SUMMARY
Hendra virus and Nipah virus (NiV), members of the Henipavirus (HNV) genus, are zoonotic paramyxoviruses known to cause severe disease across six mammalian orders, including humans. We isolated a panel of human monoclonal antibodies (mAbs) from the B cells of an individual with prior exposure to equine Hendra virus (HeV) vaccine, targeting distinct antigenic sites. The most potent class of cross-reactive antibodies achieves neutralization by blocking viral attachment to the host cell receptors ephrin-B2 and ephrin-B3, with a second class being enhanced by receptor binding. mAbs from both classes display synergistic activity in vitro. In a stringent hamster model of NiV Bangladesh (NiVB) infection, antibodies from both classes reduce morbidity and mortality and achieve synergistic protection in combination. These candidate mAbs might be suitable for use in a cocktail therapeutic approach to achieve synergistic potency and reduce the risk of virus escape.
Graphical Abstract
In brief
Doyle et al. describe two human monoclonal antibodies that target the henipavirus receptor-binding protein, HENV-103 and HENV-117, that display highly potent activity in vitro and enhanced therapeutic efficacy in vivo when delivered as a cocktail.
INTRODUCTION
Hendra virus (HeV) (Guirakhoo et al., 2002) and Nipah virus (NiV), the prototypic henipaviruses, are emerging zoonotic paramyxoviruses known to cause severe disease in humans and diverse other mammalian orders. Multiple species of pteropid bats (flying foxes) act as reservoir hosts for these negative-sense, single-stranded RNA viruses in the Paramyxoviridae family with which they are understood to have co-evolved (Chua et al., 2002; Halpin et al., 2011; Halpin et al., 2000; Vidgen et al., 2015). HeV is transmitted from flying foxes to horses and from horses to in-contact humans, causing severe respiratory and/or encephalitic disease mediated by endothelial vasculitis in both (Escaffre et al., 2013; Field, 2016; Murray et al., 1995a). HeV was identified in 1994, having caused the death of 14 of 21 infected horses and 1 of 2 infected humans in Queensland, Australia (Murray et al., 1995b; Selvey et al., 1995). Spillover has occurred sporadically with some seasonal and climatic trends since then, causing disease in 105 horses and 7 humans, with high case fatality rates (Queensland Government, 2020). NiV, which was discovered 4 years after HeV when hundreds of pig handlers fell ill with encephalitic disease (Chua et al., 1999), has continued to cause sporadic outbreaks in Bangladesh and India (Arunkumar et al., 2019; Soman Pillai et al., 2020). More direct routes of infection, including human-to-human transmission, and mortality rates approaching 100% have been observed during recent NiV outbreaks (Chadha et al., 2006; Clayton et al., 2012; Gurley et al., 2007). Anthropogenic and climatic influences on flying foxes are affecting their roosting, feeding, and migration habits as well as their susceptibility to heat-stress, disease, and injury (Kessler et al., 2018; Plowright et al., 2015). These factors together with their resultant increase in intermediate host contact (humans and domestic animals) are associated with increasing geographic range and frequency of henipavirus disease spillover (Martin et al., 2018; Walsh et al., 2017). While HeV and NiV outbreaks historically have been confined geographically to Australia and Southeast Asia, respectively, the risk of pandemic spread of these highly pathogenic agents related to regional and global population densities and the difficulty of avoiding international transmission via infected travelers have been highlighted by recent experience with SARS-CoV-2 (Morens and Fauci, 2020). Such consideration prompted the World Health Organization (WHO) to designate henipavirus infections as priority diseases requiring extensive and immediate research and development (Sweileh, 2017). The risk of a global health crisis associated with henipaviruses is exacerbated by the lack of licensed antiviral drugs or vaccines for Henipavirus (HNV) and a dearth of knowledge of the human immune response to these viruses (Escaffre et al., 2013; Gómez Román et al., 2020).
Passive immune transfer studies in both hamsters and ferrets have provided evidence that neutralizing antibodies are a correlate of immunoprotection from henipaviruses (Bossart et al., 2009; Guillaume et al., 2004, 2006). These data have been corroborated in multiple studies by investigators using murine, rabbit, or human antibody discovery technologies to isolate potently neutralizing antibodies to HeV and/or NiV (Aguilar et al., 2009; Mire et al., 2020; Zhu et al., 2006). One of these studies used phage display technology to isolate a human monoclonal antibody (hmAb), designated m102.4 (Zhu et al., 2008). This mAb potently neutralizes both HeV and NiV in vitro and protects against infection and disease in experimental henipavirus challenge models using ferrets or non-human primates (Bossart et al., 2011; Geisbert et al., 2014; Mire et al., 2016). More recently, two human mAbs, HENV-26 and HENV-32, were shown to neutralize HeV and NiV by distinct mechanisms and protect from NiV Bangladesh (NiVB) strain challenge in a ferret model (Dong et al., 2020). While these studies have laid a foundation for our understanding of how to target henipaviruses therapeutically, many questions remain regarding the antigenicity of the attachment glycoprotein, and whether escape mutations from these mAbs can develop in vivo.
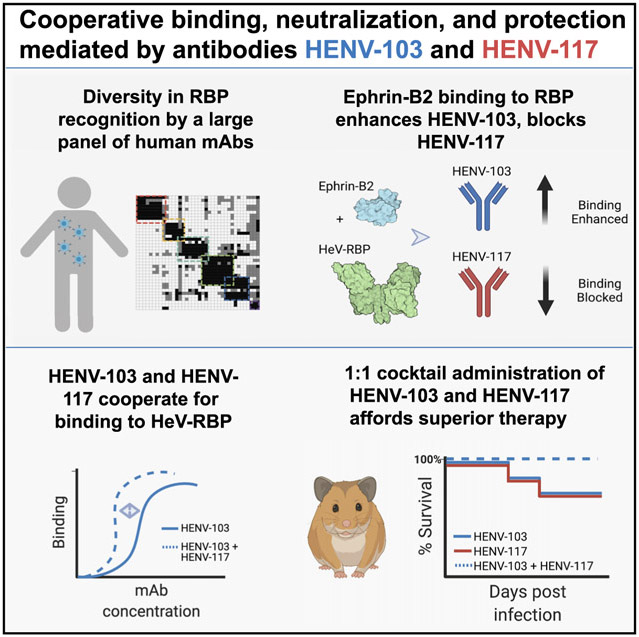
In this study, we isolated hmAbs from circulating B cells of an individual with occupation-related exposure to the equine HeV vaccine (Equivac HeV), the same individual from whom therapeutic mAbs HENV-26 and HENV-32 were previously isolated (Dong et al., 2020; Middleton et al., 2014). Members of this large panel of antibodies target diverse antigenic sites, many of which are sites of vulnerability for neutralization for at least one virus. In particular, two functional classes of antibodies that we have termed “receptor blocking” or “receptor enhanced” neutralized HeV and NiV in vitro by distinct molecular mechanisms and provided protection when used as monotherapy against lethal challenge in hamsters with the highly virulent NiVB strain. Antibodies recognizing these sites cooperate for binding to the henipavirus receptor-binding protein (RBP) glycoprotein that mediates attachment (formerly designated the G or glycosylated attachment protein, but recently renamed by the International Committee on Taxonomy of Viruses) (Rima et al., 2019). These mAbs also synergize for neutralization of both vesicular stomatitis virus (VSV)-NiVB, as well as two Cedar viruses chimerized to display the HeV or NiVB RBP and F surface glycoproteins. Cocktails of antibodies from these groups show superior therapeutic efficacy in hamsters, while bispecific antibodies bearing antigen-binding fragments from both mAbs also show therapeutic benefit. In this model, receptor-blocking mAbs induce conformational changes to the RBP that better expose the receptor-enhanced antigenic site. These results suggest that these mAbs could be used in a cocktail therapeutic approach to achieve synergistic neutralizing potency against henipavirus infections.
RESULTS
Cross-reactive, neutralizing antibodies target two distinct antigenic sites on HNV-RBP
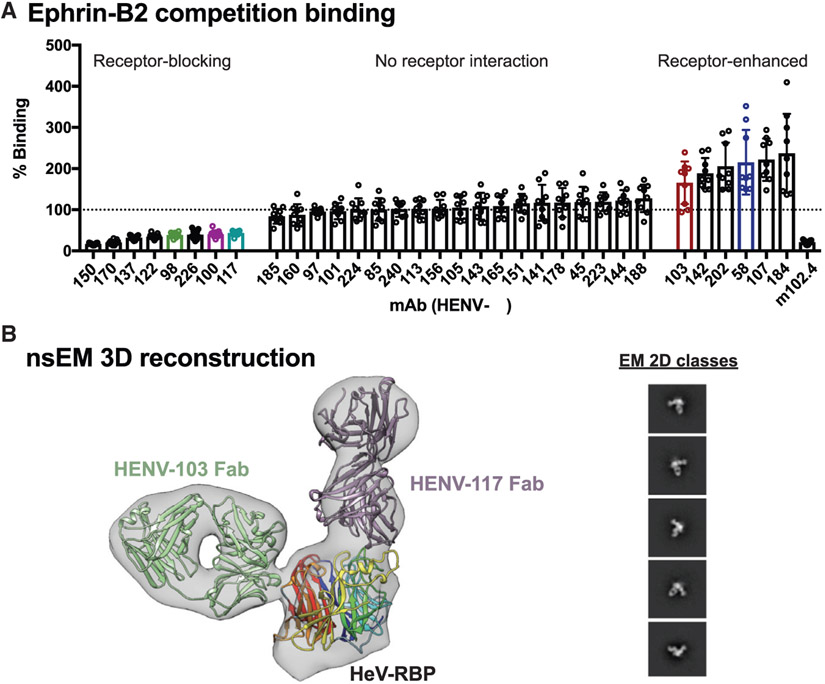
Peripheral blood mononuclear cells (PBMCs) from an Australian veterinarian with occupation-related exposure to HeV-RBP (Equivac HeV) were tested for secretion of antibodies binding to recombinant forms of the attachment (RBP) glycoproteins for NiVB, the NiV Malaysian strain (NiVM), or HeV. In total, we isolated 41 distinct new mAbs that bind henipavirus RBPs (Table S1). In order to group this large panel of mAbs rationally into those that recognized similar antigenic sites, we used a surface plasmon resonance platform to bin antibodies based on the antigenic sites they recognized on recombinant protein comprising the HeV-RBP head domain. This method immobilizes a first antibody on the surface of a gold-coated sensor chip that captures soluble antigen, and then assesses the ability of a second antibody to bind to the captured antigen. The resulting data showed that mAbs binding to HeV-RBP recognized at least six distinct major antigenic sites, designated A–F (Figures 1A and S1).
Figure 1. Identification of major antigenic sites for recognition of RBP by human mAbs.
(A) Surface plasmon resonance competition binding of human antibodies against HeV-RBP. A first antibody was applied to a gold-coated sensor chip, and recombinant HeV-RBP head domain was associated to the coupled antibody. A second antibody was applied to the sensor chip to determine binding to RBP. Black boxes indicated a pairwise interaction in which the binding of the second antibody is blocked by the first. White indicates both antibodies could bind simultaneously. Gray indicates an intermediate competition phenotype. The matrix was assembled using the Carterra Epitope software.
(B) Hydrogen-deuterium exchange mass spectrometry profiles for representative mAbs. A decrease (blue) or increase (red) in deuterium exchange on HeV-RBP in the presence of antibody is mapped onto the crystal structure of HeV-RBP (PDB: 6CMG). Structures are positioned in three orientations, with the top structure noting the ephrin-B2 binding pocket in yellow. HDX data mapped to the HeV-RBP amino acid sequence can be found at https://doi.org/10.17632/hzc4t8f4s2.1.
(C) Half-maximal binding (blue) or neutralization (green) concentrations for antibodies against recombinant proteins or live HeV or NiV, respectively.
(D) Neutralization curve plots for representative antibodies against HeV, NiV Malaysia, or NiV Bangladesh viruses. EC50 values from a single experiment representative of three independent experiments performed in technical duplicate are shown. IC50 values for neutralization are from a single independent experiment due to limitations of biosafety level 4 (BSL-4) resources. Data are represented as mean ± SD.
In tandem, we used hydrogen-deuterium exchange mass spectrometry (HDX-MS) on the head domain of HeV-RBP to map the antigenic sites of representative antibodies from each group (Figure 1B), along with binding and neutralization assays to determine cross-reactivity and functional activity (Figures 1C and 1D). Antibodies belonging to groups A and C cross-reacted with HeV, NiVM, and NiVB-RBP and neutralized the corresponding viral strains. Group A, specifically, includes the control mAb m102.4, which has been thoroughly characterized for its ability to block viral attachment to the host cell receptors ephrin-B2 and ephrin-B3, and potently neutralize both HeV and NiV (Xu et al., 2013). As expected, a representative group A mAb HENV-98 caused a decrease in deuterium exchange in a region of the HeV-RBP that corresponds to the receptor-binding site. All group A mAbs also neutralized HeV, NiVM, and NiVB strains in vitro. Notably, HENV-117 displayed exceptional potency, with half-maximal inhibitory concentration (IC50) values of 14, 8, or 15 ng/mL against HeV, NiVM, or NiVB, respectively. To date, this is the most broad and potent neutralizing mAb targeting HeV and NiV ever described, suggesting it may possess superior therapeutic activity.
Group D represents a second class of mAbs that cross-neutralize HeV, NiVM, and NiVB, albeit with roughly 10-fold less potency than group A. The group D representative mAb HENV-107 mapped to a distinct site on the HeV-RPB head domain spanning the β1 and β6 propeller blades. This region of the head domain likely lies at the interface between protomers within the dimer-of-dimers structure of the HeV-RBP tetramer, suggesting a semi-cryptic site of vulnerability on RBP (Lee and Ataman, 2011). This region has been postulated to be important in fusion triggering, as point mutations made to this region render F unable to complete its fusion cascade (Aguilar et al., 2009; Liu et al., 2013).
While mAbs in group C display limited cross-neutralization of HeV and NiV, groups B and E contain mAbs that only neutralize HeV with appreciable potency. Group F mAbs are weakly neutralizing or non-neutralizing and appear to target an antigenic site that lies on the RBP face opposite the receptor-binding domain. This epitope is likely in a site that is poorly accessible in the membrane-anchored form of RBP, lending to the poor neutralizing activity observed for these mAbs. Overall, we discovered and mapped cross-reactive, neutralizing mAbs targeting two distinct major antigenic sites that likely use distinct mechanisms to achieve virus neutralization.
Neutralizing mAbs either compete with, or are enhanced by, ephrin-B2 binding to HeV-RBP
With the knowledge that group A mAbs map to the receptor-binding domain of HeV-RBP, we sought to determine whether these antibodies could block binding of soluble ephrin-B2 to cell surface-displayed HeV-RBP. 293F cells were transiently transfected with a cDNA construct encoding full-length HeV-RBP (head, stalk, transmembrane, and cytoplasmic domains) and incubated for 72 h. These cells then were incubated with saturating concentrations of recombinant, soluble ephrin-B2, followed by addition of anti-RBP mAbs at a concentration of 2 μg/mL to assess the ability of antibodies to bind RPB in its receptor-bound state. Cells were analyzed by flow cytometry, comparing antibody binding in the presence or absence of ephrin-B2 (Figure 2A). Antibodies in group A displayed a substantial decrease in binding in the presence of ephrin-B2, supporting the hypothesis that mAbs from this group potently neutralize by blocking binding of virus to host cells. This receptor-blocking phenotype is reflected in the activity of the control mAb m102.4, which also displayed decreased signal when associated to receptor-bound RBP.
Figure 2. Receptor blocking and structural studies.
(A) Antibody binding to cell surface-displayed HeV-RBP when ephrin-B2 is bound. Cells transiently transfected with a cDNA encoding the full-length HeV-RBP were incubated with a saturating concentration of recombinantly expressed ephrin-B2. Without washing, cells were incubated with 2 μg/mL antibody, and binding was compared to binding of antibodies in the absence of ephrin-B2. The mAb m102.4 served as a control for receptor competition. Pooled data from three independent experiments are shown. Data are represented by mean ± SD.
(B) Three-dimensional reconstruction from negative stain electron microscopy (nsEM) of dimeric HeV-RBP full ectodomain bound to HENV-103 Fab and HENV-117 Fab. The EM map is shown in gray, the Fabs are in purple and green, and the RBP head domain is colored by β-propeller. 2D classes are shown, with box size of 128 at Å per pixel (Å/pix) of 3.5.
We also assessed antibodies from all other epitope binning groups for their ability to bind RBP in the presence of ephrin-B2. Antibodies from group F did not bind to surface-displayed HeV-RBP, further suggesting these antibodies cannot access this antigenic site when RBP is in its tetrameric, membrane-anchored form. Group B, C, and E mAbs bound to HeV-RBP with equal signal in the presence or absence of ephrin-B2. A second group of cross-reactive and neutralizing antibodies from group E displayed a receptor-enhanced phenotype in which binding was increased in the presence of ephrin-B2. As HDX experiments suggested that this antigenic site lies at the putative interface between protomers within the HeV-RBP dimer, it is likely that receptor binding alters the conformation of HeV-RBP, better exposing this epitope and increasing binding by mAbs to this site. This phenomenon of an antigenic site on RBP being bound better by antibodies in the presence of ephrin-B2 has been previously described (Aguilar et al., 2009; Liu et al., 2013). In summary, cross-reactive and neutralizing mAbs displayed either receptor-blocking or receptor-enhanced phenotypes, suggesting distinct neutralization mechanisms used by antibodies targeting distinct sites.
Negative stain electron microscopy (nsEM) elucidates structural determinants of recognition by receptor-blocking and receptor-enhanced mAbs
To gain insight into the structural determinants of recognition by receptor-blocking and receptor-enhanced mAbs, we performed nsEM on HeV-RBP complexed with representative Fabs based on the sequence of HENV-117 (blocking) or HENV-103 (enhanced). Initial studies with the HeV-RBP ectodomain (head and stalk domains) purified by size exclusion chromatography showed substantial structural heterogeneity of both dimeric and tetrameric complexes (data not shown). In order to generate more structurally homogeneous antigen suitable for 3D reconstruction, we purified HeV-RBP by gradient fixation ultracentrifugation using a 10%–30% glycerol gradient containing a linear 0%–0.1% glutaraldehyde gradient. This method achieved highly pure material, appropriate separation of monomeric, dimeric, and tetrameric species, and structural homogeneity induced by mild glutaraldehyde fixation (Figure S3). Dimeric HeV-RPB was complexed with a molar excess of HENV-117 and HENV-103 and assessed using nsEM. Because most RBP dimers appeared to have Fab only bound to one head domain, unbound protomer within the RBP dimer was masked to improve resolution.
Both HENV-103 and HENV-117 bind simultaneously to the HeV-RBP, further confirming that these mAbs recognize distinct antigenic sites (Figures 2B and S4C). By docking the crystal model of the head domain bound the ephrin-B2 receptor to the EM map, we observed that HENV-117 mimics the binding position of the receptor, confirming its ability to block receptor attachment (Figure S4A). Conversely, HENV-103 approaches the HeV-RBP perpendicular to the receptor binding domain at the putative interface between protomers within the RBP tetramer (Figure S4B). This antigenic site overlaps with previously published mAbs, including HENV-32 (Aguilar et al., 2009; Dong et al., 2020). In summary, HENV-103 and HENV-117 map to distinct antigenic sites by nsEM, with HENV-117 mimicking ephrin-B2 binding, while HENV-103 binds at the putative dimeric interface.
Antibodies provide therapeutic protection in a highly stringent model of Nipah Bangladesh virus challenge in Syrian golden hamsters
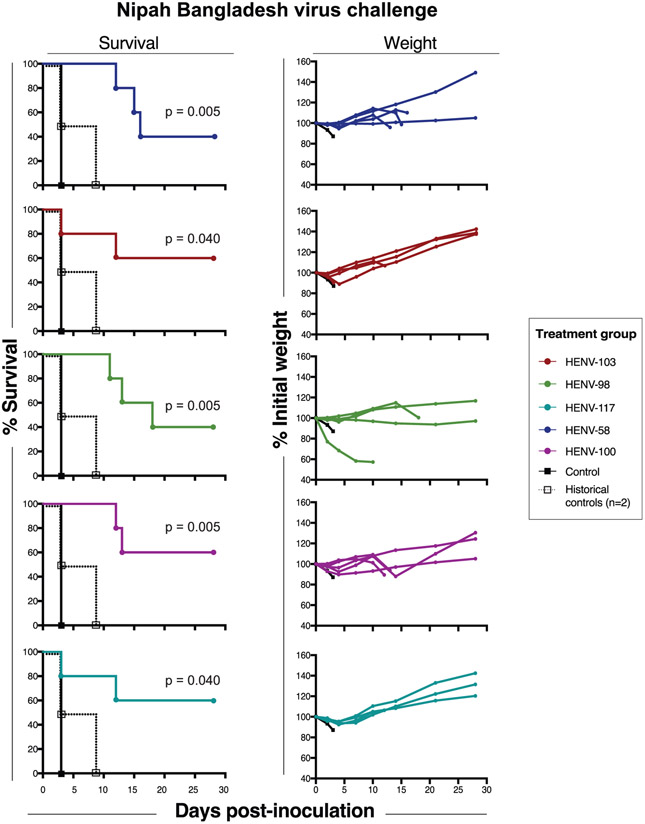
Previous studies of murine and human mAbs targeting HeV and/or NiV suggested passive immunization as a potential strategy for therapeutic intervention. To assess therapeutic activity of antibodies in this large panel, we chose five candidate mAbs representing groups A (receptor-blocking HENV-98, HENV-100, HENV-117) and D (receptor-enhanced HENV-58, HENV-103) to test in a highly stringent NiVB challenge model in hamsters (Wong et al., 2003). Disease in this model follows a two-stage disease pattern with differing sequelae: an acute respiratory distress syndrome (ARDS)-like respiratory tract component starting at day 3–4, and an encephalitic component beginning at days 8–12. On day 0, Syrian golden hamsters were challenged intranasally with 5 × 106 plaque-forming units (PFU) of NiVB. The following day, hamsters were administered a 10 mg/kg dose of antibody by the intraperitoneal (i.p.) route and monitored for 28 days after challenge. While the hamster administered a vehicle control solution succumbed at day 3, as much as 60% survival was achieved in animals administered either receptor-blocking or receptor-enhanced mAbs (Figure 3). The two most protective mAbs from each class were HENV-117 and HENV-103, for which surviving animals in each treatment group were able to maintain body weight, after a brief initial period of weight loss, throughout the study (Figure 3). HENV-117 and HENV-103 were also the two most potent mAbs from groups A and D, suggesting that in vitro potency by antibodies targeting these sites correlates with in vivo efficacy. In summary, receptor-blocking and receptor-enhanced mAbs protect hamsters from NiVB challenge, with HENV-117 and HENV-103 representing the most promising candidates targeting two distinct antigenic sites.
Figure 3. Therapeutic protection by human antibodies in hamster model of Nipah Bangladesh challenge.
Survival curves (left) and weight maintenance (right) for hamsters treated with 10 mg/kg antibody (n = 5 per group) 24 h post-inoculation with 5 × 106 PFU NiV Bangladesh by the intranasal route. An untreated control animal (n = 1) succumbed to infection 3 days post-inoculation. All weight maintenance charts include control animal in black. Two historical controls are plotted on survival curves and pooled with the experimental control to perform statistical analysis by the log rank Mantel-Cox test.
HENV-117 and HENV-103 cooperate for binding to HNV-RBP and reveal synergistic virus neutralization activity
RNA viruses, including HeV and NiV, use error-prone RNA-dependent RNA polymerase (RdRP) complexes to achieve genome replication (Welch et al., 2020). While generation of errors can lead to non-viable genomes in some cases, this process also affords viruses the ability to escape from small and large molecule antivirals by introducing amino acid substitutions in the sites recognized by these molecules (Borisevich et al., 2016). This escape pattern is of concern and has been observed in both in vitro and in vivo studies of diverse RNA viruses, showing that antibody monotherapy approaches against viral pathogens may be susceptible to failure. In order to combat escape, cocktails of antibodies targeting the same or differing antigenic sites offer a higher threshold of protection, with escape becoming statistically highly unlikely. Concurrently, studies of antibody cocktails against Ebola virus, HIV, and more recently SARS-CoV-2 show the potential for synergistic activity by neutralizing antibodies, in which one antibody potentiates the activity of another (Howell et al., 2017; Miglietta et al., 2014; Zost et al., 2020a). With this goal in mind, we sought to determine whether receptor-blocking and receptor-enhanced mAbs cooperatively bind to and neutralize henipaviruses. We hypothesized that receptor-blocking mAbs would mimic the structural rearrangements in HeV-RBP by ephrin-B2, better exposing the receptor-enhanced epitope, allowing for synergistic neutralization by combinations of these antibodies. We chose the most potent and protective candidates from each class, HENV-103 and HENV-117, for these studies.
We first tested the ability of HENV-117 to enhance the binding of HENV-103 to cell surface-displayed RBP. Using the surface-display system, we incubated HeV-RBP-transfected cells in saturating concentrations of mAbs that block ephrin-B2 binding. Without washing, we then added serial dilutions of HENV-103 chemically labeled with an Alexa Fluor 647 tag. Cells then were analyzed by flow cytometry to determine whether HENV-103 showed an increased binding signal across a dilution series in the presence of receptor-blocking mAbs. When cells were incubated with HENV-103 only, half-maximal binding was achieved at 5,289 ng/mL. When cells were first incubated with saturating concentrations of HENV-117, the half-maximal effective concentration (EC50) of HENV-103 shifted to 350 ng/mL, representing an increase in binding activity of approximately 15-fold (Figure 4A). Notably, this cooperativity is unidirectional, as HENV-103 did not increase the binding of HENV-117 (Figure 4A). This cooperative phenotype also depends on HENV-117, with increasing HENV-117 concentrations showing increased binding by a constant concentration of HENV-103 (Figure 4B). These data suggest that antibodies that bind the ephrin-B2 binding site on HeV-RBP, such as HENV-117, mimic the conformational changes induced by ephrin-B2 binding, making a semi-cryptic epitope recognized by HENV-103 more accessible.
Figure 4. Synergistic binding and neutralization.
(A) Cooperative binding by HENV-103 and HENV-117 to cell surface-displayed HeV-RBP. Cells expressing HeV-RBP were incubated with unlabeled HENV-103 or HENV-117, followed by addition of a dilution series of Alexa Fluor 647 (AF647)-labeled HENV-103 or HENV-117. Cells were analyzed by flow cytometry and gated for AF647+ cells. Data were pooled from three independent experiments performed in technical duplicate. Data are represented by mean ± SD.
(B) Dependence of HENV-117 effective concentration on HENV-103 binding enhancement. Cells were incubated with varying concentrations of unlabeled HENV-117, followed by incubation with AF647-labeled HENV-103 at 0.5 μg/mL, with enhancement calculated by comparing AF647+ cells to HENV-103 binding to HeV-RBP in the absence of HENV-117. Data from a single experiment performed in technical triplicate representative of three independent experiments are shown. Data are represented by mean ± SD.
(C) Synergistic neutralization of rCedV-HeV by HENV-103 and HENV-117 combinations. Neutralization values at each matrix concentration (top) and calculated synergy scores (bottom) are shown. Serial dilutions of HENV-103 and HENV-117 were mixed with 4,000 PFU of rCedV-HeV-GFP for 2 h, followed by addition to Vero E6 cell monolayers in 96-well plates. Formalin-fixed cells were imaged using a CTL S6 analyzer to count GFP+ cells. Neutralization was calculated by comparing treatment to virus-only control wells, with GFP+ greater than control wells considered 0% neutralization. Values were imported into SynergyFinder using a zero interactions potency (ZIP) statistical model. Delta scores >10 indicate likely synergy. Two independent experiments were performed, with data from a single representative experiment shown.
In order to show that this cooperative binding phenotype is recapitulated functionally, we performed neutralization tests using solutions containing antibody pairs to measure synergistic neutralization potential. In order to perform these multiple comparison neutralization assays quantitatively in biosafety level 2 (BSL-2) facilities, we used a non-pathogenic henipavirus chimerized with the HeV or NiVB glycoproteins. In this system, recombinant Cedar virus (rCedV) was engineered genetically to express the RPB and F from HeV or NiVB, as well as a GFP reporter. The resulting rescued, recombinant, chimeric viruses were termed rCedV-NiVB or rCedV-HeV. In this study, we used a matrix approach to test antibody pairs for neutralization synergy, in which serial dilutions of HENV-117 and HENV-103 were mixed together in a pairwise matrix, followed by incubation with rCedV-HeV. Virus/mAb mixtures then were added to Vero E6 cell monolayer cultures in 96-well plates. At approximately 22 h after inoculating cells with virus/antibody mixtures, plates were fixed and GFP+ foci were quantified to enumerate antibody neutralization values. To calculate synergy, neutralization matrix data were uploaded to the open source program SynergyFinder, and synergy scores were calculated using the zero interactions potency (ZIP) model (Ianevski et al., 2017). A score >10 suggests synergistic activity. We observed that HENV-103 and HENV-117 gave an overall ZIP score of 13.1, with selected physiologically achievable cocktail concentrations achieving synergy scores >20 (Figure 4C). This synergy was also observed when using rCedV-NiVB, as well as a VSV psuedotyped with NiVB RBP and F, a platform described in detail previously (Figures S5A and S5B) (Mire et al., 2019). Importantly, authentic NiVB was neutralized synergistically by multiple concentrations of HENV-103 and HENV-117 when titrated at equal concentrations (Figure S5C). These data together with binding studies show that antibodies from these classes cooperate for binding to RBP and synergistically neutralize chimeric and pseudotyped viruses bearing RBP and F proteins from HeV or NiVB, as well as authentic NiVB, suggesting that a combination of these mAbs may function synergistically in vivo.
Antibody cocktails and derivative bispecific mAbs provide improved therapeutic activity in hamsters
Synergy observed in vitro by HENV-103 and HENV-117 against VSV-NiVB and rCedV-NiVB suggested the potential for in vivo synergistic protection from NiVB infection. To assess this possibility, we took two separate approaches. In the first approach, we tested HENV-103 and HENV-117 as a cocktail therapy in Syrian golden hamsters. Previously, animals were treated with 10 mg/kg for each individual mAb. In this study, animals were treated with 5 mg/kg HENV-103 and 5 mg/kg HENV-117 at 24 h after intranasal inoculation with NiVB. Using monotherapy, we found that three of five animals treated with either HENV-103 or HENV-117 survived throughout the study. However, when given in combination, all animals survived (Figure 5B) and maintained/gained weight (not shown) for 28 days after infection. These data show that HENV-103 and HENV-117 provide synergistic protection in hamsters when administered together 24 h after infection with NiVB.
Figure 5. Antibody cocktail and corresponding bispecific antibody therapeutic activity in hamsters.
(A) Neutralization of VSV-NiVB by bispecific antibodies in comparison to equimolar antibody cocktail. Two independent experiments were performed in technical triplicate, with data from a single representative experiment shown. Data are represented by mean ± SD.
(B) Syrian golden hamster challenge studies with HENV-103 and HENV-117 cocktail or corresponding bispecific antibodies. Challenge studies were performed as described above. p values represent statistical significance as determined by a Mantel-Cox log rank test. n = 5 animals were included in all groups, with control animals treated with PBS at 24 h post-inoculation.
The second approach used two bispecific antibody platforms (Figure S6). The dual variable domain (DVD) construct bears two heavy and light chain variable domains in each “arm,” with the domains most Fc distal corresponding to HENV-117 (Wu et al., 2007). A similar construct, termed Bis4Ab, differs from DVD in that the Fc-distal HENV-117 component contains a full Fab fragment, whereas the HENV-103 contains only heavy and light chain variable domains in a Fc-proximal scFv format (Dimasi et al., 2019; Thanabalasuriar et al., 2017). We first tested these constructs in vitro against VSV-NiVB and found that both DVD and Bis4Ab constructs strongly neutralized VSV-NiVB with similar potency (Figure 5A). We again tested these in the Syrian golden hamster model of NiVB infection and found that in the DVD group, four of five hamster survived, while protection in the Bis4Ab group mirrored that of monotherapy, with three of five hamsters surviving. These data suggest that there may be added complexity to using bispecific antibody platforms (e.g., whether or not both antigen binding fragments can engage antigen simultaneously, serum half-life in rodents) and that combined administration of HENV-103 and HENV-117 provides superior in vivo protection in comparison to monotherapy. This feature is complemented by the added benefit of further protection from escape mutation.
DISCUSSION
Recent epidemics of Ebola, 2009 H1N1 influenza, and SARS-CoV-2 viruses highlight the need for the development of counter-measures against emerging viruses with pandemic potential before their occurrence. Pathogenic henipaviruses, particularly NiV, are emerging and highly pathogenic agents with confirmed human-to-human transmission for which licensed treatments or vaccines for human use do not yet exist (Amaya and Broder, 2020). In this study, we expanded upon our previous work and isolated a panel of mAbs specific for the henipavirus RBP glycoprotein from an individual with prior occupation-related exposure to the equine HeV-RBP subunit vaccine (Dong et al., 2020). Competition-binding and HDX-MS studies identified at least six distinct antigenic sites recognized by these mAbs. Flow cytometric studies with surface-displayed HeV-RBP showed that potently neutralizing, cross-reactive antibodies either (1) blocked HeV-RBP binding to ephrin-B2, or (2) showed enhanced binding in the presence of ephrin-B2. Antibodies that block receptor binding also induced the receptor-enhanced phenotype, showing that antibodies to these two classes cooperate for binding to HNV-RBP. In addition, these mAbs also showed synergy in neutralization of rCedV-HeV particles. As monotherapy, receptor-blocking and receptor-enhanced antibodies provided modest protection in a stringent, highly lethal NiVB challenge model in Syrian golden hamsters. In combination, these antibodies provided complete therapeutic protection in the same model of infection.
A significant concern when using antibodies as therapeutics against emerging infectious diseases due to RNA viruses is the potential for viral “mutational escape” within an infected host or immune evasion by divergent viral variants. Escape from antibody-mediated neutralization has been documented even with ultrapotent mAbs targeting conserved epitopes on viral glycoproteins (Greaney et al., 2021). Using a cocktail of mAbs provides resistance against escape, with the potential added benefit of synergistic antiviral potency, allowing for lower dosing. The potential for spillover of divergent variants of batborne paramyxoviruses (henipaviruses and rubulaviruses) is consistent with the inherent propensity of RNA viruses for rapid evolution. Furthermore, flying foxes serve as ideal reservoir hosts because of their dense community roosting patterns and relative resistance to paramyxoviral disease (Baker et al., 2012; Barr et al., 2015; Drexler et al., 2012; Luis et al., 2015; Peel et al., 2019; Sasaki et al., 2012; Vidgen et al., 2015). The discovery of protective antibodies highlighted herein, specifically HENV-103 and HENV-117, offer the opportunity to construct a cocktail of antibodies with the most desired protective properties, including against mutation escape and spillover variant viruses. Concurrently, a bispecific antibody with activity of both HENV-103 and HENV-117 is an attractive therapeutic option that endows a single therapeutic molecule with the synergistic potency of two individual mAbs. While we showed that two antibodies targeting RBP offer a synergistic benefit, the possibility exists that having antibodies targeting both RBP and F may also be of benefit. Recently, highly potent and protective anti-F antibodies have been described and may offer an ideal partner to HENV-103, HENV-117, or both as a triple antibody cocktail (Dang et al., 2019; Mire et al., 2020).
As with other paramyxoviruses, humans likely elicit highly functional antibodies against the henipavirus F glycoprotein (Merz et al., 1980). This concept is highlighted by palivizumab, an anti-F antibody used as a prophylaxis for premature infants to protect from infection by respiratory syncytial virus (Meissner et al., 1999). Although, as highlighted above, protective anti-F mAbs have been isolated, these have been uniformly of murine origin. The full antigenic landscape of the henipavirus F protein may suggest new sites of vulnerability to neutralization by mAbs and could guide the rational design of henipavirus vaccines. This opportunity is especially important considering the geographical range of henipaviruses, and the fact that a previously undescribed virus from this genus may emerge to cause a pandemic. Having knowledge of the determinants of neutralization for both RBP and F will allow for quick mobilization of platform technologies to develop vaccines, similar to what we have seen in the response to the SARS-CoV-2 pandemic (Zost et al., 2020b).
Wild-type mice are understood to be refractory to infection with henipaviruses. On the contrary, the Syrian golden hamster model of henipavirus infection has been demonstrated to recapitulate the most salient features of human disease, including the biphasic pulmonary disease followed by catastrophic neurological events (Rockx et al., 2011), making it an ideal model to screen vaccines and therapeutics. However, hamsters require an orders of magnitude higher challenge dose in order to achieve lethal disease compared to ferrets or African green monkeys (AGMs). This requirement likely contributes to a much faster disease progression and a conceivably shorter therapeutic window. Ferrets, and optimally, AGMs are likely superior animal models to further preclinical development of these promising antibody candidates. This possibility is especially true of the AGM model of NiVB infection, in which the therapeutic window for use of antibody therapies (treatment at days 3–5) is shorter than that of HeV and NiVM (treatment at days 5–7) (Mire et al., 2016). Studies in these models might further elucidate whether HENV-103 and HENV-117 are improved compared to previously described antibodies (Playford et al., 2020).
Here, and in previous studies, functional anti-henipavirus RBP-specific mAbs from multiple species have been isolated. These antibodies uniformly recognize the head domain of RBP, suggesting that this region is likely the most immunogenic domain of the RBP. Multiple studies interrogating the function of RBP, and its role in triggering the F protein to undergo significant conformational rearrangements, have pointed to the RBP stalk domain as playing a significant role in viral fusogenicity. Specifically, Aguilar and colleagues have shown that the C-terminal portion of the NiV stalk domain can trigger fusion of membranes in the absence of a head domain (Liu et al., 2013). While it is likely that the stalk domain, which is partially obstructed by the head domain, is immunogenically subdominant, it is possible that rare, circulating memory B cells harboring antibodies targeting this domain exist. Future studies interrogating the antibody response to these viruses also may shed light on the role of mAbs targeting the stalk domain of HNV-RBP, and whether these antibodies have the potential to prevent viral and host membrane fusion.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, James E. Crowe, Jr. (james.crowe@vumc.org).
Materials availability
Materials described in this paper are available for distribution for nonprofit use using templated documents from Association of University Technology Managers “Toolkit MTAs,” available at: https://autm.net/surveys-and-tools/agreements/material-transfer-agreements/mta-toolkit.
Data and code availability
All data needed to evaluate the conclusions in the paper are present in the paper or Supplemental Information. Raw HDX-MS data mapped to the sequence of HeV-RBP (in reference to Figure 1B) are publicaly available at https://doi.org/10.17632/hzc4t8f4s2.1. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
A male veterinarian residing in Australia underwent leukapharesis to obtain peripheral blood mononuclear cells (PBMCs) used for antibody discovery in this paper. Details on this donor were previously described (Dong et al., 2020).
Cell lines
Cell lines used at Vanderbilt University were tested for mycoplasma at regular intervals and only used if a negative result was achieved. Vero E6 cells were maintained in Dulbecco’s Minimal Essential Medium (DMEM) supplemented with 10% heat-inactivaated fetal bovine serum. 293F cells were maintained in suspension in FreeStyle 293F medium and incubated at 37°C in 8% CO2 while shaking at 125 RPM. Expi293F cells (Thermo Fisher, female) were maintained at the same incubation conditions as 293F cells in Expi293 medium (Thermo Fisher). ExpiCHO (Thermo Fisher, female) cells were also maintained at these conditions in ExpiCHO Expression Medium (Thermo Fisher). B95.8 (ATCC, Female) were propagated in Medium A (STEMCELL Technologies) at 37°C and were used for isolation of Epstein-Barr virus (EBV).
Viruses
NiV number 1999011924 was obtained from a patient from the 1999 outbreak in Malaysia. The isolate of NiVB used was 200401066 and was obtained from a fatal human case during the outbreak in Rajbari, Bangladesh in 2004 and passaged on Vero E6 cell monolayer cultures three times. HeV was obtained from a patient from the 1994 outbreak in Australia. All viruses were kindly provided by Dr. Thomas Ksiazek, UTMB. Each virus was propagated on Vero E6 cells in Eagle’s minimal essential medium supplemented with 10% fetal calf serum. The NiVM, NiVB and HeV challenge virus stocks were assessed for the presence of endotoxin using The Endo-safe-Portable Test System (PTS) (Charles River Laboratories, Wilmington, MA). Each virus preparation was diluted 1:10 in Limulus Amebocyte Lysate (LAL) Reagent Water per the manufacturer’s instructions, and endotoxin levels were tested in LAL Endosafe-PTS cartridges as directed by the manufacturer. Each preparation was found to be below detectable limits, whereas positive controls showed that the tests were valid. All experiments involving infectious henipaviruses were carried out at the UTMB Galveston National Laboratory under biosafety level 4 conditions. Chimeric Cedar viruses (rCedV) were rescued and handled at Uniformed Services University (unpublished). VSV-NiVB was generated at UTMB Galveston National Laboratory and handled under biosafety level 2 conditions at Vanderbilt University Medical Center.
Animal models
The animal studies were performed at the Galveston National Laboratory, University of Texas Medical Branch at Galveston (UTMB) and were approved by the UTMB Institutional Animal Care and Use Committee (IACUC). This facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Groups of 3- to 5-week old female Syrian Golden hamsters (Envigo, Indianapolis, IN) were used for in vivo studies.
METHOD DETAILS
Generation of hmAbs
Peripheral blood mononuclear cells (PBMCs) from a human subject were isolated from whole blood and transformed using Epstein-Barr virus (EBV), as previously described (Crowe, 2017). Briefly, transformed B cells were expanded and co-cultured with irradiated human PBMCs in 96-well plates. Cell supernatants were screened by ELISA using recombinant HeV-RBP or NiV-RBP head domain proteins. Wells with positive reactivity then were fused to a human-mouse heteromyeloma cell line (HMMA 2.5) and plated by limiting dilution in 384-well plates. The resulting hybridomas were cloned as single cells by fluorescence-activated cell sorting (FACS) to produce clonal hybridoma cell lines. These clonal hybridoma cells were cultured in T-225 flasks containing serum-free medium, and mAb was purified from spent medium by affinity chromatography on an ÄKTA™ pure Fast Protein Liquid Chromatography (FPLC) instrument (GE Healthcare). For large-scale, recombinant production of human mAbs, RNA was isolated from hybridoma lines, and heavy and light chain variable regions were sequenced using 5′RACE sequencing and cloned into pTwist monocistronic vectors. Constructs then were transfected into expiCHO cells and harvested 7 days post-transfection. Antibody in supernatants was purified by affinity chromatography. Material used in the combination therapy studies was produced by Mapp Biopharmaceutical, with HENV-103 containing a C46S substitution to eliminate free cysteines.
Generation of bispecific mAbs
Bispecific mAbs that combined the antigen binding domains of HENV-117 and HENV-103 into a single molecule were designed, expressed, and purified as follows. The heavy chain of the HENV-117-103 DVD combines the heavy chain variable domains of first HENV-117, then HENV-103, each separated by a flexible linker, and then followed by the IgG1 human constant heavy chain domains. Similarly, the light chain of the HENV-117-103 DVD includes the light chain variable domains of both HENV-117 and HENV-103, separated by a flexible linker and then followed by a single human kappa light chain constant domain which naturally pairs with the corresponding DVD heavy chain. The HENV-117-103 Bis4Ab was constructed by inserting a HENV-103 single-chain variable fragment (scFv) between CH1 and CH2 of the HENV-117 heavy chain. The HENV-103 scFv in the bis4Ab format contains a poly glycine-serine linker between its variable domains, and the scFv unit is also flanked by poly glycine-serine linkers. The modified heavy chain is then paired with the standard HENV-117 light chain for expression and purification (Dimasi et al., 2019). The heavy and light chains of the HENV-117-103 DVD and the HENV-117-103 Bis4Ab were cloned into pcDNA3 expression vectors. For each of the bispecific mAbs, the corresponding heavy and light chain plasmids were chemically co-transfected into ExpiCHO cells (GIBCO) and transiently expressed for 9 days. The supernatant was then clarified by centrifugation and filtration, prior to loading onto a MabSelect SuRe Protein A (GE Healthcare) affinity chromatography column using an ÄKTA™ Fast Protein Liquid Chromatography (FPLC) instrument (GE Healthcare). The column was washed with 1X PBS, and the mAbs were eluted with IgG Elution Buffer (Pierce). Following neutralization with 1 M Tris pH 8.0 to pH ~7, the eluates were concentrated to 5 mg/ml in an Amicon 30K MWCO centrifugal filter (Millipore), and then sterile-filtered using a 0.22 μM syringe filter (Millex-GP).
Neutralization assays
The virus neutralizing activity concentrations were determined for NiVM, NiVB, and HeV using a plaque reduction assay. Briefly, antibodies were diluted two-fold from 100 μg/mL to extinction and incubated with a target of ~100 plaque-forming units (pfu) of NiVM, NiVB, or HeV for 45 min at 37 °C. Virus and antibody mixtures then were added to individual wells of six-well plates of Vero 76 cell monolayer cultures. Plates were fixed and stained with neutral red two days after infection, and plaques were counted 24 h after staining. Neutralization potency was calculated based on pfu for each virus in the well without antibody. The neutralization experiments were performed in triplicate, with independent virus preparations and duplicate readings for each replicate. Mean half-maximal inhibitory concentration (IC50) values were calculated as previously described (Ferrara and Temperton, 2018).
Surface plasmon resonance (SPR) epitope binning
A continuous flow micro-spotter (CFM) instrument (Carterra) was used to generate antibody-coated SPR sensor chips (Xantec) (Abdiche et al., 2014). Briefly, mAbs were diluted to 10 μg/mL in sodium acetate pH 4.5 in a 96-well round bottom plate. A mirroring 96-well plate containing activation buffer (EDC and sulfo-NHS in 10 mM MES pH 5.5) was used first to activate the gold-plated surface of the sensor chip, followed by association of antibodies. The coated chip then was moved to an IBIS-MX96 microarray-based surface plasmon resonance imager (Carterra), where it was quenched with 1 M ethanolamine to prevent further coupling of proteins. To bin antibodies, 100 mM HeV-RBP head domain was flowed over the coated sensor chip. One-by-one, antibodies diluted to 10 μg/mL were tested for their ability to associate with antigen captured on the sensor chip. Carterra Epitope software was used to analyze data and construct competition-binding grids.
Biolayer interferometry competition binding to HeV-RBP
Competition-binding studies were performed on a biolayer interferometry instrument (Octet Red; Sartorius). Recombinant HeV-RBP was diluted to 10 μg/mL in kinetics buffer (Pall) and loaded onto HIS2 sensor tips. After an equilibration step, a first antibody was loaded onto tips at 25 μg/mL in kinetics buffer. Tips were immediated immersed in a second antibody at the same concentration to assess the ability of the second antibody to bind in the presense of a first antibody. To determine the extent to which an antibody competes with other antibodies in the panel, the maximal binding of that antibody as secondary was divided by maximal binding of the same antibody as primary (and multiplied by 100 to express as a percentage of binding). Guidelines were set as follows: 0 to 30%, no competition; 31 to 70%, partial competition; 71 to 100%, full competition.
Hydrogen-deuterium exchange mass spectrometry (HDX-MS) of Fab-HeV-RBP complexes
HDX-MS was performed as previously reported (Bennett et al., 2019). Briefly, antigen (HeV-RBP) and selected mAbs were prepared individually or in complex at a protein-concentration of 10 pmol/μL in 1 × PBS pH 7.4 and incubated for 2 h at 0°C. Deuterium labeling was performed by a 20-fold dilution of 3 μL sample in PBS pH 7.4 in D2O and incubation at 20°C for 0 s, 100 s, and 1,000 s. The reaction was quenched by a 2-fold dilution in 1 × PBS, 4 M guanidinium/HCl, 100 mM tris(2-carboxyethyl)phosphine to a final pH of 2.3 at 0°C. Samples were injected immediately into a nano-ACQUITY UPLC system controlled by an HDX manager (Waters Corporation, Milford, MA, USA). Online pepsin digestion was performed at 15°C, 10,000 psi at a flow of 100 μL/min of 0.1% formic acid in H2O using an immobilized-pepsin column. A Waters VanGuard BEH C18 1.7 μm guard column was used to trap peptides at 0°C for 6 min before separation on a Waters ACQUITY UPLC BEH C18 1.7 μm, 1 mm × 100 mm column at a flow of 40 μL/min at 0°C with a 6 min gradient of 5 to 35% acetonitrile, 0.1% formic acid in H2O. UPLC effluent was directed into a Waters Xevo G2-XS with electrospray ionization and lock-mass acquisition (human Glu-1-Fibrinopeptide B peptide, m/z = 785.8427) for peptide analysis in MSE-mode. The capillary was set to 2.8 kV, source-temperature to 80°C, desolvation temperature to 175°C, desolvation gas to 400 L/h and the instrument was scanned over a m/z-range of 50 to 2,000. All experiments were carried out in triplicate. Data analysis was accomplished using Waters ProteinLynx Global Server 3.0.3 software (non-specific protease, min fragment ion matches per peptide of three, FDR 4% and oxidation of methionine as a variable modification) for peptide identification and DynamX 3.0 software (minimum intensity of 500, minimum products 3, minimum products per amino acid 0.3 and a mass error < 15 ppm) for deuterium uptake calculations. Results are reported as an average of triplicate analyses.
Generation of VSV pseudotyped viruses bearing NiVB glycoproteins
Recombinant VSVs containing genomic inserts for expression of NiVB G and F proteins were kindly provided by Chad Mire and generated as previously described (Mire et al., 2019). Stocks of each rVSV were propagated and titrated on VSV-G transfected BHK-21 (WI-2), with viral titers determined by counting GFP+ cells using a CTL S6 Analyzer instrument (company/). To generate virus bearing both G and F glycoproteins, cells were inoculated with each VSV at MOI = 5 and incubated for 48 hours. Cell supernatants were clarified by centrifugation. Resulting VSV-NiVB was titrated on Vero cell monolayers using an xCELLigence instrument to determine the lowest virus concentration that would induce CPE as measured by cell impedance.
Cooperative binding of antibodies to antigen displayed on the surface of cells
A construct containing cDNA encoding full-length HeV-RBP protein was transfected using polyethylenimine into 293F cells, and cells were cultured at 37°C in 5% CO2 for 3 days. Cells subsequently were plated at 50,000 cells/well in V-bottom 96-well plates, washed, and incubated with either 20 μg/mL primary mAb in 30 μL or FACS buffer alone for 30 minutes at 4°C. Without washing, 30 μL serially diluted mAb labeled with Alexa Fluor 647 dye (ThermoFisher) was added to wells and incubated for 30 minutes at 4°C. Cells were washed and resuspended in FACS buffer and analyzed using an iQue Plus flow cytometer (Intellicyt). Dead cells were excluded from analysis by fluorescent staining with 4′,6-diamidino-2-phenylindole (DAPI).
Negative stain electron microscopy
For electron microscopy imaging of HeV-RBP protein and Fabs complex, we expressed the HeV-RBP full ectodomain (head domain with intact stalk domain) with a C-terminal polyhistidine tag. Expressed protein was isolated by metal affinity chromatography on HisTrap Excel columns (GE Healthcare), followed by GraFix methods using a 10% to 30% glycerol gradient and 0 to 0.1% glutaraldehyde gradient (Stark, 2010). Glutaraldehyde was quenched with 1 M Tris-Cl after fractionation. 200 μL fractions were analyzed by SDS-PAGE, with fractions corresponding to monomeric, dimeric, and tetrameric species pooled. Protein was then buffer exchanged into 50 mM Tris-Cl pH 7.5 containing 140 mM NaCl. Fabs corresponding to HENV-103 and HENV-117 were expressed and purified as previously described. Protein complexes were generated by incubation of HeV-RBPecto dimer and the two Fab in a 1:5:5 molar ratio overnight at 4°C. Approximately 3 μL of the sample at ~10 to 15 μg/mL was applied to a glow-discharged grid with continuous carbon film on 400 square mesh copper electron microscopy grids (Electron Microscopy Sciences). Grids were stained with 0.75% uranyl formate (Ohi et al., 2004). Images were recorded on a Gatan US4000 4k × 4k CCD camera using an FEI TF20 (TFS) transmission electron microscope operated at 200 keV and control with SerialEM. All images were taken at 62,000 × magnification with a pixel size of 1.757 Å per pixel in low-dose mode at a defocus of 1.5 to 1.8 μm. The total dose for the micrographs was ~35 e – per Å2. Image processing was performed using the cryoSPARC software package. Images were imported, CTF-estimated and particles were picked automatically. The particles were extracted with a box size of 256 pixels and binned to 128 pixels (pixel size of 3.514 A/pix) and 2D class averages were performed to achieve clean datasets. Classes were further classified (2D) to separated different complex variant and classes having the 2 Fab on one RBP domain were selected. Ab-initio was used to generate initial 3D volume that was further refined with a mask over one RBP-Fabs complex. The final refine volume has a resolution of ~15Å. Model docking to the EM map was done in Chimera (Pettersen et al., 2004). For the RBP head domain PDB: 6PDL was used and for the Fab PDB:12E8 or the prediction model of the Fv that was generated by SAbPred tool was used (Dunbar et al., 2016). The 3D EM map has been deposited into EMDB (EMD-23488). Chimera software was used to make all structural figures.
Neutralization synergy of VSV-NiVB
VSV-NiVB pseudotype viruses were generated as described above. In 96-well plates, serial dilutions of “receptor-blocking” and “receptor-enhanced” mAbs were mixed in a matrix arrangement, followed by addition of equal volume of VSV-NiVB diluted 1:500 in DMEM. Mixtures were incubated for 1 hour at 37°C prior to addition to Vero cell monolayers in xCELLigence 96-well E-plates containing 10,000 cells/well. Cells were incubated with virus and antibody for 1 hour at 37°C, followed by addition of DMEM + 5% FBS to wells. Plates were placed back on the xCELLigence instrument for reading of cell impedance every 15 minutes for 72 hours. Neutralization was determined by comparing cell index of treated wells versus untreated wells at a single time point (values output by xCELLigence instrument software). Neutralization values then were imported into SynergyFinder software (Ianevski et al., 2017), with delta scores calculated using the zero interactions potency (ZIP) synergy model.
Neutralization synergy of rCedV chimeric viruses
Recombinant Cedar virus (rCedV) chimeras displaying RBP and F proteins of HeV or NiVB were generated and validated as described elsewhere. Black-walled 96-well plates (Corning Life Sciences; NY, USA) were coated with 20,000 cells/well Vero E6 cells in DMEM + 10% Cosmic calf serum and incubated overnight. Approximately 24 hours later, HENV-103 and HENV-117 were diluted to indicated concentrations and incubated 1:1 with 4,000 PFU/well rCedV-HeV-GFP or rCedV-NiVB-GFP and incubated for 2 hours at 37°C. Following incubation, 90 μL virus/antibody mixtures were added to aspirated cell monolayers and were incubated at 37°C for 22 hours. Medium containing virus/antibody mixtures was aspirated, and cells were fixed with 100 μL/well 4% formalin for 20 minutes at room temperature. After aspiration, cell monolayers were gently washed 4x with DI water. Formalin-fixed plates were then scanned using the CTL S6 analyzer (Cellular Technology Limited; OH, USA). Fluorescent foci were counted using the CTL Basic Count feature and S6 software. Percent neutralization was calculated by normalizing counts to a virus only control. Matrices were then imported into SynergyFinder and analyzed as described before.
Antibody therapy in Syrian golden hamster model of Nipah Bangladesh
3 to 5 week-old Syrian golden hamsters were inoculated with 5 × 106 PFU Nipah Bangladesh (passage 3) via the intranasal route. At 24 hours post challenge, 5 animals per group were treated with 10 mg/kg antibody by intraperitoneal administration. Animals were monitored for 28 days for changes in weight, temperature, and clinical appearance. Animals were humanely euthanized at the experimental endpoint. A single untreated animal served as a control in each study highlighted.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses (except synergy calculations, as indicated below) were performed using Prism version 9 (GraphPad Software, LLC). Half maximal binding or neutralization concentrations were determined from the results of dilutional curves; data are represented as mean ± SD (Figures 1C and 1D). Antibody binding to cell-surface-displayed antigen was determined using pooled data from 3 independent experiments are shown; mean ± SD was determined (Figure 2A). Survival and weight maintenance curves including data from historical controls were plotted and used to analyze differences by the long rank Mantel-Cox test (Figure 3). Cooperative binding to cell-surface displayed antigen detected by flow cytometric analysis was determined using data pooled from 3 independent experiments performed in technical duplicate, with data calculated by mean ± SD (Figure 4A). HENV-103 binding enhancement in the presence of HENV-117 was performed in technical triplicate, with data represented by mean ± SD; a representative of 3 independent experiments is shown (Figure 4B). Neutralization synergy was calculated by comparing treatment to virus-only control wells, with GFP+ greater than control wells considered 0% neutralization. Values were imported into SynergyFinder software using a Zero Interactions Potency (ZIP) statistical model; delta scores > 10 indicate likely synergy. Two independent experiments were performed, with data from a single representative experiment shown (Figure 4C). Neutralization of cocktails and bispecific antibodies was conducted in two independent experiments with technical triplicates; data from a single representative experiment are shown as mean ± SD (Figure 5A). Animal survival curves were analyzed for differences as determined by Mantel-Cox log rank test; P values represent statistical significance (Figure 5B).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| DENV 2D22 (recombinant IgG1) | Fibriansah et al., 2015 | N/A |
| m102.4 (recombinant IgG1) | Zhu et al., 2008 | N/A |
| HENV-98 (hybridoma IgG1) | This paper | N/A |
| rHENV-98 (recombinant IgG1) | This paper | N/A |
| HENV-100 (hybridoma IgG1) | This paper | N/A |
| rHENV-100 (recombinant IgG1) | This paper | N/A |
| HENV-103 (hybridoma IgG1) | This paper | N/A |
| rHENV-103 (recombinant IgG1) | This paper | N/A |
| HENV-117 (hybridoma IgG2/4) | This paper | N/A |
| rHENV-117 (recombinant IgG1) | This paper | N/A |
| HENV-58 (hybridoma IgG1) | This paper | N/A |
| rHENV-58 (recombinant IgG1) | This paper | N/A |
| HENV-226 (hybridoma IgG1) | This paper | N/A |
| HENV-170 (hybridoma IgG1) | This paper | N/A |
| HENV-150 (hybridoma IgG1) | This paper | N/A |
| HENV-137 (hybridoma IgG1) | This paper | N/A |
| HENV-122 (hybridoma IgG, subclass ND) | This paper | N/A |
| HENV-224 (hybridoma IgG1) | This paper | N/A |
| HENV-151 (hybridoma IgG1) | This paper | N/A |
| HENV-101 (hybridoma IgG1) | This paper | N/A |
| HENV-141 (hybridoma IgG1) | This paper | N/A |
| HENV-156 (hybridoma IgG1) | This paper | N/A |
| HENV-240 (hybridoma IgG1) | This paper | N/A |
| HENV-165 (hybridoma IgG1) | This paper | N/A |
| HENV-188 (hybridoma IgG1) | This paper | N/A |
| HENV-45 (hybridoma IgG1) | This paper | N/A |
| HENV-113 (hybridoma IgG1) | This paper | N/A |
| HENV-105 (hybridoma IgG1) | This paper | N/A |
| HENV-223 (hybridoma IgG1) | This paper | N/A |
| HENV-202 (hybridoma IgG4) | This paper | N/A |
| HENV-242 (hybridoma IgG1) | This paper | N/A |
| HENV-142 (hybridoma IgG1) | This paper | N/A |
| HENV-184 (hybridoma IgG1) | This paper | N/A |
| HENV-107 (hybridoma IgG1) | This paper | N/A |
| HENV-78 (hybridoma IgG1) | This paper | N/A |
| HENV-222 (hybridoma IgG1) | This paper | N/A |
| HENV-114 (hybridoma IgG1) | This paper | N/A |
| HENV-95 (hybridoma IgG1) | This paper | N/A |
| HENV-144 (hybridoma IgG1) | This paper | N/A |
| HENV-97 (hybridoma IgG1) | This paper | N/A |
| HENV-72 (hybridoma IgG1) | This paper | N/A |
| HENV-143 (hybridoma IgG1) | This paper | N/A |
| HENV-160 (hybridoma IgG1) | This paper | N/A |
| HENV-93 (hybridoma IgG1) | This paper | N/A |
| HENV-99 (hybridoma IgG1) | This paper | N/A |
| HENV-178 (hybridoma IgG, subclass ND) | This paper | N/A |
| HENV-185 (hybridoma IgG1) | This paper | N/A |
| HENV-83 (hybridoma IgG1) | This paper | N/A |
| HENV-103-117-DVD | This paper | N/A |
| HENV-103-117-Bis4Ab | This paper | N/A |
| Goat Anti-Human IgG-PE | Southern Biotech | Cat# 2040-09 |
| Goat Anti-Human Lambda-HRP | Southern Biotech | Cat# 2070-05 |
| Goat Anti-Human Kappa-HRP | Southern Biotech | Cat# 2060-05 |
| Mouse Anti-Human IgG1 Fc-AP | Southern Biotech | Cat# 9054-04 |
| Mouse Anti-Human IgG2 Fc-AP | Southern Biotech | Cat# 9070-04 |
| Mouse Anti-Human IgG3 Hinge-AP | Southern Biotech | Cat# 9210-04 |
| Mouse Anti-Human IgG4 Fc-AP | Southern Biotech | Cat# 9200-04 |
| Goat Anti-Human IgG-AP | Meridian Life Sciences | Cat# W90088A |
| Bacterial and virus strains | ||
| Hendra virus | Dr. Tom Ksiazek, UTMB | Mire et al., 2014; Geisbert et al., 2021 |
| Nipah Malaysia virus | Dr. Tom Ksiazek, UTMB | Bossart et al., 2012; Geisbert et al., 2014 |
| Nipah Bangladesh virus | Dr. Tom Ksiazek, UTMB | Mire et al., 2016; Geisbert et al., 2021 |
| VSV-NiVB-G virus | Dr. Chad Mire, UTMB | Mire et al., 2019 |
| VSV-NiVB-F virus | Dr. Chad Mire, UTMB | Mire et al., 2019 |
| VSV-NiVB (G+F) virus | Dr. Chad Mire, UTMB | Mire et al., 2019 |
| rCedV-HeV virus | This paper | N/A |
| rCedV-NiVB virus | This paper | N/A |
| Biological samples | ||
| PBMCs from HENV-immune donor | Dong et al., 2020 | Vanderbilt Vaccine Center Biorepository ID #1124 |
| Chemicals, peptides, and recombinant proteins | ||
| Cyclosporin A (CSA) | Sigma-Aldrich | Cat# C1832 |
| CpG | Sigma-Aldrich | Cat# C3742-25MG |
| Chk2 inhibitor | Sigma-Aldrich | Cat# C3742 |
| ClonaCell-HY Medium A | STEMCELL Technologies | Cat# 3801 |
| ClonalCell-HY Medium E | STEMCELL Technologies | Cat# 3805 |
| 384-well plate | Nunc | Cat# 164688 |
| 384-well plate (non-treated) | Thermo Fisher/Nunc | Cat# 262203 |
| 96-well flat plate | Thermo Fisher Scientific | Cat# 167008 |
| 96-well plate | Falcon | Cat# 353072 |
| 96-well plate, V-bottom | Corning | Cat# 3894 |
| ExpiFectamine 293 Transfection Kit | Thermo Fisher Scientific | Cat# A14525 |
| ClonalCell-HY Medium E | STEMCELL Technologies | Cat# 3805 |
| 384-well plate | Nunc | Cat# 164688 |
| Ouabain | Sigma-Aldrich | Cat# O3125 |
| Hypoxanthine | Sigma-Aldrich | Cat# H0137 |
| HAT | Sigma-Aldrich | Cat# H0262 |
| G-rex | Wilson Wolf | Cat# 80240M |
| Hybridoma Serum Free Media | GIBCO | Cat# 12045 |
| HiTrap MabSelectSure | GE Health Life Sciences | Cat# 11003494 |
| ExpiCHO Expression Medium | Thermo Fisher Scientific | Cat# A2910001 |
| Ouabain | Sigma-Aldrich | Cat# O3125 |
| Hypoxanthine | Sigma-Aldrich | Cat# H0137 |
| HAT | Sigma-Aldrich | Cat# H0262 |
| G-rex | Wilson Wolf | Cat# 80240M |
| Opti-MEM 1 Reduced Serum Medium | GIBCO | Cat# 31985088 |
| Fetal Bovine Serum, ultra-low IgG | Thermo Fisher Scientific | Cat# 16250078 |
| Trypsin EDTA (0.25%), Phenol Red | Thermo Fisher Scientific | Cat# 25200114 |
| DMEM, high glucose | Thermo Fisher Scientific | Cat# 11965118 |
| FBS, heat inactivated | Thermo Fisher Scientific | Cat# A3840102 |
| 96 well clear V-bottom plate | Corning | Cat# 3894 |
| D-PBS | Corning | Cat# 21031CM |
| DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride) | Thermo Fisher Scientific | Cat# D1306 |
| Histopaque(R)-1077, sterile-filtered, density: 1.077 g/mL | Sigma-Aldrich | Cat# 10771-500ML |
| AP substrate | Sigma-Aldrich | Cat# SO942-200AAB |
| Alexa Fluor 647 NHS Ester (Succinimidyl Ester) | Thermo Fisher Scientific | Cat# A37573 |
| Polyethylenimine, linear | Polysciences, Inc. | Cat# 23966 |
| Uranyl formate | EMS | Cat# CF400-CU-50 |
| Critical commercial assays | ||
| AlexaFluor ™ 647 Antibody Labeling Kit | Thermo Fisher Scientific | Cat# A20186 |
| Expifectamine ™ 293 Transfection Kit | Thermo Fisher Scientific | Cat# A14526 |
| Experimental models: Cell lines | ||
| B95.8 cells | ATCC | Cat# VR-1492 |
| Expi293F cells | Thermo Fisher Scientific | Cat# A14527 |
| 293F cells | Thermo Fisher Scientific | Cat# R79007 |
| ExpiCHO cells | Thermo Fisher Scientific | Cat# A29127 |
| Vero E6 cells | ATCC | Cat# CRL-1586 |
| Human-mouse HMMA 2.5 cells | Dr. L. Cavacini | N/A |
| Experimental models: Organisms/strains | ||
| Syrian Golden Hamster | Envigo (formerly Harlan Sprague Dawley) | N/A |
| Recombinant DNA | ||
| Plasmid: HeV-G full length | Dong et al., 2020 | GenScript Biotech |
| Plasmid: HeV-G head domain | Dong et al., 2020 | GenScript Biotech |
| Plasmid: NiV-G (Malaysia) head domain | Dong et al., 2020 | GenScript Biotech |
| Plasmid: NiV-G (Bangladesh) head domain | Dong et al., 2020 | GenScript Biotech |
| Plasmid: human ephrinB2 (aa 28-165) | Dong et al., 2020 | Twist Biosciences Inc. |
| Plasmid: HeV-G full ectodomain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-58 Heavy Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-58 Light Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-98 Heavy Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-98 Light Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-100 Heavy Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-100 Light Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-103 Heavy Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-103 Light Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-117 Heavy Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-117 Light Chain | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-103 Fab | This paper | Twist Biosciences Inc. |
| Plasmid: HENV-117 Fab | This paper | Twist Biosciences Inc. |
| Software and algorithms | ||
| GraphPad Prism 9.1 Software | GraphPad Software, Inc. | https://www.graphpad.com |
| iQue® Forecyt® Software | Sartorius | N/A |
| Octet Data Acquisition 10.0.3.12 Software | ForteBio | https://www.sartorius.com/en/products/protein-analysis |
| Octet Data Analysis 11.0.2.5 Software | ForteBio | https://www.sartorius.com/en/products/protein-analysis |
| PyMOL Software | Schrödinger | https://pymol.org/2/ |
| SynergyFinder Software | Ianevski et al., 2017 | https://synergyfinder.fimm.fi |
| CompuSyn Software | Chou and Martin, 2005 | https://www.combosyn.com/ |
| UCSF Chimera 1.14 Software | Pettersen et al., 2004 | RRID:SCR_004097 |
| Cryosparc 3.2 Software | Punjani et al., 2017 | RRID:SCR_016501 |
| SerialEM 3.7 Software | Mastronarde, 2005 | RRID:SCR_017293 |
| Other | ||
| ÄKTA pure chromatography system | Cytiva | N/A |
| HiTrap Protein G High Performance | Cytiva | Cat# 17040501 |
| HiTrap MabSelect SuRe | Cytiva | Cat# 11003494 |
| HisTrap HP His Tag column | Cytiva | Cat# 17524701 |
| Intellicyt iQue Screener Plus | Sartorius | N/A |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare | Cat# 28989335 |
| xCELLigence RTCA Analyzer | Agilent | N/A |
| Wasatch SPR | Carterra | N/A |
| ImmunoSpot S6 Plate Imager | CTL | N/A |
| FEI TF20 electron microscope with Gatan US4000 4k × 4k CCD camera | TFS | N/A |
| 400 mesh copper EM grids | EMS | Cat# 22451 |
Highlights.
Antibodies targeting the henipavirus receptor-binding protein (RBP) were isolated
Two potently neutralizing mAbs, HENV-103 and HENV-117, bind to distinct sites on the RPB
HENV-103 and HENV-117 delivered as a cocktail display enhanced protection in hamsters
ACKNOWLEDGMENTS
We thank Chris Gainza and Rachel Nargi from Vanderbilt in the Crowe laboratory for technical support with antibody production and purification; Joseph Reidy, Andrew Trivette, and Robert Carnahan for intellectual contributions to antibody production and sequencing; and Ian Setliff and Ivelin Georgiev for statistical support. We thank Noah Ditto of Carterra for assistance in SPR data analysis. We acknowledge BioRender for use in generating the graphical abstract. This work was supported by a grant from the National Institutes of Health (NIH) U19 AI142764 and departmental funds from UTMB to T.W.G. and UC7AI094660 for BSL-4 operations support of the Galveston National Laboratory. The work of M.P.D. was supported by NIH fellowship grant F31 AI152332. The project described was supported by CTSA award no. UL1 TR002243 from the National Center for Advancing Translational Sciences (NCATS). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIAID or NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109628.
DECLARATION OF INTERESTS
J.E.C. has served as a consultant for Luna Biologics, is a member of the Scientific Advisory Board of Meissa Vaccines, and is Founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received unrelated sponsored research agreements from Takeda, IDBiologics, and AstraZeneca. E.A.K., Z.A.B., and B.R.W. are employees and shareholders of Mapp. L.Z. is an employee, shareholder, and co-owner of Mapp. C.C.B. is a US federal employee, and C.C.B. and M.A. are co-inventors on US and foreign patents pertaining to Cedar virus and methods of use and recombinant Cedar virus chimeras, whose assignees are the US as represented by the Henry M. Jackson Foundation for the Advancement of Military Medicine (Bethesda, MD, USA). The remaining authors declare no competing interests.
SUPPORTING CITATIONS
The following reference appears in the supplemental information: Bowden et al. (2010).
REFERENCES
- Abdiche YN, Miles A, Eckman J, Foletti D, Van Blarcom TJ, Yeung YA, Pons J, and Rajpal A (2014). High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS ONE 9, e92451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar HC, Ataman ZA, Aspericueta V, Fang AQ, Stroud M, Negrete OA, Kammerer RA, and Lee B (2009). A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F). J. Biol. Chem 284, 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya M, and Broder CC (2020). Vaccines to emerging viruses: Nipah and Hendra. Annu. Rev. Virol 7, 447–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunkumar G, Chandni R, Mourya DT, Singh SK, Sadanandan R, Sudan P, and Bhargava B; Nipah Investigators People and Health Study Group (2019). Outbreak investigation of Nipah virus disease in Kerala, India, 2018. J. Infect. Dis 219, 1867–1878. [DOI] [PubMed] [Google Scholar]
- Baker KS, Todd S, Marsh G, Fernandez-Loras A, Suu-Ire R, Wood JLN, Wang LF, Murcia PR, and Cunningham AA (2012). Co-circulation of diverse paramyxoviruses in an urban African fruit bat population. J. Gen. Virol 93, 850–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J, Smith C, Smith I, de Jong C, Todd S, Melville D, Broos A, Crameri S, Haining J, Marsh G, et al. (2015). Isolation of multiple novel paramyxoviruses from pteropid bat urine. J. Gen. Virol 96, 24–29. [DOI] [PubMed] [Google Scholar]
- Bennett MR, Bombardi RG, Kose N, Parrish EH, Nagel MB, Petit RA, Read TD, Schey KL, Thomsen IP, Skaar EP, and Crowe JE (2019). Human mAbs to Staphylococcus aureus IsdA provide protection through both heme-blocking and Fc-mediated mechanisms. J. Infect. Dis 219, 1264–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisevich V, Lee B, Hickey A, DeBuysscher B, Broder CC, Feldmann H, and Rockx B (2016). Escape from monoclonal antibody neutralization affects henipavirus fitness in vitro and in vivo. J. Infect. Dis 213, 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Zhu Z, Middleton D, Klippel J, Crameri G, Bingham J, McEachern JA, Green D, Hancock TJ, Chan YP, et al. (2009). A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute Nipah virus infection. PLoS Pathog. 5, e1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Geisbert TW, Feldmann H, Zhu Z, Feldmann F, Geisbert JB, Yan L, Feng YR, Brining D, Scott D, et al. (2011). A neutralizing human monoclonal antibody protects african green monkeys from Hendra virus challenge. Sci. Transl. Med 3, 105ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Rockx B, Feldmann F, Brining D, Scott D, LaCasse R, Geisbert JB, Feng YR, Chan YP, Hickey AC, et al. (2012). A Hendra virus G glycoprotein subunit vaccine protects African green monkeys from Nipah virus challenge. Sci. Transl. Med 4, 146ra107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden TA, Crispin M, Harvey DJ, Jones EY, and Stuart DI (2010). Dimeric architecture of the Hendra virus attachment glycoprotein: Evidence for a conserved mode of assembly. J. Virol 84, 6208–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ, Ksiazek TG, and Mishra A (2006). Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis 12, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC, and Martin N (2005). CompuSyn for Drug Combinations: PC Software and User’s Guide: A Computer Program for Quantitation of Synergism and Antagonism in Drug Combinations, and the Determination of IC50and ED50 and LD50 Values (Paramus, NJ: ComboSyn Inc; ). [Google Scholar]
- Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PS, Ksiazek TG, Zaki SR, Paul G, Lam SK, and Tan CT (1999). Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354, 1257–1259. [DOI] [PubMed] [Google Scholar]
- Chua KB, Koh CL, Hooi PS, Wee KF, Khong JH, Chua BH, Chan YP, Lim ME, and Lam SK (2002). Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 4, 145–151. [DOI] [PubMed] [Google Scholar]
- Clayton BA, Middleton D, Bergfeld J, Haining J, Arkinstall R, Wang L, and Marsh GA (2012).Transmission routes for Nipah virus from Malaysia and Bangladesh. Emerg. Infect. Dis 18, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JE Jr. (2017). Principles of broad and potent antiviral human antibodies: insights for vaccine design. Cell Host Microbe 22, 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang HV, Chan YP, Park YJ, Snijder J, Da Silva SC, Vu B, Yan L, Feng YR, Rockx B, Geisbert TW, et al. (2019). An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections. Nat. Struct. Mol. Biol 26, 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimasi N, Fleming R, Wu H, and Gao C (2019). Molecular engineering strategies and methods for the expression and purification of IgG1-based bispecific bivalent antibodies. Methods 154, 77–86. [DOI] [PubMed] [Google Scholar]
- Dong J, Cross RW, Doyle MP, Kose N, Mousa JJ, Annand EJ, Borisevich V, Agans KN, Sutton R, Nargi R, et al. (2020). Potent henipavirus neutralization by antibodies recognizing diverse sites on Hendra and Nipah virus receptor binding protein. Cell 183, 1536–1550.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, et al. (2012). Bats host major mammalian paramyxoviruses. Nat. Commun 3, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar J, Krawczyk K, Leem J, Marks C, Nowak J, Regep C, Georges G, Kelm S, Popovic B, and Deane CM (2016). SAbPred: A structure-based antibody prediction server. Nucleic Acids Res. 44 (W1), W474–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaffre O, Borisevich V, Carmical JR, Prusak D, Prescott J, Feldmann H, and Rockx B (2013). Henipavirus pathogenesis in human respiratory epithelial cells. J. Virol 87, 3284–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara F, and Temperton N (2018). Pseudotype neutralization assays: From laboratory bench to data analysis. Methods Protoc. 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Ibarra KD, Ng T-S, Smith SA, Tan JL, Lim X-N, Ooi JSG, Kostyuchenko VA, Wang J, and de Silva AM (2015). DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HE (2016). Hendra virus ecology and transmission. Curr. Opin. Virol 16, 120–125. [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Mire CE, Geisbert JB, Chan YP, Agans KN, Feldmann F, Fenton KA, Zhu Z, Dimitrov DS, Scott DP, et al. (2014). Therapeutic treatment of Nipah virus infection in nonhuman primates with a neutralizing human monoclonal antibody. Sci. Transl. Med 6, 242ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Bobb K, Borisevich V, Geisbert JB, Agans KN, Cross RW, Prasad AN, Fenton KA, Yu H, Fouts TR, et al. (2021). A single dose investigational subunit vaccine for human use against Nipah virus and Hendra virus. NP J Vaccines 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez Román R, Wang LF, Lee B, Halpin K, de Wit E, Broder CC, Rahman M, Kristiansen P, and Saville M (2020). Nipah@20: Lessons learned from another virus with pandemic potential. MSphere 5, e00602–e00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, et al. (2021). Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29, 44–57.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume V, Contamin H, Loth P, Georges-Courbot MC, Lefeuvre A, Marianneau P, Chua KB, Lam SK, Buckland R, Deubel V, and Wild TF (2004). Nipah virus: Vaccination and passive protection studies in a hamster model. J. Virol 78, 834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume V, Contamin H, Loth P, Grosjean I, Courbot MC, Deubel V, Buckland R, and Wild TF (2006). Antibody prophylaxis and therapy against Nipah virus infection in hamsters. J. Virol 80, 1972–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirakhoo F, Pugachev K, Arroyo J, Miller C, Zhang ZX, Weltzin R, Georgakopoulos K, Catalan J, Ocran S, Draper K, and Monath TP (2002). Viremia and immunogenicity in nonhuman primates of a tetravalent yellow fever-dengue chimeric vaccine: Genetic reconstructions, dose adjustment, and antibody responses against wild-type dengue virus isolates. Virology 298, 146–159. [DOI] [PubMed] [Google Scholar]
- Gurley ES, Montgomery JM, Hossain MJ, Bell M, Azad AK, Islam MR, Molla MA, Carroll DS, Ksiazek TG, Rota PA, et al. (2007). Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis 13, 1031–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin K, Young PL, Field HE, and Mackenzie JS (2000). Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol 81, 1927–1932. [DOI] [PubMed] [Google Scholar]
- Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, Rahman SA, Hughes T, Smith C, Field HE, and Daszak P; Henipavirus Ecology Research Group (2011). Pteropid bats are confirmed as the reservoir hosts of henipaviruses: A comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg 85, 946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KA, Brannan JM, Bryan C, McNeal A, Davidson E, Turner HL, Vu H, Shulenin S, He S, Kuehne A, et al. (2017). Cooperativity enables non-neutralizing antibodies to neutralize ebolavirus. Cell Rep. 19, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianevski A, He L, Aittokallio T, and Tang J (2017). SynergyFinder: A web application for analyzing drug combination dose-response matrix data. Bioinformatics 33, 2413–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MK, Becker DJ, Peel AJ, Justice NV, Lunn T, Crowley DE, Jones DN, Eby P, Sánchez CA, and Plowright RK (2018). Changing resource landscapes and spillover of henipaviruses. Ann. N Y Acad. Sci 1429, 78–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, and Ataman ZA (2011). Modes of paramyxovirus fusion: A Henipavirus perspective. Trends Microbiol. 19, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, and Aguilar HC (2013). Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 9, e1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis AD, O’Shea TJ, Hayman DTS, Wood JLN, Cunningham AA, Gilbert AT, Mills JN, and Webb CT (2015). Network analysis of host-virus communities in bats and rodents reveals determinants ofcross-species transmission. Ecol. Lett 18, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Yanez-Arenas C, Chen C, Plowright RK, Webb RJ, and Skerratt LF (2018). Climate change could increase the geographic extent of Hendravirus spillover risk. EcoHealth 15, 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Meissner HC, Welliver RC, Chartrand SA, Law BJ, Weisman LE, Dorkin HL, and Rodriguez WJ (1999). Immunoprophylaxis with palivizumab, a humanized respiratory syncytial virus monoclonal antibody, for prevention of respiratory syncytial virus infection in high risk infants: a consensus opinion. Pediatr. Infect. Dis. J 18, 223–231. [DOI] [PubMed] [Google Scholar]
- Merz DC, Scheid A, and Choppin PW (1980). Importance of antibodies to the fusion glycoprotein of paramyxoviruses in the prevention of spread of infection. J. Exp. Med 151, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D, Pallister J, Klein R, Feng YR, Haining J, Arkinstall R, Frazer L, Huang JA, Edwards N, Wareing M, et al. (2014). Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg. Infect. Dis 20, 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miglietta R, Pastori C, Venuti A, Ochsenbauer C, and Lopalco L (2014). Synergy in monoclonal antibody neutralization of HIV-1 pseudoviruses and infectious molecular clones. J. Transl. Med 12, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire CE, Geisbert JB, Agans KN, Feng YR, Fenton KA, Bossart KN, Yan L, Chan YP, Broder CC, and Geisbert TW (2014). A recombinant Hendra virus G glycoprotein subunit vaccine protects nonhuman primates against Hendra virus challenge. J. Virol 88, 4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire CE, Satterfield BA, Geisbert JB, Agans KN, Borisevich V, Yan L, Chan YP, Cross RW, Fenton KA, Broder CC, and Geisbert TW (2016). Pathogenic differences between Nipah virus Bangladesh and Malaysia strains in primates: Implications for antibody therapy. Sci. Rep 6, 30916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire CE, Geisbert JB, Agans KN, Versteeg KM, Deer DJ, Satterfield BA, Fenton KA, and Geisbert TW (2019). Use of single-injection recombinant vesicular stomatitis virus vaccine to protect nonhuman primates against lethal Nipah virus disease. Emerg. Infect. Dis 25, 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire CE, Chan YP, Borisevich V, Cross RW, Yan L, Agans KN, Dang HV, Veesler D, Fenton KA, Geisbert TW, and Broder CC (2020). A cross-reactive humanized monoclonal antibody targeting fusion glycoprotein function protects ferrets against lethal Nipah virus and Hendra virus infection. J. Infect. Dis 221 (Suppl 4), S471–S479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, and Fauci AS (2020). Emerging pandemic diseases: How we got to COVID-19. Cell 182, 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K, Rogers R, Selvey L, Selleck P, Hyatt A, Gould A, Gleeson L, Hooper P, and Westbury H (1995a). A novel morbillivirus pneumonia of horses and its transmission to humans. Emerg. Infect. Dis 1, 31–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, Westbury H, Hiley L, Selvey L, Rodwell B, et al. (1995b). A morbillivirus that caused fatal disease in horses and humans. Science 268, 94–97. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, and Walz T (2004). Negative staining and image classification— Powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel AJ, Wells K, Giles J, Boyd V, Burroughs A, Edson D, Crameri G, Baker ML, Field H, Wang LF, et al. (2019). Synchronous shedding of multiple bat paramyxoviruses coincides with peak periods of Hendra virus spillover. Emerg. Microbes Infect 8, 1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Playford EG, Munro T, Mahler SM, Elliott S, Gerometta M, Hoger KL, Jones ML, Griffin P, Lynch KD, Carroll H, et al. (2020). Safety, tolerability, pharmacokinetics, and immunogenicity of a human monoclonal antibody targeting the G glycoprotein of henipaviruses in healthy adults: A first-in-human, randomised, controlled, phase 1 study. Lancet Infect. Dis 20, 445–454. [DOI] [PubMed] [Google Scholar]
- Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, Middleton D, Reid PA, McFarlane RA, Martin G, et al. (2015). Ecological dynamics of emerging bat virus spillover. Proc. Biol. Sci 282, 20142124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryo-SPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Queensland Government (2020). Summary of Hendravirus incidents in horses. https://www.business.qld.gov.au/industries/service-industries-professionals/service-industries/veterinary-surgeons/guidelines-hendra/incident-summary.
- Rima B, Balkema-Buschmann A, Dundon WG, Duprex P, Easton A, Fouchier R, Kurath G, Lamb R, Lee B, Rota P, and Wang L; Ictv Report Consortium (2019). ICTV virus taxonomy profile: Paramyxoviridae. J. Gen. Virol 100, 1593–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B, Brining D, Kramer J, Callison J, Ebihara H, Mansfield K, and Feldmann H (2011). Clinical outcome of henipavirus infection in hamsters is determined by the route and dose of infection. J. Virol 85, 7658–7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Setiyono A, Handharyani E, Rahmadani I, Taha S, Adiani S, Subangkit M, Sawa H, Nakamura I, and Kimura T (2012). Molecular detection of a novel paramyxovirus in fruit bats from Indonesia. Virol. J 9, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvey LA, Wells RM, McCormack JG, Ansford AJ, Murray K, Rogers RJ, Lavercombe PS, Selleck P, and Sheridan JW (1995). Infection of humans and horses by a newly described morbillivirus. Med. J. Aust 162, 642–645. [DOI] [PubMed] [Google Scholar]
- Soman Pillai V, Krishna G, and Valiya Veettil M (2020). Nipah virus: Past outbreaks and future containment. Viruses 12, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H (2010). GraFix: Stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126. [DOI] [PubMed] [Google Scholar]
- Sweileh WM (2017). Global research trends of World Health Organization’s top eight emerging pathogens. Global. Health 13, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanabalasuriar A, Surewaard BG, Willson ME, Neupane AS, Stover CK, Warrener P, Wilson G, Keller AE, Sellman BR, DiGiandomenico A, and Kubes P (2017). Bispecific antibody targets multiple Pseudomonas aeruginosa evasion mechanisms in the lung vasculature. J. Clin. Invest 127, 2249–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidgen ME, de Jong C, Rose K, Hall J, Field HE, and Smith CS (2015). Novel paramyxoviruses in Australian flying-fox populations support host-virus co-evolution. J. Gen. Virol 96, 1619–1625. [DOI] [PubMed] [Google Scholar]
- Walsh MG, Wiethoelter A, and Haseeb MA (2017). The impact of human population pressure on flying fox niches and the potential consequences for Hendra virus spillover. Sci. Rep 7, 8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch SR, Tilston NL, Lo MK, Whitmer SLM, Harmon JR, Scholte FEM, Spengler JR, Duprex WP, Nichol ST, and Spiropoulou CF (2020). Inhibition of Nipah virus by defective interfering particles. J. Infect. Dis 221 (Suppl 4), S460–S470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KT, Grosjean I, Brisson C, Blanquier B, Fevre-Montange M, Bernard A, Loth P, Georges-Courbot MC, Chevallier M, Akaoka H, et al. (2003). A golden hamster model for human acute Nipah virus infection. Am. J. Pathol 163, 2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ying H, Grinnell C, Bryant S, Miller R, Clabbers A, Bose S, McCarthy D, Zhu RR, Santora L, et al. (2007). Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat. Bio-technol 25, 1290–1297. [DOI] [PubMed] [Google Scholar]
- Xu K, Rockx B, Xie Y, DeBuysscher BL, Fusco DL, Zhu Z, Chan YP, Xu Y, Luu T, Cer RZ, et al. (2013). Crystal structure of the Hendra virus attachment G glycoprotein bound to a potent cross-reactive neutralizing human monoclonal antibody. PLoS Pathog. 9, e1003684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Dimitrov AS, Bossart KN, Crameri G, Bishop KA, Choudhry V, Mungall BA, Feng YR, Choudhary A, Zhang MY, et al. (2006). Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J. Virol 80, 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Bossart KN, Bishop KA, Crameri G, Dimitrov AS, McEachern JA, Feng Y, Middleton D, Wang LF, Broder CC, and Dimitrov DS (2008). Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J. Infect. Dis 197, 846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, et al. (2020a). Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost SJ, Gilchuk P, Chen RE, Case JB, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Chen EC, et al. (2020b). Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med 26, 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or Supplemental Information. Raw HDX-MS data mapped to the sequence of HeV-RBP (in reference to Figure 1B) are publicaly available at https://doi.org/10.17632/hzc4t8f4s2.1. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.