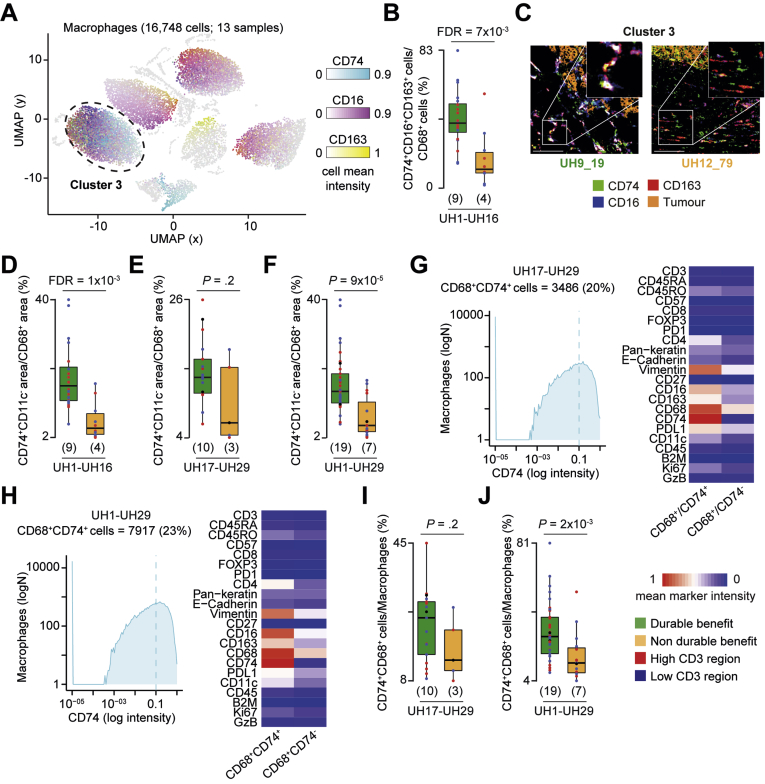
Figure 4.
Difference in CD74+ macrophages between DB- and nDB-CRCs. (A) Uniform Manifold Approximation and Projection (UMAP) map of 16,748 macrophages in 30 regions from 13 hypermutated CRCs in the discovery cohort. Cells were grouped in 9 clusters based on the expression of 11 phenotypic markers using Seurat9 (Supplementary Table 8) and colored according to the mean intensities of representative markers. The circle indicates the cluster enriched in DB-CRCs. (B) Proportions of cluster 3 (CD68+CD74+ cells) over the total macrophages in DB- and nDB-CRCs. Distributions were compared using the 2-sided Wilcoxon’s rank sum test. Benjamini-Hochberg false discovery rate (FDR) correction was applied for testing over 9 clusters. (C) IMC-derived images of CD74, CD16, and CD163 and tumor-associated markers (E-cadherin and pan-keratin) in 2 representative samples. Scale bar = 100 μm. (D) Comparisons of normalized CD74+ area between DB- and nDB-CRCs in the discovery, (E) validation, and (F) both cohorts using the 2-sided Wilcoxon’s rank sum test. For the discovery cohort, Benjamini-Hochberg FDR correction was applied for testing over 9 combinations of macrophage markers (Supplementary Table 6). (G) CD74+ macrophages in the validation and (H) combined cohorts were identified by applying a threshold of 0.1 CD74 expression to all macrophages after IMC image histologic inspection. Mean marker intensities in CD74+ and CD74− macrophages are reported and normalized across all markers and cells. (I) Comparison of normalized of CD74+ macrophages between DB- and nDB-CRCs in the validation and (J) combined cohorts. Distributions were compared using the 2-sided Wilcoxon’s rank sum test. The number of patients in each tumor group is reported in brackets. The horizontal line in the middle of each box indicates the median; the top and bottom borders of the box mark the 75th and 25th percentiles, respectively, and the vertical lines mark minimum and maximum of all the data.