The global pandemic of dengue/dengue haemorrhagic fever: current status and prospects for the future. Ann Acad Med Singap. 1998;27:227–234. [PubMed] [Google Scholar]
- 11.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 12.Henchal E A, Polo S L, Vorndam V, Yaemsiri C, Innis B L, Hoke C H. Sensitivity and specificity of a universal primer set for the rapid diagnosis of dengue virus infections by polymerase chain reaction and nucleic acid hybridization. Am J Trop Med Hyg. 1991;45:418–428. doi: 10.4269/ajtmh.1991.45.418. [DOI] [PubMed] [Google Scholar]
- 13.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igarashi A. Impact of dengue virus infection and its control. FEMS Immunol Med Microbiol. 1997;18:291–300. doi: 10.1111/j.1574-695X.1997.tb01058.x. [DOI] [PubMed] [Google Scholar]
- 15.Kanesa-thasan N, Chang G J, Smoak B L, Magill A, Burrous M J, Hoke C H., Jr Molecular and epidemiologic analysis of dengue virus isolates from Somalia. Emerg Infect Dis. 1998;4:299–303. doi: 10.3201/eid0402.980220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanciotti R S, Calisher C H, Gubler D J, Chang G-J, Vorndam A V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis J G, Chang G J, Lanciotti R S, Trent D W. Direct sequencing of large flavivirus PCR products for analysis of genome variation and molecular epidemiological investigations. J Virol Methods. 1992;38:11–23. doi: 10.1016/0166-0934(92)90165-a. [DOI] [PubMed] [Google Scholar]
- 18.Mangada M N, Igarashi A. Molecular and in vitro analysis of eight dengue type 2 viruses isolated from patients exhibiting different disease severities. Virology. 1998;244:458–466. doi: 10.1006/viro.1998.9093. [DOI] [PubMed] [Google Scholar]
- 19.Meiyu F, Huosheng C, Cuihua C, Xiaodong T, Lianhua J, Yifei P, Weijun C, Huiyu G. Detection of flaviviruses by reverse transcriptase-polymerase chain reaction with the universal primer set. Microbiol Immunol. 1997;41:209–213. doi: 10.1111/j.1348-0421.1997.tb01192.x. [DOI] [PubMed] [Google Scholar]
- 20.Monath T P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monath T P. Early indicators in acute dengue infection. Lancet. 1997;350:1719–1720. doi: 10.1016/S0140-6736(05)63567-2. [DOI] [PubMed] [Google Scholar]
- 22.Morita K, Maemoto T, Honda S, Onishi K, Murata M, Tanaka M, Igarashi A. Rapid detection of virus genome from imported dengue fever and dengue hemorrhagic fever patients by direct polymerase chain reaction. J Med Virol. 1994;44:54–58. doi: 10.1002/jmv.1890440111. [DOI] [PubMed] [Google Scholar]
- 23.Pierre V, Drouet M T, Deubel V. Identification of mosquito-borne flavivirus sequences using universal primers and reverse transcription/polymerase chain reaction. Res Virol. 1994;145:93–104. doi: 10.1016/s0923-2516(07)80011-2. [DOI] [PubMed] [Google Scholar]
- 24.Pinheiro F P, Corber S J. Global situation of dengue and dengue haemorrhagic fever, and its emergence in the Americas. World Health Stat Q. 1997;50:161–169. [PubMed] [Google Scholar]
- 25.Rico-Hesse R, Harrison L M, Salas R A, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa M T, Nogueira R M, da Rosa A T. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 26.Rigau-Pèrez J G, Clark G G, Gubler D J, Reiter P, Sanders E J, Vorndam A V. Dengue and dengue haemorrhagic fever. Lancet. 1998;352:971–977. doi: 10.1016/s0140-6736(97)12483-7. [DOI] [PubMed] [Google Scholar]
- 27.Rossi C A, Drabick J J, Gambel J M, Sun W, Lewis T E, Henchal E A. Laboratory diagnosis of acute dengue fever during the United Nations mission in Haiti, 1995–1996. Am J Trop Med Hyg. 1998;59:275–278. doi: 10.4269/ajtmh.1998.59.275. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz H, Emmerich P. Detection of specific immunoglobulin M antibody to different flaviviruses by use of enzyme-labeled antigens. J Clin Microbiol. 1984;19:664–667. doi: 10.1128/jcm.19.5.664-667.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz H, Emmerich P, ter Meulen J. Imported tropical virus infections in Germany. Arch Virol Suppl. 1996;11:67–74. doi: 10.1007/978-3-7091-7482-1_8. [DOI] [PubMed] [Google Scholar]
- 30.Sudiro T M, Ishiko H, Green S, Vaughn D W, Nisalak A, Kalayanarooj S, Rothman A L, Raengsakulrach B, Janus J, Kurane I, Ennis F A. Rapid diagnosis of dengue viremia by reverse transcriptase-polymerase chain reaction using 3′-noncoding region universal primers. Am J Trop Med Hyg. 1997;56:424–429. doi: 10.4269/ajtmh.1997.56.424. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka M. Rapid identification of flavivirus using the polymerase chain reaction. J Virol Methods. 1993;41:311–322. doi: 10.1016/0166-0934(93)90020-r. [DOI] [PubMed] [Google Scholar]
- 32.Thayan R, Morita K, Vijayamalar B, Zainah S, Chew T K, Oda K, Sinniah M, Igarashi A. Comparison of sequences of E/NS1 gene junction of dengue type 3 virus following culture subpassage in C6/36 cells to study the possible occurrence of mutations. Southeast Asian J Trop Med Public Health. 1997;28:380–386. [PubMed] [Google Scholar]