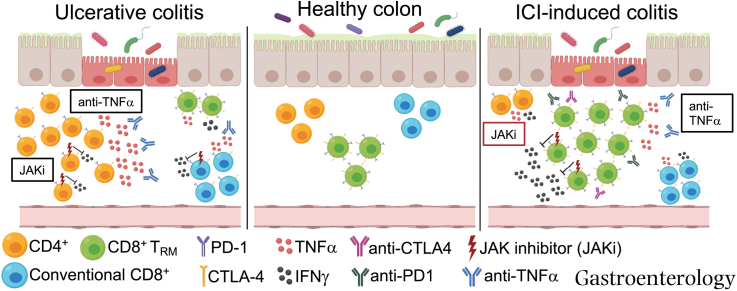
Abstract
Background & Aims
The pathogenesis of immune checkpoint inhibitor (ICI)–colitis remains incompletely understood. We sought to identify key cellular drivers of ICI-colitis and their similarities to idiopathic ulcerative colitis, and to determine potential novel therapeutic targets.
Methods
We used a cross-sectional approach to study patients with ICI-colitis, those receiving ICI without the development of colitis, idiopathic ulcerative colitis, and healthy controls. A subset of patients with ICI-colitis were studied longitudinally. We applied a range of methods, including multiparameter and spectral flow cytometry, spectral immunofluorescence microscopy, targeted gene panels, and bulk and single-cell RNA sequencing.
Results
We demonstrate CD8+ tissue resident memory T (TRM) cells are the dominant activated T cell subset in ICI-colitis. The pattern of gastrointestinal immunopathology is distinct from ulcerative colitis at both the immune and epithelial-signaling levels. CD8+ TRM cell activation correlates with clinical and endoscopic ICI-colitis severity. Single-cell RNA sequencing analysis confirms activated CD8+ TRM cells express high levels of transcripts for checkpoint inhibitors and interferon-gamma in ICI-colitis. We demonstrate similar findings in both anti–CTLA-4/PD-1 combination therapy and in anti–PD-1 inhibitor-associated colitis. On the basis of our data, we successfully targeted this pathway in a patient with refractory ICI-colitis, using the JAK inhibitor tofacitinib.
Conclusions
Interferon gamma–producing CD8+ TRM cells are a pathological hallmark of ICI-colitis and a novel target for therapy.
Keywords: Immunotherapy Colitis, Checkpoint Colitis, Ulcerative Colitis, Tofacitinib
Abbreviations used in this paper: cDNA, complementary DNA; DCC, anti–CTLA-4/PD-1 dual checkpoint inhibitor colitis; DCNC, anti–CTLA-4/PD-1 dual checkpoint inhibitor no colitis; FMT, fecal microbiota transplantation; HV, healthy volunteer; ICI, immune checkpoint inhibitor; IFNG, interferon gamma; irAE, immune-related adverse event; PD-1, programmed cell death protein 1; PDC, anti–PD-1-monotherapy colitis; RNASeq, RNA sequencing; scRNA, single-cell RNA; TRM, tissue resident memory T cell; UC, ulcerative colitis; UCEIS, Ulcerative Colitis Endoscopic Index of Severity
Graphical abstract
We present an analysis of the immunopathology in checkpoint-inhibitor colitis, a common adverse effect of cancer immunotherapy. We used our findings to successfully identify a novel therapy for a case of refractory colitis.
See Covering the Cover synopsis on page 1079; See editorial on page 1106.
What You Need to Know.
Background and Context
ICI-colitis is a common adverse effect of checkpoint inhibitors, can mimic IBD, and currently has empirically derived treatment guidelines.
New Findings
We identify both unique and overlapping immunopathology in ICI-colitis and UC. CD8+ TRM cells are the key effector cells in ICI-colitis. TRM cells strongly express checkpoint proteins and IFNG. We present the first PD1-inhibitor–associated colitis single-cell analysis that demonstrates consistent TRM cell activation. IFNG-JAK-STAT activation identified tofacitinib as a potential therapy, although IFNG blockade could negatively affect oncological response.
Limitations
This is a small human cohort study. Further investigation will be required to understand the role of the microbiome in TRM cell activation and the safety of JAK inhibition.
Impact
This analysis of CD8+ TRM cells and associated immune pathways in ICI-colitis provides a basis for targeted therapy development. We provide an immunologic rationale for the use of JAK inhibitor therapy in refractory ICI-colitis.
Immune checkpoint inhibitors (ICIs) are revolutionizing the treatment of melanoma and other cancers but come at the cost of immune-related adverse events (irAEs). These irAEs commonly affect the gastrointestinal tract, with those receiving combination anti–CTLA-4 and PD-1 therapy displaying increased rates of ICI-colitis (32%–37%) compared to those treated with anti–PD-1 monotherapy (4%–6%).1,2 There is a higher incidence of ICI-diarrhea (44%),1 probably due to unconfirmed colitis. ICI-colitis results in the greatest overall mortality of irAE, although other rarer toxicities (eg, myocarditis) have lower individual survival rates.3
Current management for ICI-colitis includes systemic corticosteroids and subsequent anti-TNFα therapy (infliximab) for inadequately controlled disease.4 Alternative therapies include anti-α4β7 integrin (vedolizumab)5 and fecal microbiota transplantation (FMT).6 These therapeutic approaches are empirically derived from the treatment of idiopathic inflammatory bowel disease (IBD), without an understanding of how analogous this newer entity is to more classical forms of colitis. Refractory cases of ICI-colitis occur, resulting in steroid toxicity and, on occasion, colectomy. It is anticipated that greater insight into the mechanisms underlying ICI-colitis will lead to more targeted treatments. ICI-colitis is heterogeneous, but can mimic UC and, less commonly, Crohn’s disease.7 We opted to use UC as the external disease control, as both conditions typically affect the rectum and/or sigmoid colon and are amenable to flexible sigmoidoscopy.
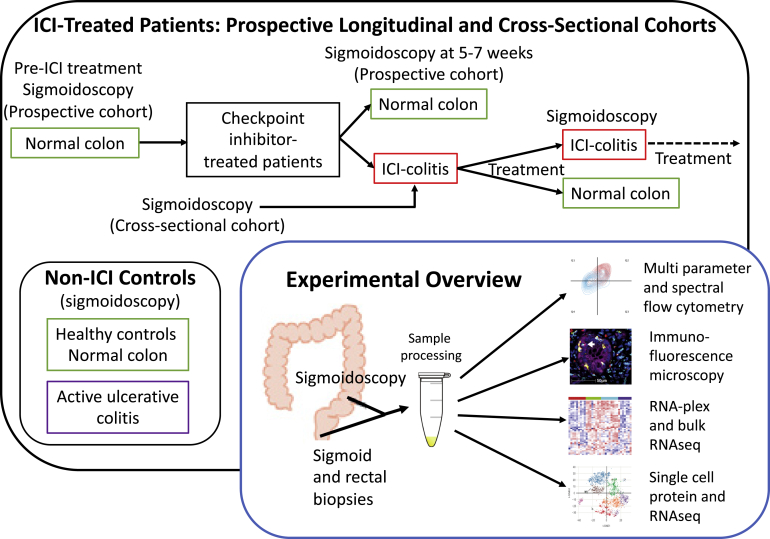
At the outset of this study, little was known about the cellular and molecular pathogenesis of ICI-colitis. The available data suggested that anti–CTLA-4–associated colitis is associated with CD8+ T cells8 and an up-regulation of Th1 and Th17 effector pathways, including interferon-gamma (IFNG).9 We previously demonstrated that anti–CTLA-4/PD-1 colitis is associated with high levels of activated (HLA-DR+CD38+) memory CD8+ T cells,10 and lower proportions of regulatory T cells compared with UC.10 We hypothesized that CD8+ TRM cells are implicated in the pathogenesis of ICI-colitis, postulating that they would become activated in an off-target consequence, and sought to understand the signaling pathways involved (Figure 1).
Figure 1.
Study design.
TRM cells are specialist lymphocytes enriched at mucosal sites, including the gut,11 and display minimal circulation. CD8+ TRM cells classically express CD69 and CD103, and play an important role in mucosal immunity (reviewed in Sasson et al12). They are implicated in the pathogenesis of autoimmune skin conditions.13 TRM-like tumor infiltrating lymphocytes also mediate anti-tumor responses,14,15 and a higher proportion of TRM-like tumor infiltrating lymphocytes correlates with disease-free survival.16,17 TRM-like tumor infiltrating lymphocytes express high levels of checkpoint proteins,18, 19, 20 which appear to act primarily as negative regulators rather than markers of exhaustion.
Recent data utilized single-cell technology to identify cytotoxic CD8+ T cells as the main pathogenic gastrointestinal population in anti–CTLA-4/PD-1 colitis.21 It was inferred through T cell receptor sequence analysis that these may derive, in part, from TRM cells. Up-regulation of both IFNG and TNFA signaling pathways was identified in the CD4+ and CD8+ T cells of patients with ICI-colitis, acting on the myeloid cellular compartment.21 Our study supports and extends the prior analysis via a broader range of experimental techniques. We also provide a direct comparison with idiopathic UC and include analysis of anti–PD-1 monotherapy colitis and anti–CTLA-4/PD-1 gastritis. Our data taken together directly implicate IFNG-expressing CD8+ TRM cells as a major pathogenic T cell population in the colon.
Finally, on the basis of our laboratory data, we predicted that tofacitinib would be an effective therapy in a case of refractory anti–PD-1 colitis and used this successfully. These data, consistent with the subsequently published case reports of successful tofacitinib therapy,22,23 provide compelling insights into the underlying pathogenic mechanisms in this clinical setting and novel pathways to target therapeutically.
Materials and Methods
Subjects
We studied patients with anti–CTLA-4/PD-1 colitis (dual checkpoint inhibitor colitis [DCC]; n = 15), anti–CTLA-4/PD-1 treated with no colitis (DCNC; n = 10), anti–PD-1 colitis (PDC; n = 6), anti–PD-1–treated with no colitis (n = 5), active UC (disease flare assessment; n = 10), and healthy volunteers (HVs; n = 22). The patients with UC and ICI-colitis are reasonably matched in terms of inflammation severity and previous and current therapy but do have a longer median acute flare duration in the UC cohort (see Supplementary Table 1), and the usual caveats must apply when interpreting real-world human data from subjects that are not perfectly aligned. All patients provided written informed consent for participation (Oxford GI Illness Biobank 16/YH/0247 or PRISE [Predicting Immunotherapy Side Effects] study, London-Surrey Research Ethics Committee: REC18/LO/0412). This consent enabled invitation for sigmoidoscopy pre-ICI, at week 5–7 post ICI treatment and at the development of colitis symptoms (see Figure 1). Colitis disease activity is comparable across UC and ICI-colitis. The duration of active colitis and the treatment at the time of sampling are detailed in Supplementary Table 1. Rectal and sigmoid biopsies were protocolized. Individual patient samples studied in each experiment are shown in Supplementary Table 2.
The clinical characteristics of patients with anti–CTLA-4/PD-1–associated gastritis (n = 4) and HV controls (n = 7) are shown in Supplementary Table 3.
Isolation of Mononuclear Cells From Gastrointestinal Tissue
Colonic biopsies were placed in RPMI media containing penicillin and streptomycin and 10% fetal calf serum. Biopsies underwent enzymatic and mechanical digestion with 1 mg/mL Collagenase D (Sigma-Aldrich) and 100 μg/mL DNAse I (ThermoFisher) and were shaken at 37°C for 1 hour. Biopsies were dissociated using gentleMACS Dissociator (Miltenyi Biotech) and passed through a 70-μm strainer.
Flow Cytometry
Flow cytometry was performed on freshly isolated mononuclear cells using a near-infrared live/dead stain (Invitrogen) and an initial monoclonal antibody panel (see Supplementary Methods) was performed on a 3-laser LSR Fortessa X-20 (BD Biosciences). An extended TRM cell phenotyping monoclonal antibody panel (see Supplementary Methods) was performed on an Aurora spectral analyzer (Cytek).
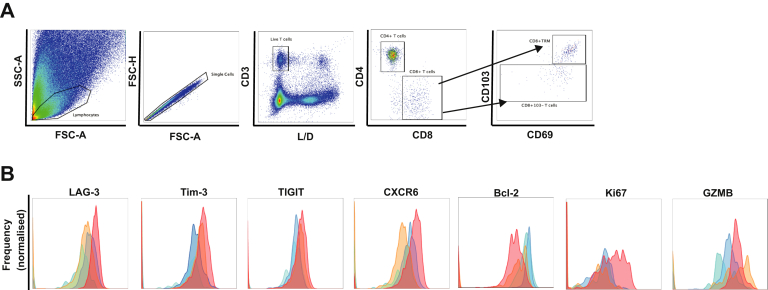
Data were analyzed using FACSDIVA software, version 8.0.1 (BD Biosciences). Gating strategies are shown in Supplementary Figure 2A. Lymphocyte populations are reported as a proportion of parent populations.
Supplementary Figure 2.
(A) Flow cytometric gating strategy. (B) CD8+CD69+CD103+ TRM cells in anti-CTLA-4/PD-1 colitis (red) displayed high levels of checkpoint molecules LAG-3, Tim-3, TIGIT, and chemokine receptor CXCR6. Intracellularly they had high Ki-67 and granzyme-B and low levels of anti-apoptotic protein Bcl-2. Anti-CTLA-4/PD-1–associated colitis non-TRM cells (orange), CTLA-4/PD-1 treatment with no colitis CD8+ TRM cells (dark blue) and non-TRM cells (light blue). Histograms based on live CD45+CD8+ T cells. Data are representative of 5 experiments.
Statistics
Differences between groups were determined using the unpaired nonparametric Mann-Whitney test. Correlation analysis was performed using the nonparametric Spearman test. All analyses were performed using SPSS software (IBM, Armonk, NY). Medians and interquartile ranges are reported throughout. A P value < .05 was considered statistically significant. When multiple comparisons were performed, a Bonferroni correction was made (see figure legends).
Multiplex Immunofluorescence Microscopy
Multiplex immunofluorescence staining was carried out on 4-μm formalin-fixed paraffin embedded sections using the OPAL protocol (AKOYA Biosciences) on the Leica BOND RXm autostainer (Leica Microsystems). Six consecutive staining cycles were performed using primary antibody-Opal fluorophore pairings. Whole slide scans and multispectral images were obtained on the AKOYA Biosciences Vectra Polaris. Batched analyzed multispectral images were fused in HALO (Indica Labs) to produce a spectrally unmixed reconstructed whole tissue image.
Nanostring RNA Plex
Targeted gene expression was measured using 150 μg of RNA extracted from pinch biopsies and a 770-gene human autoimmune profiling panel with a custom 10-gene spike in set (Supplementary Methods). Samples were analyzed on an nCounter Sprint profiler with downstream analysis using nSolver freeware (Nanostring), Gene Set Enrichment Analysis (Broad Institute), and R studio (Boston).
Bulk RNA Sequencing
Bulk RNA sequencing (RNASeq) analysis was performed using 900 ng per sample of RNA extracted from pinch biopsies and the GRCH37.EBVB95-8wt reference genome. Total RNA was converted to complementary DNA (cDNA) with second-strand cDNA incorporating a 2'-deoxyuridine 5'-triphosphate. cDNA was end-repaired with PolyA tails and was adapter-ligated. Sequencing was performed on a NovaSeq6000 (Illumina). Bulk RNASeq was analyzed using Partek Flow software. Library generation and sequencing were performed at the Oxford Genomics Centre. Data analysis was performed according to published standards.24, 25, 26, 27
10X Genomics Library Preparation and Sequencing
Single-cell (sc)RNASeq libraries were generated using 10X Genomics Chromium scRNA Reagents Kits (v1 Chemistry). Live CD45+ cells were sorted using a FACSAriaIII cell sorter (BD Biosciences) and resuspended in phosphate-buffered saline with 0.04% bovine serum albumin at approximately 1000 cells/μL and loaded onto 2 lanes of the Chromium Controller. Captured cell number was 5876. Library quality and concentration was determined using a TapeStation (Agilent) and Qubit 2.0 Fluorometer (Thermo Fisher). Libraries were sequenced on an Illumina HiSeq 4000 to a mean depth of 64,000 mean reads/cell. Library generation and sequencing were performed at the Oxford Genomics Centre.
Droplet-Based (10X Genomics) Single-Cell RNA Sequencing Data Analysis
FastQ generation, read alignment, barcode counting, and UMI counting was performed using the Cell Ranger Pipeline, version 2.2.0. Downstream processing steps were performed using Seurat, version 2.3.4. Genes expressed in fewer than 10 cells were removed. Cells with a local minimum of the UMI distribution to the left of the mode UMI count, <500 genes, and >10,000 UMIs, > 2500 genes, and/or > 10% mitochondrial reads were removed. Data were log-normalized and scaled, with cell–cell variation due to UMI counts, percent mitochondrial reads, and S and G2M cell cycle scores regressed out.
Genetic De-Multiplexing Single-Cell RNA Sequencing
Demultiplexing scRNASeq was run with the inferred genotypes from the bulk RNASeq data that we have sequenced as part of this same project. We used the GATK variant calling pipeline on the samples included in each pool (GX06/GX18) and fed that to demuxlet as described in Kang et al.28
Single-Cell RNA Sequencing Data Processing and Quality Control
Cellranger (version 3.0.2) mkfastq was applied to the Illumina BCL output to produce FASTQ files. Cellranger count was then applied to each FASTQ file to produce a feature barcoding and gene expression library. Cellranger aggr was used to combine samples for merged analysis.
We applied scater package to filter out single-cell profiles that were outliers for any metric, as low-quality libraries.29 Data analysis was performed according to published standards.30, 31, 32, 33 All datasets used and additional scripts are available online (https://bitbucket.org/Fairfaxlab/prise-sarah-sassion/src/master/).
Identification T Cell Clusters
We used the area under the curve to calculate whether a T cell reference gene set was enriched within the expressed genes for each cell.34 We used the Human Protein Atlas database reference gene list for T cells, downloaded from https://www.proteinatlas.org/humanproteome/blood/blood+cells+summary, cell type group enriched genes. the repository is provided as a supplementary file (repository_reference_gene_sets.txt).
Single-Cell Protein and RNA Sequencing Expression
Live CD45+CD3+ T cells were sorted using a FACSAriaIII cell sorter (BD Biosciences). A total of 46,000 T cells were sorted and stained with a cocktail of 70 oligo-conjugated AbSeq antibodies (BD Biosciences; see Supplementary Table 4) for 45 minutes at 4ºC. Cells were then washed to remove residual unbound AbSeq antibodies and loaded onto 3 BD Rhapsody cartridges (BD Biosciences) for single-cell capture.
Complementary DNA Library Preparation and Sequencing
Single-cell capture and cDNA library preparation were performed using the BD Rhapsody Express single-cell analysis system (BD Biosciences) and a customized T cell expression panel (Supplementary Table 5), according to the manufacturer’s instructions (for further details including data analysis and quality control, see Supplementary Methods).
Results
The Majority of Activated Colon CD8+ T Cells in Anti–CTLA-4/PD-1 Colitis Are Tissue Resident Memory T Cells
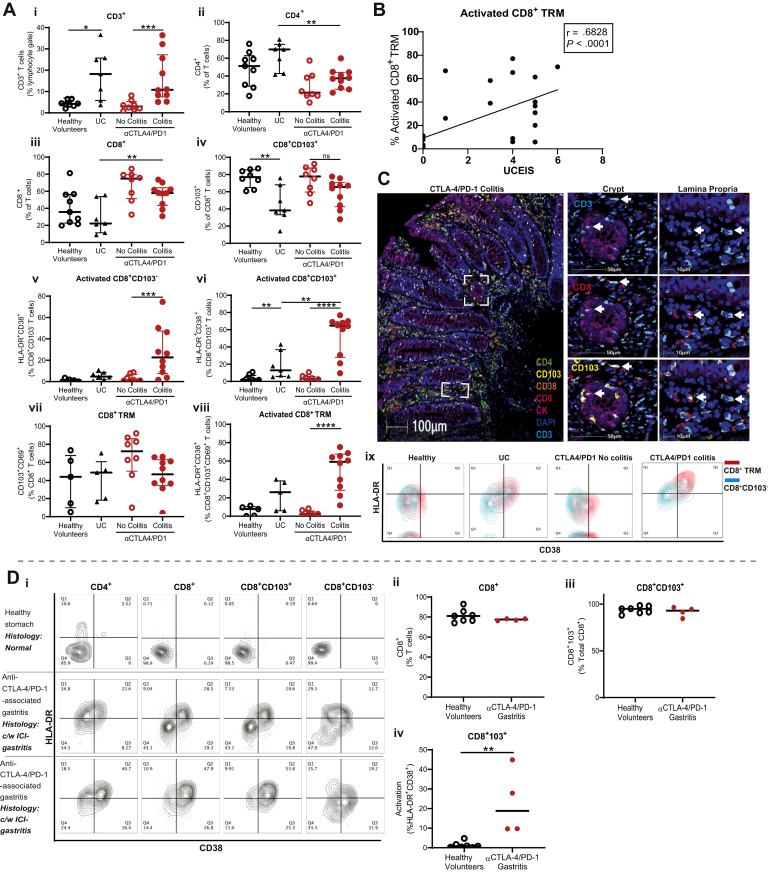
We previously demonstrated that the majority of T cells in the colon of ICI-colitis were CD8+.10 We sought to determine whether these CD8+ T cells had a TRM cell phenotype. We found that both UC and DCC groups are associated with increased proportions of CD3+ T cells in the affected tissue compared with healthy gut and DCNC groups, respectively (Figure 2Ai). Compared with UC, DCC is associated with proportionately fewer CD4+ T cells and more CD8+ T cells (Figure 2Aii–iii). The proportion of CD8+CD103+ T cells is lower in active UC compared with HVs (Figure 2Aiv). Patients with DCC have a very high proportion of activated CD8+CD103+ T cells, as defined by the co-expression of HLA-DR and CD38 (median 65% of CD8+CD103+ T cells), and this is higher than in DCNC (3%; P < .0001) and UC (13%; P < .01) groups (Figure 2Avi). There is some activation of CD8+103– “non–tissue-resident” T cells in DCC compared with DCNC; however, the proportion is much lower than in the CD103+ subset (Figure 2Av). We used a more stringent definition of CD8+ TRM cells, that is, co-expression of CD69 and CD103 to confirm high levels of cellular activation of CD8+ TRM cells in DCC compared with DCNC (Figure 2Avii–viii).
Figure 2.
CD8+ TRM cells predominate in ICI-colitis and their activation correlates with endoscopic and histologic findings. Mononuclear cells from colonic biopsies from HVs (n = 8), active UC (n = 7), DCC (n = 12), and DCNC (n = 8). Flow cytometry demonstrates (Ai–iii) DCC is associated with a CD3+ T cell lymphocytosis, and CD8+ T cells predominance. (iv–vi) The majority of CD8+ T cells express tissue-residency marker CD103, with higher activation in colitis than CD8+103– counterparts. (vii–viii) The proportion of CD8+CD69+CD103+ TRM cells does not significantly differ across disease states; however, activation is highest in the DCC group. ∗P < .02; ∗∗P < .01; ∗∗∗P < .001; and ∗∗∗∗P < .0001 by Mann-Whitney test with Bonferroni correction. (ix) Co-expression of activation markers CD38 and HLA-DR are highest in patients with UC and DCC with CD8+CD69+CD103+ TRM cells (red) displaying higher expression of these markers than CD8+CD103– nonresident T cells (blue). Live CD45+CD8+ T cells are shown. (B) Proportion of activated CD8+ TRM cells positively correlates with anti–CTLA-4/PD-1 colitis severity and measured by UCEIS (Spearman correlation). (C) Multiplexed spectral microscopy of a patient with DCC. Colocalization of CD3, CD8, and CD103 is demonstrated in both gastrointestinal crypts and in the lamina propria. CK, cytokeratin; DAPI, 4′,6-diamidino-2-phenylindole nuclear stain. Data representative of 3 experiments. (D) Live, singlet CD45+CD3+ T cells are displayed. (I) Cellular activation of T cells (top right quadrant) in healthy stomach and patients with anti–CTLA-4/PD-1 gastritis. In both health and anti–CTLA-4/PD-1 gastritis the majority of T cells are (ii) CD8+ with (iii) TRM cell phenotype. (iv) Increased cellular activation is present in anti–CTLA-4/PD-1 gastritis compared with healthy stomach.
Higher cellular activation of CD103+CD69+ CD8+ TRM cells compared to CD103– CD8+ T cells was detected across all patient groups and is represented in Figure 1Aix.
The Proportion of Activated Colon CD8+ Tissue Resident Memory T Cells Correlates Longitudinally With Clinical and Endoscopic Findings
We investigated whether the proportion of activated CD8+ TRM cells in the colon was an accurate biomarker of the presence of DCC and response to therapy. Overall, CD8+ TRM cell activation positively correlates with endoscopic severity of DCC using the Ulcerative Colitis Endoscopic Index of Severity (UCEIS) score (Figure 2B). UCEIS score details are provided in Supplementary Figure 1. We previously reported that UCEIS can be used as an objective endoscopic marker of ICI-colitis clinical outcomes.2 We confirmed by multiplexed spectral fluorescence microscopy that in DCC, CD8+ TRM cells reside both in gastrointestinal crypts and in the lamina propria (Figure 2C).
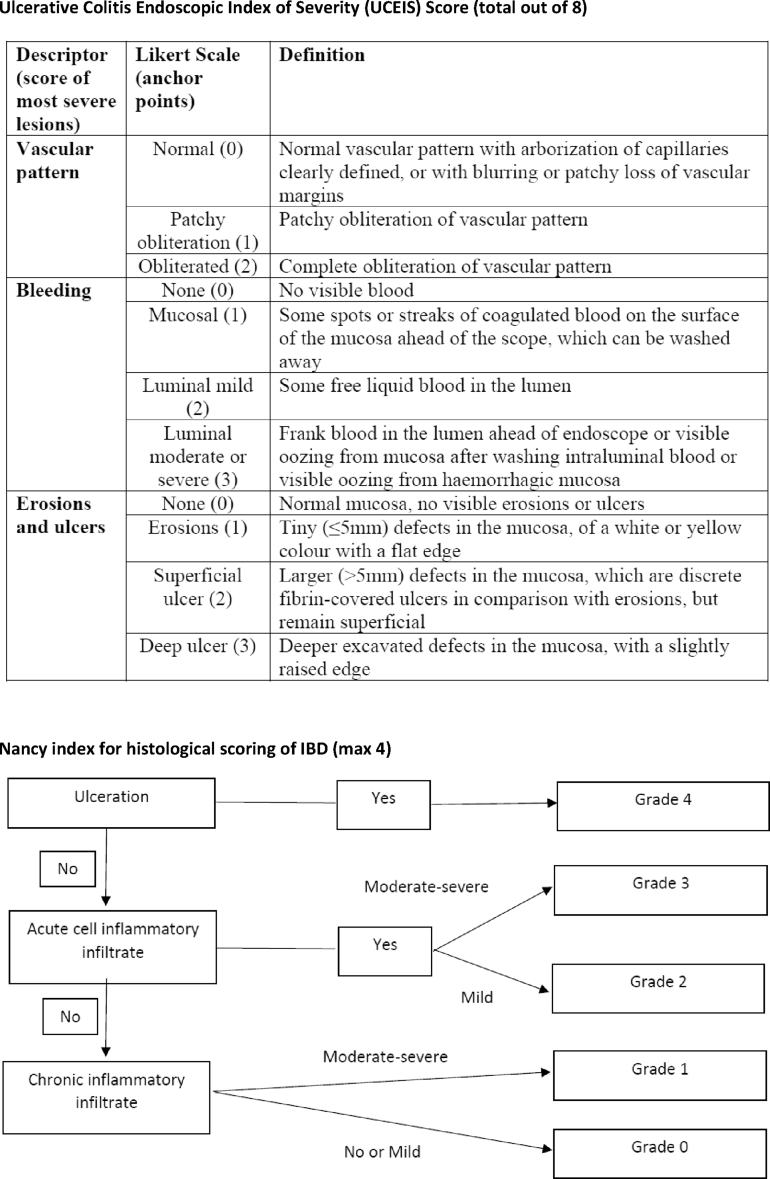
Supplementary Figure 1.
UCEIS and Nancy Index system for the scoring of UC.
We investigated whether activation of CD8+ TRM cells was a phenomenon specific to ICI-colitis or was evident in other forms of irAEs, such as ICI-gastritis (Figure 2Di). CD8+ TRM comprise the majority of T cells in the gastric mucosa in both health and anti–CTLA-4/PD-1 gastritis (Figure 2Dii–iii). The proportion of activated CD8+ TRM cells was low in health (<1%; Figure 2Div) and increased in anti–CTLA-4/PD-1 gastritis (30%–51%; Figure 2Div).
We performed an extended flow cytometric panel to further characterize the CD8+ TRM cells in anti–CTLA-4/PD-1 colitis (Supplementary Figure 2B).
Anti–CTLA-4/PD-1 Colitis Has a Transcriptome Distinct From Ulcerative Colitis With Up-Regulated Interferon-Gamma Signaling
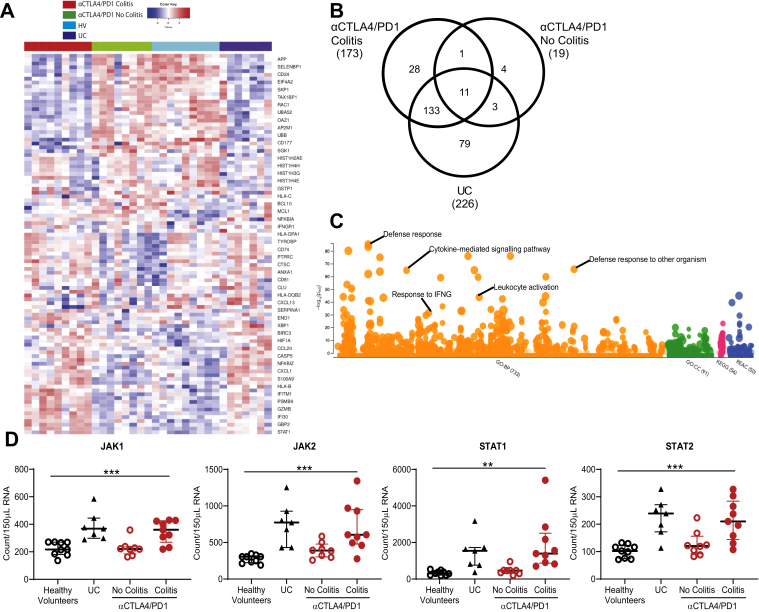
We performed a 780-gene autoimmune profiling panel using colonic RNA to determine unique and common features among DCC, DCNC, and UC groups. A heatmap of the top 50 defining features demonstrates that DCC is associated with up-regulated STAT1, GBP2, IFI30, GZMB, PSMB9, IFITM1, HLAB, S100A9, and CXCL1 (Figure 3A).
Figure 3.
Targeted gene panel analysis of ICI-colitis includes unique and common features compared with UC and high expression of the IFNG signaling pathway. A 780-gene Nanostring analysis of colonic biopsy RNA from HVs (n = 8), patients with active UC (n = 5), DCC (n = 9), and DCNC (n = 8). (A) Heatmap of top 50 differentially expressed genes. (B) Number of genes up-regulated ≥2-fold compared with HVs demonstrates 173 of up-regulated genes in DCC, only 12 of 173 are also up-regulated in the DCNC group (limited on-treatment effect); 144 of 173 genes are common between DCC and UC, 28 of 173 genes are unique to DCC. (C) Manhattan plot indicating significantly up-regulated pathways, including response to IFNG. The complete list is provided in Supplementary Table 4. (D) RNA expression of canonical markers of IFNG signaling JAK1, JAK2, STAT1, and STAT2 are higher in DCC and UC groups compared with healthy controls and DCNC groups. ∗∗P < .01 and ∗∗∗P < .001 by 1-way analysis of variance.
We identified 259 genes that are up-regulated more than 2-fold across the DCC, DCNC, and UC groups compared to HVs (Figure 3B). Of the 173 genes up-regulated in DCC, only 12 of 173 are common to the DCNC group, indicating these changes are not simply an on-treatment effect. Of the 173 genes up-regulated in DCC, 144 of 173 are in common with UC, and 29 are unique (Figure 3B).
Exploration of the 173 up-regulated genes in DCC using g:Profiler pathway analysis highlights a number of biological pathways (Figure 3C). The top 30 pathways include defense response and response to external biotic stimulus and cytokine-mediated pathways. The most pathway-specific results include a response/cellular response to IFNG (Figure 3C; Supplementary Table 6). We confirm that the canonical JAK/STAT components of IFNG signaling are up-regulated in anti–CTLA-4/PD-1 colitis compared to controls (Figure 3D).
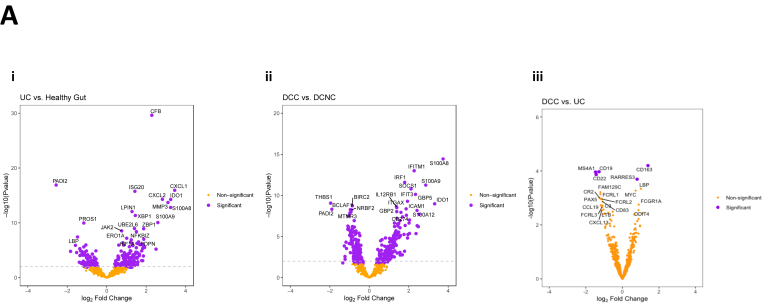
Volcano plots showing differentially expressed genes in UC vs HV, DCC vs DCNC, and DCC vs UC are shown in Supplementary Figure 3. Up-regulated genes common to DCC and UC include S100A8, S100A9, and IDO1. DCC is associated with lower expression of canonical B cell markers CD19, MS4A1(CD20), and CD22 compared to UC.
Supplementary Figure 3.
Data generated from a 780-gene Nanostring experiment analyzing RNA extracted from the gastrointestinal tract of HVs (n = 8), patients with active UC (n = 5), anti–CTLA-4/PD-1 colitis (DCC; n = 9) and anti–CTLA-4/PD-1 treated with no colitis (DCNC; n = 8) are shown. Plots show differentially expressed genes in (i) UC vs HV and (ii) DCC vs DCNC and DCC vs UC.
Bulk RNA Sequencing Analysis Confirms a Distinct Transcriptome for Anti–CTLA-4/PD-1–Associated Colitis Enriched for Interferon-Gamma Signaling
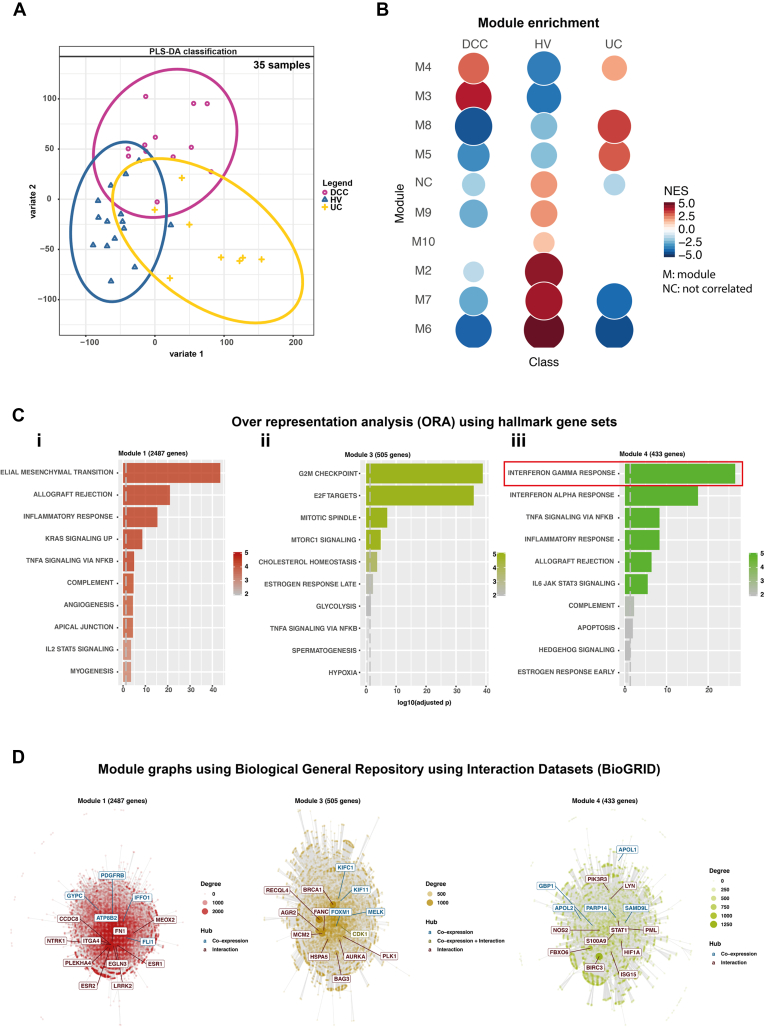
RNASeq analysis from bulk RNA extracted from colonic biopsies confirms the transcriptomic signature associated with DCC is distinct from UC (Figure 4A and B). As opposed to the Nanostring panel, which selects for genes expressed by lymphocytes, the bulk RNASeq analysis is predominated by epithelial signals. Analysis of modular hallmark gene sets demonstrates patients with DCC have highly expressed IFNG response, in excess of TNFA signaling (Figure 4C).
Figure 4.
Bulk RNASeq analysis confirms anti–CTLA-4/PD-1 colitis has a transcriptome distinct from UC with IFNG signaling stronger than TNFα signaling. Bulk RNASeq data generated from total RNA extracted from patients with DCC, those with active UC and HVs are shown. (A) Partial least squares-discriminate analysis (PLS-DA) demonstrate the divergent transcriptome of DCC and UC. (B) Module enrichment analysis demonstrated overexpression of hallmark gene “modules” 3 and 4 in DCC. (C) Over-representation analysis demonstrates the over-represented genesets in modules 1, 3, and 4. Over-expressed pathways include IFNG signaling (box),which was more highly expressed than TNFα signaling. (D) Co-expression and interaction of genes in modules 1, 3, and 4 as determined by Biological General Repository using Interaction Datasets (BioGRID). Blue indicates co-expressed genes; brown indicates gene interaction; and green indicates gene co-expression and interaction.
Single-Cell RNA Sequencing Confirms Anti–CTLA-4/PD-1 Colitis Is Associated With High Proportions of Activated CD8+ Tissue Resident Memory T Cells That Express Transcripts for CTLA-4, PD-1, TIGIT, TIM-3, LAG-3, and Interferon-Gamma
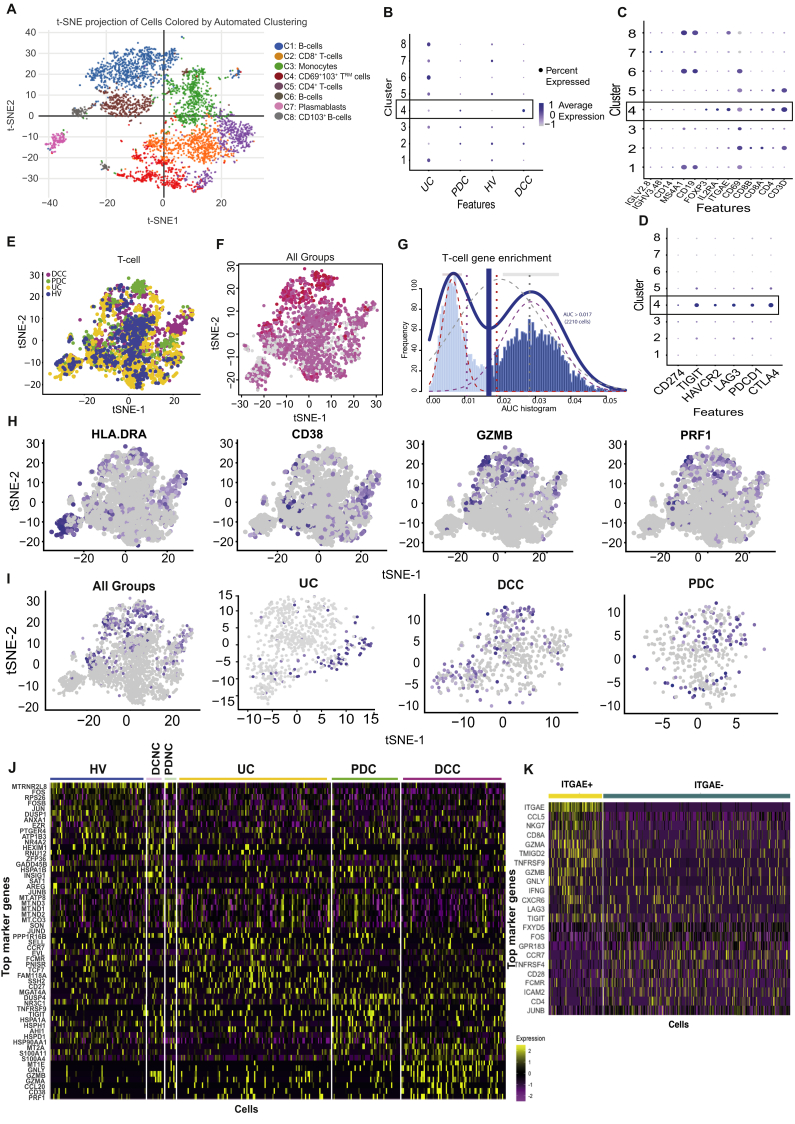
Single-cell analysis formed 8 main clusters (Figure 5A) that include T cells (clusters 2, 4, and 5), B cells (clusters 1, 6, and 8), plasmablasts (cluster 7), and monocytes (cluster 3). UC has a higher proportion of B cells and plasmablasts compared with ICI-colitis groups and HV (Figure 5B). The defining features of each cluster are shown in Figure 5C and Supplementary Table 7. Co-expression of ITGAE (CD103) and CD69 is strongest in cluster 4 and identifies cells with a TRM cell phenotype. The DCC group has the highest proportion of cells with a TRM cell phenotype (cluster 4; Figure 5B). These cells have a very high expression of immune checkpoint transcripts, including CTLA4, PDCD1 (PD-1), TIGIT, HAVCR2(TIM-3), and LAG3, which are minimally detected or absent in the other clusters (Figure 5D).
Figure 5.
CD8+ TRM cells in ICI-colitis express high proportions of checkpoint inhibitors, cellular activation/cytotoxicity markers and IFNG. scRNASeq analysis of 5876 cells from HVs (n = 3), active UC (n = 2), DCC (n = 3), DCNC (n = 3), PD-1 colitis (PDC; n = 2), and PD-1 treated with no colitis (PDNC; n = 3). (A) t-stochastic neighbor embedding (t-SNE) projection of live CD45+ lymphocytes formed 8 transcriptionally distinct clusters. (B) Proportion of clusters formed from cells from each disease state, with cluster 4 (box) most common in DCC. (C) Canonical gene markers of each clusters used to define annotation with cluster 4 (box) expressing CD3, CD8, CD69, and CD103 consistent with TRM cells. (D) High expression of immune checkpoint molecules on (cluster 4) CD8+ TRM cells (box). (E) t-SNE projection of T cells, highlighted by patient group. (F) Distribution of CD8+ TRM cells as shown by cells co-expressing CD8, CD69, and ITGAE(CD103) in pink (low expression) and red (high expression). (G) Histogram showing cells that express a canonical gene-set list for T cells (dark blue) were selected from the total data for analysis in E, F, G, H, I, and K. (H) Expression of activation markers HLADR, GZMB, PRF1, and CD38 (to a lesser extent) overlap with the CD8+ TRM cell zone. (I) Expression of IFNG overlaps with the CD8+ TRM cell zone with IFNG being detected in UC, DCC, and PDC groups. (J) Heatmap based on all CD45+ cells showing the most differentially expressed genes in each patient group. (K) Heatmap based on T cells only showing differential expression between ITGAE(CD103)+ and ITGAE– cells.
We further investigated the T cell component of the scRNAseq dataset (Figure 5E and G). CD8+ T cells with a TRM cell phenotype can be identified as cells co-expressing transcripts for CD8A, CD8B, ITGAE, and CD69 (Figure 5F), and there is evidence for these cells expressing high amounts of HLADR, GZMB, and PRF1 (Figure 5H). We have demonstrated by Nanostring and bulk RNASeq that IFNG signaling pathways are enriched in anti–CTLA-4/PD-1 colitis, but the source of IFNG message cannot be determined at a bulk RNASeq level. Using scRNASeq, we are able to confirm IFNG production in T cells that overlap with the TRM cell zones (Figure 5F and I) and that IFNG production is detected in all UC, DCC, and anti–PD-1 colitis (PDC) groups (Figure 5I).
Heatmap analysis demonstrates that each group has a distinct set of up-regulated genes (Figure 5J). T cells from patients with DCC have significantly higher expression of MT2A, S100A11, GNLY, MT1E, GZMB, and CCL20. T cells from patients with PDC have significantly higher expression of DUSP4, NR3C1, HLADPB1, HLADR85, and HSPA1A. T cells from patients with UC have significantly higher expression of SELL, CCR7, IFITM3, FAM118A, and FCMR.
The defining characteristics of ITGAE+(CD103+) colonic T cells (compared to ITGAE– T cells) are shown in Figure 5K. These cells have significantly higher expression of CCL5, NKG7, CD8A, GZMA, TMIGD2, TNFRSF9, GZMB, GNLY, IFNG, CXCR6, LAG3, and TIGIT, and lower expression of CCR7, CD28, and CD4.
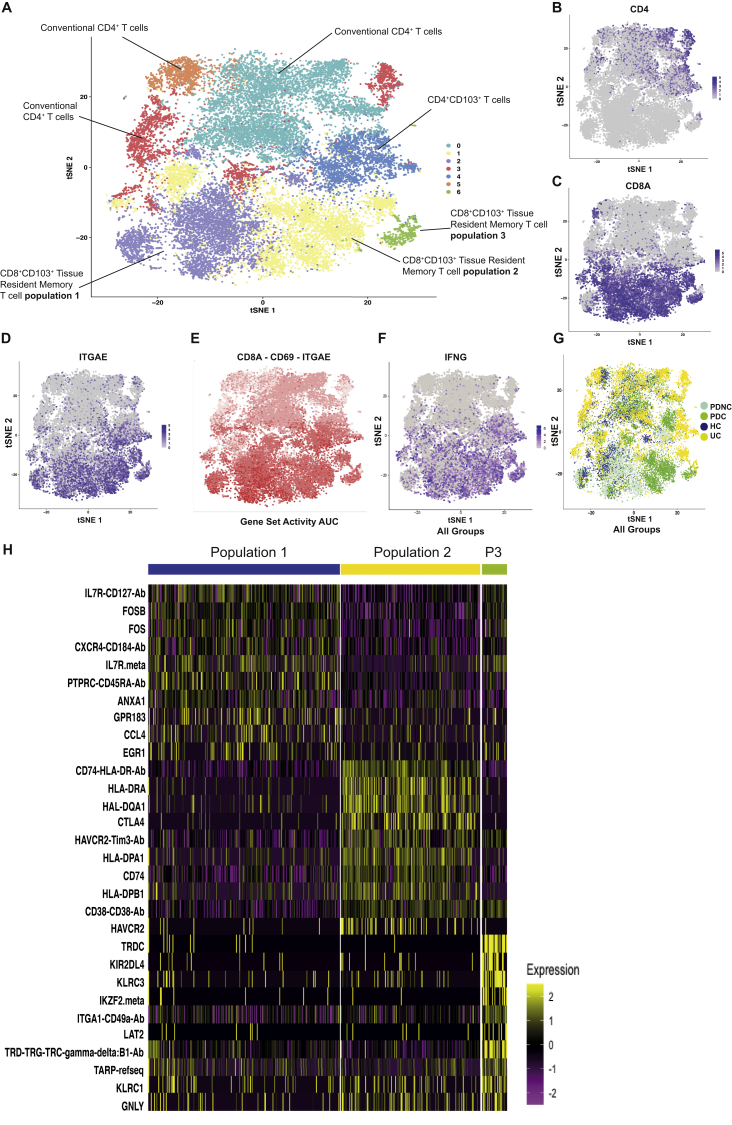
To confirm our finding that CD8+ TRM cells display high expression of IFNG, including in patients with PDC, we performed a second single-cell protein and RNASeq experiment, this time sorting on live CD45+CD3+ T cells (Figure 6). The cells form 7 clusters (Figure 6A and Supplementary Table 8), including 3 clusters of CD8+ TRM cells (clusters 1, 2, and 6), a tissue-resident CD4+ T cell population (cluster 4), and 3 populations of nonresident classical T cells (clusters 0, 3, and 5). There is a clear separation of CD4+ (Figure 6B) and CD8+ T cell populations (Figure 6C), with CD103(ITGAE) predominantly overlapping with CD8+ T cells (Figure 6D). Co-expression of CD8, CD103(ITGAE), and CD69 defines the CD8+ TRM cell clusters (Figure 6E). Expression of IFNG overlaps with CD8+ TRM cell populations and, to a lesser degree, the CD4+CD103+ T cells (Figure 6F). T cells from patients with PD-1 colitis are predominantly in the CD8+ TRM cell populations (clusters 1 and 6) and are distinct from T cells from active UC, which are predominantly in the conventional and CD4+ T cell zones (Figure 6G).
Figure 6.
CD8+ TRM cells express high levels of IFNG in PD-1–associated colitis. Data from a single cell protein and RNASeq analysis of 23,265 gut-derived T cells from HVs (n = 4), patients with active UC (n = 3), PD-1 colitis (PDC; n = 5) and PD-1 treated with no colitis (PDNC; n = 2). (A) t-stochastic neighbor embedding (t-SNE) projection of live T cells formed 7 distinct clusters. t-SNE plots of all groups showing expression of (B) CD4, (C) CD8A, and (D) ITGAE(CD103). (E) Distribution of CD8+ TRM cells as shown by cells co-expressing CD8, CD69, and ITGAE(CD103) in pink (low expression) and red (high expression). (F) Expression of IFNG is shown in all groups, displaying overlap with CD8+ TRM cell zones. (G) Distribution of cells based on patient groups demonstrates T cells from patients with PDC are found predominantly in the CD8+ TRM cell zones (clusters 1 and 6). (H) Heatmap based on CD8+ TRM cell populations 1–3 only displaying top differentially expressed genes.
We extracted data pertaining to the 3 CD8+ TRM cell populations (Figure 6H) and found that the CD8+ TRM cell population 2, comprised mostly cells from patients with PD-1 colitis, has markedly high expression of activation markers (HLADR, CD38) and checkpoint molecules (CTLA4, TIM3). This transcriptome is distinct from CD8+ TRM cell population 1, which has a greater representation in health and expresses high levels of IL7R and CCL4. The smaller CD8+ TRM cell population 3 again contains unique features, including KIR2DL4, KLRC3, and includes a γδ T cell component.
Tofacitinib Results in Rapid Resolution of Treatment-Refractory Anti–PD-1–Associated Colitis and Reversal of CD8+ Tissue Resident Memory T Cell Activation and Interferon-Gamma Signaling
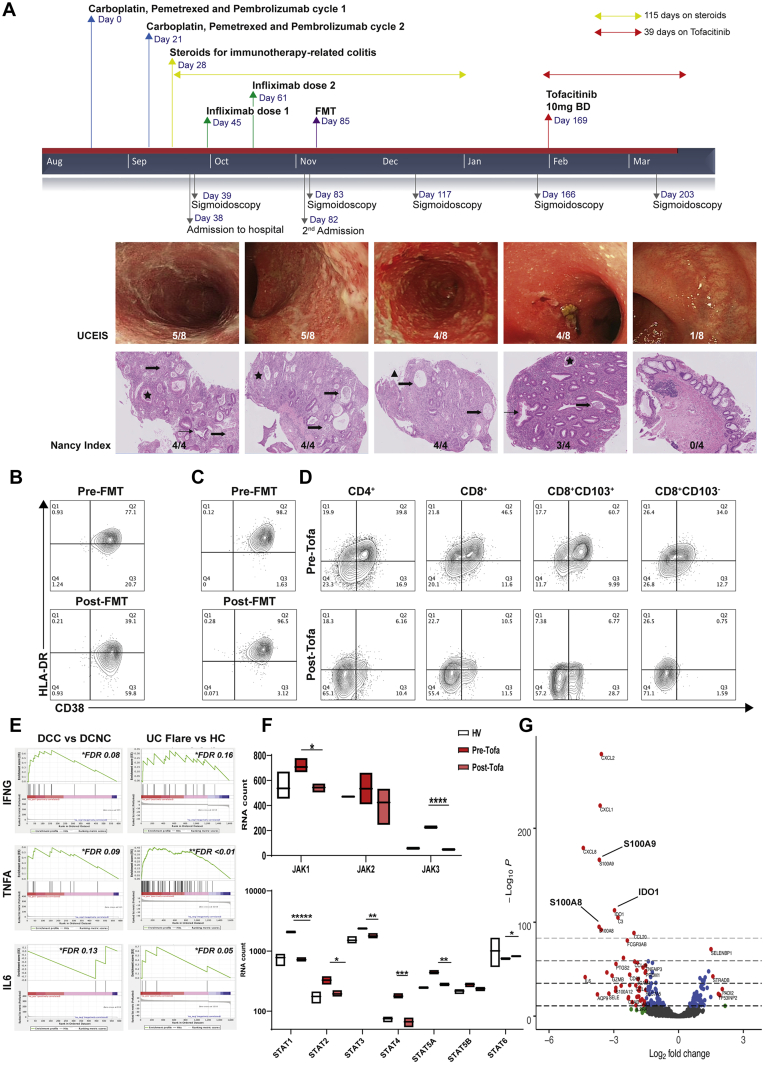
A 61-year-old man with metastatic non-small-cell lung cancer was treated with combination chemotherapy (carboplatin and pemetrexed) and pembrolizumab (see Figure 7A). After 2 cycles, he developed symptoms of ICI-colitis, which was confirmed endoscopically and histologically (UCEIS and Nancy score details are in Supplementary Figure 1). He did not respond to intravenous steroids or 2 doses of infliximab. We had previously seen a rapid resolution of refractory anti–CTLA-4/PD-1 colitis with FMT treatment in a different patient, with corresponding decreases in CD8+ TRM cell activation (Figure 7B). We prioritized FMT over vedolizumab as he required rapid induction therapy. There was a modest initial clinical response to FMT; however, on follow-up sigmoidoscopies over the subsequent 12 weeks, he continued to have refractory colitis of an equivalent endoscopic and histologic severity. This was confirmed by flow cytometry (Figure 7C). Our data collectively provided us with evidence that tofacitinib, a JAK inhibitor, was a potential therapeutic option. After discussion with the patient, tofacitinib was prescribed at 10 mg twice per day, with concomitant venous thromboembolism prophylaxis.35 He made an immediate clinical response and achieved endoscopic and histologic remission by 5 weeks (Figure 7A), commensurate with resolution of activated CD8+ TRM cells (61%–7%; Figure 7D). Tofacitinib was ceased after 6 weeks, and he restarted chemotherapy. He has made a good oncological response and 10 months later colitis has not recurred.
Figure 7.
Tofacitinib results in rapid resolution of treatment-refractory ICI-colitis, and correlates with resolution of CD8+ TRM cell activation and down-regulation of JAK/STAT signaling. (A) Clinical time course of a 61-year-old man with non–small-cell lung cancer treated with carboplatin, pemetrexed, and pembrolizumab. The anti–PD-1 colitis was refractory to multiple therapies. Tofacitinib resulted in prompt resolution of clinical symptoms, and endoscopic and histopathology inflammation. Tofacitinib was continued for 6 weeks. Star, crypt abscess; thick arrow, attenuated crypt; thin arrow, crypt architectural distortion; triangle, erosion. (B) FMT response in a previous patient with ICI-colitis, where clinical resolution was associated with normalization of CD8+ TRM cell activation. Flow cytometry gated on live CD3+CD8+CD69+CD103+ TRM cells. (C) Using the same donor stool, FMT did not result in resolution of clinical symptoms or resolution of CD8+ TRM cell activation in this 61-year-old man who subsequently received tofacitinib. (D) Flow cytometry plots gated on live single CD45+CD3+ T cells are shown. Before tofacitinib, widespread activation of CD4+ and CD8+ T cells is evident, with the highest level of activation in CD8+CD103+ TRM cell subset (61%). After 6 weeks of tofacitinib, there is resolution of T cell activation, including in the CD8+CD103+ TRM cell subset (7%). (E) Gene set enrichment analysis of bulk RNASeq data demonstrates IFNG signaling pathway enrichment in ICI-colitis. (F) Data from Nanostring RNAplex assay from the tofacitinib-treated patient and 3 HVs. Tofacitinib results in significant down-regulation of JAK1, JAK3, STAT1, STAT2, STAT3, STAT4, and STAT5A. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; and ∗∗∗∗∗P < .00001 by Mann-Whitney test. (G) Volcano plot depicting pre and post tofacitinib.
An RNAplex assay of colonic mucosal RNA pre- and post-tofacitinib therapy shows down-regulation of key JAK-STAT signaling components known to be down-stream of IFNG signaling (Figure 7F). The overall transcriptional response to tofacitinib therapy demonstrates down-regulation of transcripts including S100A8, IDO1, and S100A9 (Figure 7G).
Discussion
The profound success of ICIs has resulted in broader applications, and an increasing incidence of ICI-colitis that is frequent and has the greatest overall irAE mortality.3
Here we present a comprehensive analysis of ICI-colitis using multiparameter and spectral flow cytometry, RNAplex, and bulk and single-cell analysis. We find that in all patient groups, the majority of colon-derived CD8+ T cells are TRM cells, and that in anti–CTLA-4/PD-1 colitis, the highest activation levels are seen in TRM cells. CD8+ TRM cell activation anti–CTLA-4/PD-1 colitis correlates with clinical, endoscopic, and histopathologic findings, and the response to treatment over time. In addition, CD8+ TRM cell activation is also present in ICI gastritis, which involves a distinct epithelium and microenvironment from the colon, and this may have implications for the pathogenesis of extragastrointestinal irAEs.
We have made a direct comparison with active UC and find that the level of CD8+CD103+ T cell activation is significantly greater in the DCC group. We identify both the immunologic commonalities with UC and disparate features. In addition to differences in T cell populations, we demonstrate that B lineage populations are over-represented in the UC samples and comparatively absent in ICI groups, indicating that pathogenic B cells likely play a smaller role in ICI-colitis. The IFNG-response pathway is up-regulated in both UC and DCC, but with an enhanced activation in both DCC and PDC indicating that targeting this pathway may be even more effective in ICI-colitis.
Our scRNASeq experiments confirm that CD8+ TRM cells are enriched in ICI-colitis, and display the highest proportion of immune checkpoint transcripts, including CTLA4 and PCDC1 (PD-1). This supports recently published data21 and provides a likely mechanism by which these cells become disproportionally and rapidly activated after ICI administration. We confirm that the production of IFNG clusters in the same region as CD8+ TRM cells, extending the data from Luoma et al,21 which identified IFNG and TNFA up-regulation in the CD4+ and CD8+ T cells, and acting on the myeloid cellular compartment. Our analysis of ITGAE(CD103)+ T cells reveals an expression profile similar to CD8+ TRM cells in vitiligo,13 with 5 up-regulated genes that largely relate to cytotoxicity featuring in both datasets: GZMB, GNLY, NKG7, CCL5, and IFNG. Our data suggest that in health, CD8+ TRM cells express a homeostatic signature, including IL7R, but that in ICI-associated colitis, there is significant up-regulation of activation molecules and checkpoint molecules.
We find compelling evidence for up-regulated IFNG signaling in ICI-colitis, more so than TNFA, which is the current target of ICI-colitis rescue therapy. We present a patient with refractory ICI-colitis who we treated successfully with tofacitinib, and with robust resolution of CD8+ TRM cell activation. This aligns with the recent case reports of successful tofacitinib therapy for treatment-refractory ICI-colitis.22,23 We acknowledge that a series of cases cannot conclusively determine a treatment’s large-scale efficacy or safety. IFNG signaling is a well-established pathway in tumor control. Early work in murine models found that neutralizing IFNG interferes with tumor rejection in immunocompetent hosts.36 Mice lacking IFNG signaling components IFNGR1 or STAT1 develop a higher percentage of tumors at a faster rate.37 We recognize concern that use of JAK inhibition in ICI-colitis may deactivate not only colonic but also intra-tumoral CD8+ TRM cells, which are a key therapeutic target.18,38 In human melanoma, IL-7 signaling,39 T cell infiltration, and IFNG signaling signatures40 have a high association with tumor response to immune checkpoint inhibitors, and conversely defects in IFNG signaling, including loss-of function mutations in IFNGR1, JAK1, JAK2, and STAT1, are associated with resistance to checkpoint blockade.41, 42, 43, 44, 45 Although larger clinical trials are needed to establish the safety and efficacy of tofacitinib for ICI-colitis, our analysis provides a complete bench-to-bedside cycle: from a novel disease entity, hypothesis, and mechanistic study, to intervention with a rationally repurposed therapeutic. Our data suggest that tofacitinib may be a useful therapy in patients with refractory ICI-colitis as a salvage therapy.
Our study has limitations. We acknowledge our relatively small cohort of patients and, given the real-world nature of our study, these patients are not perfectly matched. In addition, the relatively low cell number in some of the experiments, notably the initial scRNASeq, mean that the UC and ICI-colitis comparison could bias toward commonalities. Comparison of a chronic disease (UC) with longstanding inflammatory response against an acute iatrogenic colitis may also confound the data. Although the patients with UC do have a longer median flare duration, the cohorts are otherwise well-matched in relation to inflammation severity, and previous and current therapy (see Supplementary Table 1). Future studies could aim to reduce the heterogenicity of enrolled ICI-colitis and comparator where possible, and this may require larger multicenter studies. In addition, although we attempted to study ICI-colitis CD8+ TRM cells ex vivo, the high rates of apoptosis, in keeping with down-regulated Bcl-2 expression (see Supplementary Figure 2), made this challenging. We suggest cluster 4 in our scRNASeq experiment is likely numerically under-represented, given our flow cytometry results were conducted on fresh tissue.
There remain many unanswered questions, including what factors drive a subset of patients to develop ICI-colitis, leaving others unaffected. We postulate that activated CD8+ TRM cells in ICI-colitis are responding to commensal or pathogenic microbes, and that this results in high levels of cellular activation and IFNG signaling that propagates downstream and widespread tissue activation. Furthermore, the use of FMT for treatment of ICI-colitis (for which we describe one treatment success) implies that replacement of the microbiome may remove the instigating antigen.6
Our work has identified that IFNG-producing CD8+ TRM cells are a cellular hallmark of ICI-colitis. This has important implications for targeted therapy for ICI-colitis, as evidenced by the successful application of a JAK inhibitor. These findings also suggest that medications that specifically target CD103 may prove effective therapy, and note monoclonal antibody targeting the β7 integrin chain. Finally, our data on CD8+ TRM cell activation in both colon and gastric epithelium may have broader relevance for other extra-gastrointestinal irAEs.
Acknowledgments
The authors thank the patients for their generous participation in this study. Thanks also to James Chivenga and the Oxford Translational Gastroenterology Unit Biobanking team for assistance with biopsy collection. Thank you to Dr Helen Ferry and Liam Hardy for assistance with cell-sorting experiments, and to Dr Joanna Hester and Jim White for assistance and expertise with Nanostring. The authors acknowledge Dr Rubeta Matin, who provides dermatological care to the melanoma patients. Stephanie M. Slevin, Vincent T. F. Cheung, and Isar Nassiri contributed equally to this work.
CRediT Authorship Contributions
Sarah C. Sasson, PhD (Conceptualization: Equal; Investigation: Lead; Writing – original draft: Equal; Writing – review & editing: Equal).
Stephanie Slevin, DPhil (Formal analysis: Supporting; Investigation: Supporting).
Vincent T. F. Cheung, MRCP (Funding acquisition: Supporting; Investigation: Supporting; Project administration: Equal).
Isar Nassiri, PhD (Formal analysis: Equal; Methodology: Supporting; Software: Supporting).
Anna Olsson-Brown, DPhil (Investigation: Supporting).
Eve Fryer, FRCPath (Validation: Supporting).
Ricardo C. Ferreira, PhD (Formal analysis: Supporting).
Dominik Trzupek, PhD (Formal analysis: Supporting).
Gupta Tarun, MRCP (Investigation: Supporting; Project administration: Supporting).
Lulia Al-Hillawi, MRCP (Investigation: Supporting).
Mari-lenna Issaias, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting).
Easton Alistair, FRCPath (Investigation: Supporting; Methodology: Supporting).
Leticia Campo, PhD (Investigation: Supporting; Methodology: Supporting).
Michael Fitzpatrick, DPhil (Investigation: Supporting; Methodology: Supporting).
Adams Joss, FRCP (Clinical management: Supporting).
Meenali Chitnis, FRCP (Clinical management: Supporting).
Andrew Protheroe, FRCP (Investigation: Supporting; Clinical management: Supporting).
Mark Tuthill, FRCP (Project administration: Supporting; Clinical management: Supporting).
Nicholas Coupe, FRCP (Project administration: Supporting; Clinical management: Supporting).
Alison Simmons, DPhil, FRCP (Methodology: Supporting; Resources: Supporting).
Miranda Payne, FRCP (Investigation: Supporting; Project administration: Supporting; Clinical management: Supporting).
Mark R. Middleton, FRCP (Conceptualization: Supporting; Investigation: Supporting).
Simon PL Travis, DPhil, FRCP (Investigation: Supporting; Writing – review & editing: Supporting).
Benjamin P. Fairfax, FRCP (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting).
Paul Klenerman, DPhil, FRCP (Conceptualization: Equal; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Equal; Resources: Equal; Supervision: Equal; Writing – review & editing: Equal).
Oliver Brain, DPhil, FRCP (Conceptualization: Lead; Formal analysis: Equal; Investigation: Equal; Methodology: Supporting; Supervision: Lead; Writing – original draft: Equal).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Sarah C. Sasson and Stephanie M. Slevin are supported by an Oxford-Bristol Myers Squibb Postdoctoral Fellowship. Vincent T. F. Cheung is supported by a Norman Collisson Foundation Fellowship and an Oxford Health Services Research Committee grant. Ricardo C. Ferreira and Dominik Trzupek are supported by a strategic award and grant from the Juvenile Diabetes Research Foundation (4-SRA-2017-473-A-A and 1-SRA-2019-657-A-N) and the Wellcome Trust (107212/A/15/Z). Paul Klenerman is supported by the Wellcome Trust (WT109965MA), National Insitutes of Health (U192U19AI082630) and is a National Institute for Health Research senior fellow. Oliver Brain, Benjamin P. Fairfax, and Mark R. Middleton receive support from Oxford National Institute for Health Research Biomedical Research Centre. The Translational Histopathology Laboratory is funded by the Experimental Cancer Medicines Centres. Reagents for the Nanostring experiments were provided as part of a Nanostring grant. The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://doi.org/10.1053/j.gastro.2021.06.025.
Supplementary Methods
Illustrative Figures
Illustrative figures were created with the assistance of BioRender.com
Flow Cytometry
Flow cytometry on freshly isolated gut mononuclear cells was performed using a near infra-red live/dead stain (Invitrogen) and the following monoclonal antibodies: CD3 PE-CF594(UCHT1), CD4-BV650(SK3), CD8-AF700(SK1), CD38-FITC(HIT2) (all, BD Biosciences), CD45RO-BV510(UCHL1), CD69-BV785(FN50), CD103-BV605(Ber-ACT8), and HLA-DR-BV711(L243) (Biolegend). Isotype control stains were performed using mouse IgG2aκ and IgG1κ (BD Biosciences). This work was performed on a 3-laser LSR Fortessa X-20 (BD Biosciences).
An extended TRM cell phenotyping panel was run using the aforementioned near infra-red live/dead stain and following antibodies: CD4-BV650(OKT4), CD38-AF488(HIT2), CD45RO-BV510(UCHL1), CD69-BV785(FN50), CD103-BV605(Ber-ACT8), Bcl-2-AF647(100), CTLA-4-PE(BN13), CXCR6-APC(KO41E5), HLA-DR-BV570(L243), Ki-67-PerCP-Cy5.5(Ki-67), LAG3-BV421(11C3C65), PD-1-AF647(EH12.2H7) (Biolegend), CD8-AF532(RPA-T8), TIGIT-PERCP-EF710(46-9500-42) (ThermoFisher Scientific), CD3-PECF594(UCHT1), granzyme B-BV421(GB11), and TIM-3-BB515(7D3) (BD Biosciences). Granzyme B, Ki-67, and Bcl-2 staining was performed using an intracellular and intranuclear permeabilization kit (ThermoFisher). The extended panel was performed on an Aurora spectral analyzer (Cytek, Freemont, CA).
Data were analyzed using FACSDIVA software, version 8.0.1 (BD Biosciences). Compensation was calculated before each experiment. Approximately 150,000 lymphocytes (50,000 live T cells) were analyzed per colon biopsy. Gating strategies are shown in Figure 1Aix–x. Lymphocyte populations are reported as a proportion of parent populations. Specifically, live CD3+ T cells are reported as percentage of total live lymphocytes (as defined by side scatter vs forward scatter, CD45+, and exclusion of viability dye). CD4+ and CD8+ T cells are reported as a percentage of live T cells. CD8+CD103+CD69+ TRM cells are reported as a percentage of total CD8+ T cells. Activated (HLA-DR+CD38+) cells are reported as percentage of memory CD4+ or CD8+ T cell populations.
Nanostring
Custom 10-gene spike in set: CD69, ITGAE(CD103), RUNX3, NR4A1, MKI67, LAMP1, TNFSF9(CD137), CXCL16, HAVCR2(TIM-3), and IL2RA.
Single-Cell Protein and RNA Sequencing Expression
Viably cryopreserved gut-derived mononuclear cells were thawed in a 37°C water bath before washing and quantitation. The number of cells were normalized across all subjects. Individual patient samples were labeled with oligo-conjugated sample tag antibodies (Sample Multiplexing Kit; BD Biosciences) by incubating for 15 minutes at room temperature followed by the addition of anti-human CD45-APC (Clone HI3-; BD Biosciences) and CD3-PE-CF594 (Clone UCHT1; BD Biosciences) for 15 minutes at room temperature. Cells were washed twice with 15 mL phosphate-buffered saline and centrifuged at 500g before combining into a single tube and staining with SyberGreen viability dye (Sigma).
Single-Cell Protein and RNA Sequencing Complementary DNA Library Preparation and Sequencing
cDNA was amplified for 10 cycles using the predesigned T cell expression panel (BD Biosciences) in combination with supplementary custom primer panel. In total, the primer set used in this assay contained 565 probes targeting 534 different genes. The resulting PCR1 products were purified using AMPure XP magnetic beads (Beckman Coulter) and the respective messenger RNA (mRNA) and AbSeq/Sample tag products were separated based on size-selection, using different bead ratios (0.7X and 1.2X, respectively). The purified mRNA and Sample tag PCR1 products were further amplified (10 cycles), and the resulting PCR2 products purified by size selection (0.8X and 1.2X for the mRNA and sample tag libraries, respectively). The concentration, size, and integrity of the resulting polymerase chain reaction products was assessed using both Qubit (High Sensitivity dsDNA kit; Thermo Fisher) and the Agilent 4200 Tapestation system (High Sensitivity D1000 screentape; Agilent). The final products were normalized to 2.5 ng/μL (mRNA), 1 ng/μL (sample tag), and 0.2 ng/μL (AbSeq) and underwent a final round of amplification (6 cycles for mRNA and sample tag and 7 cycles for AbSeq) using indexes for Illumina sequencing to prepare the final libraries. Final libraries were quantified using Qubit and Agilent Tapestation and pooled (approximately 23:73:4% mRNA/AbSeq/Sample tag ratio) to achieve a final concentration of 5 nM. Final pooled libraries were spiked with 15% PhiX control DNA to increase sequence complexity and sequenced (150-bp paired-end) on a NovaSeq sequencer (Illumina).
Single-Cell RNA and Protein: Data Analysis and Quality Control
After filtering the PhiX reads, we achieved a final coverage of 567 × 106 reads. The FASTQ files obtained from sequencing were analyzed following the BD Biosciences Rhapsody pipeline (BD Biosciences), as described previously.1 The distribution-based error correction–adjusted molecule counts were imported and the expression matrices analyzed using R package.
We applied scater package to filter out single-cell profiles that were outliers for any metric, as low-quality libraries.2 Technical noise was modeled using the scran package3 based on the optimal number of principal components.4
Sub-setting was performed to select cells expressing CD8A, CD8B, and CD4, ITGAE, and IFNG. A principle component analysis was run with 7 canonical clusters selected for downstream analysis using the Seurat package.5 An integrated analysis of all merged data was performed using defined canonical clusters. Plots were generated using ggpubr (version 0.2) and customizing ggplot2.6
We applied area under the curve and bimodal distribution to separate the distributions and evaluate the strength of enrichment of each reference cell with genes in an indicated cell.
Supplementary Table 1.
Clinical Characteristics of DCC, DCNC, UC, PDC, PDNC, and HV Groups
| Variable | DCC | DCNC | UC | PD-1 checkpoint inhibitor-related colitis | PD-1 checkpoint inhibitor no colitis | HVs |
|---|---|---|---|---|---|---|
| n | 15 | 10 | 10 | 7 | 5 | 22 |
| Age, y, median (IQR) | 69 (59–70) | 60 (54–70) | 44 (34–49) | 64 (61–71) | 72 (70–74) | 67 (50–73) |
| Sex, male, n (%) | 10 (67) | 3 (30) | 6 (60) | 6 (86) | 4 (80) | 12 (55) |
| Cancer | NA | NA | ||||
| Melanoma stage, n | ||||||
| IIIB | 1 | 1 | 0 | 2 | ||
| IIIC | 1 | 2 | 0 | 0 | ||
| IIID | 0 | 0 | 1 | 1 | ||
| IV | 11 | 7 | 0 | 0 | ||
| Renal | 2 | 0 | 3 | 1 | ||
| Lung | 0 | 0 | 3 | 1 | ||
| Serum LDH,aU/L, median (IQR) | 259 (222–301) | 289 (259–331) | NA | 216 (210–235) | 210 (205–227) | NA |
| Distribution of UC | NA | NA | 4 Pancolitis 6 Left-sided colitis |
NA | NA | NA |
| Time from UC diagnosis to endoscopy, mo, median (IQR) | NA | NA | 50 (41–161) | NA | NA | NA |
| Time from diarrhoea onset to endoscopy, d, median (IQR) | 10 (6–29) | 9 (3–26) | 27 (1–49) | 16 (9–17) | 12 (11–13) | NA |
| Patients on steroids at endoscopy, n (%) | 12 (80) | 5 (50) | 3 (30) | 2 (29) | 0 (0) | NA |
| Time on steroids at endoscopy, d, median (IQR) | 21 (28–63) | 16 (2–27) | 45 (40–54) | 32 (19–45) | 0 (0) | NA |
| Colitis treatment, n | NA | NA | NA | |||
| Mesalazine | 0 | 7 | 0 | |||
| Prednisolone | 15 | 6 | 7 | |||
| Intravenous methylprednisone | 11 | 0 | 0 | |||
| Azathioprine | 0 | 3 | 0 | |||
| Mycophenolate mofetil | 1 | 0 | 0 | |||
| Infliximab | 8b | 3c | 2d | |||
| Adalimumab | 0 | 1 | 0 | |||
| Vedolizumab | 4d | 3e | 0 | |||
| FMT | 1d | 0 | 1d | |||
| Tofacitinib | 0 | 0 | 1d | |||
| Colectomy | 0 | 0 | 0 | |||
| Endoscopic findings (UCEIS score), n | ||||||
| 0 | 0 | 10 | 0 | 1 | 5 | 22 |
| 1 | 4 | 0 | 2 | 0 | 0 | 0 |
| 2 | 1 | 0 | 0 | 2 | 0 | 0 |
| 3 | 3 | 0 | 2 | 0 | 0 | 0 |
| 4 | 4 | 0 | 1 | 3 | 0 | 0 |
| 5 | 1 | 0 | 5 | 1 | 0 | 0 |
| 6 | 2 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 |
| Histologic findings, n | NA | |||||
| Focal active colitis | 2 | 0 | 0 | 0 | 0 | |
| Collagenous colitis | 1 | 0 | 0 | 2 | 0 | |
| Lymphocytic colitis | 1 | 0 | 0 | 1 | 0 | |
| IBD-like colitis | 7 | 0 | 10 (UC) | 1 | 0 | |
| Infectious/NSAID-like colitis | 3 | 0 | 0 | 3 | 0 | |
| Normal | 1 | 10 | 0 | 0 | 5 | |
| Nancy score, n | NA | |||||
| 0 | 0 | 10 | 0 | 1 | 5 | |
| 1 | 3 | 0 | 1 | 1 | 0 | |
| 2 | 5 | 0 | 3 | 3 | 0 | |
| 3 | 5 | 0 | 4 | 2 | 0 | |
| 4 | 2 | 0 | 2 | 0 | 0 |
IQR, interquartile range; LDH, lactate dehydrogenase; NA, not applicable; NSAID, nonsteroidal anti-inflammatory drug.
gene Nanostring experiment analyzing RNA extracted from the gastrointestinal tract of HVs (n = 8), patients with active UC (n = 5), anti–CTLA-4/PD-1 colitis (DCC; n = 9) and anti–CTLA-4/PD-1 treated with no colitis (DCNC; n = 8) are shown. Plots show differentially expressed genes in (i) UC vs HV and (ii) DCC vs DCNC and DCC vs UC.
Normal range, 90–235 U/L.
2 of 8 patients were on infliximab at the time of endoscopy and 6 had it subsequently.
2 of 3 patients were on infliximab at the time of endoscopy and 1 had had it prior.
Biologic (or FMT) was given after the endoscopy in patients.
2 of 3 patients were on vedolizumab at the time of endoscopy and 1 had had it prior.
Supplementary Table 2.
Patient Samples and Experiment Allocations
| Sample | Category | Flow cytometry |
Fluorescence microscopy | Bulk RNASeq | Nanostring | scRNASeq | Single-cell protein-RNASeq | |
|---|---|---|---|---|---|---|---|---|
| Basic panel | Extended panel | |||||||
| HV3664 | HV | • | ||||||
| HV3536 | HV | • | ||||||
| HV3665 | HV | • | • | |||||
| HV3684 | HV | • | • | |||||
| HV3523 | HV | • | • | • | ||||
| HV3947 | HV | • | • | |||||
| HV3379 | HV | • | • | |||||
| HV3132 | HV | • | • | • | ||||
| HV3530 | HV | • | ||||||
| HV3085 | HV | • | • | |||||
| HV3667 | HV | • | ||||||
| HV3384 | HV | • | • | |||||
| HV3127 | HV | • | • | |||||
| HV3452 | HV | • | ||||||
| HV3687 | HV | • | ||||||
| HV3859 | HV | • | • | |||||
| HV3947 | HV | • | • | |||||
| HV4166 | HV | • | • | • | ||||
| HV3367 | HV | • | ||||||
| HV4407 | HV | • | ||||||
| HV2885 | HV | • | ||||||
| HV4116 | HV | • | ||||||
| IBDU3087 | UC | • | • | • | • | |||
| UC1-2865 | UC | • | • | • | • | |||
| UC7 | UC | • | • | • | ||||
| UC8 | UC | • | • | • | • | |||
| UC9 | UC | • | • | |||||
| UC2-3105 | UC | • | ||||||
| UC4-3814 | UC | • | • | • | • | • | • | |
| UC6-3817 | UC | • | • | • | • | • | ||
| UC1975 | UC | • | ||||||
| IBD240 | UC | • | ||||||
| DCC4051 | CTLA-4/PD-1 colitis | • | • | • | • | • | ||
| PRISE3 | CTLA-4/PD-1 colitis | • | • | |||||
| DCC4442 | CTLA-4/PD-1 colitis | • | • | |||||
| PRISE14 | CTLA-4/PD-1 colitis | • | • | |||||
| DCC3188 | CTLA-4/PD-1 colitis | • | • | • | • | |||
| DCC3237 | CTLA-4/PD-1 colitis | • | • | • | • | |||
| DCC3700 | CTLA-4/PD-1 colitis | • | • | • | • | |||
| DCC4158 | CTLA-4/PD-1 colitis | • | • | • | • | |||
| PRISE7 | CTLA-4/PD-1 colitis | • | • | • | • | • | ||
| DCC4054 | CTLA-4/PD-1 colitis | • | • | • | • | |||
| DCC4055 | CTLA-4/PD-1 colitis | • | • | |||||
| DCC4059 | CTLA-4/PD-1 colitis | • | • | • | ||||
| LP11 | CTLA-4/PD-1 colitis | • | ||||||
| LP13 | CTLA-4/PD-1 colitis | • | ||||||
| LP16 | CTLA-4/PD-1 colitis | • | ||||||
| 2PRISE3 | CTLA-4/PD-1 TX no colitis | • | • | • | • | • | ||
| DCNC4149 | CTLA-4/PD-1 TX no colitis | • | • | • | • | • | ||
| DCNC3662 | CTLA-4/PD-1 TX no colitis | • | • | • | • | • | ||
| DCNC4158 | CTLA-4/PD-1 TX no colitis | • | ||||||
| DCNC4159 | CTLA-4/PD-1 TX no colitis | • | • | • | ||||
| DCNC3723 | CTLA-4/PD-1 TX no colitis | • | • | • | • | |||
| PRISE1 | CTLA-4/PD-1 TX no colitis | • | • | • | • | |||
| PRISE11 | CTLA-4/PD-1 TX no colitis | • | • | |||||
| PRISE12 | CTLA-4/PD-1 TX no colitis | • | • | • | • | |||
| DCNC-3237 | CTLA-4/PD-1 TX no colitis | • | • | |||||
| CC2-3126 | PD-1 colitis | • | • | |||||
| CC8-3660 | PD-1 colitis | • | ||||||
| GI4378 | PD-1 colitis | • | ||||||
| GI4303 | PD-1 colitis | • | ||||||
| PRISE5 | PD-1 colitis | • | ||||||
| GI4445 | PD-1 colitis | • | ||||||
| 3592 | PD-1 TX no colitis | • | ||||||
| PRISE13 | PD-1 TX no colitis | • | ||||||
| PRISE2 | PD-1 TX no colitis | • | ||||||
| GI4069 | PD-1 TX no colitis | • | ||||||
| PRISE15 | PD-1 TX no colitis | • | ||||||
Supplementary Table 3.
Clinical Characteristics of DCG and HV Groups
| Characteristic | DCG | HV |
|---|---|---|
| n | 4 | 7 |
| Age, y, median (IQR) | 69 (65–73) | 66 (51–72) |
| Sex, male, n (%) | 3 (75) | 3 (43) |
| Cancer, n | NA | |
| Melanoma stage IV | 3 | |
| Renal | 1 | |
| Serum LDH,aU/L, median (IQR) | 256 (230–309) | NA |
| Time from symptom onset to endoscopy, d, median (IQR) | 18 (14–33) | NA |
| Patients on steroids at endoscopy, n (%) | 2 (50) | NA |
| Time on steroids at endoscopy, d, median (IQR) | 11 (10–12) | NA |
| Gastritis treatment, n | NA | |
| Prednisolone | 4 | |
| Infliximab | 1b |
DCG, dual checkpoint inhibitor-related gastritis; IQR, interquartile range; LDH, lactate dehydrogenase; NA, not applicable.
Normal range, 90–235 U/L.
Biologic was given after the endoscopy in patient.
Supplementary Material
References
- 1.Larkin J., Hodi F.S., Wolchok J.D. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:1270–1271. doi: 10.1056/NEJMc1509660. [DOI] [PubMed] [Google Scholar]
- 2.Cheung V.T.F., Gupta T., Olsson-Brown A. Immune checkpoint inhibitor-related colitis assessment and prognosis: can IBD scoring point the way? Br J Cancer. 2020;123:207–215. doi: 10.1038/s41416-020-0882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D.Y., Salem J.E., Cohen J.V. Fatal toxic effects associated with immune checkpoint inhibitors: a systematic review and meta-analysis. JAMA Oncol. 2018;4:1721–1728. doi: 10.1001/jamaoncol.2018.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmer J.R., Lacchetti C., Schneider B.J. Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2018;36:1714–1768. doi: 10.1200/JCO.2017.77.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu-Sbeih H., Ali F.S., Wang X. Early introduction of selective immunosuppressive therapy associated with favorable clinical outcomes in patients with immune checkpoint inhibitor-induced colitis. J Immunother Cancer. 2019;7:93. doi: 10.1186/s40425-019-0577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Wiesnoski D.H., Helmink B.A. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. 2018;24:1804–1808. doi: 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soularue E., Lepage P., Colombel J.F. Enterocolitis due to immune checkpoint inhibitors: a systematic review. Gut. 2018;67:2056–2067. doi: 10.1136/gutjnl-2018-316948. [DOI] [PubMed] [Google Scholar]
- 8.Arriola E., Wheater M., Lopez M.A. Evaluation of immune infiltration in the colonic mucosa of patients with ipilimumab-related colitis. Oncoimmunology. 2016;5 doi: 10.1080/2162402X.2016.1209615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamias G., Delladetsima I., Perdiki M. Immunological characteristics of colitis associated with anti-CTLA-4 antibody therapy. Cancer Invest. 2017;35:443–455. doi: 10.1080/07357907.2017.1324032. [DOI] [PubMed] [Google Scholar]
- 10.Sasson S.C., Zaunders J.J., Nahar K. Mucosal-associated invariant T (MAIT) cells are activated in the gastrointestinal tissue of patients with combination ipilimumab and nivolumab therapy-related colitis in a pathology distinct from ulcerative colitis. Clin Exp Immunol. 2020;202:335–352. doi: 10.1111/cei.13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thome J.J., Yudanin N., Ohmura Y. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159:814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasson S.C., Gordon C.L., Christo S.N. Local heroes or villains: tissue-resident memory T cells in human health and disease. Cell Mol Immunol. 2020;17:113–122. doi: 10.1038/s41423-019-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheuk S., Schlums H., Gallais Sérézal I. CD49a Expression defines tissue-resident CD8. Immunity. 2017;46:287–300. doi: 10.1016/j.immuni.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik B.T., Byrne K.T., Vella J.L. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci Immunol. 2017;2(10) doi: 10.1126/sciimmunol.aam6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park S.L., Buzzai A., Rautela J. Tissue-resident memory CD8. Nature. 2019;565(7739):366–371. doi: 10.1038/s41586-018-0812-9. [DOI] [PubMed] [Google Scholar]
- 16.Ganesan A.P., Clarke J., Wood O. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. 2017;18:940–950. doi: 10.1038/ni.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards J., Wilmott J.S., Madore J. CD103. Clin Cancer Res. 2018;24:3036–3045. doi: 10.1158/1078-0432.CCR-17-2257. [DOI] [PubMed] [Google Scholar]
- 18.Savas P., Virassamy B., Ye C. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24:986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 19.Boddupalli C.S., Bar N., Kadaveru K. Interlesional diversity of T cell receptors in melanoma with immune checkpoints enriched in tissue-resident memory T cells. JCI Insight. 2016;1(21) doi: 10.1172/jci.insight.88955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke J., Panwar B., Madrigal A. Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J Exp Med. 2019;216:2128–2149. doi: 10.1084/jem.20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luoma A.M., Suo S., Williams H.L. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell. 2020;182:655–671. doi: 10.1016/j.cell.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esfahani K., Hudson M., Batist G. Tofacitinib for refractory immune-related colitis from PD-1 therapy. N Engl J Med. 2020;382:2374–2375. doi: 10.1056/NEJMc2002527. [DOI] [PubMed] [Google Scholar]
- 23.Bishu S., Melia J., Sharfman W. Efficacy and outcome of tofacitinib in immune checkpoint inhibitor colitis. Gastroenterology. 2021;160:932–934. doi: 10.1053/j.gastro.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohart F., Gautier B., Singh A. mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13(11) doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo P.S.T., Ferreira G.R., Cardozo L.E. CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinformatics. 2018;19(1):56. doi: 10.1186/s12859-018-2053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A., Birger C., Thorvaldsdóttir H. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oughtred R., Stark C., Breitkreutz B.J. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang H.M., Subramaniam M., Targ S. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018;36:89–94. doi: 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy D.J., Campbell K.R., Lun A.T. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lun A.T., Bach K., Marioni J.C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber W., Carey V.J., Gentleman R. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lun A.T., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlin J.S., Hamey F.K., Pijuan-Sala B. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood. 2018;131(21):e1–e11. doi: 10.1182/blood-2017-12-821413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aibar S., González-Blas C.B., Moerman T. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mease P., Charles-Schoeman C., Cohen S. Incidence of venous and arterial thromboembolic events reported in the tofacitinib rheumatoid arthritis, psoriasis and psoriatic arthritis development programmes and from real-world data. Ann Rheum Dis. 2020;79:1400–1413. doi: 10.1136/annrheumdis-2019-216761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dighe A.S., Richards E., Old L.J. Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity. 1994;1:447–456. doi: 10.1016/1074-7613(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan D.H., Shankaran V., Dighe A.S. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. 1998;95:7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards J., Wilmott J.S., Madore J. CD103(+) Tumor-resident CD8(+) T cells are associated with improved survival in immunotherapy-naïve melanoma patients and expand significantly during anti-PD-1 treatment. Clin Cancer Res. 2018;24:3036–3045. doi: 10.1158/1078-0432.CCR-17-2257. [DOI] [PubMed] [Google Scholar]
- 39.Shi L.Z., Fu T., Guan B. Interdependent IL-7 and IFN-gamma signalling in T-cell controls tumour eradication by combined alpha-CTLA-4+alpha-PD-1 therapy. Nat Commun. 2016;7:12335. doi: 10.1038/ncomms12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grasso C.S., Tsoi J., Onyshchenko M. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2020;38:500–515. doi: 10.1016/j.ccell.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao J., Shi L.Z., Zhao H. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167:397–404. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaretsky J.M., Garcia-Diaz A., Shin D.S. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manguso R.T., Pope H.W., Zimmer M.D. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547(7664):413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sade-Feldman M., Yizhak K., Bjorgaard S.L. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998–1013. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalbasi A., Tariveranmoshabad M., Hakimi K. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci Transl Med. 2020;12(565) doi: 10.1126/scitranslmed.abb0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Trzupek D., Dunstan M., Cutler A.J. Discovery of CD80 and CD86 as recent activation markers on regulatory T cells by protein-RNA single-cell analysis. Genome Med. 2020;12:55. doi: 10.1186/s13073-020-00756-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy D.J., Campbell K.R., Lun A.T. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huber W., Carey V.J., Gentleman R. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lun A.T., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuart T., Butler A., Hoffman P. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida A., Loy A., Hofmann H. ggplot2 Compatible quantile-quantile plots in R. R J. 2018;10:248–261. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.